Pan-Domain Analysis of ZIP Zinc Transporters
Abstract
:1. Introduction
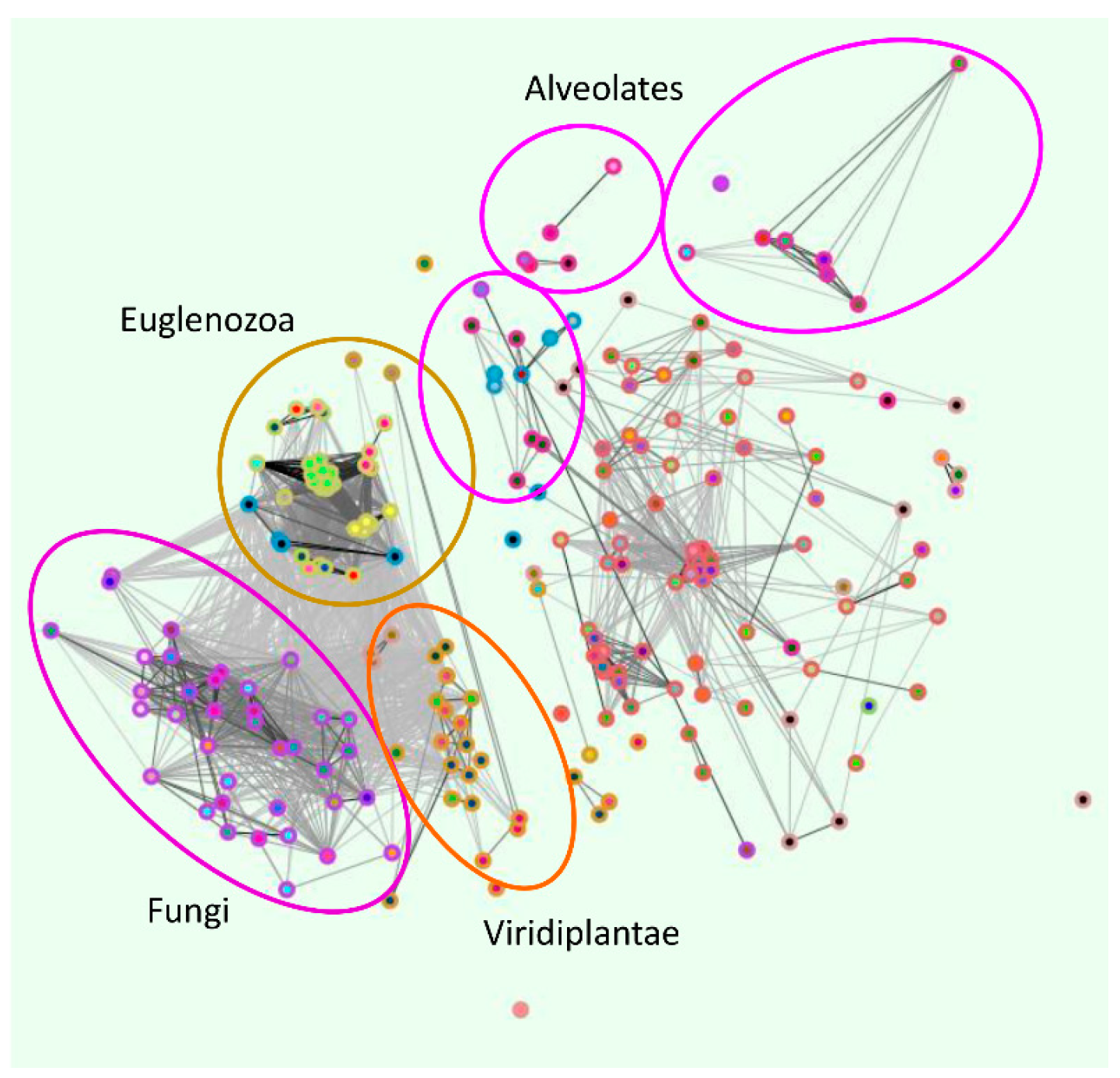
2. Phylogenetic Analysis of ZIP Transporter
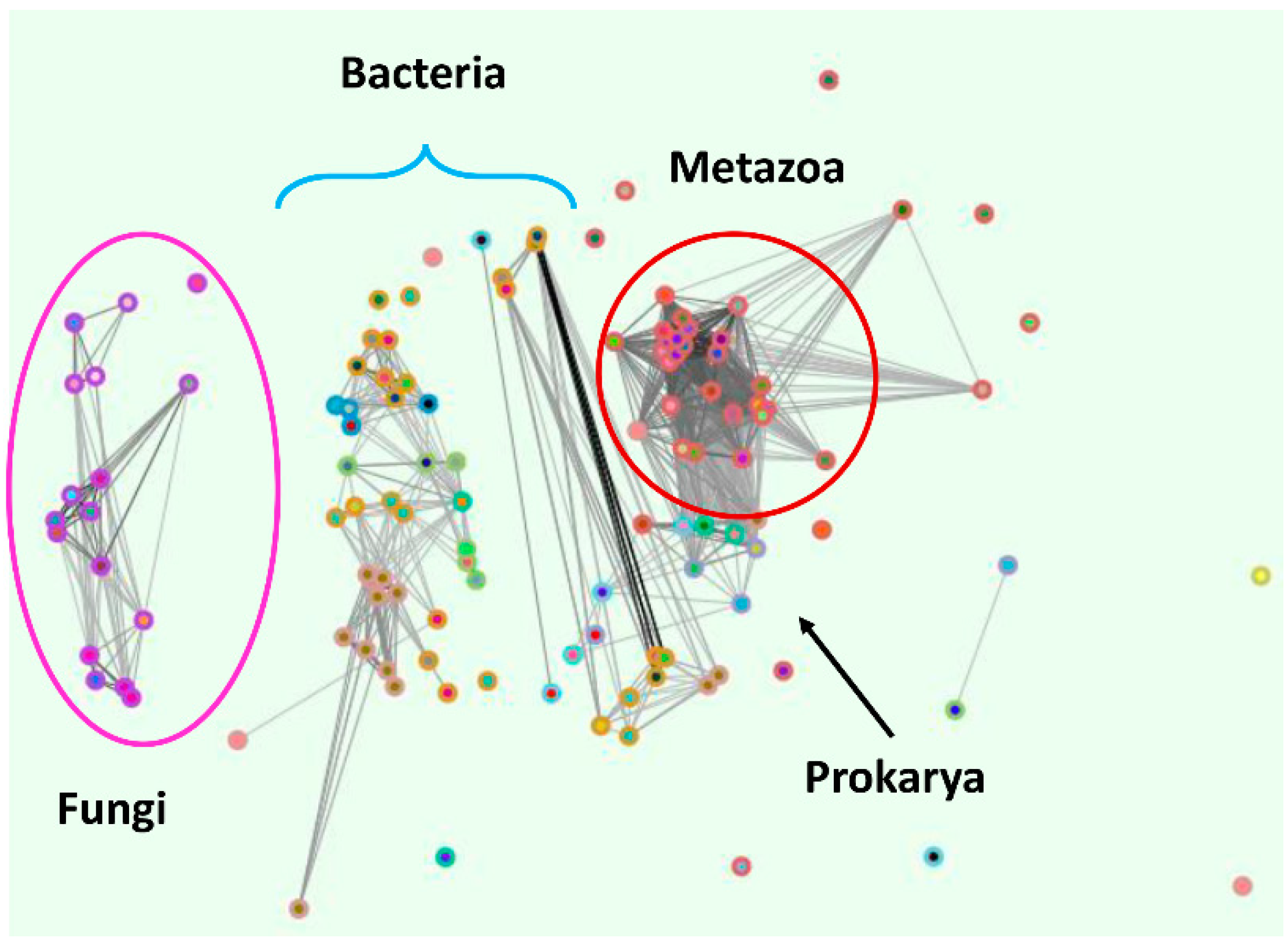
3. Conserved Plasma Membrane Importer Cluster (OG5_126707)
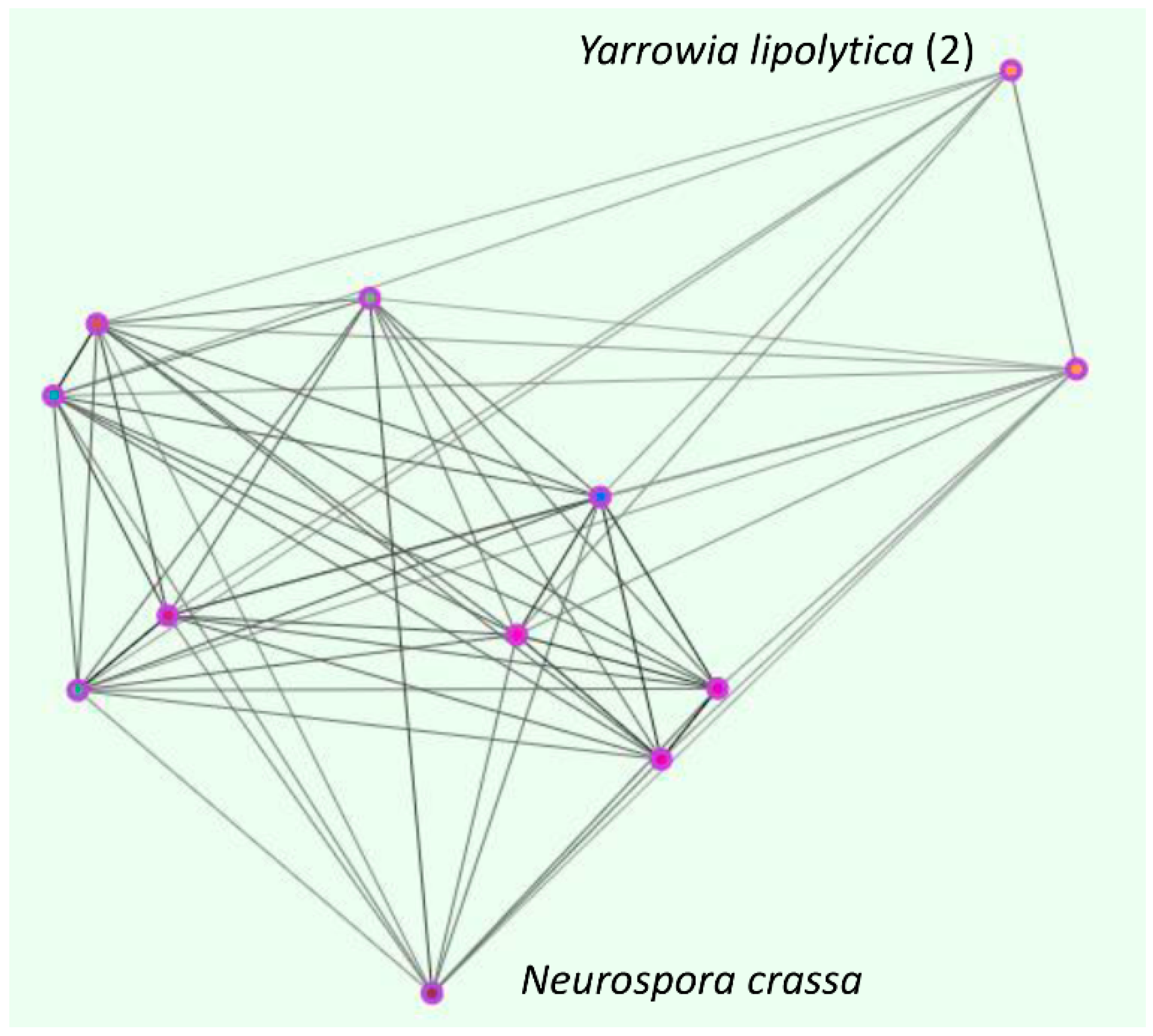
4. The Fungal Zincophore Locus Cluster (OG5_141027)
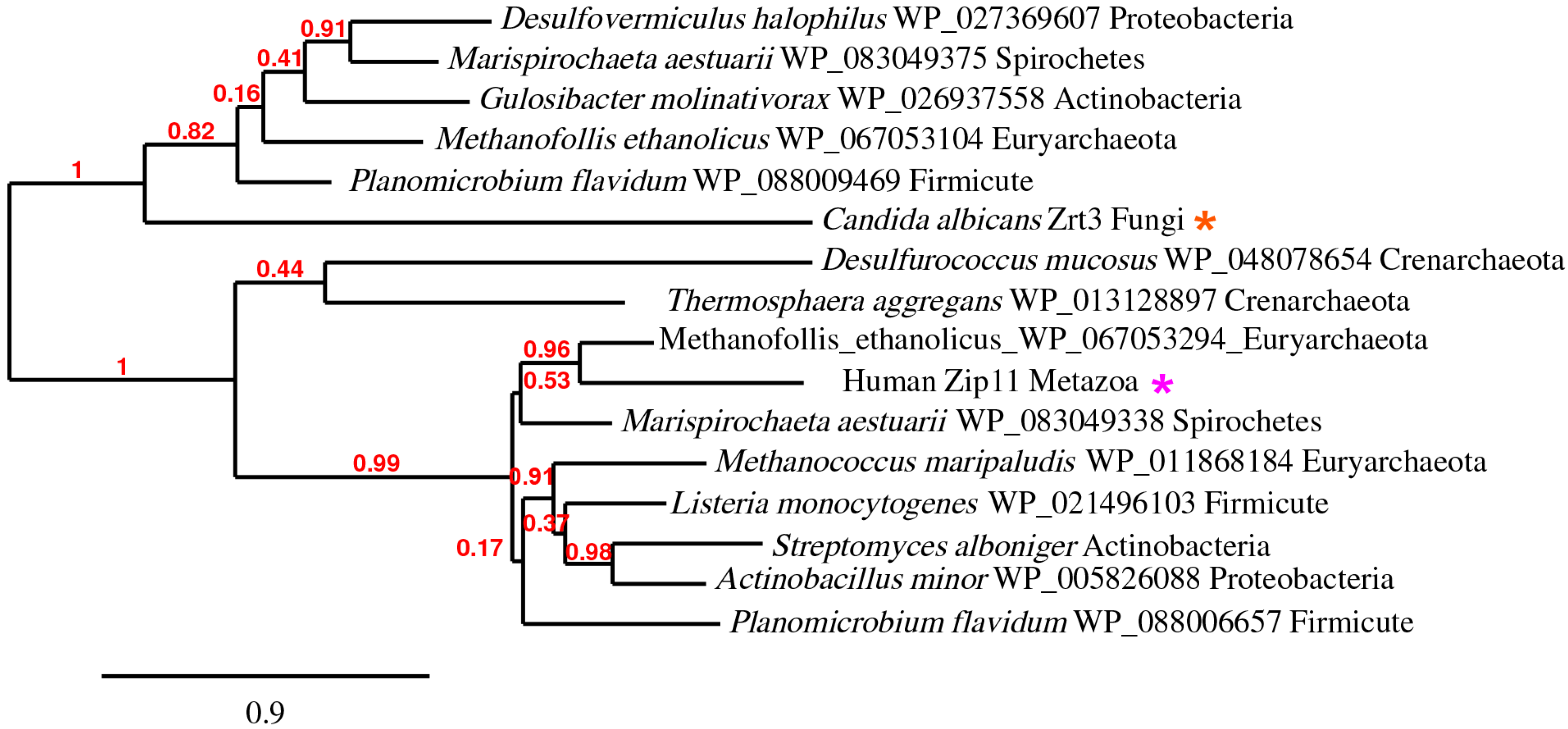
5. The ZupT/ZIP11/Zrt3 Pan-Domain Supercluster (OG5_127397)
Supplementary Materials
Author Contributions
Conflicts of Interest
Funding
References
- Botella, H.; Peyron, P.; Levillain, F.; Poincloux, R.; Poquet, Y.; Brandli, I.; Wang, C.; Tailleux, L.; Tilleul, S.; Charriere, G.M.; et al. Mycobacterial P1-type atpases mediate resistance to zinc poisoning in human macrophages. Cell Host Microbe 2011, 10, 248–259. [Google Scholar] [CrossRef] [PubMed]
- Hood, M.I.; Skaar, E.P. Nutritional immunity: Transition metals at the pathogen-host interface. Nat. Rev. Microbiol. 2012, 10, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Capdevila, D.A.; Wang, J.; Giedroc, D.P. Bacterial strategies to maintain zinc metallostasis at the host-pathogen interface. J. Biol. Chem. 2016, 291, 20858–20868. [Google Scholar] [CrossRef] [PubMed]
- Eide, D.J. Zinc transporters and the cellular trafficking of zinc. Biochim. Biophys. Acta 2006, 1763, 711–722. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Eide, D. The yeast zrt1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc. Natl. Acad. Sci. USA 1996, 93, 2454–2458. [Google Scholar] [CrossRef] [PubMed]
- Eide, D.; Broderius, M.; Fett, J.; Guerinot, M.L. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl. Acad. Sci. USA 1996, 93, 5624–5628. [Google Scholar] [CrossRef] [PubMed]
- Karlinsey, J.E.; Maguire, M.E.; Becker, L.A.; Crouch, M.L.; Fang, F.C. The phage shock protein pspa facilitates divalent metal transport and is required for virulence of Salmonella enterica sv. Typhimurium. Mol. Microbiol. 2010, 78, 669–685. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.J.; Culotta, V.C. Suppression of oxidative damage by Saccharomyces cerevisiae atx2, which encodes a manganese-trafficking protein that localizes to golgi-like vesicles. Mol. Cell. Biol. 1996, 16, 6303–6312. [Google Scholar] [CrossRef] [PubMed]
- Amich, J.; Calera, J.A. Zinc acquisition: A key aspect in Aspergillus fumigatus virulence. Mycopathologia 2014, 178, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Schneider Rde, O.; Diehl, C.; Dos Santos, F.M.; Piffer, A.C.; Garcia, A.W.; Kulmann, M.I.; Schrank, A.; Kmetzsch, L.; Vainstein, M.H.; Staats, C.C. Effects of zinc transporters on Cryptococcus gattii virulence. Sci. Rep. 2015, 5, 10104. [Google Scholar] [CrossRef] [PubMed]
- Do, E.; Hu, G.; Caza, M.; Kronstad, J.W.; Jung, W.H. The zip family zinc transporters support the virulence of Cryptococcus neoformans. Med. Mycol. 2016, 54, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Dade, J.; DuBois, J.C.; Pasula, R.; Donnell, A.M.; Caruso, J.A.; Smulian, A.G.; Deepe, G.S., Jr. Hczrt2, a zinc responsive gene, is indispensable for the survival of Histoplasma capsulatum in vivo. Med. Mycol. 2016, 54, 865–875. [Google Scholar] [CrossRef] [PubMed]
- Fischer, S.; Brunk, B.P.; Chen, F.; Gao, X.; Harb, O.S.; Iodice, J.B.; Shanmugam, D.; Roos, D.S.; Stoeckert, C.J., Jr. Using orthomcl to assign proteins to orthomcl-db groups or to cluster proteomes into new ortholog groups. Curr. Protoc. Bioinform. 2011. [Google Scholar] [CrossRef]
- Gaither, L.A.; Eide, D.J. The human zip1 transporter mediates zinc uptake in human k562 erythroleukemia cells. J. Biol. Chem. 2001, 276, 22258–22264. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Dufner-Beattie, J.; Kim, B.E.; Petris, M.J.; Andrews, G.; Eide, D.J. Zinc-stimulated endocytosis controls activity of the mouse zip1 and zip3 zinc uptake transporters. J. Biol. Chem. 2004, 279, 24631–24639. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Eide, D. The zrt2 gene encodes the low affinity zinc transporter in Saccharomyces cerevisiae. J. Biol. Chem. 1996, 271, 23203–23210. [Google Scholar] [CrossRef] [PubMed]
- Vicentefranqueira, R.; Moreno, M.A.; Leal, F.; Calera, J.A. The zrfa and zrfb genes of Aspergillus fumigatus encode the zinc transporter proteins of a zinc uptake system induced in an acid, zinc-depleted environment. Eukaryot. Cell 2005, 4, 837–848. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, S.; Barreira da Silva, R.; Shawki, A.; Castro, H.; Lamy, M.; Eide, D.; Costa, V.; Mackenzie, B.; Tomas, A.M. Lizip3 is a cellular zinc transporter that mediates the tightly regulated import of zinc in Leishmania infantum parasites. Mol. Microbiol. 2015, 96, 581–595. [Google Scholar] [CrossRef] [PubMed]
- Crawford, A.; Lehtovirta-Morley, L.E.; Alamir, O.; Niemiec, M.J.; Alawfi, B.; Alsarraf, M.; Skrahina, V.; Costa, A.C.B.P.; Anderson, A.; Yellagunda, S.; et al. Biphasic zinc compartmentalisation in a human fungal pathogen. Unpublished work. 2017. [Google Scholar]
- Amich, J.; Vicentefranqueira, R.; Leal, F.; Calera, J.A. Aspergillus fumigatus survival in alkaline and extreme zinc-limiting environments relies on the induction of a zinc homeostasis system encoded by the zrfc and aspf2 genes. Eukaryot. Cell 2010, 9, 424–437. [Google Scholar] [CrossRef] [PubMed]
- Citiulo, F.; Jacobsen, I.D.; Miramon, P.; Schild, L.; Brunke, S.; Zipfel, P.; Brock, M.; Hube, B.; Wilson, D. Candida albicans scavenges host zinc via pra1 during endothelial invasion. PLoS Pathog. 2012, 8, e1002777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, D. An evolutionary perspective on zinc uptake by human fungal pathogens. Metallomics 2015, 7, 979–985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grass, G.; Wong, M.D.; Rosen, B.P.; Smith, R.L.; Rensing, C. Zupt is a zn(Ⅱ) uptake system in Escherichia coli. J. Bacteriol. 2002, 184, 864–866. [Google Scholar] [CrossRef] [PubMed]
- Herzberg, M.; Bauer, L.; Nies, D.H. Deletion of the zupt gene for a zinc importer influences zinc pools in Cupriavidus metallidurans ch34. Metallomics 2014, 6, 421–436. [Google Scholar] [CrossRef] [PubMed]
- Grass, G.; Franke, S.; Taudte, N.; Nies, D.H.; Kucharski, L.M.; Maguire, M.E.; Rensing, C. The metal permease zupt from Escherichia coli is a transporter with a broad substrate spectrum. J. Bacteriol. 2005, 187, 1604–1611. [Google Scholar] [CrossRef] [PubMed]
- MacDiarmid, C.W.; Gaither, L.A.; Eide, D. Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J. 2000, 19, 2845–2855. [Google Scholar] [CrossRef] [PubMed]
- Alamir, O.; Wilson, D. Zrt3 mediates vacuolar zinc efflux in Candida albicans. Unpublished work. 2017. [Google Scholar]
- Kelleher, S.L.; Velasquez, V.; Croxford, T.P.; McCormick, N.H.; Lopez, V.; MacDavid, J. Mapping the zinc-transporting system in mammary cells: Molecular analysis reveals a phenotype-dependent zinc-transporting network during lactation. J. Cell. Physiol. 2012, 227, 1761–1770. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Wu, A.; Zhang, Z.; Yan, G.; Zhang, F.; Zhang, L.; Shen, X.; Hu, R.; Zhang, Y.; Zhang, K.; et al. Characterization of the gufa subfamily member SLC39A11/Zip11 as a zinc transporter. J. Nutr. Biochem. 2013, 24, 1697–1708. [Google Scholar] [CrossRef] [PubMed]
- Baldauf, S.L.; Roger, A.J.; Wenk-Siefert, I.; Doolittle, W.F. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 2000, 290, 972–977. [Google Scholar] [CrossRef] [PubMed]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.Fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dereeper, A.; Audic, S.; Claverie, J.M.; Blanc, G. Blast-explorer helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 2010, 10, 8. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lehtovirta-Morley, L.E.; Alsarraf, M.; Wilson, D. Pan-Domain Analysis of ZIP Zinc Transporters. Int. J. Mol. Sci. 2017, 18, 2631. https://doi.org/10.3390/ijms18122631
Lehtovirta-Morley LE, Alsarraf M, Wilson D. Pan-Domain Analysis of ZIP Zinc Transporters. International Journal of Molecular Sciences. 2017; 18(12):2631. https://doi.org/10.3390/ijms18122631
Chicago/Turabian StyleLehtovirta-Morley, Laura E., Mohammad Alsarraf, and Duncan Wilson. 2017. "Pan-Domain Analysis of ZIP Zinc Transporters" International Journal of Molecular Sciences 18, no. 12: 2631. https://doi.org/10.3390/ijms18122631
APA StyleLehtovirta-Morley, L. E., Alsarraf, M., & Wilson, D. (2017). Pan-Domain Analysis of ZIP Zinc Transporters. International Journal of Molecular Sciences, 18(12), 2631. https://doi.org/10.3390/ijms18122631



