Membrane-Bound Class III Peroxidases: Unexpected Enzymes with Exciting Functions
Abstract
1. Introduction
2. Membrane-Bound Class III Peroxidases
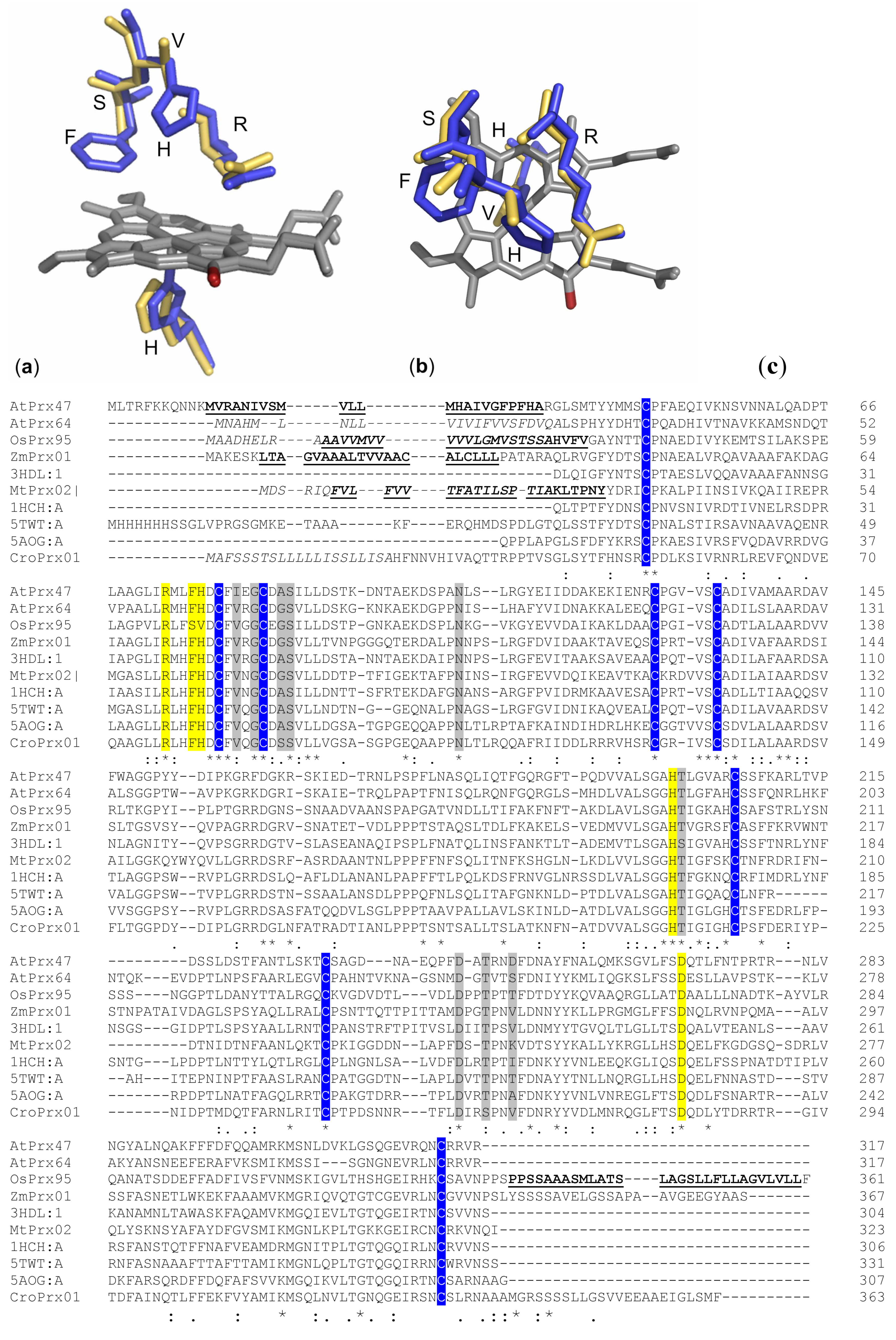
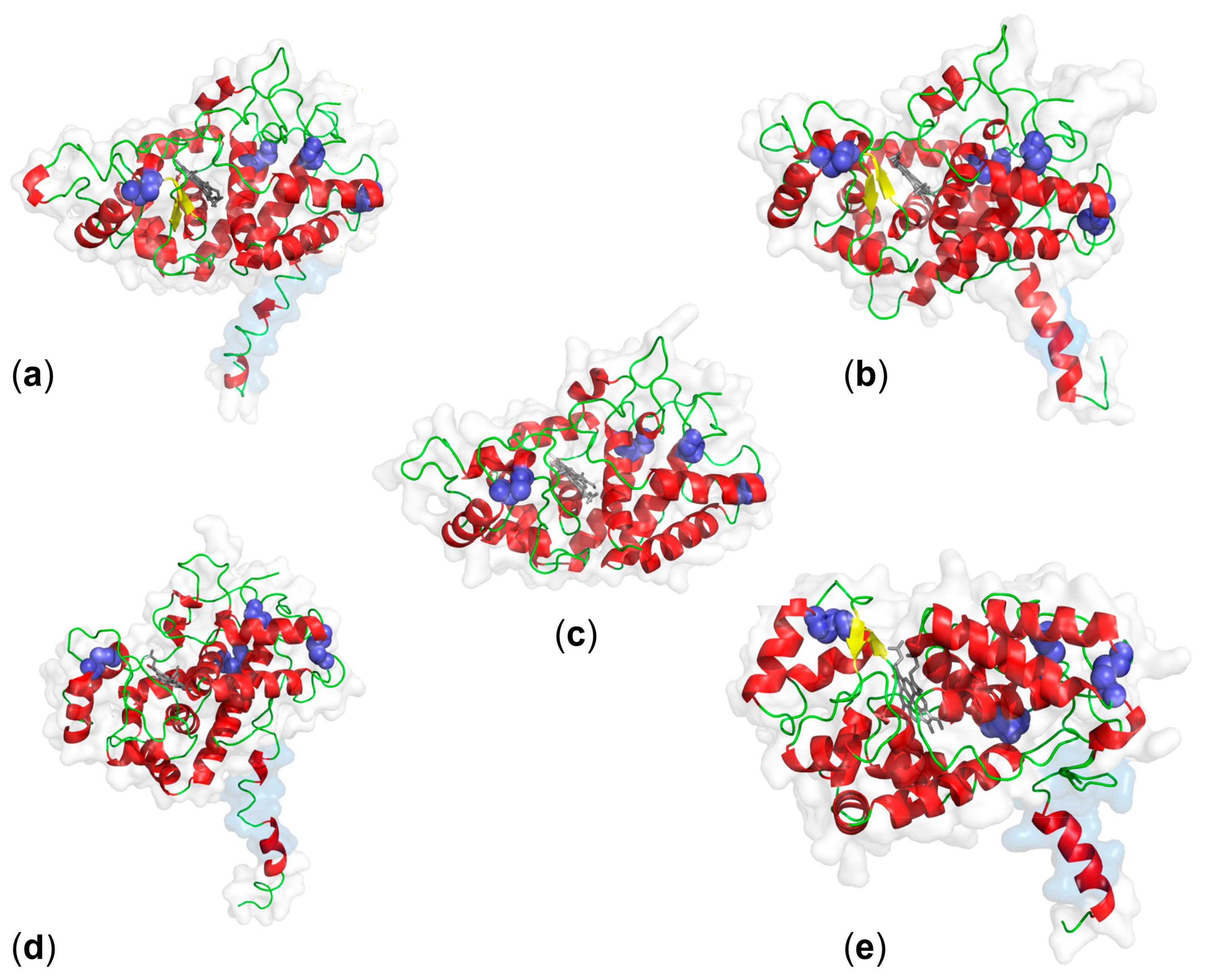
3. Structure
4. Topology and Spatiotemporal Organization
4.1. Transmembrane Spanning Domains
4.2. Protein–Protein Interaction
4.3. Spatiotemporal Organization
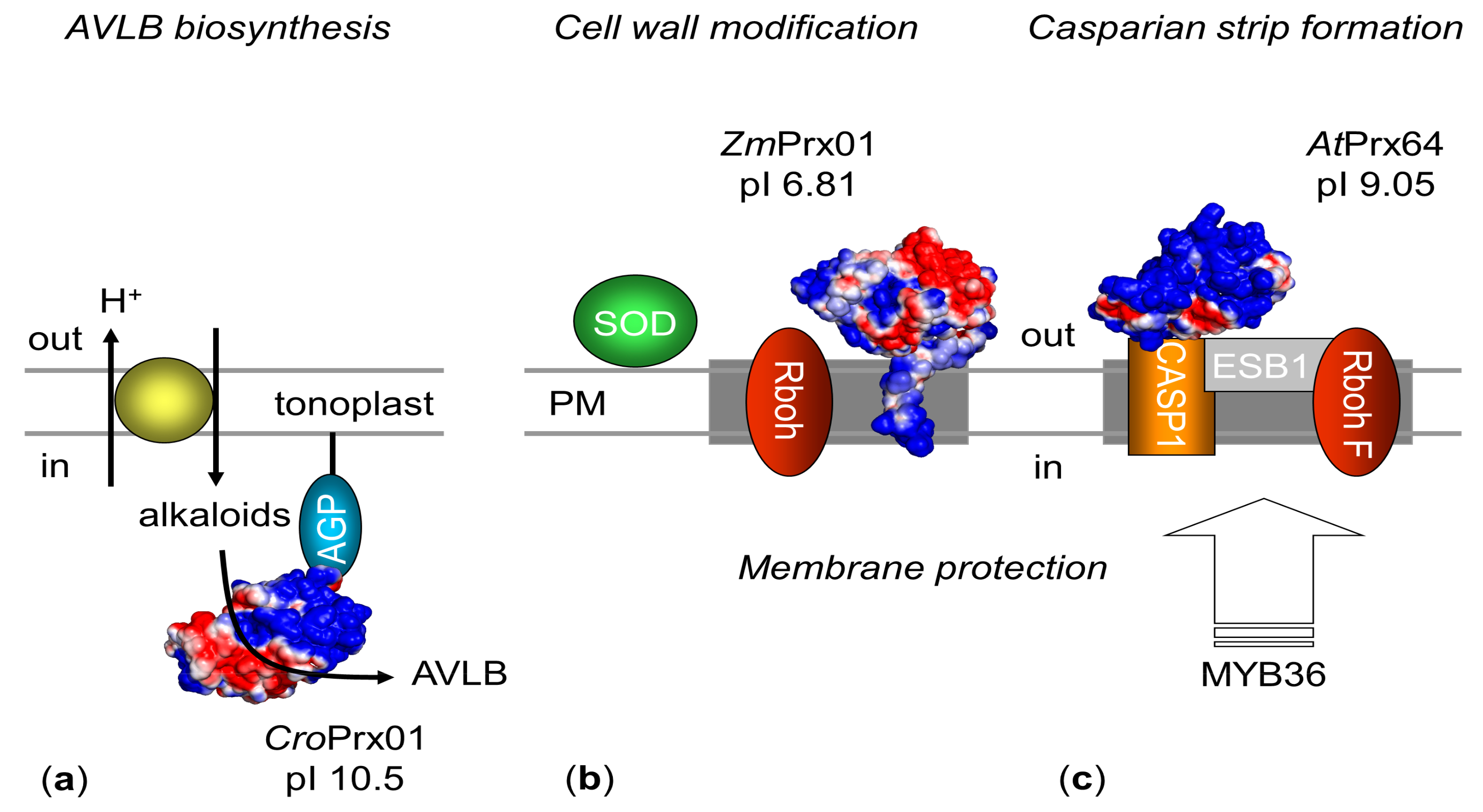
5. Function
5.1. Molecular Function
Substrates
5.2. Biological Functions
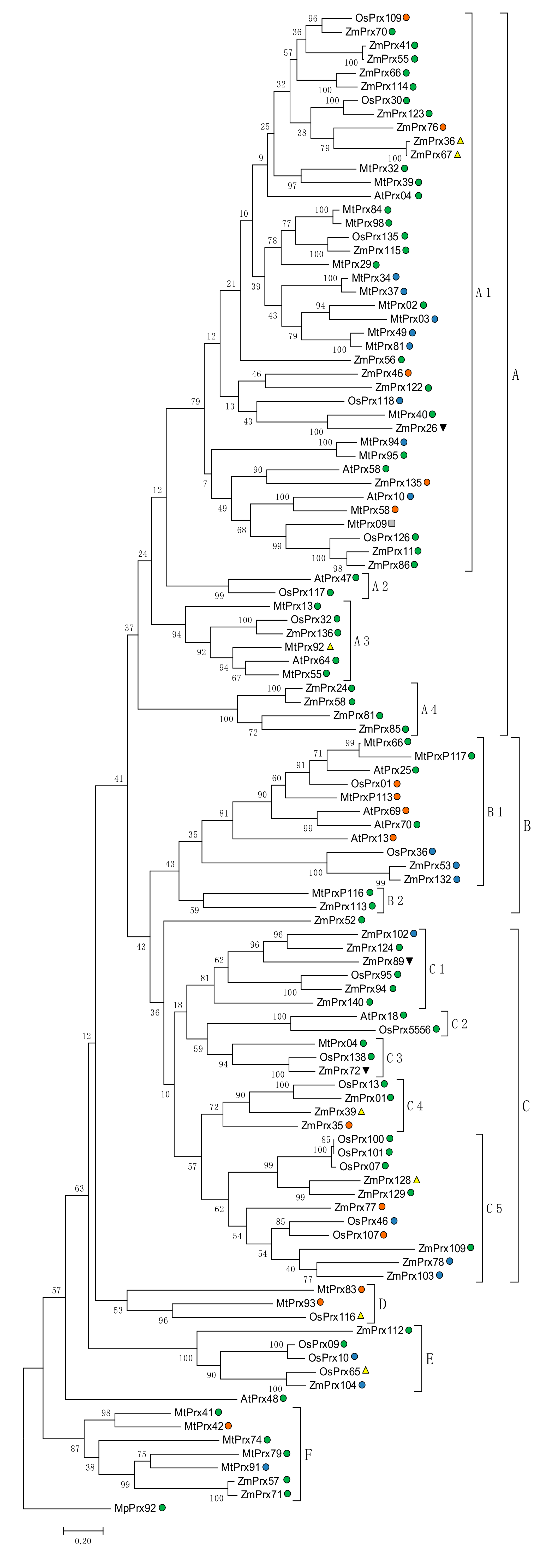
5.2.1. Orthologous
5.2.2. Phylogeny
6. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| AGP | Arabinogalactan protein |
| ARE | Antioxidant-responsive elements |
| AtPrx47 | Arabidopsis thaliana peroxidase 47 |
| AtPrx64 | AtPer64, Arabidopsis thaliana peroxidase 64 |
| AVLB | -3′,4′-anhydrovinblastine |
| BLAST | Basic local alignment search tool |
| BvPrx12 | Beta vulgaris peroxidase 12 |
| CASPs | Casparian strip domain proteins |
| CroPrx01 | CrPrx1; Catharanthus roseus peroxidase 1 |
| DRM | Detergent resistant membrane |
| ER | Endoplasmic reticulum |
| ESB1 | Enhanced Suberin 1 |
| Gaertn. | Gärtner, Joseph |
| GFP | Green fluorescence protein |
| hrCNE | High resolution clear native electrophoresis |
| Heynh. | Heynhold, Gustav, |
| HRP | Horseradish peroxidase |
| IAA | Indol-3-acetic acid |
| l-DOPA | l-3,4-dihydroxyphenylalanine |
| MEGA7 | Molecular Evolutionary Genetics Analysis Version 7.0 |
| B.May. | Bernhard. Meyer |
| MiM | Mitochondrial inner membrane |
| MoM | Mitochondrial outer membrane |
| MS | Mass spectrometry |
| MtPrx02 | Medicago truncatula peroxidase 2 |
| MW | Molecular weight |
| MYB36 | MYB domain protein 36 |
| NADH | Nicotinamide adenine dinucleotide |
| NADPH | Nicotinamide adenine dinucleotide phosphate |
| OsPrx95 | Oryza sativa peroxidase 95 |
| PCA | Pyrrolidone carboxylic acid |
| pI | Point isoelectric |
| PM | Plasma membrane |
| pmPOX2a | Plasma membrane peroxidase 2a |
| PSORT | protein subcellular localization prediction tool |
| PsPrx13 | Pisum sativum peroxidase 13 |
| PyMOL | Python-enhanced molecular graphics tool |
| Rboh | Respiratory burst oxidase homolouge |
| ROS | Reactive oxygen species |
| Scherb. | Scherbius, Johannes |
| SOD | Superoxide dismutase |
| TMH | Transmembrane helix |
| TMHMM | Trans Membrane Helix Markov Model |
| Vac | Vacuole |
| ZmPrx01 | Zea mays peroxidase 1 |
References
- Welinder, K.G. Superfamily of plant, fungal and bacterial peroxidases. Curr. Opin. Struct. Biol. 1992, 2, 388–393. [Google Scholar] [CrossRef]
- Zámocký, M.; Hofbauer, S.; Schaffner, I.; Gasselhuber, B.; Nicolussi, A.; Soudi, M.; Pirker, K.F.; Furtmüller, P.G.; Obinger, C. Independent evolution of four heme peroxidase superfamilies. Arch. Biochem. Biophys. 2015, 574, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Fawal, N.; Li, Q.; Savelli, B.; Brette, M.; Passaia, G.; Fabre, M.; Mathé, C.; Dunand, C. PeroxiBase: A database for large-scale evolutionary analysis of peroxidases. Nucleic Acids Res. 2013, 41, D441–D444. [Google Scholar] [CrossRef] [PubMed]
- Duroux, L.; Welinder, K.G. The peroxidase gene family in plants: A phylogenetic overview. J. Mol. Evol. 2003, 57, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Tognolli, M.; Penel, C.; Greppin, H.; Simon, P. Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene 2002, 288, 129–138. [Google Scholar] [CrossRef]
- Welinder, K.G.; Justesen, A.F.; Kjaersgard, I.V.; Jensen, R.B.; Rasmussen, S.K.; Jespersen, H.M.; Duroux, L. Structural diversity and transcription of class III peroxidases from Arabidopsis thaliana. Eur. J. Biochem. 2002, 269, 6063–6081. [Google Scholar] [CrossRef] [PubMed]
- Passardi, F.; Cosio, C.; Penel, C.; Dunand, C. Peroxidases have more functions than a swiss army knife. Plant Cell Rep. 2005, 24, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Shigeto, J.; Tsutsumi, Y. Diverse functions and reactions of class III peroxidases. New Phytol. 2016, 209, 1395–1402. [Google Scholar] [CrossRef] [PubMed]
- Pandey, V.P.; Awasthi, M.; Singh, S.; Tiwari, S.; Dwivedi, U.N. A comprehensive review on function and application of plant peroxidases. Biochem. Anal. Biochem. 2017, 6. [Google Scholar] [CrossRef]
- Díaz, J.; Pomar, F.; Bernal, Á.; Merino, F. Peroxidases and the metabolism of capsaicin in Capsicum annuum L. Phytochem. Rev. 2004, 3, 141–157. [Google Scholar] [CrossRef]
- Hiraga, S.; Sasaki, K.; Ito, H.; Ohashi, Y.; Matsui, H. A large family of class III plant peroxidases. Plant Cell Physiol. 2001, 42, 462–468. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Q.; Zhao, Y.; Han, G.; Zhu, S. Systematic analysis of maize class III peroxidase gene family reveals a conserved subfamily involved in abiotic stress response. Gene 2015, 566, 95–108. [Google Scholar] [CrossRef] [PubMed]
- De Gara, L. Class III peroxidases and ascorbate metabolism in plants. Phytochem. Rev. 2004, 3, 195–205. [Google Scholar] [CrossRef]
- Lüthje, S.; Meisrimler, C.N.; Hopff, D.; Möller, B. Phylogeny, topology, structure and functions of membrane-bound class III peroxidases in vascular plants. Phytochemistry 2011, 72, 1124–1135. [Google Scholar] [CrossRef] [PubMed]
- Narendra, S.; Venkataramani, S.; Shen, G.; Wang, J.; Pasapula, V.; Lin, Y.; Kornyeyev, D.; Holaday, A.S.; Zhang, H. The arabidopsis ascorbate peroxidase 3 is a peroxisomal membrane-bound antioxidant enzyme and is dispensable for arabidopsis growth and development. J. Exp. Bot. 2006, 57, 3033–3042. [Google Scholar] [CrossRef] [PubMed]
- Laloue, H.; Weber-Lofti, F.; Lucau-Danila, A.; Guillemaut, P. Identification of ascorbate and guaiacol peroxidase in needle chloroplasts of spruce trees. Plant Physiol. Biochem. 1997, 35, 341–346. [Google Scholar]
- Ros Barceló, A.; Ferrer, M.A.; Florenciano, G.E.; Muñoz, R. The tonoplast localization of two basic isoperoxidases of high pI in Lupinus. Bot. Acta 1991, 104, 272–278. [Google Scholar] [CrossRef]
- Andrews, J.; Adams, S.R.; Burton, K.S.; Evered, C.E. Subcellular localization of peroxidase in tomato fruit skin and possible implications for the regulation of fruit growth. J. Exp. Bot. 2002, 53, 2185–2191. [Google Scholar] [CrossRef] [PubMed]
- Andrews, J.; Adams, S.R.; Burton, K.S.; Edmondson, R.N. Partial purification of tomato fruit peroxidase and its effect on the mechanical properties of tomato fruit skin. J. Exp. Bot. 2002, 53, 2393–2399. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Calvino, L.; Faulkner, C.; Walshaw, J.; Saalbach, G.; Bayer, E.; Benitez-Alfonso, Y.; Maule, A. Arabidopsis plasmodesmal proteome. PLoS ONE 2011, 6, e18880. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.M.R.; Hilliou, F.; Duarte, P.; Pereira, L.G.; Almeida, I.; Leech, M.; Memelink, J.; Barcelo, A.R.; Sottomayor, M. Molecular cloning and characterization of a vacuolar class III peroxidases involved in the metabolism of anticancer alkaloids in Catharanthus roseus. Plant Physiol. 2008, 146, 403–417. [Google Scholar] [CrossRef] [PubMed]
- Mika, A.; Buck, F.; Lüthje, S. Membrane-bound class III peroxidases: Identification, biochemical properties and sequence analysis of isoenzymes purified from maize (Zea mays L.) roots. J. Proteom. 2008, 71, 412–424. [Google Scholar] [CrossRef] [PubMed]
- Sottomayor, M.; Ros Barceló, A. Peroxidase from Catharanthus roseus (L.) G. Don and the biosynthesis of a-30-40-anhydrovinlastine: A specific role for a multifunctional enzyme. Protoplasma 2003, 222, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Mika, A.; Lüthje, S. Properties of guaiacol peroxidase activities isolated from corn root plasma membranes. Plant Physiol. 2003, 132, 1489–1498. [Google Scholar] [CrossRef] [PubMed]
- Lüthje, S.; Möller, B.; Perrineau, F.C.; Wöltje, K. Plasma membrane electron pathways and oxidative stress. Antioxid. Redox Signal. 2013, 18, 2163–2183. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Qi, Y.; Zhu, Q.; Chen, X.; Wang, N.; Zhao Chen, H.; Cui, X.; Xu, L.; Zhang, W. New changes in the plasma-membrane-associated proteome of rice roots under salt stress. Proteomics 2009, 9, 3100–3114. [Google Scholar] [CrossRef] [PubMed]
- Meisrimler, C.N.; Planchon, S.; Renaut, J.; Sergeant, K.; Lüthje, S. Alteration of plasma membrane-bound redox systems of iron deficient pea roots by chitosan. J. Proteom. 2011, 74, 1437–1449. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Rubio, M.C.; Alassimone, J.; Geldner, N. A mechanism for localized lignin deposition in the endodermis. Cell 2013, 153, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, T.; Borghi, M.; Wang, P.; Danku, J.M.; Kalmbach, L.; Hosmani, P.S.; Naseer, S.; Fujiwara, T.; Geldner, N.; Salt, D.E. The MYB36 transcription factor orchestrates Casparian strip formation. Proc. Natl. Acad. Sci. USA 2015, 112, 10533–10538. [Google Scholar] [CrossRef] [PubMed]
- Tokunaga, N.; Kaneta, T.; Sato, S.; Sato, Y. Analysis of expression profiles of three peroxidase genes associated with lignification in Arabidopsis thaliana. Physiol. Plantarum 2009, 136, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Budworth, P.; Han, B.; Brown, D.; Chang, H.S.; Zou, G.; Wang, X. Toward elucidating the global gene expression patterns of developing Arabidopsis: Parallel analysis of 8300 genes by a high-density oligonucleotide probe array. Plant Physiol. Biochem. 2001, 39, 221–242. [Google Scholar] [CrossRef]
- Wu, Y.; Yang, Z.; How, J.; Xu, H.; Chen, L.; Li, K. Overexpression of a peroxidase gene (AtPrx64) of Arabidopsis thaliana in tobacco improves plant’s tolerance to aluminum stress. Plant Mol. Biol. 2017, 95, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, B.; Furt, F.; Hartmann, M.A.; Michaelson, L.V.; Carde, J.P.; Sargueil-Boiron, F.; Rossignol, M.; Napier, J.A.; Cullimore, J.; Bessoule, J.J.; et al. Characterization of lipid rafts from Medicago truncatula root plasma membranes: A proteomic study reveals the presence of a raft–associated redox system. Plant Physiol. 2007, 144, 408–418. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Carbonell, E.; Takahashi, D.; Lüthje, S.; González-Reyes, J.A.; Mongrand, S.; Contreras-Moreira, B.; Abadía, A.; Uemura, M.; Abadía, J.; López-Millán, A.F. A shotgun proteomic approach reveals that Fe deficiency causes marked changes in the protein profiles of plasma membrane and detergent-resistant microdomain preparations from Beta vulgaris roots. J. Proteome Res. 2016, 15, 2510–2524. [Google Scholar] [CrossRef] [PubMed]
- Sottomayor, M.; Duarte, P.; Figueiredo, R.; Ros Barceló, A. A vacuolar class III peroxidase and the metabolism of anticancer indole alkaloids in Catharanthus roseus. Plant Signal. Behav. 2008, 3, 899–901. [Google Scholar] [CrossRef] [PubMed]
- Berglund, G.I.; Carlsson, G.H.; Smith, A.T.; Szöke, H.; Henriksen, A.; Hajdu, J. The catalytic pathway of horseradish peroxidase at high resolution. Nature 2003, 417, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Poulos, T.L.; Edwards, S.L.; Wariishi, H.; Gold, M.H. Crystallographic refinement of lignin peroxidase at 2 A. J. Biol. Chem. 1993, 268, 4429–4440. [Google Scholar] [PubMed]
- Newmyer, S.L.; Ortiz de Montellano, P.R. Horseradish peroxidase His-42-->Ala, His-42-->Val, and Phe-41-->Ala mutants. Histidine catalysis and control of substrate access to the heme iron. J. Biol. Chem. 1995, 270, 19430–19438. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, I.J.; Ishikaea, H.; Kim, S.; Massari, A.M.; Fayer, M.D. Substrate binding and protein conformational dynamics measured by 2D-IR vibrational echo spectroscopy. Proc. Natl. Acad. Sci. USA 2007, 104, 2637–2642. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, L.; de Moura, P.R.; Bleicher, L.; Nascimento, A.S.; Zamorano, L.S.; Calvete, J.J.; Sanz, L.; Perez, A.; Bursakov, S.; Roig, M.G.; et al. Crystal structure of highly glycosylated peroxidase from Royal Palm Tree. J. Struct. Biol. 2010, 169, 226–242. [Google Scholar] [CrossRef] [PubMed]
- Moural, T.W.; Lewis, K.M.; Barnaba, C.; Zhu, F.; Palmer, N.A.; Sarath, G.; Scully, E.D.; Jones, J.P.; Sattler, S.E.; Kang, C. Characterization of Class III Pperoxidases from switchgrass. Plant Physiol. 2017, 173, 417–433. [Google Scholar] [CrossRef] [PubMed]
- Nnamchi, C.I.; Parkin, G.; Efimov, I.; Basran, J.; Kwon, H.; Svistunenko, D.A.; Agirre, J.; Okolo, B.N.; Moneke, A.; Nwanguma, B.C.; et al. Structural and spectroscopic characterisation of a heme peroxidase from Sorghum. J. Biol. Inorg. Chem. 2016, 21, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, H.; Krogh, A. Prediction of signal peptides and signal anchors by a hidden Markov model. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998, 6, 122–130. [Google Scholar] [PubMed]
- Blom, N.; Sicheritz-Ponten, T.; Gupta, R.; Gammeltoft, S.; Brunak, S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 2004, 4, 1633–1649. [Google Scholar] [CrossRef] [PubMed]
- Nakai, K.; Kanehisa, M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 1992, 14, 897–911. [Google Scholar] [CrossRef]
- Newman, T.; De Bruijn, F.J.; Green, P.; Keegstra, K.; Kende, H.; McIntosh, L.; Ohlrogge, J.; Raikhel, N.; Somerville, S.; Thomashow, M. Genes galore: A summary of methods for accessing results from large-scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol. 1994, 106, 1241–1255. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Stemberg, M. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Wass, M.N.; Kelley, L.A.; Sternberg, M.J. 3DLigandSite: Predicting ligand-binding sites using similar structures. NAR 2010, 38, 469–473. [Google Scholar] [CrossRef] [PubMed]
- Lüthje, S.; Meisrimler, C.N.; Hopff, D.; Schütze, T.; Köppe, J.; Heino, K. Class III peroxidases. Meth. Mol. Biol. 2014, 1072, 687–706. [Google Scholar] [CrossRef]
- Asada, K.; Miyake, C.; Ogawa, K.; Hossain, M.A. Microcompartmentation of ascorbate peroxidase and regeneration of ascorbate from ascorbate radical: Its dual role in chloroplasts. In Proceedings of the IV. International Symposium on Plant Peroxidases: Biochemistry and Physiology, Vienna, Austria, 6–10 July 1996; Obinger, C., Burner, U., Ebermann, R., Penel, C., Greppin, H., Eds.; pp. 163–167, ISBN 2881640087. [Google Scholar]
- Jespersen, H.M.; Kjærsgard, V.H.; Østergaard, L.; Welinder, K.G. From sequence analysis of three novel ascorbate peroxidases from Arabidopsis thaliana to structure, function and evolution of seven types of ascorbate peroxidase. Biochem. J. 1997, 326, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Kieselbach, T.; Bystedt, M.; Hynds, P.; Robinson, C.; Schroder, W.P. A peroxidase homologue and novel plastocyanin located by proteomics to the Arabidopsis chloroplast thylakoid lumen. FEBS Lett. 2000, 480, 271–276. [Google Scholar] [CrossRef]
- Yabuta, Y.; Motoki, T.; Yoshimura, K.; Takeda, T.; Ishikawa, T.; Shigeoka, S. Thylakoid membrane-bound ascorbate peroxidase is a limiting factor of antioxidative systems under photo-oxidative stress. Plant J. 2002, 32, 915–925. [Google Scholar] [CrossRef] [PubMed]
- Bunkelmann, J.; Trelease, R.N. Ascorbate peroxidase. A prominent membrane protein in oilseed glyoxysomes. Plant Physiol. 1996, 110, 589–598. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, T.; Yoshimura, K.; Sakai, K.; Tamoi, M.; Takeda, T.; Shigeoka, S. Molecular characterization and physiological role of a glyoxysome-bound ascorbate peroxidase from spinach. Plant Cell Physiol. 1998, 39, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Nito, K.; Yamaguchi, K.; Kondo, M.; Hayashi, M.; Nishimura, M. Pumpkin peroxisomal ascorbate peroxidase is localized on peroxisomal membranes and unknown membranous structures. Plant Cell Physiol. 2001, 42, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, K.; Mori, H.; Nishimura, M. A novel isoenzyme of ascorbate peroxidase localized on glyoxysomal and leaf peroxisomal membranes in pumpkin. Plant Cell Physiol. 1995, 36, 1157–1162. [Google Scholar] [CrossRef] [PubMed]
- Lüthje, S.; Hopff, D.; Schmitt, A.; Meisrimler, C.N.; Menckhoff, L. Hunting for low abundant redox proteins in plant plasma membranes. J. Proteom. 2009, 72, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Welinder, K.G.; Larsen, Y.B. Covalent structure of soybean seed coat peroxidase. Biochim. Biophys. Acta 2004, 1698, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Sigrist, C.J.A.; de Castro, E.; Cerutti, L.; Cuche, B.A.; Hulo, N.; Bridge, A.; Bougueleret, L.; Xenarios. New and continuing developments at PROSITE. Nucleic Acids Res. 2012, 21, D344–D347. [Google Scholar] [CrossRef]
- Mika, A.; Boenisch, M.J.; Hopff, D.; Lüthje, S. Membrane-bound guaiacol peroxidases are regulated by methyl jasmonate, salicylic acid, and pathogen elicitors. J. Exp. Bot. 2010, 61, 831–841. [Google Scholar] [CrossRef] [PubMed]
- Mika, A.; (University of Hamburg, Hamburg, Germany); Lüthje, S.; (University of Hamburg, Hamburg, Germany). Personal communication, 2007.
- Morel, J.; Claverol, S.; Mongrand, S.; Furt, F.; Fromentin, J.; Bessoule, J.J.; Blein, J.P.; Simon-Plas, F. Proteomics of plant detergent-resistant membranes. Mol. Cell. Proteom. 2006, 5, 1396–1411. [Google Scholar] [CrossRef] [PubMed]
- Simon-Plas, F.; Perraki, A.; Bayer, E.; Gerbeau-Pissot, P.; Mongrand, S. An update on plant membrane rafts. Curr. Opin. Plant Biol. 2011, 14, 642–649. [Google Scholar] [CrossRef] [PubMed]
- Showalter, A.M. Arabinogalactan-proteins: Structure, expression and function. Cell Mol. Life Sci. 2001, 58, 1399–1417. [Google Scholar] [CrossRef] [PubMed]
- Wink, M. The plant vacuole: A multifunctional compartment. J. Exp. Bot. 1993, 44, 231–246. [Google Scholar]
- Carqueijeiro, I.; Noronha, H.; Duarte, P.; Gerós, H.; Sottomayor, M. Vacuolar transport of the medicinal alkaloids from Catharanthus roseus is mediated by a proton-driven antiport. Plant Physiol. 2013, 162, 1486–1496. [Google Scholar] [CrossRef] [PubMed]
- Lüthje, S. Plasma membrane redox systems: Lipid rafts and protein assemblies. Prog. Bot. 2008, 69, 169–200. [Google Scholar] [CrossRef]
- Roppolo, D.; De Rybel, B.; Tendon, V.D.; Pfister, A.; Alassimone, J.; Vermeer, J.E.; Yamazaki, M.; Stierhof, Y.D.; Beeckman, T.; Geldner, N. A novel protein family mediates Casparian strip formation in the endodermis. Nature 2011, 473, 380–383. [Google Scholar] [CrossRef] [PubMed]
- Halpin, C. Cell Biology: Up Against the Wall. Curr. Biol. 2013, 23, R1050. [Google Scholar] [CrossRef] [PubMed]
- Christensen, J.H.; Bauw, G.; van Montagu, M.; Boerjan, W. Towards the identification of lignin specific peroxidases in poplar. In Proceedings of the IV. International Symposium on Plant Peroxidases: Biochemistry and Physiology, Vienna, Austria, 6–10 July 1996; Obinger, C., Burner, U., Ebermann, R., Penel, C., Greppin, H., Eds.; pp. 113–117, ISBN 2881640087. [Google Scholar]
- Cosio, C.; Dunand, C. Specific functions of individual class III peroxidase genes. J. Exp. Bot. 2009, 60, 391–409. [Google Scholar] [CrossRef] [PubMed]
- Mika, A.; Minibayeva, F.; Beckett, R.; Lüthje, S. Possible functions of extracellular peroxidases in stress-induced generation and detoxification of active oxygen species. Phytochem. Rev. 2004, 3, 173–193. [Google Scholar] [CrossRef]
- Sies, H. Hydrogen peroxide as a central redox signaling molecule in physiological oxidative stress: Oxidative eustress. Redox Biol. 2017, 11, 613–619. [Google Scholar] [CrossRef] [PubMed]
- Noctor, G.; Reichheld, J.P.; Foyer, C.H. ROS-related redox regulation and signaling in plants. Semin. Cell. Dev. Biol. 2018, 80, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Schraudner, M.; Langebartels, C.; Sandermann, H., Jr. Plant defence systems and ozone. Biochem. Soc. Trans. 1996, 24, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Bolwell, G.P.; Bindschedler, L.V.; Blee, K.A.; Butt, V.S.; Davies, D.R.; Gardner, S.L.; Gerrish, C.; Minbayeva, F. The apoplastic oxidative burst in response to biotic stress in plants: A three-component system. J. Exp. Bot. 1996, 53, 1367–1376. [Google Scholar] [CrossRef]
- Schützendübel, A.; Polle, A. Plant responses to abiotic stresses: Heavy metal-induced oxidative stress and protection by mycorrhization. J. Exp. Bot. 2002, 53, 1351–1365. [Google Scholar] [CrossRef] [PubMed]
- Minibayeva, F.; Lüthje, S.; Kolesnikov, O.; Chasov, A.; Beckett, R.P.; Vylegzhanina, N.; Buck, F.; Böttger, M. Wound-induced apoplastic peroxidase activities: Their roles in the production and detoxification of reactive oxygen species. Plant Cell Environ. 2009, 32, 497–508. [Google Scholar] [CrossRef] [PubMed]
- Cakmak, I.; van de Wetering, D.A.M.; Marschner, H.; Bienfait, H.F. Involvement of superoxide radical in extracellular ferric reduction by iron-deficient bean roots. Plant Physiol. 1987, 85, 310–314. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Q.S.; Liang, H.G. Lipid peroxidation caused by the redox system of plasma membranes from wheat roots. J. Plant Physiol. 1995, 145, 261–265. [Google Scholar] [CrossRef]
- Rawyler, A.; Arpagaus, S.; Braendle, R. Impact of oxygen stress and energy availability on membrane stability of plant cells. Ann. Bot. 2002, 90, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Marjamaa, K.; Kukkola, E.M.; Fagerstedt, K. The role of xylem class III peroxidases in lignification. J. Exp. Bot. 2009, 60, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Vanholme, R.; Demedts, B.; Morreel, K.; Ralph, J.; Boerjan, W. Lignin biosynthesis and structure. Plant Physiol. 2010, 153, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin biosynthesis. Ann. Rev. Plant Biol. 2003, 54, 519–546. [Google Scholar] [CrossRef] [PubMed]
- Novo-Uzal, E.; Fernández-Pérez, F.; Herrero, J.; Gutiérrez, J.; Gómez-Ros, L.V.; Bernal, M.Á.; Díaz, J.; Cuello, J.; Pomar, F.; Pedreño, M.A. From Zinnia to Arabidopsis: Approaching the involvement of peroxidases in lignification. J. Exp. Bot. 2013, 64, 3499–3518. [Google Scholar] [CrossRef] [PubMed]
- Vishwanath, S.J.; Delude, C.; Domergue, F.; Rowland, O. Suberin: Biosynthesis, regulation, and polymer assembly of a protective extracellular barrier. Plant Cell Rep. 2015, 34, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Takahama, U. Hydrogen peroxide scavenging systems in vacuoles of mesophyll cells of Vicia faba. Phytochemistry 1992, 31, 1127–1133. [Google Scholar] [CrossRef]
- Soares, A.R.; Marchiosi, R.; Siqueira-Soares Rde, C.; Barbosa de Lima, R.; Dantas dos Santos, W.; Ferrarese-Filho, O. The role of l-DOPA in plants. Plant Signal. Behav. 2014, 9, e28275. [Google Scholar] [CrossRef] [PubMed]
- Kawano, T. Roles of the reactive oxygen species-generating peroxidase reactions in plant defense and growth induction. Plant Cell Rep. 2003, 21, 829–837. [Google Scholar] [CrossRef] [PubMed]
- Ranocha, P.; Dima, O.; Nagy, R.; Felten, J.; Corratgé-Faillie, C.; Novák, O.; Morreel, K.; Lacombe, B.; Martinez, Y.; Pfrunder, S.; et al. Arabidopsis WAT1 is a vacuolar auxin transport facilitator required for auxin homoeostasis. Nat. Commun. 2013, 4, 2625. [Google Scholar] [CrossRef] [PubMed]
- Journet, E.P. MtPrx02, Unpublished, PeroxiBase.
- Takahama, U.; Oniki, T. Effects of ascorbate on oxidation of hydroxycinnamic acid derivatives and the mechanism of oxidation of sinapic acid by cell wall-bound peroxidases. Plant Cell Phys. 1994, 35, 593–600. [Google Scholar] [CrossRef]
- Pomar, F.; Merino, F.; Ros Barceló, A. O-4-linked coniferyl and sinapyl aldehyde in lignifying cell walls are the main targets of the Wiesner (phloroglucinol-HCl) reaction. Protoplasma 2002, 220, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Lamb, C.; Dixon, R.A. The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 251–275. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.T.; Wu, P.; Wang, H.; Zhou, Q. Response of soybean seed germination to cadmium and acid rain. Biol. Trace Elem. Res. 2011, 144, 1186–1196. [Google Scholar] [CrossRef] [PubMed]
- Steffens, B. The role of ethylene and ROS in salinity, heavy metal, and flooding responses in rice. Front. Plant Sci. 2014, 5, 685. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, X.L.; Zhang, R.X.; Yuan, H.Y.; Wang, M.M.; Yang, H.Y.; Ma, H.Y.; Liu, D.; Jiang, C.J.; Liang, Z.W. Root damage under alkaline stress is associated with reactive oxygen species accumulation in rice (Oryza sativa L.). Front. Plant Sci. 2017, 8, 1580. [Google Scholar] [CrossRef] [PubMed]
- Sergio, L.; Cardinali, A.; De Paola, A.; Di Venere, D. Biochemical properties of soluble and bound peroxidases from artichoke heads and leaves. Food Technol. Biotechnol. 2009, 47, 32–38. [Google Scholar]
- Martínez-Cortés, T.; Pomar, F.; Espiñeira, J.M.; Merino, F.; Novo-Uzal, E. Purification and kinetic characterization of two peroxidases of Selaginella martensii Spring. involved in lignification. Plant Physiol. Biochem. 2012, 52, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.K.; Zhu, D.F.; Zhang, Y.P.; Chen, H.Z.; Xiang, J.; Lin, X.Q. Low pH-Induced changes of antioxidant enzyme and ATPase activities in the roots of rice (Oryza sativa L.) seedlings. PLoS ONE 2015, 10, e0116971. [Google Scholar] [CrossRef] [PubMed]
- Haslekås, C.; Grini, P.E.; Nordgard, S.H.; Thorstensen, T.; Viken, M.K.; Nygaard, V. ABI3 mediates expression of the peroxiredoxin antioxidant AtPER1 gene and induction by oxidative stress. Plant Mol. Biol. 2003, 53, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Ferreres, F.; Figueiredo, R.; Bettencourt, S.; Carqueijeiro, I.; Oliveira, J.; Gil-Izquierdo, A.; Pereira, D.M.; Valentao, P.; Andrade, P.B.; Duarte, P.; et al. Identification of phenolic compounds in isolated vacuoles of the medicinal plant Catharanthus roseus and their interaction with vacuolar class III peroxidase: An H2O2 affair? J. Exp. Bot. 2011, 62, 2841–2854. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Jaggi, M.; Taneja, J.; Sinha, A.K. Cloning and characterization of two new Class III peroxidase genes from Catharanthus roseus. Plant Physiol. Biochem. 2011, 49, 404–412. [Google Scholar] [CrossRef] [PubMed]
- Baum, S.F.; Tran, P.N.; Silk, W.K. Effects of salinity on xylem structure and water use in growing leaves of sorghum. New Phytol. 2000, 146, 119–127. [Google Scholar] [CrossRef]
- Karahara, I.; Ikeda, A.; Kodo, T.; Uetake, Y. Development of the Casparian strip in primary roots of maize under salt stress. Planta 2004, 219, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Downie, J.A. Legume nodulation. Curr. Biol. 2014, 24, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Ngoune Tandzi, L.; Shelton Mutengwa, C.; Mangaptche Ngonkeu, E.L.; Gracen, V. Breeding maize for tolerance to acidic soils: A review. Agronomy 2018, 8, 84. [Google Scholar] [CrossRef]
- Sekhon, R.S.; Lin, H.; Childs, K.L.; Hansey, C.N.; Buell, C.R.; de Leon, N.; Kaeppler, S.M. Genome-wide atlas of transcription during maize development. Plant J. 2011, 66, 553–563. [Google Scholar] [CrossRef] [PubMed]
| Species | Acc. No. 1 | Protein 2 | MW 3 | pI 3 | SignalP 4 | N-Glyco 5 | NetPhos6 | PCA 7 | PSORT 8 | Response to |
|---|---|---|---|---|---|---|---|---|---|---|
| A. thaliana | Q9LE15 | AtPrx04 | 34.28 | 7.87 | 1-19 | 1 | 39 | + | PM | senescence |
| Q9FX85 | AtPrx10 | 38.03 | 6.17 | --- | 2 | 29 | --- | Vac | hormones and stress | |
| O49293 | AtPrx13 | 36.88 | 5.15 | --- | 1 | 23 | + | ER | --- | |
| Q9SK52 | AtPrx18 | 35.50 | 5.03 | 1-29 | 2 | 41 | --- | PM | floral organ development | |
| O80822 | AtPrx25 | 37.45 | 8.48 | --- | 2 | 38 | --- | PM | etiolation and drought | |
| Q9SZB9 | AtPrx47 MS | 35.97 | 8.57 | --- | 1 | 30 | --- | PM | lignification of vessels | |
| O81755 | AtPrx48 | 44.79 | 4.99 | --- | 2 | 42 | --- | PM | --- | |
| P59120 | AtPrx58 | 35.29 | 4.92 | 1-23 | 2 | 30 | + | PM | --- | |
| Q43872 | AtPrx64 MS | 34.71 | 9.05 | 1-22 | 3 | 30 | --- | PM | etiolation and drought | |
| Q96511 | AtPrx69 | 35.68 | 9.47 | 1-23 | 3 | 29 | --- | ER | etiolation and cold stress | |
| Q9FMI7 | AtPrx70 | 36.00 | 6.97 | 1-23 | 2 | 25 | --- | PM | --- | |
| M. truncatula | G7KFK6 | MtPrx02 MS | 36.02 | 9.20 | 1-23 | 1 | 27 | --- | PM | nitrogen starvation, wounding and pathogen |
| A0A072UYJ4 | MtPrx03 | 35.99 | 5.35 | 1-25 | 3 | 28 | --- | Vac | rhizobium inoculation | |
| G8A179 | MtPrx04 | 35.48 | 8.32 | 1-22 | 5 | 34 | --- | PM | nitrogen starvation and pathogen | |
| A0A072URQ9 | MtPrx09 | 35.92 | 6.51 | 1-25 | 6 | 41 | + | Golgi | phosphate starvation and pathogen | |
| G7IC23 | MtPrx13 | 35.26 | 8.01 | 1-25 | 2 | 22 | --- | PM | nodulation and pathogen | |
| G7KFM2 | MtPrx29 | 35.79 | 9.41 | 1-28 | 1 | 27 | + | PM | drought, wounding, nitrogen starvation, phosphate starvation, pathogen, methyl jasmonate and ultraviolet (UV) irradiation | |
| A0A072UJD7 | MtPrx32 | 36.88 | 8.62 | 1-23 | 1 | 35 | --- | PM | nitrogen starvation and pathogen | |
| Q1RXM7 | MtPrx34 | 34.38 | 9.11 | 1-22 | 3 | 32 | --- | Vac | drought, insect damage and pathogen | |
| I3S041 | MtPrx37 | 34.90 | 7.15 | 1-22 | 1 | 30 | --- | Vac | drought, insect damage, phosphate starvation and pathogen | |
| G7IF04 | MtPrx39 | 36.19 | 6.5 | 1-25 | 5 | 31 | --- | PM | pathogen | |
| A0A072UQ08 | MtPrx40 | 37.48 | 4.73 | 1-28 | 2 | 33 | + | PM | pathogen | |
| A0A072TWY1 | MtPrx41 | 34.60 | 5.03 | 1-23 | 1 | 30 | + | PM | methyl jasmonate, nodulation and pathogen | |
| Q1SC11 | MtPrx42 | 36.08 | 9.09 | 1-25 | 1 | 27 | + | ER | phosphate starvation and pathogen | |
| A0A072UHR9 | MtPrx49 | 35.30 | 9.72 | 1-25 | --- | 31 | --- | Vac | nodulation and pathogen | |
| M. truncatula | G7I8C1 | MtPrx55 | 34.47 | 9.45 | 1-23 | 2 | 33 | --- | PM | nodulation and pathogen |
| G7K822 | MtPrx58 | 36.06 | 6.87 | 1-25 | 2 | 31 | + | ER | nodulation and pathogen | |
| G7JCW8 | MtPrx66 | 34.64 | 8.21 | 1-19 | 1 | 27 | + | PM | nitrogen starvation and pathogen | |
| A0A072UYL4 | MtPrx74 | 38.49 | 9.28 | 1-29 | 2 | 28 | + | PM | nodulation and pathogen | |
| I3T3G6 | MtPrx79 | 36.97 | 8.82 | --- | 2 | 48 | --- | PM | nodulation * | |
| A0A072UIE4 | MtPrx81 | 35.06 | 8.14 | 1-25 | --- | 30 | --- | Vac | pathogen | |
| G7JZ11 | MtPrx83 | 41.78 | 5.78 | 1-23 | --- | 29 | --- | ER | --- | |
| G7J9S0 | MtPrx84 | 33.82 | 9.17 | 1-26 | 2 | 44 | + | PM | --- | |
| Q1SAT8 | MtPrx91 | 36.52 | 7.22 | 1-21 | 3 | 35 | --- | Vac | --- | |
| G7LB60 | MtPrx92 | 34.76 | 8.81 | 1-25 | 1 | 40 | --- | MoM | nodulation and pathogen | |
| G7JJA7 | MtPrx93 | 35.84 | 5.79 | 1-23 | 2 | 36 | + | ER | --- | |
| G7JKU3 | MtPrx94 | 46.33 | 9.21 | --- | 3 | 49 | --- | Vac | methyl jasmonate | |
| G7JKV1 | MtPrx95 | 38.06 | 6.14 | --- | 5 | 36 | --- | PM | methyl jasmonate * | |
| G7J9S2 | MtPrx98 | 33.99 | 8.06 | 1-26 | 3 | 48 | + | PM | --- | |
| G7INU9 | MtPrx[P]113 | 12.74 | 5.56 | 1-24 | --- | 10 | --- | ER | cadmium treatment, nodulation, nematode | |
| A2Q4B7 | MtPrx[P]116 | 33.77 | 7.98 | 1-14 | 1 | 23 | --- | PM | infection and pathogen * | |
| G7JCW9 | MtPrx[P]117 | 17.60 | 6.23 | 1-19 | --- | 13 | --- | PM | nitrogen and phosphate starvation and pathogen * | |
| O. sativa | Q5VR15 | OsPrx01 | 34.78 | 6.88 | 1-24 | 1 | 30 | --- | ER | --- |
| Q5U1T6 | OsPrx07 | 31.93 | 5.32 | 1-20 | 1 | 27 | --- | PM | --- | |
| Q5U1T4 | OsPrx09 | 35.50 | 5.67 | 1-11 | 5 | 31 | --- | PM | etiolation and drought * | |
| Q5U1T3 | OsPrx10 | 36.20 | 5.38 | --- | 5 | 34 | --- | Vac | --- | |
| Q5U1T0 | OsPrx13 | 36.18 | 5.16 | 1-21 | 8 | 18 | --- | PM | --- | |
| Q6ER51 | OsPrx30 | 34.43 | 7.52 | 1-26 | 1 | 33 | + | PM | --- | |
| Q5U1R1 | OsPrx32 | 34.06 | 8.42 | 1-30 | 2 | 30 | --- | PM | pathogen * | |
| Q5U1Q7 | OsPrx36 | 50.76 | 4.85 | 1-17 | 2 | 45 | + | Vac | --- | |
| Q5U1P7 | OsPrx46 | 35.95 | 7.02 | 1-29 | 1 | 20 | --- | Vac | nitrogen and phosphate starvation * | |
| Q7XUL1 | OsPrx5556 | 51.48 | 9.15 | 1-26 | 5 | 86 | + | PM | gibberellic acid and heat * | |
| Q6AVZ8 | OsPrx65 | 37.61 | 7.20 | 1-24 | 2 | 21 | --- | MoM | drought * | |
| Q8L3W2 | OsPrx95 MS | 37.58 | 5.63 | 1-28 | 6 | 38 | --- | PM | --- | |
| Q6Z121 | OsPrx100 | 32.78 | 5.68 | 1-20 | 2 | 27 | --- | PM | --- | |
| Q6Z121 | OsPrx101 | 32.78 | 5.68 | 1-20 | 2 | 27 | --- | PM | --- | |
| Q8GVG6 | OsPrx106-1 | 40.15 | 7.22 | 1-19 | 2 | 47 | --- | Vac | --- | |
| A2YP47 | OsPrx106-2 | 40.15 | 7.22 | 1-19 | 2 | 45 | --- | Vac | --- | |
| Q8GVG0 | OsPrx107 | 34.23 | 9.19 | 1-22 | 3 | 15 | --- | ER | nitrogen and phosphate starvation * | |
| Q5U1I4 | OsPrx109 | 33.02 | 8.26 | 1-21 | 4 | 32 | + | ER | anaerobic stress, brassinolide and giberellic acid treatments and nematode infection * | |
| O. sativa | Q6Z3Y8 | OsPrx116 | 36.05 | 7.69 | 1-19 | 1 | 29 | + | MoM | pH and oxidative stress * |
| Q5U1H6 | OsPrx117 | 33.51 | 6.65 | 1-25 | 1 | 30 | --- | PM | pH, oxidative stress, nitrogen starvation and pathogen * | |
| Q6UU25 | OsPrx118 | 36.93 | 4.96 | 1-25 | 2 | 23 | --- | Vac | --- | |
| Q7XHB1 | OsPrx126 | 35.48 | 4.45 | 1-27 | 1 | 33 | + | PM | pH, oxidative stress, nematode infection and pathogen * | |
| Q5U1F8 | OsPrx135 | 34.83 | 8.79 | 1-31 | 3 | 38 | + | PM | --- | |
| Q5U1F5 | OsPrx138 | 35.66 | 8.40 | --- | 2 | 17 | --- | PM | nitrogen starvation * | |
| Z. mays | A5H8G4 | ZmPrx01 MS | 38.36 | 6.81 | --- | 5 | 35 | --- | PM | cell wall modification, wounding and pathogen |
| A0A1D6KUF1 | ZmPrx11 | 35.44 | 5.14 | 1-28 | 3 | 28 | + | PM | pH, oxidative stress and pathogen * | |
| B4FHG3 | ZmPrx24 MS | 37.82 | 5.9 | 1-25 | 2 | 36 | --- | PM | pH, oxidative stress, abscissic and ethylene stress, etiolation and heat shock * | |
| B4FD28 | ZmPrx26 | 37.50 | 5.01 | 1-38 | 3 | 34 | --- | MiM | pH, drought and pathogen * | |
| B6T173 | ZmPrx35 | 36.80 | 6.05 | 1-22 | 2 | 24 | --- | ER | --- | |
| A0A1D6H655 | ZmPrx36 | 33.09 | 4.90 | 1-24 | 2 | 29 | + | MoM | heat and oxidative stress * | |
| B4FVT1 | ZmPrx39 | 37.91 | 8.30 | 1-21 | 8 | 34 | --- | MoM | pathogen | |
| B4FJX1 | ZmPrx41 | 35.75 | 4.88 | --- | 1 | 30 | --- | PM | pH, drought and etiolation * | |
| A0A1D6FBJ6 | ZmPrx46 | 38.65 | 6.19 | 1-23 | 1 | 33 | --- | ER | --- | |
| Q9ZTS6 | ZmPrx52 | 34.59 | 8.96 | 1-27 | 4 | 29 | --- | PM | pathogen * | |
| K7VQB0 | ZmPrx53 | 46.55 | 4.98 | 1-24 | 1 | 42 | + | Vac | --- | |
| B6TU39 | ZmPrx55 | 34.96 | 4.85 | 1-42 | 1 | 33 | + | PM | cold stress | |
| A0A1D6IMZ0 | ZmPrx56 | 34.57 | 4.35 | 1-25 | 3 | 36 | --- | PM | pathogen * | |
| B6T5R9 | ZmPrx57 | 37.58 | 5.73 | 1-19 | 3 | 26 | --- | PM | pathogen * | |
| B4FH68 | ZmPrx58 MS | 36.91 | 6.35 | 1-25 | 1 | 30 | --- | PM | pH, oxidative stress, abscissic and ethylene stress, etiolation and heat shock * | |
| A5H454-1 | ZmPrx66 MS | 33.42 | 8.39 | 1-29 | 4 | 70 | + | PM | drought | |
| A0A1D6H655 | ZmPrx67 | 32.77 | 4.89 | --- | 4 | 26 | --- | MoM | heat, drought, pathogen and oxidative stress * | |
| A5H452 | ZmPrx70 MS | 33.41 | 9.08 | 1-25 | 4 | 34 | + | ER/PM | drought | |
| B4FMF8 | ZmPrx71 | 35.71 | 9.64 | 1-28 | 3 | 30 | --- | PM | --- | |
| B4F7T9 | ZmPrx72 | 36.67 | 9.31 | --- | 2 | 22 | --- | MiM | nitrogen starvation * | |
| B6SNF9 | ZmPrx76 | 33.30 | 8.64 | 1-24 | --- | 33 | --- | ER | drought | |
| B4FH35 | ZmPrx77 | 35.71 | 9.64 | 1-32 | 6 | 20 | + | ER | --- | |
| A0A1D6IKW2 | ZmPrx78 | 41.42 | 5.86 | 1-21 | 3 | 40 | + | Vac | pH and drought * | |
| B4FG39 | ZmPrx81 | 36.42 | 7.66 | 1-29 | 3 | 35 | --- | PM | drought | |
| A0A1D6E530 | ZmPrx85 | 35.48 | 5.35 | 1-23 | 1 | 32 | --- | PM | pathogen | |
| Z. mays | Q9ZTS8 | ZmPrx86 | 35.51 | 4.37 | 1-21 | 3 | 34 | + | PM | pathogen |
| A0A1D6HQQ9 | ZmPrx89 | 36.08 | 4.87 | 1-32 | 10 | 23 | + | MiM | pH, anoxia, ethylene and gibberellic acid * | |
| A0A1D6HQQ8 | ZmPrx94 | 37.24 | 5.82 | 1-29 | 6 | 30 | --- | PM | drought | |
| B6TWB1 | ZmPrx102 | 37.11 | 6.00 | 1-26 | 4 | 24 | --- | Vac | pH and pathogen * | |
| A0A1D6IKX3 | ZmPrx103 | 57.26 | 5.06 | 1-27 | 4 | 106 | --- | Vac | --- | |
| B4FBH0 | ZmPrx104 | 38.04 | 8.09 | 1-27 | 2 | 23 | --- | Vac | etiolation * | |
| B4FYH1 | ZmPrx109 | 37.94 | 6.00 | 1-22 | 3 | 40 | --- | PM | pathogen * | |
| A0A1D6J1L2 | ZmPrx112 | 41.37 | 7.03 | 1-27 | 6 | 64 | --- | PM | etiolation, drought and pathogen * | |
| A0A1D6KQI0 | ZmPrx113 | 38.28 | 10.26 | --- | 3 | 27 | --- | PM | --- | |
| C0PPB6 | ZmPrx114 | 32.71 | 5.20 | 1-23 | 5 | 38 | + | PM | pH, drought and pathogen * | |
| B6U6W0 | ZmPrx115 | 37.59 | 8.31 | --- | 3 | 40 | + | PM | etiolation, cell wall modification, wounding and pathogen * | |
| A0A1D6H652 | ZmPrx122 | 34.37 | 6.88 | 1-28 | 2 | 29 | + | PM | drought * | |
| B4FA32 | ZmPrx123 | 34.01 | 6.50 | 1-25 | 1 | 25 | + | PM | etiolation, abscisic acid and pathogen * | |
| A0A1D6N9N5 | ZmPrx124 | 37.42 | 4.84 | 1-23 | 2 | 25 | --- | PM | pH and sulfur deficiency * | |
| K7TMB0 | ZmPrx128 | 35.80 | 8.05 | 1-36 | 3 | 30 | --- | MoM | --- | |
| K7U151 | ZmPrx129 | 33.75 | 6.80 | 1-26 | 1 | 20 | --- | PM | --- | |
| A0A1D6JNY2 | ZmPrx132 | 46.64 | 4.99 | 1-22 | 1 | 40 | + | Vac | --- | |
| K7VFH6 | ZmPrx135 | 35.67 | 4.98 | 1-31 | --- | 22 | --- | ER | abscisic acid and heat * | |
| C4IZ20 | ZmPrx136 | 34.48 | 8.45 | 1-22 | 2 | 21 | --- | PM | etiolation, drought and pathogen * | |
| A0A1D6JF04 | ZmPrx140 | 37.88 | 8.88 | 1-21 | 3 | 31 | --- | PM | pH * |
| Substrate | CroPrx01 | AtPrx64 | AtPrx47 | MtPrx02 | OsPrx95 | ZmPrx01 | HRP |
|---|---|---|---|---|---|---|---|
| ascorbic acid | −5.47 | −5.39 | −5.94 | −5.48 | −5.56 | −5.29 | −5.88 |
| l-DOPA 1 | −8.23 | −8.39 | 0.24 | 12.62 | 7.11 | 12.34 | −0.34 |
| indole acetic acid | −15.41 | −13.66 | −12.95 | −17.24 | −13.05 | −13.6 | −11.79 |
| NADH 2 | −9.26 | −8.6 | −12.82 | −12.42 | −13.32 | −9.89 | −6.29 |
| NADPH 3 | −12.93 | −13.69 | −13.78 | −11.69 | −11.95 | −12.49 | −5.44 |
| cinnamyl alcohol | −7.95 | −7.98 | −7.67 | −8.74 | −7.67 | −6.39 | −5.85 |
| coniferyl alcohol | −4.56 | −4.33 | −3.77 | −3.73 | −4.85 | −4.54 | −3.81 |
| sinalpyl alcohol | −7.31 | −7.88 | −5.23 | −8.02 | −6.51 | −7.26 | −3.35 |
| ferulic acid | −6.8 | −7.18 | −8.14 | −9.04 | −6.11 | −5.22 | −6.86 |
| caffeic acid | −5.35 | −5.34 | −5.06 | −5.98 | −5.91 | −4.88 | −2.07 |
| p-coumaric acid | −4.96 | −5.77 | −5.32 | −5.46 | −6.08 | −6.4 | −5.54 |
| Protein | Z. mays | O. sativa | M. truncatula | A. thaliana | C. roserus |
|---|---|---|---|---|---|
| CroPrx01 | ZmPrx16 (55%) | OsPrx23 (53%) | MtPrx48 (54%) | AtPrx12 (59%) | CroPrx03 (84%) |
| AtPrx64 | ZmPrx136 (65%) | OsPrx32 (60%) | MtPrx55 (72%) | AtPrx66(52%) | CroPrx40 (72%) CroPrx43 (72%) |
| AtPrx47 | ZmPrx15 (57%) | OsPrx117 (60%) | MtPrx19 (71%) | AtPrx66 (47%) AtPrx64 (46%) | CroPrx09 (75%) |
| MtPrx02 | ZmPrx120 (52%) | OsPrx40 (59%) | MtPrx70 (96%) | AtPrx52 (46%) | CroPrx04 (55%) |
| OsPrx95 | ZmPrx94 (73%) | OsPrx97 (50%) OsPrx134 (50%) | MtPrx07 (43%) | AtPrx56 (46%) | CroPrx49 (48%) CroPrx59 (47%) |
| ZmPrx1 | ZmPrx101 (68%) | OsPrx12 (74%) | MtPrx07(52%) | AtPrx39 (46%) | CroPrx14 (48%) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lüthje, S.; Martinez-Cortes, T. Membrane-Bound Class III Peroxidases: Unexpected Enzymes with Exciting Functions. Int. J. Mol. Sci. 2018, 19, 2876. https://doi.org/10.3390/ijms19102876
Lüthje S, Martinez-Cortes T. Membrane-Bound Class III Peroxidases: Unexpected Enzymes with Exciting Functions. International Journal of Molecular Sciences. 2018; 19(10):2876. https://doi.org/10.3390/ijms19102876
Chicago/Turabian StyleLüthje, Sabine, and Teresa Martinez-Cortes. 2018. "Membrane-Bound Class III Peroxidases: Unexpected Enzymes with Exciting Functions" International Journal of Molecular Sciences 19, no. 10: 2876. https://doi.org/10.3390/ijms19102876
APA StyleLüthje, S., & Martinez-Cortes, T. (2018). Membrane-Bound Class III Peroxidases: Unexpected Enzymes with Exciting Functions. International Journal of Molecular Sciences, 19(10), 2876. https://doi.org/10.3390/ijms19102876