De Novo Assembly and Analysis of Polygonatum sibiricum Transcriptome and Identification of Genes Involved in Polysaccharide Biosynthesis
Abstract
:1. Introduction
2. Results
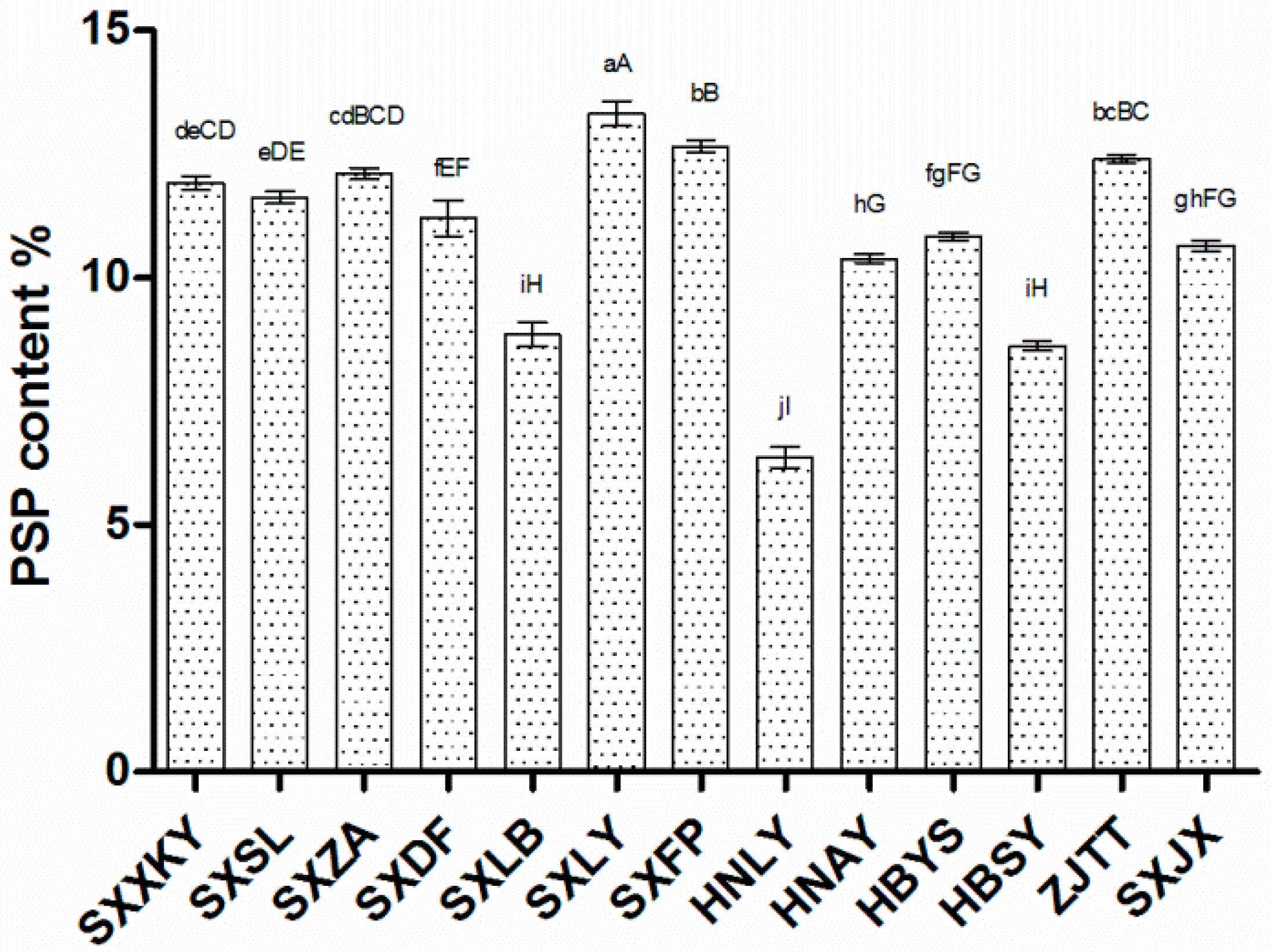
2.1. PSP Content Varies among Polygonatum sibiricum Germplasms
2.2. Illumina Sequencing, De Novo Assembly, and Assessment of Assembly Program
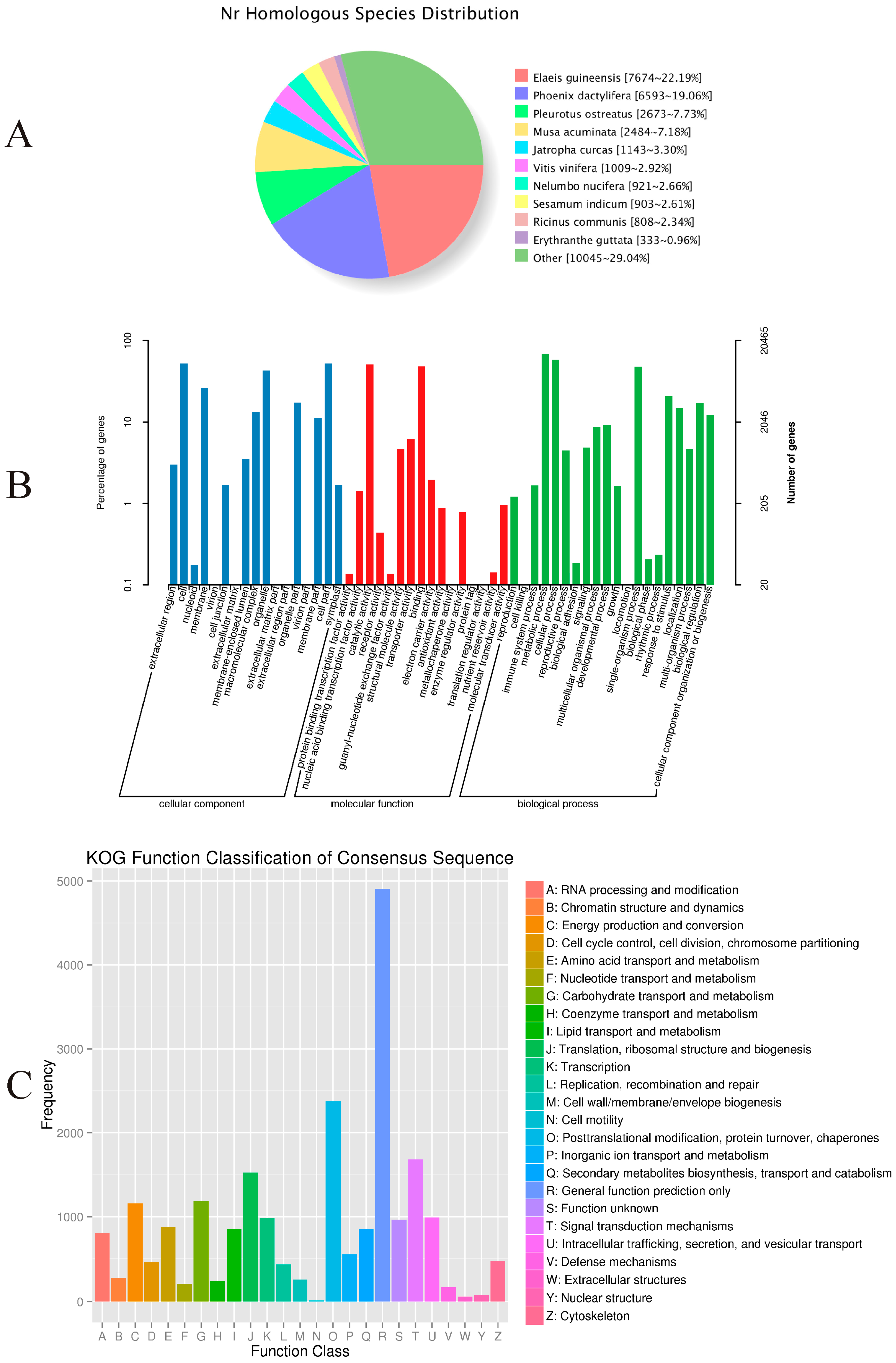
2.3. Functional Annotations
2.4. Kyoto Encyclopedia of Genes and Genomes Pathway Analysis
2.5. Candidate Genes Involved in PSP Biosynthesis
2.6. Analysis of Differential Gene Expression in Lueyang, Shaanxi (SXLY) and Luoyang, Henan (HNLY) Germplasms
2.7. Analysis of PSP Biosynthetic Pathway
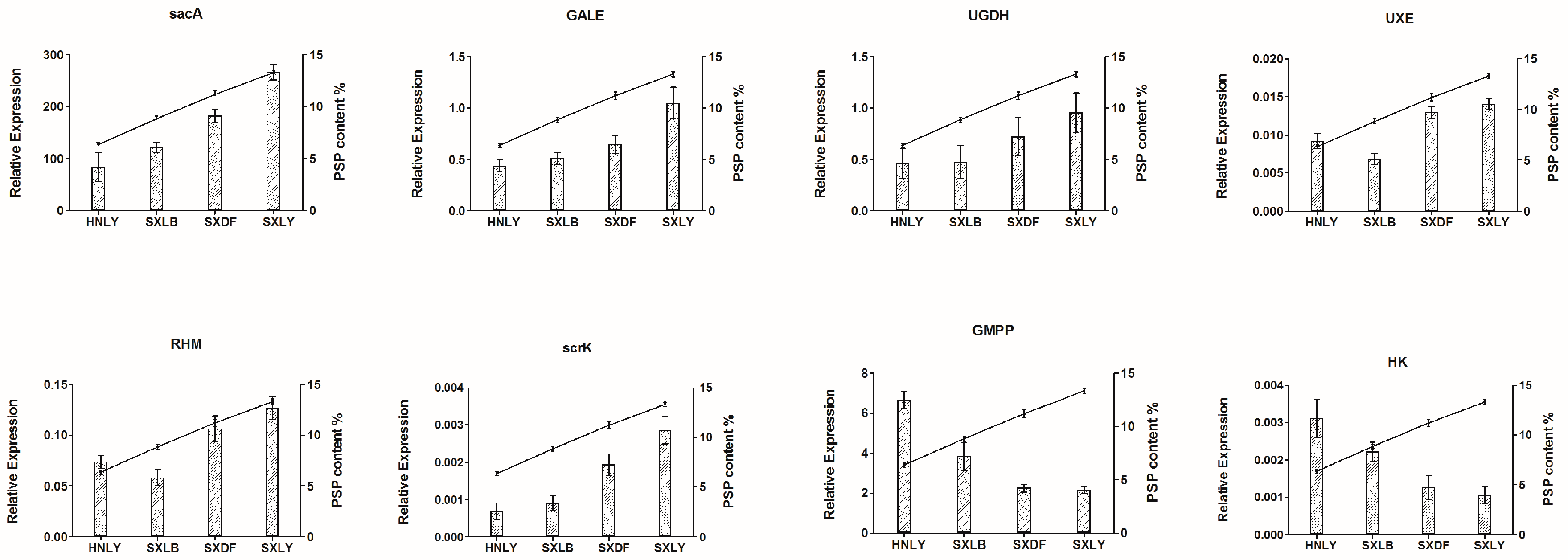
2.8. Real-Time PCR Analysis
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Plant Materials
4.3. Isolation and Detection of Polysaccharides
4.4. Library Preparation and Sequencing (mRNA-Seq)
4.5. De Novo Assembly and Functional Annotation
4.6. Analysis of Differential Gene Expression
4.7. Identification of Genes Involved in PSP Biosynthesis
4.8. Real-Time PCR
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Zhao, X.; Li, J. Chemical constituents of the genus Polygonatum and their role in medicinal treatment. Nat. Prod. Commun. 2015, 10, 683–688. [Google Scholar] [PubMed]
- Virk, J.K.; Kumar, S.; Singh, R.; Tripathi, A.C.; Saraf, S.K.; Gupta, V.; Bansal, P. Isolation and characterization of quinine from Polygonatum verticillatum: A new marker approach to identify substitution and adulteration. J. Adv. Pharm. Technol. Res. 2016, 7, 153–158. [Google Scholar] [PubMed]
- Liu, L.; Dong, Q.; Dong, X.T.; Fang, J.N.; Ding, K. Structural investigation of two neutral polysaccharides isolated from rhizome of Polygonatum sibiricum. Carbohydr. Polym. 2007, 70, 304–309. [Google Scholar]
- Zong, S.; Zeng, G.; Zou, B.; Li, K.; Fang, Y.; Lu, L.; Xiao, D.; Zhang, Z. Effects of Polygonatum sibiricum polysaccharide on the osteogenic differentiation of bone mesenchymal stem cells in mice. Int. J. Clin. Exp. Pathol. 2015, 8, 6169–6180. [Google Scholar] [PubMed]
- Zhang, H.; Cao, Y.; Chen, L.; Wang, J.; Tian, Q.; Wang, N.; Liu, Z.; Li, J.; Wang, N.; Wang, X.; et al. A polysaccharide from Polygonatum sibiricum attenuates amyloid-β-induced neurotoxicity in PC12 cells. Carbohydr. Polym. 2015, 117, 879–886. [Google Scholar] [CrossRef] [PubMed]
- Zeng, G.F.; Zhang, Z.Y.; Lu, L.; Xiao, D.Q.; Xiong, C.X.; Zhao, Y.X.; Zong, S.H. Protective effects of Polygonatum sibiricum polysaccharide on ovariectomy-induced bone loss in rats. J. Ethnopharmacol. 2011, 136, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.X.; Wu, S.; Huang, X.L.; Hu, X.Q.; Zhang, Y. Hypolipidemic activity and antiatherosclerotic effect of polysaccharide of Polygonatum sibiricum in rabbit model and related cellular mechanisms. Evid. Based Complement. Altern. Med. 2015, 2015, 391065. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.H.; Lv, G.Y.; Li, B.; Zhang, Y.L.; Su, J.; Chen, S.H. Study on effect of Polygonatum sibiricum on Yin deficiency model rats induced by long-term overload swimming. Zhongguo Zhong Yao Za Zhi 2014, 39, 1886–1891. (In Chinese) [Google Scholar] [PubMed]
- Xiao, M.; Zhang, Y.; Chen, X.; Lee, E.J.; Barber, C.J.; Chakrabarty, R.; Desgagné-Penix, I.; Haslam, T.M.; Kim, Y.B.; Liu, E.; et al. Transcriptome analysis based on next-generation sequencing of non-model plants producing specialized metabolites of biotechnological interest. J. Biotechnol. 2013, 166, 122–134. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Gereltu, B.; Zhaorigetu; Narisu. Determination of relative molecular mass and composition for Polygonatum sibiricum polysaccharide by high performance liquid chromatography. Chin. J. Chromatogr. 2005, 23, 394–396. [Google Scholar] [PubMed]
- Tang, Q.; Ma, X.; Mo, C.; Wilson, I.W.; Song, C.; Zhao, H.; Yang, Y.; Fu, W.; Qiu, D. An efficient approach to finding Siraitia grosvenorii triterpene biosynthetic genes by RNA-seq and digital gene expression analysis. BMC Genom. 2011, 12, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Alonso, A.P.; Wilkerson, C.G.; Keegstra, K. Deep EST profiling of developing fenugreek endosperm to investigate galactomannan biosynthesis and its regulation. Plant Mol. Biol. 2012, 79, 243–258. [Google Scholar] [CrossRef] [PubMed]
- Pauly, M.; Gille, S.; Liu, L.; Mansoori, N.; de Souza, A.; Schultink, A.; Xiong, G. Hemicellulose biosynthesis. Planta 2013, 238, 627–642. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Zhou, H.; Liu, C.; Zhang, J.; Li, N.; Zhao, Z.; Sun, G.; Zhong, Y. A molasses habitat-derived fungus Aspergillus tubingensis XG21 with high β-fructofuranosidase activity and its potential use for fructooligosaccharides production. AMB Express 2017, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Uematsu, K.; Suzuki, N.; Iwamae, T.; Inui, M.; Yukawa, H. Expression of Arabidopsis plastidial phosphoglucomutase in tobacco stimulates photosynthetic carbon flow into starch synthesis. J. Plant Physiol. 2012, 169, 1454–1462. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhen, L.; Tan, X.; Li, L.; Wang, X. The involvement of hexokinase in the coordinated regulation of glucose and gibberellin on cell wall invertase and sucrose synthesis in grape berry. Mol. Biol. Rep. 2014, 41, 7899–7910. [Google Scholar] [CrossRef] [PubMed]
- Perez-Cenci, M.; Salerno, G.L. Functional characterization of Synechococcus amylosucrase and fructokinase encoding genes discovers two novel actors on the stage of cyanobacterial sucrose metabolism. Plant Sci. 2014, 224, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Park, J.I.; Ishimizu, T.; Suwabe, K.; Sudo, K.; Masuko, H.; Hakozaki, H.; Nou, I.S.; Suzuki, G.; Watanabe, M. UDP-glucose pyrophosphorylase is rate limiting in vegetative and reproductive phases in Arabidopsis thaliana. Plant Cell Physiol. 2010, 51, 981–996. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, P.; Zetsche, K. A close temporal and spatial correlation between cell growth, cell wall synthesis and the activity of enzymes of mannan synthesis in Acetabularia mediterranea. Planta 1979, 145, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Huang, J.; Gu, X.; Bar-Peled, M.; Xu, Y. Evolution of plant nucleotide-sugar interconversion enzymes. PLoS ONE 2011, 6, e27995. [Google Scholar] [CrossRef] [PubMed]
- Soares, J.S.; Gentile, A.; Scorsato, V.; Lima, A.C.; Kiyota, E.; Dos, S.M.L.; Piattoni, C.V.; Huber, S.C.; Aparicio, R.; Menossi, M. Oligomerization, membrane association, and in vivo phosphorylation of sugarcane UDP-glucose pyrophosphorylase. J. Biol. Chem. 2014, 289, 33364–33377. [Google Scholar] [CrossRef] [PubMed]
- Mamoon, R.H.; Amjad, N.M.; Bao, L.; Hussain, S.Z.; Lee, J.M.; Ahmad, M.Q.; Chung, G.; Yang, S.H. Genome-wide analysis of family-1 UDP-glycosyltransferases in soybean confirms their abundance and varied expression during seed development. J. Plant Physiol. 2016, 206, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Gachon, C.M.; Langlois-Meurinne, M.; Saindrenan, P. Plant secondary metabolism glycosyltransferases: The emerging functional analysis. Trends Plant. Sci. 2005, 10, 542–549. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.B.; Liu, J.H.; Xiao, Y.; Zhang, F.; Chen, J.F.; Ji, Q.; Tan, H.X.; Huang, X.; Feng, H.; Huang, B.K.; et al. Deep sequencing reveals the effect of MeJA on scutellarin biosynthesis in Erigeron breviscapus. PLoS ONE 2015, 10, e0143881. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Allan, A.C.; Li, C.; Wang, Y.; Yao, Q. De novo assembly and characterization of the transcriptome of the chinese medicinal herb, Gentiana rigescens. Int. J. Mol. Sci. 2015, 16, 11550–11573. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, K.; Li, J.; Zhao, X.; Que, Q.; Li, P.; Huang, H.; Deng, X.; Singh, S.K.; Wu, A.M.; Chen, X. Transcriptomic analysis of multipurpose timber yielding tree Neolamarckia cadamba during xylogenesis using RNA-Seq. PLoS ONE 2016, 11, e0159407. [Google Scholar] [CrossRef] [PubMed]
- Keller, R.; Renz, F.S.; Kossmann, J. Antisense inhibition of the GDP-mannose pyrophosphorylase reduces the ascorbate content in transgenic plants leading to developmental changes during senescence. Plant J. 1999, 19, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, C.; Xie, Y.; Li, N.; Ning, Z.; Du, N.; Huang, X.; Zhong, Y. Enhancing fructooligosaccharides production by genetic improvement of the industrial fungus Aspergillus niger ATCC 20611. J. Biotechnol. 2017, 249, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Li, N.N.; Qian, W.J.; Wang, L.; Cao, H.L.; Hao, X.Y.; Yang, Y.J.; Wang, X.C. Isolation and expression features of hexose kinase genes under various abiotic stresses in the tea plant (Camellia sinensis). J. Plant Physiol. 2017, 209, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Gao, J.; Xue, F.; Yu, X.; Shao, T. Extraction optimization, purification and physicochemical properties of polysaccharides from Gynura medica. Molecules 2016, 21, 397. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Jianqi, L.I.; Songfeng, W.U.; Zhu, Y.; Chen, Y.; Fuchu, H.E. Integrated nr database in protein annotation system and its localization. Comput. Eng. 2006, 32, 71–74. [Google Scholar]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M.; et al. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2004, 32, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Krylov, D.M.; Makarova, K.S.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; Rao, B.S.; et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004, 5, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, 277–280. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. Profile hidden Markov models. Bioinformatics. 1998, 14, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Hwang, D.S.; Lee, B.Y.; Park, J.C.; Lee, Y.H.; Lee, J.S. De novo assembly and annotation of the marine mysid (Neomysis awatschensis) transcriptome. Mar. Genom. 2016, 28, 41–43. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000, 132, 365–386. [Google Scholar] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆ΔCt Method. Methods. 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
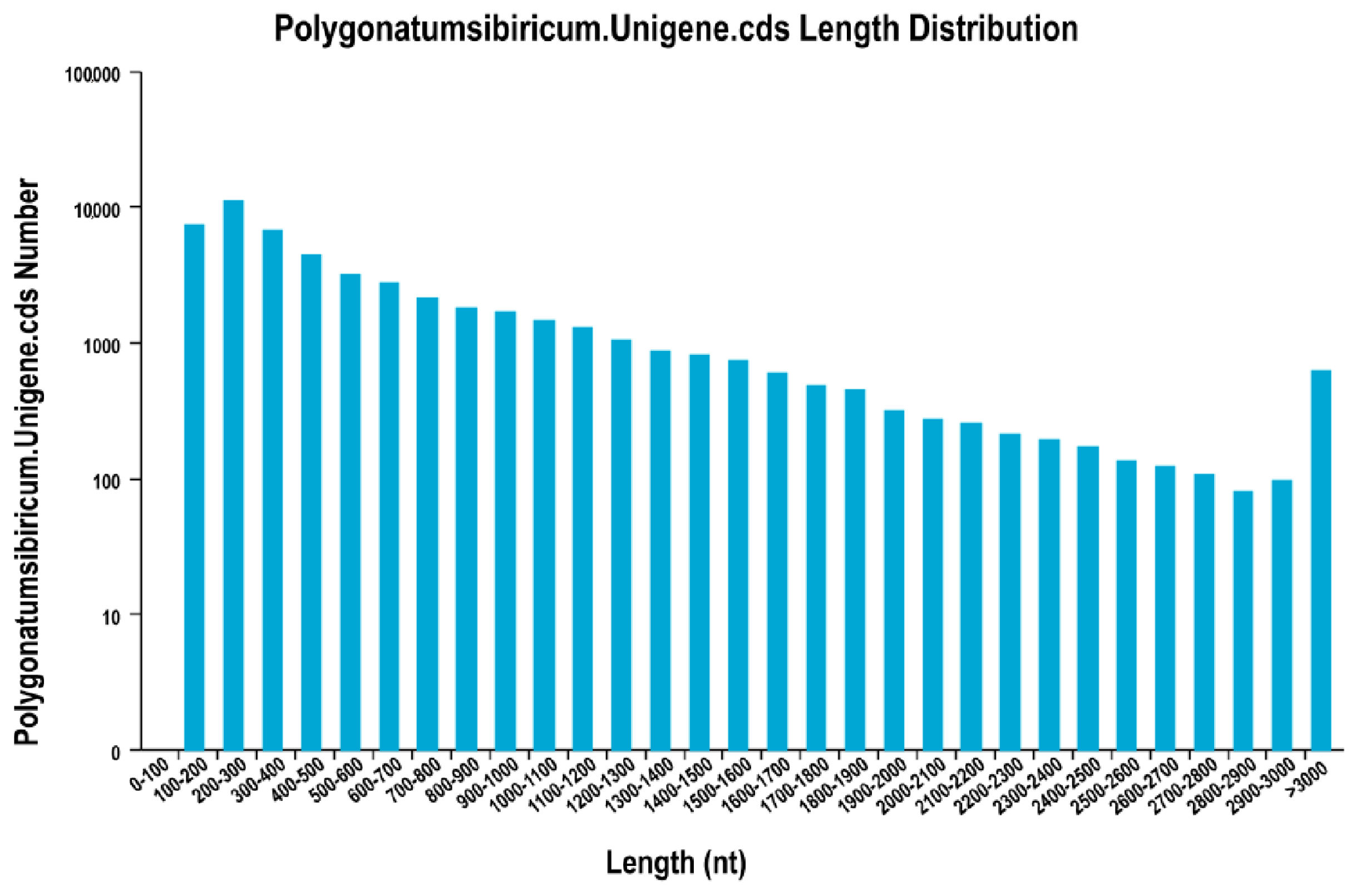
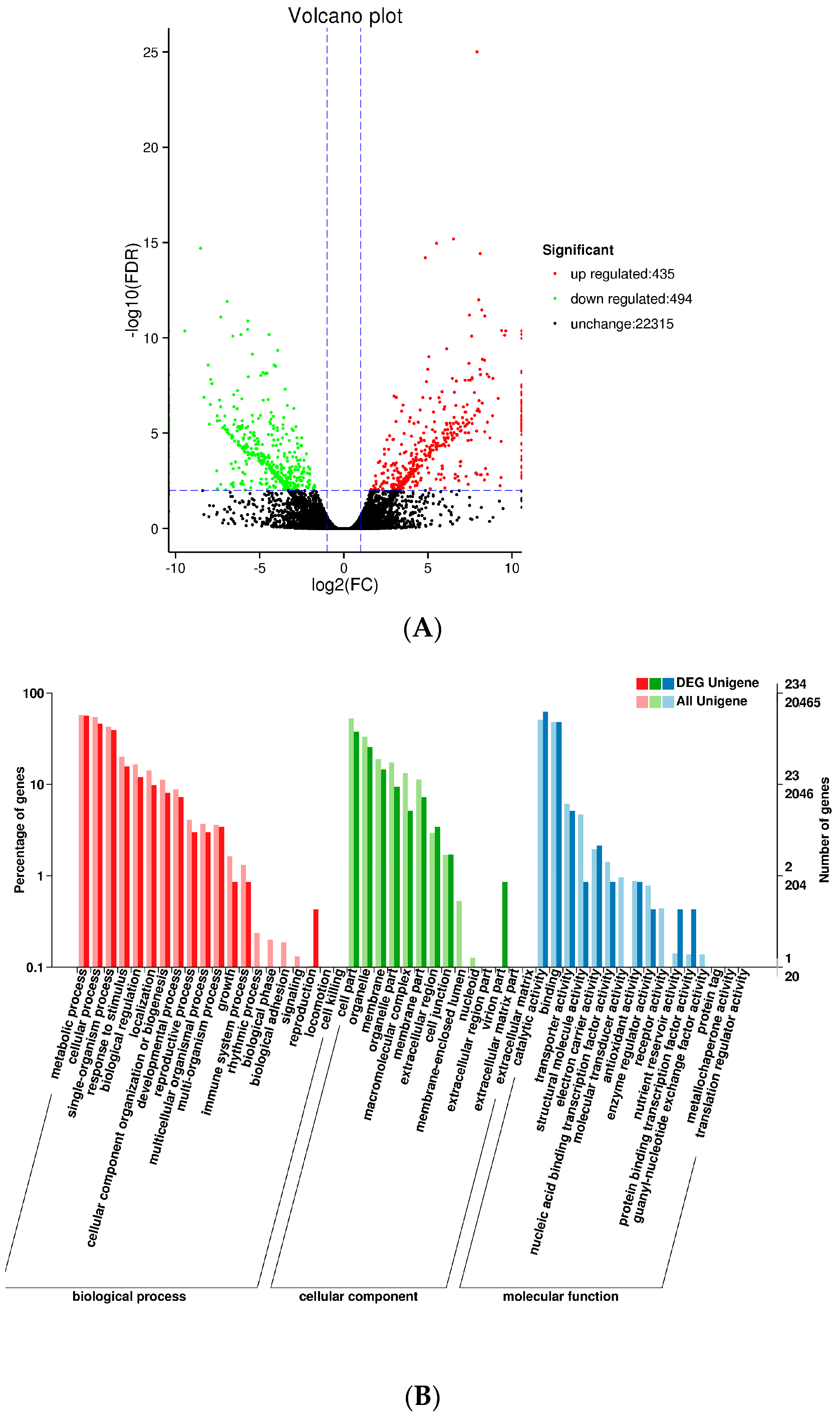
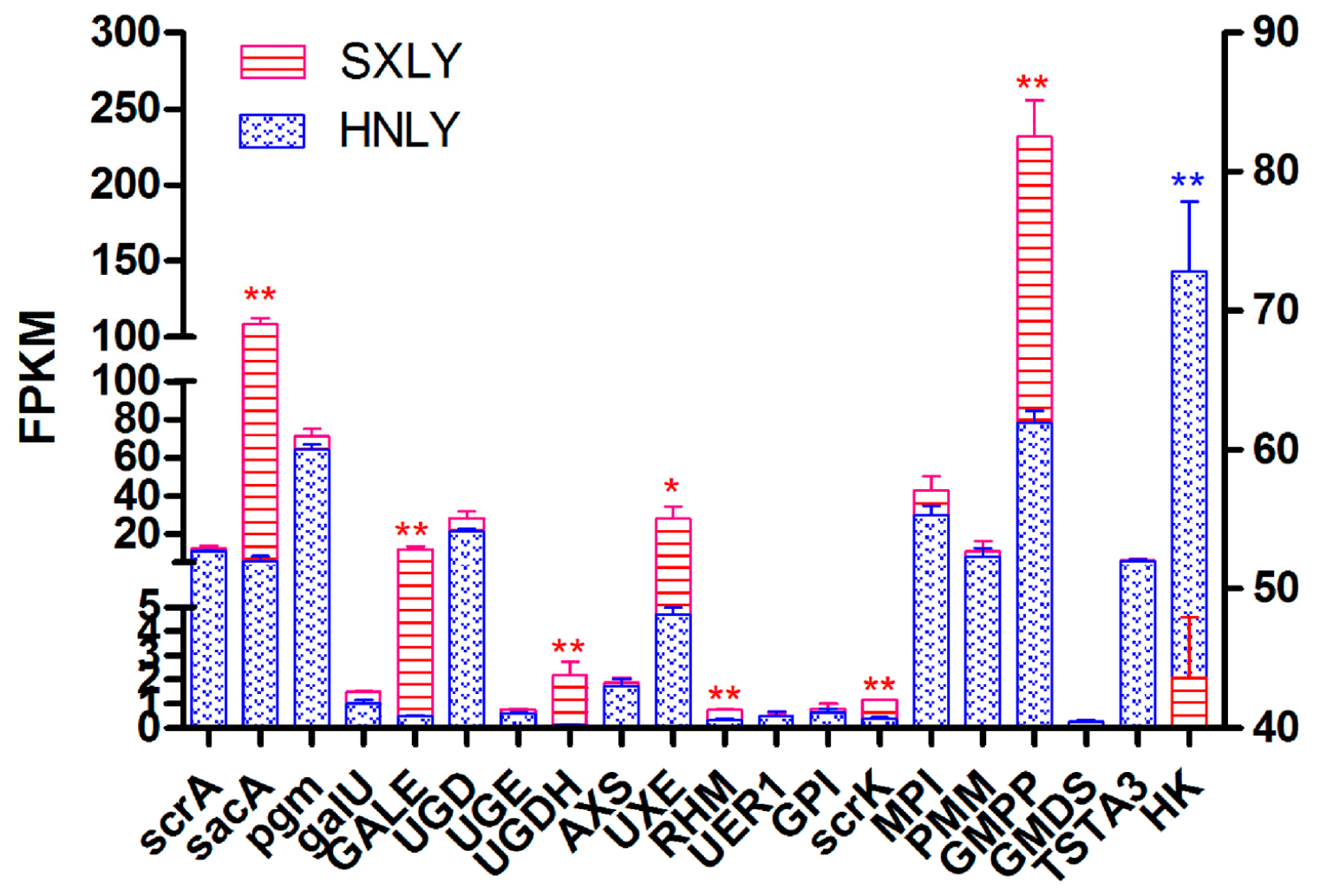
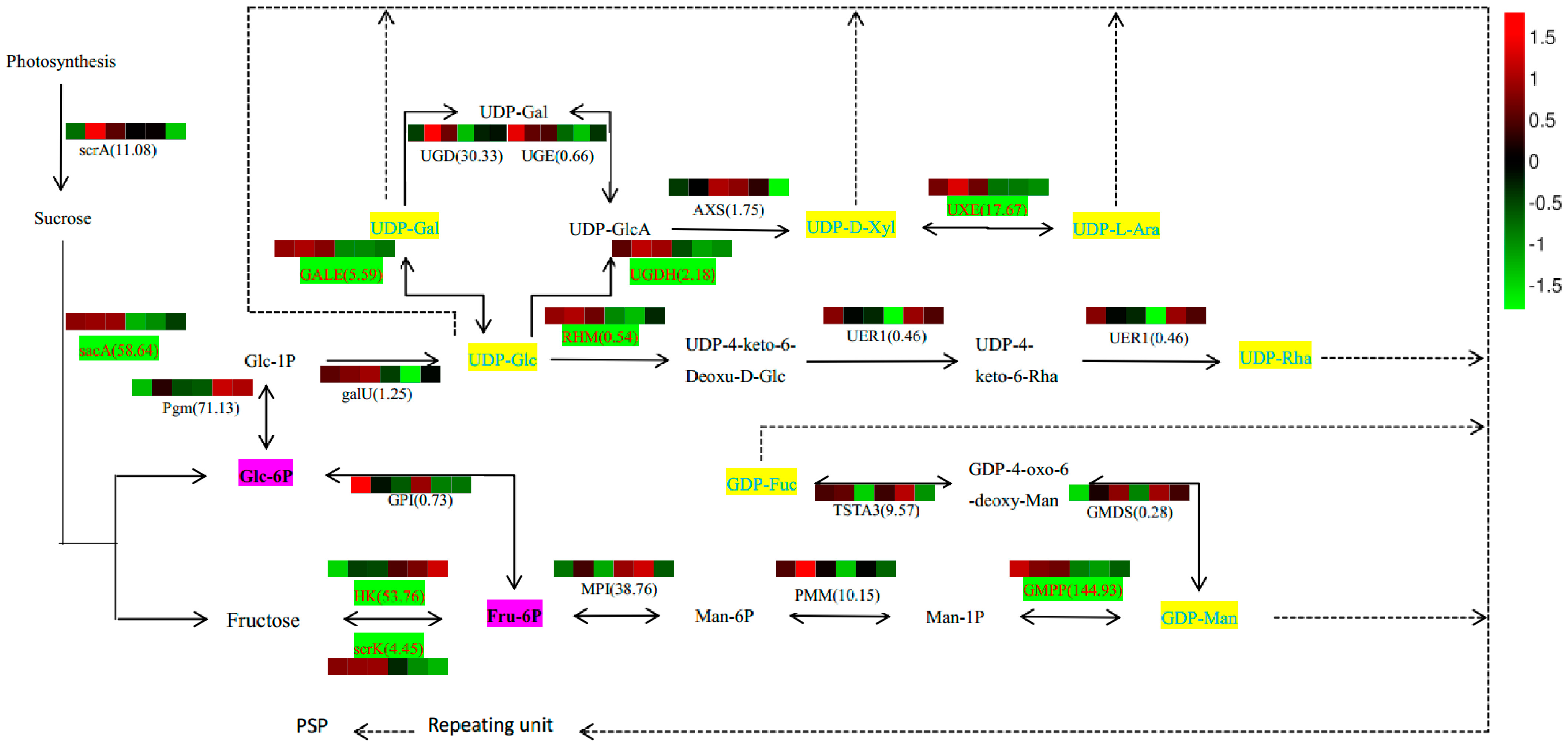

| ID 1 | Number of Reads | Number of Bases | Guanine-Cytosine (GC) Content | N Percentage | % ≥Q30 |
|---|---|---|---|---|---|
| PSABC | 48,703,742 | 14,514,388,398 | 50.21% | 0.00 | 93.57 |
| PS01 | 29,810,908 | 8,876,080,296 | 49.81% | 0.00 | 93.40 |
| PS02 | 30,941,196 | 9,196,437,754 | 49.21% | 0.00 | 93.44 |
| PS03 | 36,392,600 | 10,740,869,676 | 49.37% | 0.00 | 93.55 |
| PS04 | 40,885,773 | 12,098,862,416 | 49.70% | 0.00 | 93.31 |
| PS05 | 22,762,445 | 6,786,142,412 | 48.66% | 0.00 | 94.31 |
| PS06 | 32,982,501 | 9,849,781,232 | 48.96% | 0.00 | 93.35 |
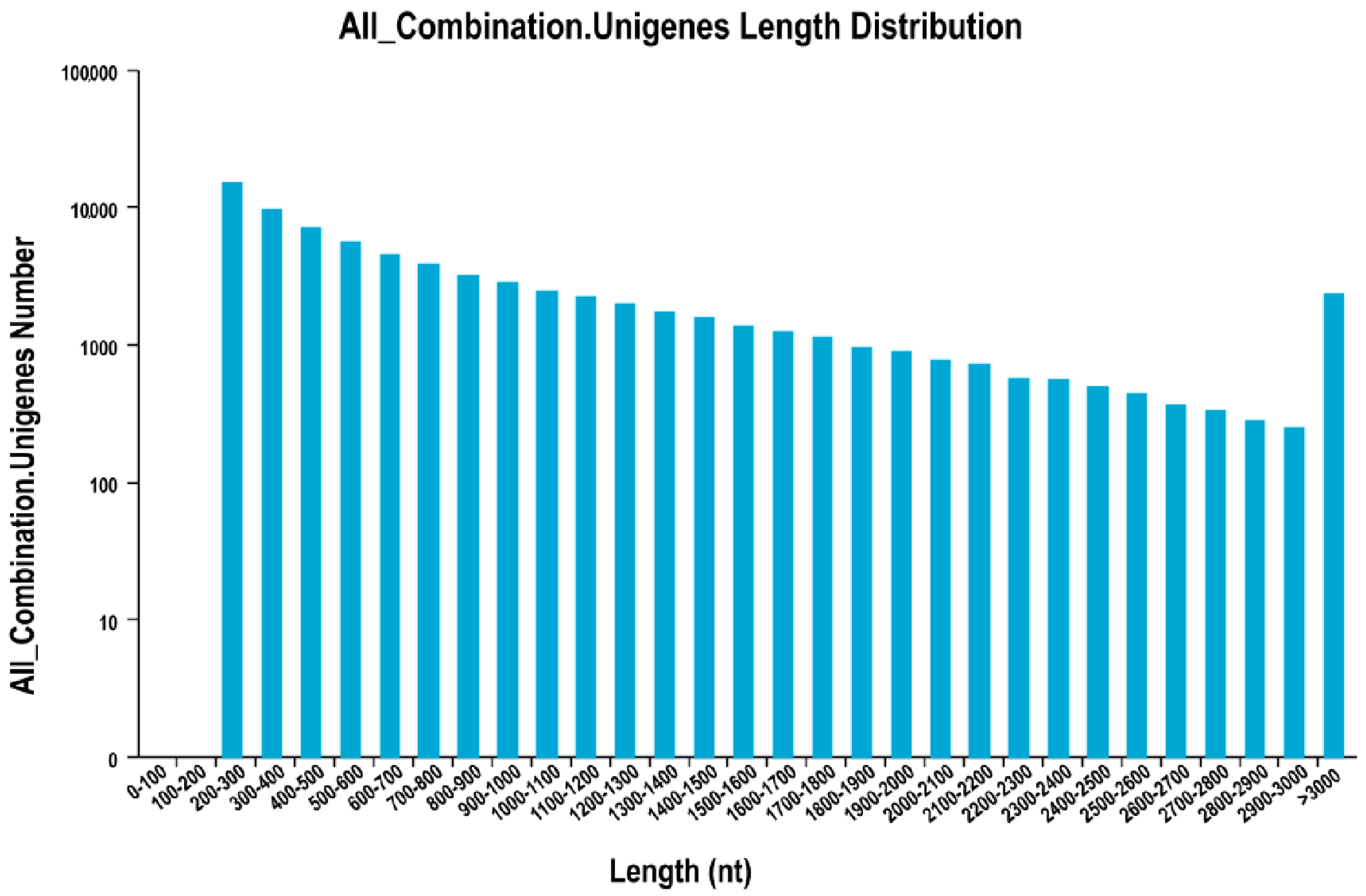
| Length Range (bp) | Number of Contigs | Transcript Abundance | Number of Unigenes |
|---|---|---|---|
| 200–300 | 23,592,360 (99.41%) | 19,417 (14.59%) | 15,057 (20.31%) |
| 300–500 | 72,139 (0.30%) | 26,138 (19.63%) | 16,631 (22.43%) |
| 500–1000 | 42,020 (0.18%) | 37,052 (27.83%) | 19,930 (26.89%) |
| 1000–2000 | 19,425 (0.08%) | 33,660 (25.29%) | 15,422 (20.80%) |
| >2000 | 7128 (0.03%) | 16,854 (12.66%) | 7090 (9.56%) |
| Total number | 23,733,072 | 133,121 | 74,130 |
| Total length | 1,025,259,786 | 138,445,867 | 66,783,395 |
| N50 length | 45 | 1532 | 1364 |
| Mean length | 43.20 | 1040.00 | 900.90 |
| Database | Number Annotated | 300 bp < Length Number of Unigenes < 1000 bp | Number Longer than 1000 bp |
|---|---|---|---|
| Clusters of Orthologous Groups (COG) | 12,175 (16.42%) | 4011 (5.41%) | 6892 (9.30%) |
| Gene Ontology (GO) | 20,465 (27.61%) | 8253 (11.13%) | 10,444 (14.09%) |
| Kyoto Encyclopedia of Genes and Genomes (KEGG) | 14,267 (19.25%) | 5748 (7.75%) | 6797 (9.17%) |
| euKaryotic Orthologous Groups (KOG) | 20,240 (27.30%) | 7785 (10.50%) | 10,681 (14.41%) |
| Protein family (Pfam) | 24,732 (33.36%) | 8700 (11.74%) | 14,264 (19.24%) |
| Swiss-Prot | 21,965 (29.63%) | 8574 (11.57%) | 11,727 (15.82%) |
| non-redundant (NR) | 34,601 (46.68%) | 14,135 (19.07%) | 17,043 (22.99%) |
| All annotated | 35,793 948.28%) | 14,618 (19.72%) | 17,140 (23.12%) |
| Pathway | Number of Unigenes | KO Entry |
|---|---|---|
| Ribosome | 891 (6.25%) | KO03010 |
| Carbon metabolism | 657 (4.61%) | KO01200 |
| Biosynthesis of amino acids | 578 (4.05%) | KO01230 |
| Protein processing in endoplasmic reticulum | 473 (3.32%) | KO04141 |
| Spliceosome | 402 (2.82%) | KO03040 |
| Oxidative phosphorylation | 397 (2.78%) | KO00190 |
| RNA transport | 365 (2.56%) | KO03013 |
| Glycolysis/Gluconeogenesis | 304 (2.13%) | KO00010 |
| Plant hormone signal transduction | 295 (2.07%) | KO04075 |
| Starch and sucrose metabolism | 284 (1.99%) | KO00500 |
| Plant-pathogen interaction | 282 (1.98%) | KO04626 |
| RNA degradation | 245 (1.72%) | KO03018 |
| Purine metabolism | 243 (1.70%) | KO00230 |
| Carbon fixation in photosynthetic organisms | 236 (1.65%) | KO00710 |
| Phagosome | 233 (1.63%) | KO04145 |
| Ubiquitin-mediated proteolysis | 232 (1.63%) | KO04120 |
| Endocytosis | 229 (1.61%) | KO04144 |
| Pyruvate metabolism | 227 (1.59%) | KO00620 |
| mRNA surveillance pathway | 224 (1.57%) | KO03015 |
| Cysteine and methionine metabolism | 215 (1.51%) | KO00270 |
| Pathway | Number of Unigenes | KO Entry |
|---|---|---|
| Glycolysis/Gluconeogenesis | 304 | KO00010 |
| Starch and sucrose metabolism | 284 | KO00500 |
| Pyruvate metabolism | 227 | KO00620 |
| Amino sugar and nucleotide sugar metabolism | 215 | KO00520 |
| Citrate cycle (Tricarboxylic Acid (TCA) cycle) | 170 | KO00020 |
| Pentose phosphate pathway | 128 | KO00030 |
| Galactose metabolism | 128 | KO00052 |
| Fructose and mannose metabolism | 113 | KO00051 |
| Pentose and glucuronate interconversions | 98 | KO00040 |
| N-Glycan biosynthesis | 74 | KO00510 |
| Other glycan degradation | 65 | KO00511 |
| Glycosylphosphatidylinositol(GPI)-anchor biosynthesis | 31 | KO00563 |
| Glycosaminoglycan degradation | 23 | KO00531 |
| Enzyme Code | Enzyme Name | Abbreviation | Number | FPKM |
|---|---|---|---|---|
| 2.7.1.211 | Phosphotransferase System | scrA | 3 | 11.08 |
| 3.2.1.26 | β-fructofuranosidase | sacA | 31 | 58.64 |
| 5.4.2.2 | Phosphoglucomutase | pgm | 5 | 71.13 |
| 2.7.7.9 | Uridine-diphosphate glucose pyrophosphorylase | galU | 8 | 1.25 |
| 5.1.3.2 | UDP-glucose 4-epimerase | GALE | 4 | 5.59 |
| 1.1.1.- | UDP-D-galactose dehydrogenase | UGD | 4 | 30.33 |
| 5.1.3.6 | UDP-glucuronate 4-epimerase | UGE | 8 | 0.66 |
| 1.1.1.22 | UDP-glucose 6-dehydrogenase | UGDH | 9 | 2.18 |
| AXS | UDP-apiose/xylose synthase | AXS | 3 | 1.75 |
| 5.1.3.5 | UDP-arabinose 4-epimerase | UXE | 5 | 17.67 |
| 4.2.1.76 | UDP-glucose 4,6-dehydratase | RHM | 10 | 0.54 |
| 5.1.3.- , 1.1.1.- | 3,5-epimerase-4-reductase | UER1 | 4 | 0.46 |
| 5.3.1.9 | Glucose-6-phosphate isomerase | GPI | 7 | 0.73 |
| 2.7.1.1 | Hexokinase | HK | 13 | 53.76 |
| 2.7.1.4 | Fructokinase | scrK | 8 | 4.45 |
| 5.3.1.8 | Mannose-6-phosphate isomerase | MPI | 3 | 38.76 |
| 5.4.2.8 | Phosphomannomutase | PMM | 1 | 10.15 |
| 2.7.7.13 | Mannose-1-phosphate guanylyltransferase | GMPP | 8 | 144.93 |
| 4.2.1.47 | GDP-mannose 4,6-dehydratase | GMDS | 3 | 0.28 |
| 1.1.1.271 | GDP-L-fucose synthase | TSTA3 | 1 | 9.57 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Wang, B.; Hua, W.; Niu, J.; Dang, K.; Qiang, Y.; Wang, Z. De Novo Assembly and Analysis of Polygonatum sibiricum Transcriptome and Identification of Genes Involved in Polysaccharide Biosynthesis. Int. J. Mol. Sci. 2017, 18, 1950. https://doi.org/10.3390/ijms18091950
Wang S, Wang B, Hua W, Niu J, Dang K, Qiang Y, Wang Z. De Novo Assembly and Analysis of Polygonatum sibiricum Transcriptome and Identification of Genes Involved in Polysaccharide Biosynthesis. International Journal of Molecular Sciences. 2017; 18(9):1950. https://doi.org/10.3390/ijms18091950
Chicago/Turabian StyleWang, Shiqiang, Bin Wang, Wenping Hua, Junfeng Niu, Kaikai Dang, Yi Qiang, and Zhezhi Wang. 2017. "De Novo Assembly and Analysis of Polygonatum sibiricum Transcriptome and Identification of Genes Involved in Polysaccharide Biosynthesis" International Journal of Molecular Sciences 18, no. 9: 1950. https://doi.org/10.3390/ijms18091950
APA StyleWang, S., Wang, B., Hua, W., Niu, J., Dang, K., Qiang, Y., & Wang, Z. (2017). De Novo Assembly and Analysis of Polygonatum sibiricum Transcriptome and Identification of Genes Involved in Polysaccharide Biosynthesis. International Journal of Molecular Sciences, 18(9), 1950. https://doi.org/10.3390/ijms18091950