Mapping the Contact Sites of the Escherichia coli Division-Initiating Proteins FtsZ and ZapA by BAMG Cross-Linking and Site-Directed Mutagenesis
Abstract
1. Introduction
2. Results
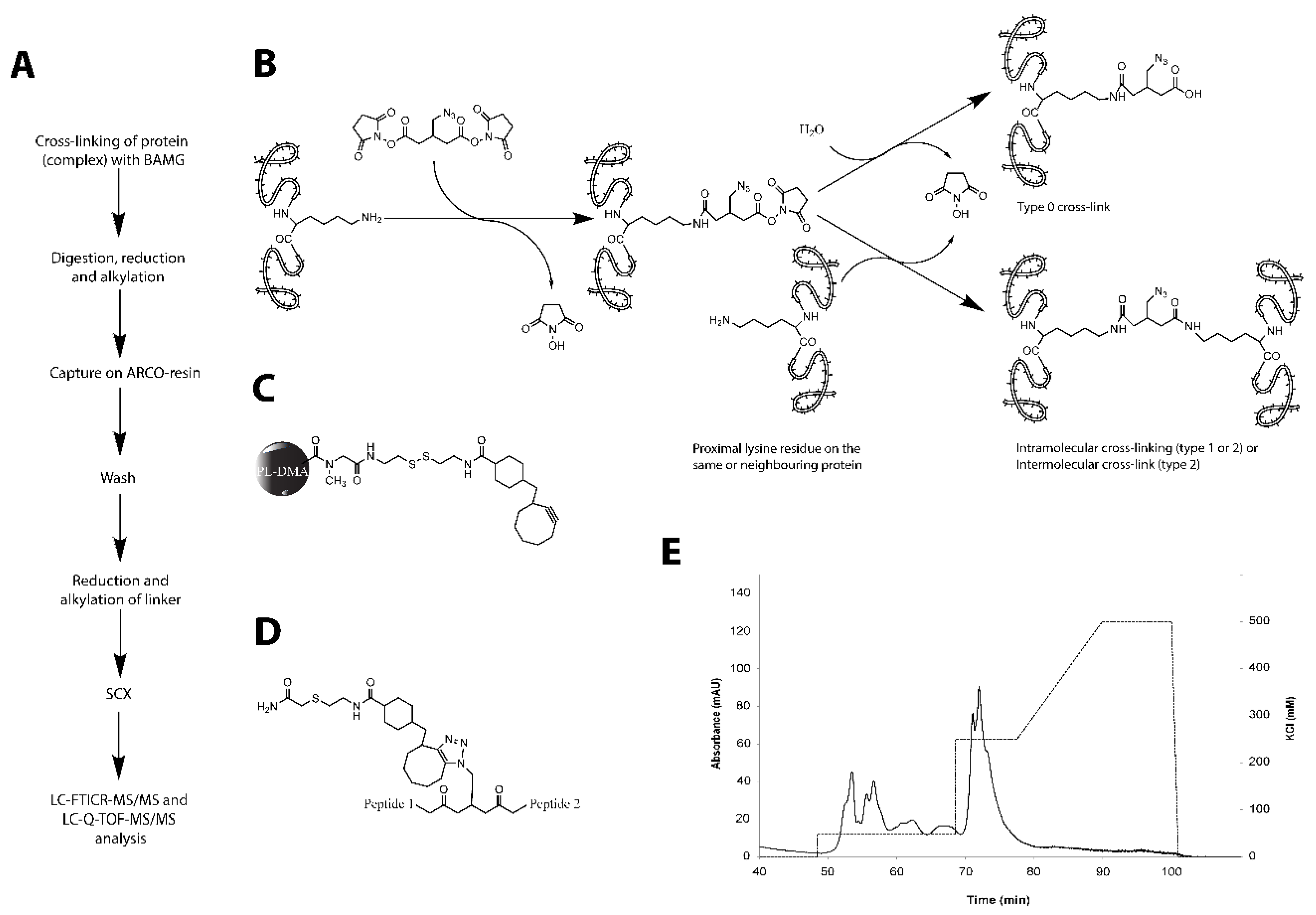
2.1. Optimization of Cross-Linking Conditions for Mass Spectrometric Analysis
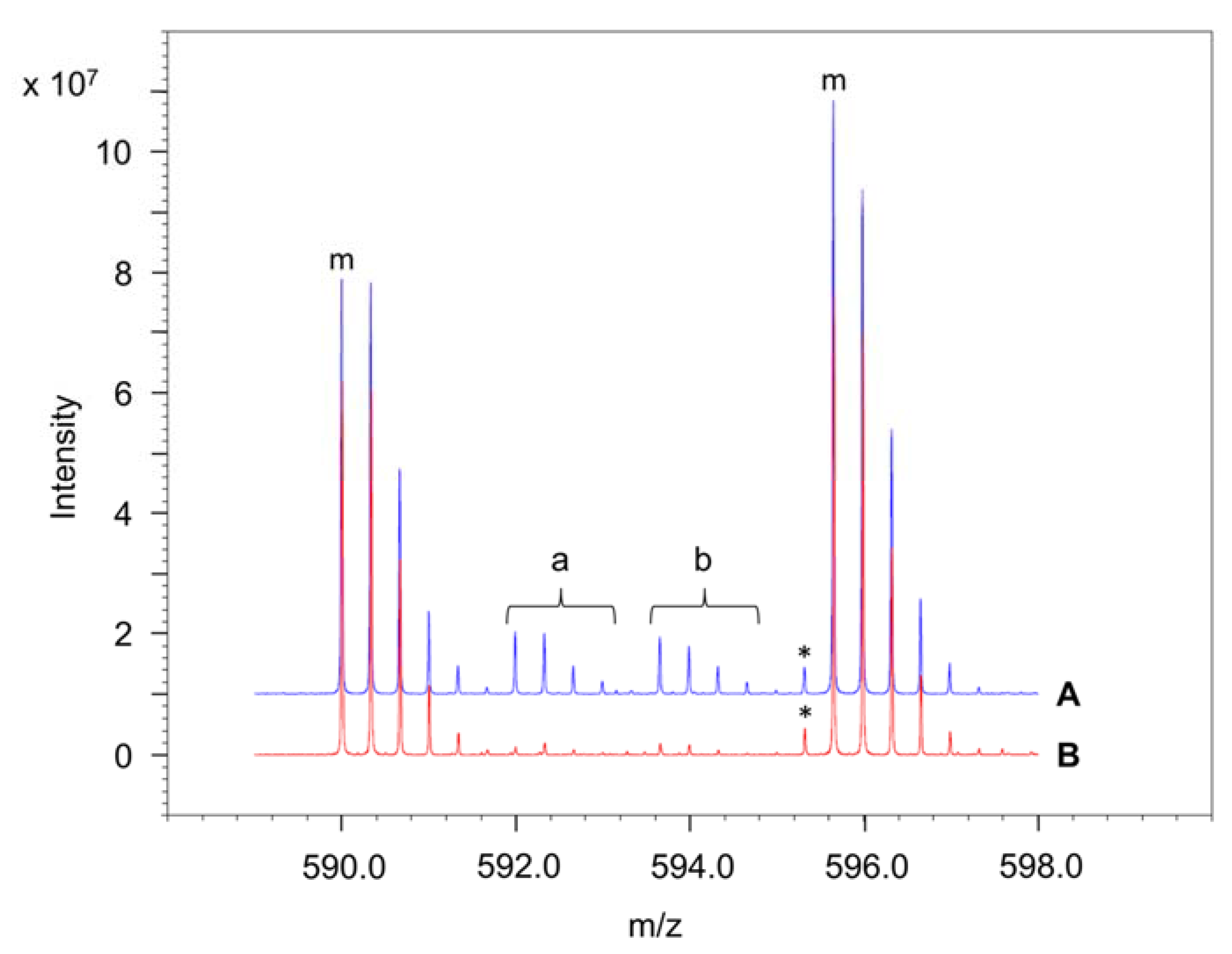
2.2. Cross-Links are Identified at a Low False Discovery Rate
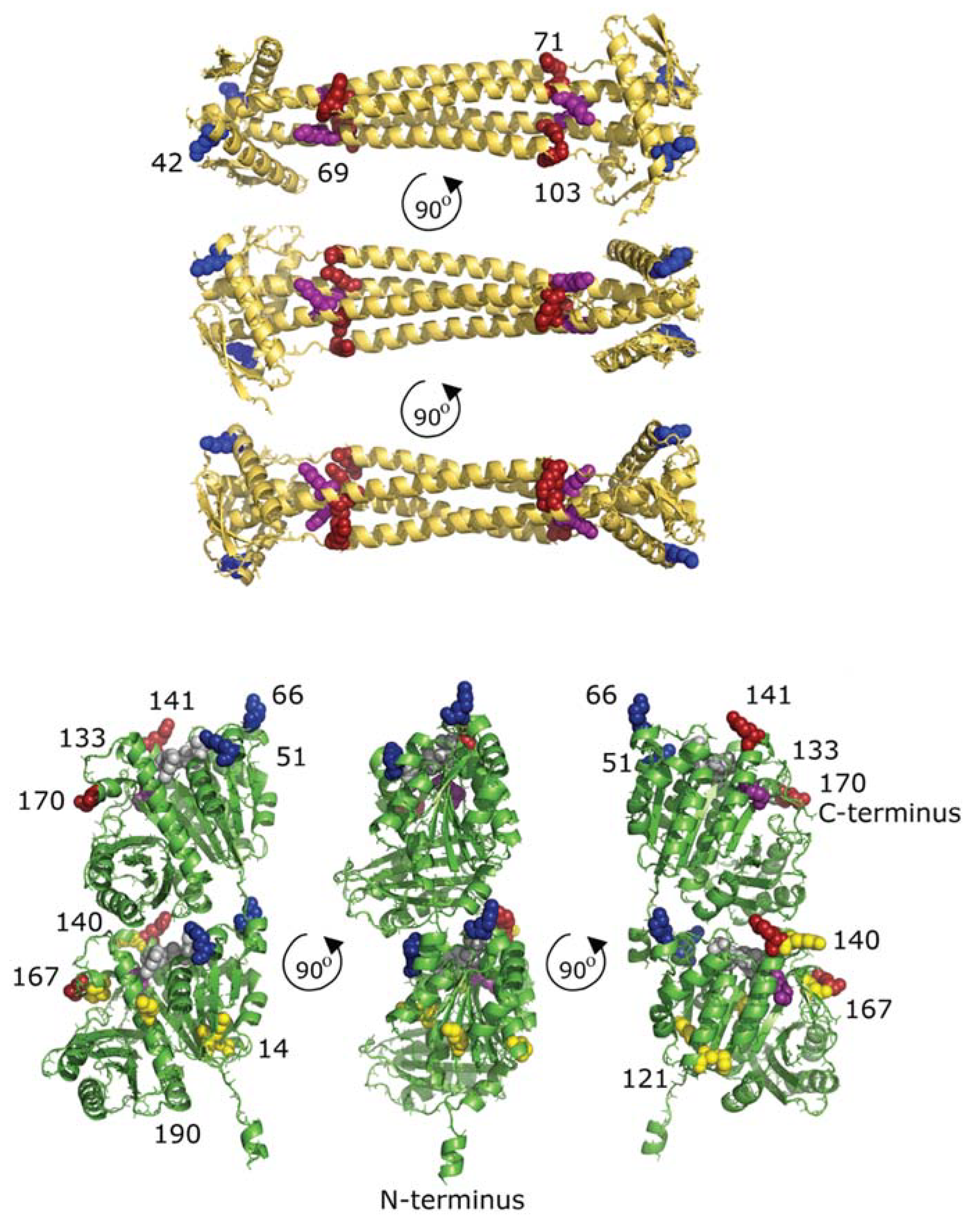
2.3. Cross-Links between FtsZ Molecules
2.4. Interprotein Cross-Links of ZapA
2.5. Cross-Links between ZapA and FtsZ
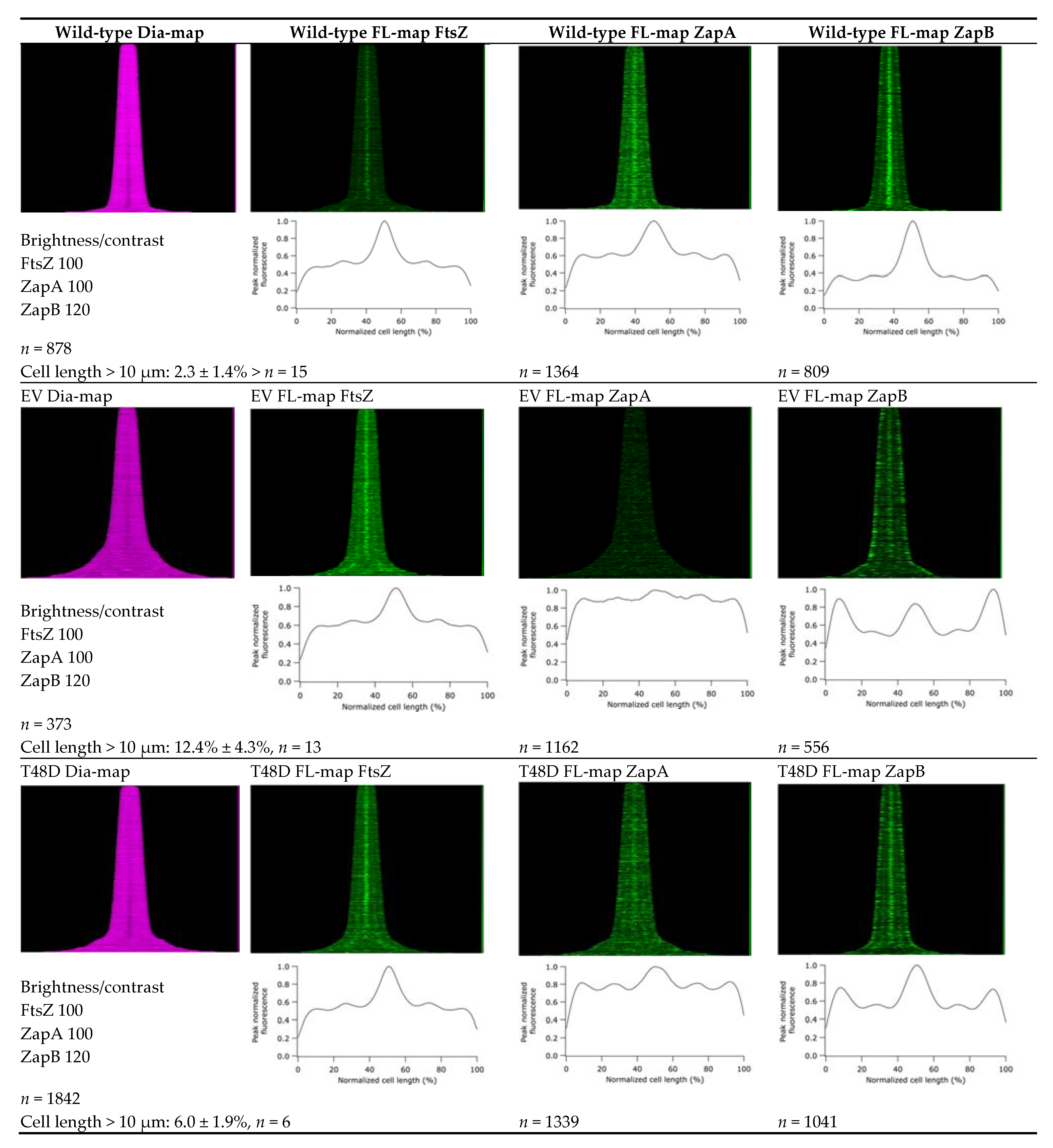
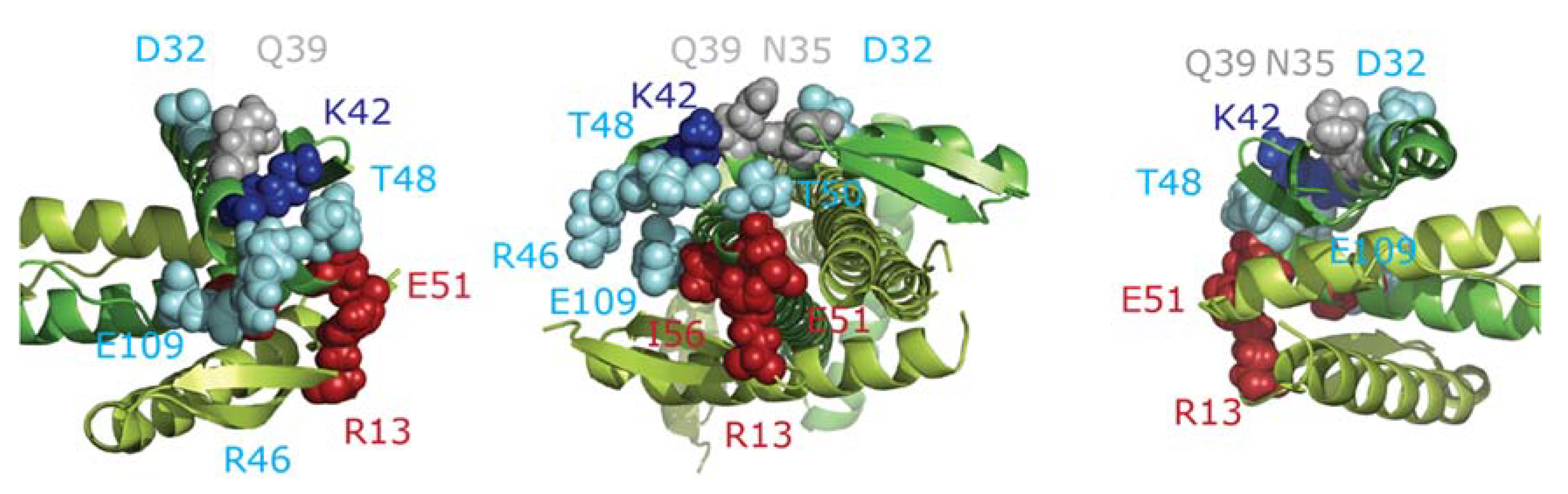
2.6. ZapA Mutants
2.7. Docking of FtsZ and ZapA
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2.Site-Directed Mutagenesis and Plasmid Construction
4.3. Microscopy and Image Analysis
4.4. Cross-Linking
4.5. Isolation of Cross-Linked Peptides
4.6. Determination of the Amounts of Interprotein and Intraprotein Cross-Links Using 14N- and 15N-Labelled Peptides
4.7. Identification of Type 2 Cross-Linked Peptides
4.8. Determination of the False Discovery Rate (FDR)
4.9. Docking
5. Conclusions
- BAMG X-linking yields inter and intracross-links very accurately.
- ZapA keeps FtsZ protofilaments apart as no X-links between two different FtsZ molecules were found in the presence of ZapA. In contrast, under protofilament bundling conditions, i.e., in the presence of Ca2+, a number of cross-links between different FtsZ molecules was found.
- The structurally disordered C-terminal 55 amino acids of FtsZ occupied a limited space and are likely not extended in the absence of other cell division proteins.
- Cross-links confirm the tetrameric structure of ZapA in solution.
- The FtsZ filament binds to the front of the globular domain of ZapA. Both proteins are almost in the same plane at an angle of about 70°. Sufficient ZapA is present in the cells to cross-link most FtsZ protofilaments.
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Den Blaauwen, T.; Hamoen, L.W.; Levin, P.A. The divisome at 25: The road ahead. Curr. Opin. Microbiol. 2017, 36, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Den Blaauwen, T.; Buddelmeijer, N.; Aarsman, M.E.; Hameete, C.M.; Nanninga, N. Timing of FtsZ assembly in Escherichia coli. J. Bacteriol. 1999, 181, 5167–5175. [Google Scholar] [PubMed]
- Van der Ploeg, R.; Verheul, J.; Vischer, N.O.E.; Alexeeva, S.; Hoogendoorn, E.; Postma, M.; Banzhaf, M.; Vollmer, W.; den Blaauwen, T. Colocalization and interaction between elongasome and divisome during a preparative cell division phase in Escherichia coli. Mol. Microbiol. 2013, 87, 1074–1087. [Google Scholar] [CrossRef] [PubMed]
- Den Blaauwen, T.; Aarsman, M.E.G.; Vischer, N.O.E.; Nanninga, N. Penicillin-binding protein PBP2 of Escherichia coli localizes preferentially in the lateral wall and at mid-cell in comparison with the old cell pole. Mol. Microbiol. 2003, 47, 539–547. [Google Scholar] [CrossRef] [PubMed]
- De Pedro, M.A.; Quintela, J.C.; Höltje, J.V.; Schwarz, H. Murein segregation in Escherichia coli. J. Bacteriol. 1997, 179, 2823–2834. [Google Scholar] [CrossRef] [PubMed]
- Pichoff, S.; Lutkenhaus, J. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 2002, 21, 685–693. [Google Scholar] [CrossRef] [PubMed]
- Monahan, L.G.; Robinson, A.; Harry, E.J. Lateral FtsZ association and the assembly of the cytokinetic Z ring in bacteria. Mol. Microbiol. 2009, 74, 1004–1017. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, T.; Ploeger, G.E.J.; Verheul, J.; Comvalius, A.D.; Martos, A.; Alfonso, C.; van Marle, J.; Rivas, G.; den Blaauwen, T. The GTPase activity of Escherichia coli FtsZ determines the magnitude of the FtsZ polymer bundling by ZapA in vitro. Biochemistry 2009, 48, 11056–11066. [Google Scholar] [CrossRef] [PubMed]
- Dajkovic, A.; Pichoff, S.; Lutkenhaus, J.; Wirtz, D. Cross-linking FtsZ polymers into coherent Z rings. Mol. Microbiol. 2010, 78, 651–668. [Google Scholar] [CrossRef] [PubMed]
- Galli, E.; Gerdes, K. Spatial resolution of two bacterial cell division proteins: ZapA recruits ZapB to the inner face of the Z-ring. Mol. Microbiol. 2010, 76, 1514–1526. [Google Scholar] [CrossRef] [PubMed]
- Buss, J.A.; Peters, N.T.; Xiao, J.; Bernhardt, T.G. ZapA and ZapB form an FtsZ-independent structure at midcell. Mol. Microbiol. 2017, 104, 652–663. [Google Scholar] [CrossRef]
- Ebersbach, G.; Galli, E.; Møller-Jensen, J.; Löwe, J.; Gerdes, K. Novel coiled-coil cell division factor ZapB stimulates Z ring assembly and cell division. Mol. Microbiol. 2008, 68, 720–735. [Google Scholar] [CrossRef] [PubMed]
- Buss, J.; Coltharp, C.; Shtengel, G.; Yang, X.; Hess, H.; Xiao, J. A Multi-layered Protein Network Stabilizes the Escherichia coli FtsZ-ring and Modulates Constriction Dynamics. PLoS Genet. 2015, 11, e1005128. [Google Scholar] [CrossRef] [PubMed]
- Espeli, O.; Borne, R.; Dupaigne, P.; Thiel, A.; Gigant, E.; Mercier, R.; Boccard, F. A MatP-divisome interaction coordinates chromosome segregation with cell division in E. coli. EMBO J. 2012, 31, 3198–3211. [Google Scholar] [CrossRef] [PubMed]
- Mercier, R.; Petit, M.-A.; Schbath, S.; Robin, S.; El Karoui, M.; Boccard, F.; Espéli, O. The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cell 2008, 135, 475–485. [Google Scholar] [CrossRef] [PubMed]
- Coltharp, C.; Buss, J.; Plumer, T.M.; Xiao, J. Defining the rate-limiting processes of bacterial cytokinesis. Proc. Natl. Acad. Sci. USA 2016, 113, E1044–E1053. [Google Scholar] [CrossRef] [PubMed]
- Low, H.H.; Moncrieffe, M.C.; Löwe, J. The crystal structure of ZapA and its modulation of FtsZ polymerisation. J. Mol. Biol. 2004, 341, 839–852. [Google Scholar] [CrossRef] [PubMed]
- Roach, E.J.; Kimber, M.S.; Khursigara, C.M. Crystal structure and site-directed mutational analysis reveals key residues involved in Escherichia coli ZapA function. J. Biol. Chem. 2014, 289, 23276–23286. [Google Scholar] [CrossRef] [PubMed]
- Galli, E.; Gerdes, K. FtsZ-ZapA-ZapB Interactome of Escherichia coli. J. Bacteriol. 2012, 194, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Oliva, M.A.; Trambaiolo, D.; Löwe, J. Structural insights into the conformational variability of FtsZ. J. Mol. Biol. 2007, 373, 1229–1242. [Google Scholar] [CrossRef] [PubMed]
- Elsen, N.L.; Lu, J.; Parthasarathy, G.; Reid, J.C.; Sharma, S.; Soisson, S.M.; Lumb, K.J. Mechanism of action of the cell-division inhibitor PC190723: Modulation of FtsZ assembly cooperativity. J. Am. Chem. Soc. 2012, 134, 12342–12345. [Google Scholar] [CrossRef] [PubMed]
- Buske, P.J.; Levin, P.A. A flexible C-terminal linker is required for proper FtsZ assembly in vitro and cytokinetic ring formation in vivo. Mol. Microbiol. 2013, 89, 249–263. [Google Scholar] [CrossRef] [PubMed]
- Sundararajan, K.; Miguel, A.; Desmarais, S.M.; Meier, E.L.; Casey Huang, K.; Goley, E.D. The bacterial tubulin FtsZ requires its intrinsically disordered linker to direct robust cell wall construction. Nat. Commun. 2015, 6, 7281. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, M.A.; Huang, K.-H.; Zeng, W.; Janakiraman, A. Structure of the Z Ring-associated Protein, ZapD, Bound to the C-terminal Domain of the Tubulin-like Protein, FtsZ, Suggests Mechanism of Z Ring Stabilization through FtsZ Cross-linking. J. Biol. Chem. 2017, 292, 3740–3750. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Mukherjee, A.; Lutkenhaus, J. Recruitment of ZipA to the division site by interaction with FtsZ. Mol. Microbiol. 1999, 31, 1853–1861. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Margolin, W. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J. Bacteriol. 1999, 181, 7531–7544. [Google Scholar] [PubMed]
- Haney, S.A.; Glasfeld, E.; Hale, C.; Keeney, D.; He, Z.; de Boer, P. Genetic analysis of the Escherichia coli FtsZ.ZipA interaction in the yeast two-hybrid system. Characterization of FtsZ residues essential for the interactions with ZipA and with FtsA. J. Biol. Chem. 2001, 276, 11980–11987. [Google Scholar] [CrossRef] [PubMed]
- Mosyak, L.; Zhang, Y.; Glasfeld, E.; Haney, S.; Stahl, M.; Seehra, J.; Somers, W.S. The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J. 2000, 19, 3179–3191. [Google Scholar] [CrossRef] [PubMed]
- Pacheco-Gómez, R.; Cheng, X.; Hicks, M.R.; Smith, C.J.I.; Roper, D.I.; Addinall, S.; Rodger, A.; Dafforn, T.R. Tetramerization of ZapA is required for FtsZ bundling. Biochem. J. 2013, 449, 795–802. [Google Scholar] [PubMed]
- Kasper, P.T.; Back, J.W.; Vitale, M.; Hartog, A.F.; Roseboom, W.; de Koning, L.J.; van Maarseveen, J.H.; Muijsers, A.O.; de Koster, C.G.; de Jong, L. An aptly positioned azido group in the spacer of a protein cross-linker for facile mapping of lysines in close proximity. ChemBioChem 2007, 8, 1281–1292. [Google Scholar] [PubMed]
- Buncherd, H.; Nessen, M.A.; Nouse, N.; Stelder, S.K.; Roseboom, W.; Dekker, H.L.; Arents, J.C.; Smeenk, L.E.; Wanner, M.J.; van Maarseveen, J.H.; et al. Selective enrichment and identification of cross-linked peptides to study 3-D structures of protein complexes by mass spectrometry. J. Proteom. 2012, 75, 2205–2215. [Google Scholar] [CrossRef] [PubMed]
- Sossong, T.M.; Brigham-Burke, M.R.; Hensley, P.; Pearce, K.H. Self-Activation of Guanosine Triphosphatase Activity by Oligomerization of the Bacterial Cell Division Protein FtsZ. Biochemistry 1999, 38, 14843–14850. [Google Scholar] [CrossRef] [PubMed]
- González, J.M.; Vélez, M.; Jiménez, M.; Alfonso, C.; Schuck, P.; Mingorance, J.; Vicente, M.; Minton, A.P.; Rivas, G. Cooperative behavior of Escherichia coli cell-division protein FtsZ assembly involves the preferential cyclization of long single-stranded fibrils. Proc. Natl. Acad. Sci. USA 2005, 102, 1895–1900. [Google Scholar] [CrossRef] [PubMed]
- Caplan, M.R.; Erickson, H.P. Apparent cooperative assembly of the bacterial cell division protein FtsZ demonstrated by isothermal titration calorimetry. J. Biol. Chem. 2003, 278, 13784–13788. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Bjornson, K.; Redick, S.D.; Erickson, H.P. A rapid fluorescence assay for FtsZ assembly indicates cooperative assembly with a dimer nucleus. Biophys. J. 2004, 88, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Romberg, L.; Levin, P.A. Assembly dynamics of the bacterial cell division protein FTSZ: Poised at the edge of stability. Annu. Rev. Microbiol. 2003, 57, 125–154. [Google Scholar] [CrossRef] [PubMed]
- Schilling, B.; Row, R.H.; Gibson, B.W.; Guo, X.; Young, M.M. MS2Assign, automated assignment and nomenclature of tandem mass spectra of chemically crosslinked peptides. J. Am. Soc. Mass Spectrom. 2003, 14, 834–850. [Google Scholar] [CrossRef]
- Mendieta, J.; Rico, A.I.; López-Viñas, E.; Vicente, M.; Mingorance, J.; Gómez-Puertas, P. Structural and Functional Model for Ionic (K+/Na+) and pH Dependence of GTPase Activity and Polymerization of FtsZ, the Prokaryotic Ortholog of Tubulin. J. Mol. Biol. 2009, 390, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Buske, P.J.; Mittal, A.; Pappu, R.V.; Levin, P.A. An intrinsically disordered linker plays a critical role in bacterial cell division. Semin. Cell Dev. Biol. 2015, 37, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Gardner, K.A.J.A.; Moore, D.A.; Erickson, H.P. The C-terminal linker of Escherichia coli FtsZ functions as an intrinsically disordered peptide. Mol. Microbiol. 2013, 89, 264–275. [Google Scholar] [CrossRef] [PubMed]
- Scheffers, D.-J.; de Wit, J.G.; den Blaauwen, T.; Driessen, A.J.M. GTP hydrolysis of cell division protein FtsZ: Evidence that the active site is formed by the association of monomers. Biochemistry 2002, 41, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Small, E.; Marrington, R.; Rodger, A.; Scott, D.J.; Sloan, K.; Roper, D.; Dafforn, T.R.; Addinall, S.G. FtsZ polymer-bundling by the Escherichia coli ZapA orthologue, YgfE, involves a conformational change in bound GTP. J. Mol. Biol. 2007, 369, 210–221. [Google Scholar] [CrossRef] [PubMed]
- Van Zundert, G.C.P.; Rodrigues, J.P.G.L.M.; Trellet, M.; Schmitz, C.; Kastritis, P.L.; Karaca, E.; Melquiond, A.S.J.; van Dijk, M.; de Vries, S.J.; Bonvin, A.M.J.J. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, C.; Boelens, R.; Bonvin, A.M.J.J. HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003, 125, 1731–1737. [Google Scholar] [CrossRef] [PubMed]
- Vedyaykin, A.D.; Vishnyakov, I.E.; Polinovskaya, V.S.; Khodorkovskii, M.A.; Sabantsev, A.V. New insights into FtsZ rearrangements during the cell division of Escherichia coli from single-molecule localization microscopy of fixed cells. Microbiologyopen 2016, 5, 378–386. [Google Scholar] [CrossRef] [PubMed]
- Buss, J.; Coltharp, C.; Huang, T.; Pohlmeyer, C.; Wang, S.-C.; Hatem, C.; Xiao, J. In vivo organization of the FtsZ-ring by ZapA and ZapB revealed by quantitative super-resolution microscopy. Mol. Microbiol. 2013, 89, 1099–1120. [Google Scholar] [CrossRef] [PubMed]
- Vischer, N.O.E.; Verheul, J.; Postma, M.; van den Berg van Saparoea, B.; Galli, E.; Natale, P.; Gerdes, K.; Luirink, J.; Vollmer, W.; Vicente, M.; et al. Cell age dependent concentration of Escherichia coli divisome proteins analyzed with ImageJ and ObjectJ. Front. Microbiol. 2015, 6, 586. [Google Scholar] [CrossRef] [PubMed]
- Läppchen, T.; Hartog, A.F.; Pinas, V.A.; Koomen, G.-J.; den Blaauwen, T. GTP analogue inhibits polymerization and GTPase activity of the bacterial protein FtsZ without affecting its eukaryotic homologue tubulin. Biochemistry 2005, 44, 7879–7884. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Buddelmeier, N.; Aarsman, M.E.G.; den Blaauwen, T. Immunolabeling of Proteins in Situ in Escherichia coli K12 Strains. Bio-Protocol 2013, 3, 1–4. [Google Scholar]
- Koppelman, C.-M.; Aarsman, M.E.G.; Postmus, J.; Pas, E.; Muijsers, A.O.; Scheffers, D.-J.; Nanninga, N.; den Blaauwen, T. R174 of Escherichia coli FtsZ is involved in membrane interaction and protofilament bundling, and is essential for cell division. Mol. Microbiol. 2004, 51, 645–657. [Google Scholar] [CrossRef] [PubMed]
- Läppchen, T.; Pinas, V.A.; Hartog, A.F.; Koomen, G.-J.; Schaffner-Barbero, C.; Andreu, J.M.; Trambaiolo, D.; Löwe, J.; Juhem, A.; Popov, A.V.; et al. Probing FtsZ and tubulin with C8-substituted GTP analogs reveals differences in their nucleotide binding sites. Chem. Biol. 2008, 15, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Taverner, T.; Hall, N.E.; O’Hair, R.A.J.; Simpson, R.J. Characterization of an antagonist interleukin-6 dimer by stable isotope labeling, cross-linking, and mass spectrometry. J. Biol. Chem. 2002, 277, 46487–46492. [Google Scholar] [CrossRef] [PubMed]
- Maiolica, A.; Cittaro, D.; Borsotti, D.; Sennels, L.; Ciferri, C.; Tarricone, C.; Musacchio, A.; Rappsilber, J. Structural analysis of multiprotein complexes by cross-linking, mass spectrometry, and database searching. Mol. Cell. Proteom. 2007, 6, 2200–2211. [Google Scholar] [CrossRef] [PubMed]
- Panchaud, A.; Singh, P.; Shaffer, S.A.; Goodlett, D.R. xComb: A cross-linked peptide database approach to protein-protein interaction analysis. J. Proteome Res. 2010, 9, 2508–2515. [Google Scholar] [CrossRef] [PubMed]
- Buncherd, H.; Roseboom, W.; Ghavim, B.; Du, W.; de Koning, L.J.; de Koster, C.G.; de Jong, L. Isolation of cross-linked peptides by diagonal strong cation exchange chromatography for protein complex topology studies by peptide fragment fingerprinting from large sequence databases. J. Chromatogr. A 2014, 1348, 34–46. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, J.P.G.L.M.; Trellet, M.; Schmitz, C.; Kastritis, P.; Karaca, E.; Melquiond, A.S.J.; Bonvin, A.M.J.J. Clustering biomolecular complexes by residue contacts similarity. Proteins 2012, 80, 1810–1817. [Google Scholar] [CrossRef] [PubMed]


| Protein(s) | Linked Residues | A Peptide | B Peptide | (Å) | ZapA, No Ca2+ | No ZapA, Ca2+ | ||
|---|---|---|---|---|---|---|---|---|
| Spectral Counts b | Type | Spectral Counts | Type | |||||
| FtsZ | 141–66 | KR | TAVGQTIQIGSGITKGLGAGANPEVGR | 24.8 | 11 | Intra | 6 | Intra |
| FtsZ | 141–133 | KR | DLGILTVAVVTKPFNFEGK | 12.3 | 2 | Intra | – | – |
| FtsZ-FtsZ | 141–367 | KR | VVNDNAPQTAKEPDYLDIPAFLR | u | 3 | Intra | 4 | Intra |
| FtsZ-FtsZ | 141–380 | KR | KQAD | u | 3 | Intra | 9 | Mix |
| FtsZ-FtsZ | 170–367 | LLKVLGR | VVNDNAPQTAKEPDYLDIPAFLR | u | - | - | 3 | Mix |
| FtsZ-FtsZ | 380–51 | KQAD | KTAVGQTIQIGSGITK | u | 5 | Intra | 13 | Mix |
| FtsZ | 380–66 | KQAD | TAVGQTIQIGSGITKGLGAGANPEVGR | u | 4 | Intra | - | - |
| FtsZ-FtsZ | 380–170 | KQAD | LLKVLGR | u | 3 | Intra | 5 | Mix |
| FtsZ-FtsZ | 380–367 | KQAD | VVNDNAPQTAKEPDYLDIPAFLR | u | 5 | Intra | 10 | Intra |
| ZapA-FtsZ | 42–51 | LQDLKER | KTAVGQTIQIGSGITK | 15.4 a | 5 | Inter | ||
| ZapA-FtsZ | 42–66 | LQDLKER | TAVGQTIQIGSGITKGLGAGANPEVGR | 16.8 a | 4 | Inter | ||
| ZapA-ZapA | 42–42 | LQDLKER | LQDLKER | 26.3 | 2 | Inter | ||
| ZapA-ZapA | 42–103 | LQDLKER | ITEKTNQNFE | 23.9 | 3 | Inter | ||
| ZapA-ZapA | 71–103 | AKTR | ITEKTNQNFE | 8.4 | 8 | Inter | ||
| ZapA-ZapA | 71–69 | AKTR | VTNEQLVFIAALNISYELAQEKAK | 9.0 | 2 | Inter | ||
| ZapA-ZapA | 103–103 | ITEKTNQNFE | ITEKTNQNFE | 22.3 | 1 | Inter |
| ZapA Mutants | Cell Length ± S.E.M. (μm) | n a | Cell Length >10 μm ± S.E.M. (%) | ZapA Fluorescence b |
|---|---|---|---|---|
| TB28 | 4.13 ± 0.05 | (2)4 | 0.1 ± 0.1 | 100 |
| Empty vector | 6.87 ± 0.87 | (4)13 | 12.4 ± 4.3 | 38 |
| Wild-type | 5.02 ± 0.39 | (5)15 | 2.3 ± 1.4 | 100 |
| R13D | 5.70 ± 0.70 | (3)9 | 7.3 ± 3.2 | 95 |
| D32A | 4.81 ± 0.17 | (2)3 | 1.0 ± 0.4 | 92 |
| D32K | 5.11 ± 0.04 | (2)4 | 2.8 ± 1.5 | 106 |
| N35A | 4.54 ± 0.05 | (2)3 | 0.6 ± 0.3 | 79 |
| N35D | 5.05 ± 0.09 | (2)3 | 1.4 ± 1.0 | 68 |
| Q39A | 4.67 ± 1.23 | (2)3 | 0.8 ± 0.3 | 82 |
| Q39E | 4.94 ± 0.12 | (1)3 | 2.4 ± 0.6 | 104 |
| Q39K | 4.96 ± 0.08 | (2)3 | 1.4 ± 0.8 | 88 |
| K42A | 5.22 ± 0.26 | (2)5 | 3.2 ± 1.0 | 87 |
| K42E | 5.20 ± 0.35 | (2)4 | 2.8 ± 1.7 | 98 |
| R46E | 5.85 ± 0.17 | (2)5 | 5.8 ± 0.7 | 115 |
| T48D | 5.86 ± 0.37 | (2)6 | 6.0 ± 1.9 | 67 |
| T48R | 4.60 ± 0.6 | (2)5 | 2.3 ± 1.2 | 70 |
| T50R, E51D | 5.28 ± 0.16 | (2)6 | 3.0 ± 0.7 | 75 |
| E51K | 6.74 ± 0.35 | (3)7 | 10.9 ± 2.9 | 96 |
| I56K | 7.08 ± 0.41 | (2)7 | 12.9 ± 2.3 | 99 |
| E109K | 5.58 ± 0.37 | (3)7 | 4.7 ± 2.3 | 67 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roseboom, W.; Nazir, M.G.; Meiresonne, N.Y.; Mohammadi, T.; Verheul, J.; Buncherd, H.; Bonvin, A.M.J.J.; De Koning, L.J.; De Koster, C.G.; De Jong, L.; et al. Mapping the Contact Sites of the Escherichia coli Division-Initiating Proteins FtsZ and ZapA by BAMG Cross-Linking and Site-Directed Mutagenesis. Int. J. Mol. Sci. 2018, 19, 2928. https://doi.org/10.3390/ijms19102928
Roseboom W, Nazir MG, Meiresonne NY, Mohammadi T, Verheul J, Buncherd H, Bonvin AMJJ, De Koning LJ, De Koster CG, De Jong L, et al. Mapping the Contact Sites of the Escherichia coli Division-Initiating Proteins FtsZ and ZapA by BAMG Cross-Linking and Site-Directed Mutagenesis. International Journal of Molecular Sciences. 2018; 19(10):2928. https://doi.org/10.3390/ijms19102928
Chicago/Turabian StyleRoseboom, Winfried, Madhvi G. Nazir, Nils Y. Meiresonne, Tamimount Mohammadi, Jolanda Verheul, Hansuk Buncherd, Alexandre M. J. J. Bonvin, Leo J. De Koning, Chris G. De Koster, Luitzen De Jong, and et al. 2018. "Mapping the Contact Sites of the Escherichia coli Division-Initiating Proteins FtsZ and ZapA by BAMG Cross-Linking and Site-Directed Mutagenesis" International Journal of Molecular Sciences 19, no. 10: 2928. https://doi.org/10.3390/ijms19102928
APA StyleRoseboom, W., Nazir, M. G., Meiresonne, N. Y., Mohammadi, T., Verheul, J., Buncherd, H., Bonvin, A. M. J. J., De Koning, L. J., De Koster, C. G., De Jong, L., & Den Blaauwen, T. (2018). Mapping the Contact Sites of the Escherichia coli Division-Initiating Proteins FtsZ and ZapA by BAMG Cross-Linking and Site-Directed Mutagenesis. International Journal of Molecular Sciences, 19(10), 2928. https://doi.org/10.3390/ijms19102928







