Implementation of Replica-Averaged Restraints from Nuclear Magnetic Resonance Measurement with UNRES Coarse Grained Model of Polypeptide Chains
Abstract
1. Introduction
2. Results and Discussion
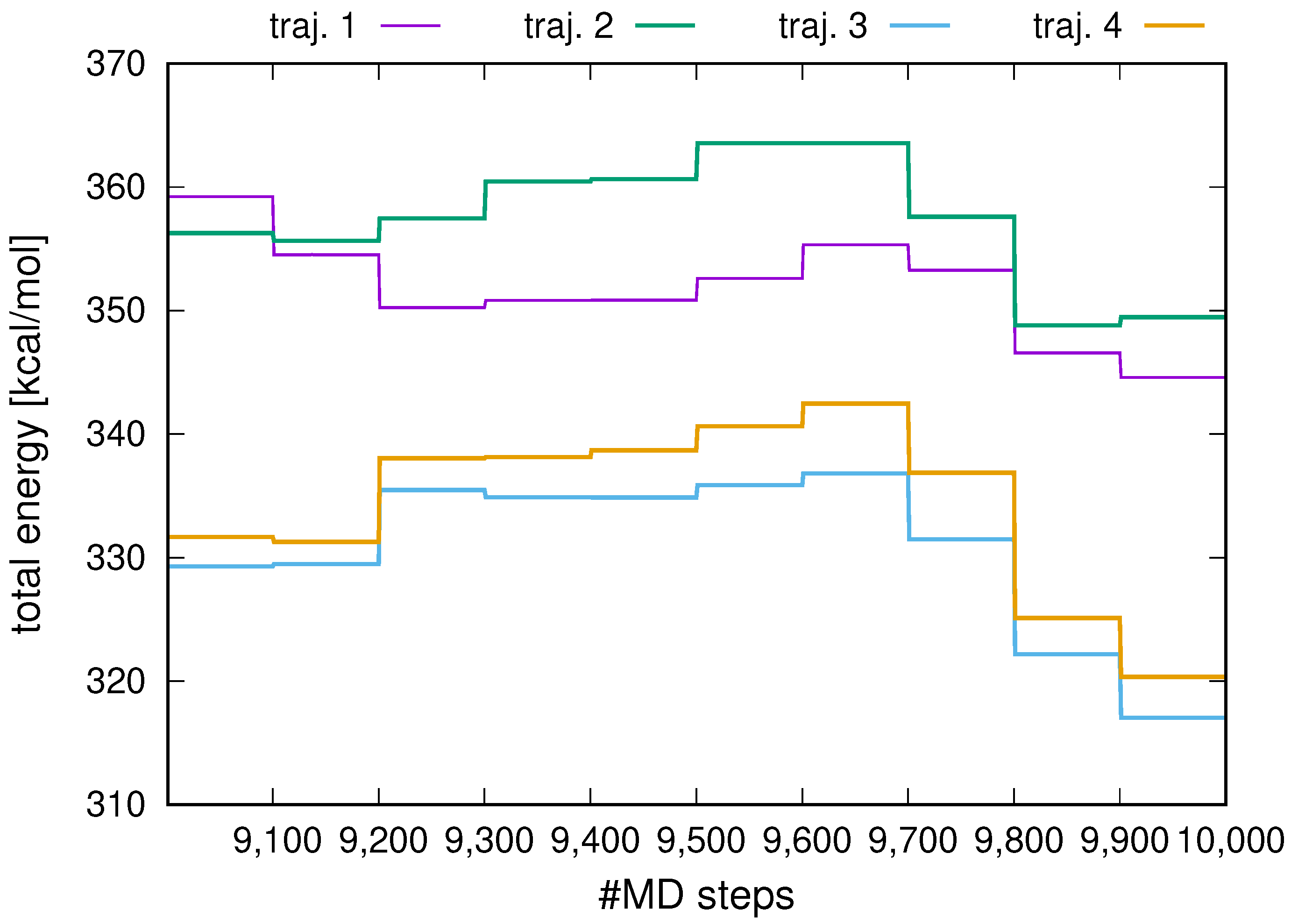
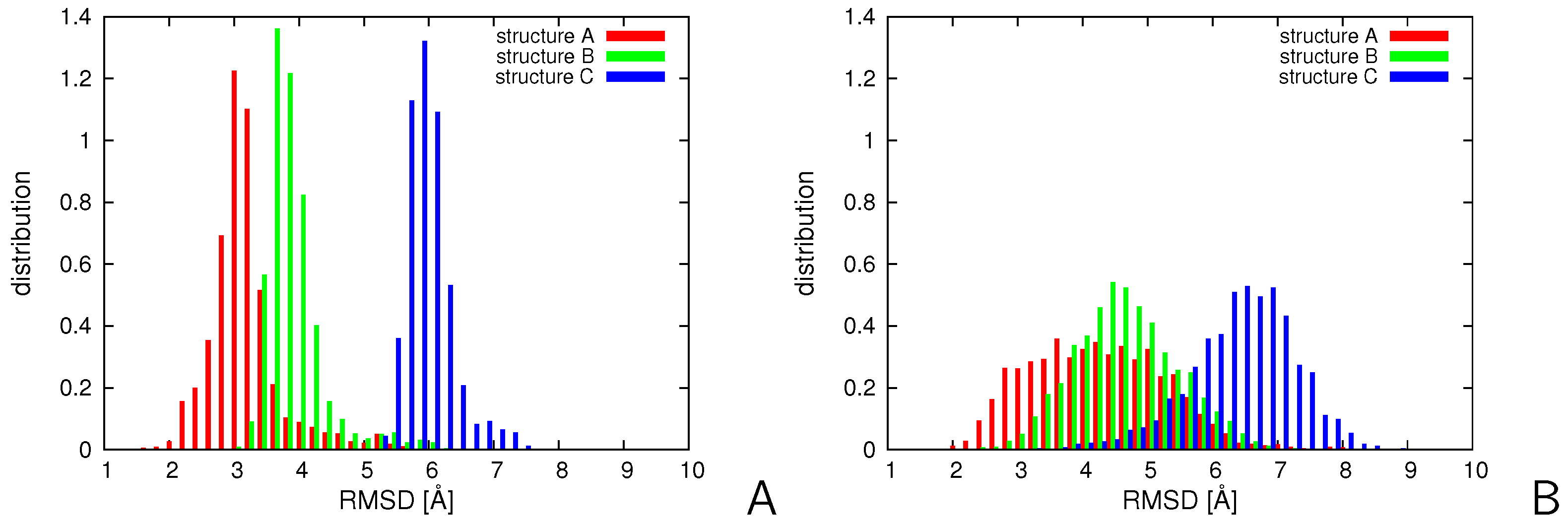
2.1. Stability of Replica-Averaged Simulations
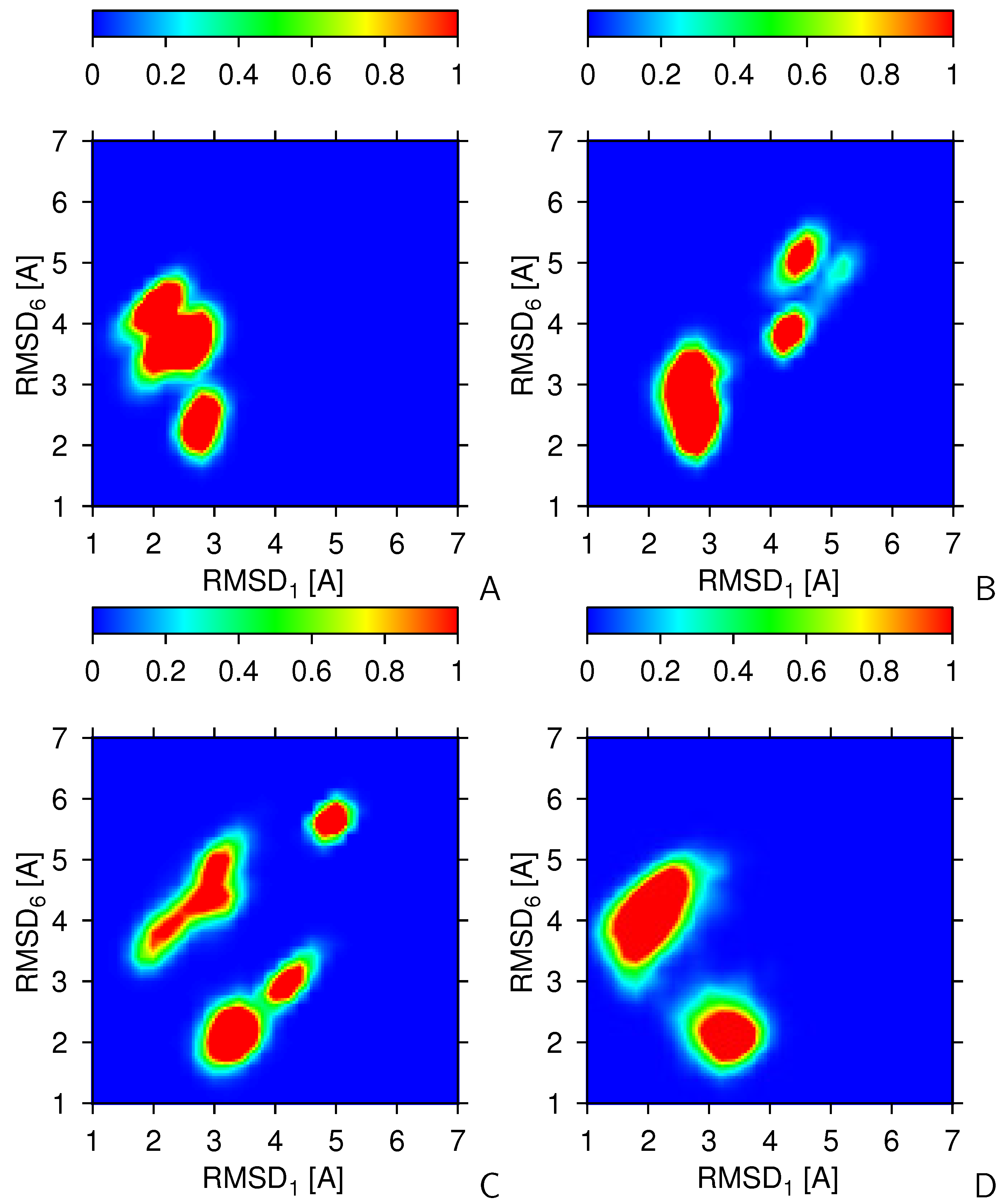
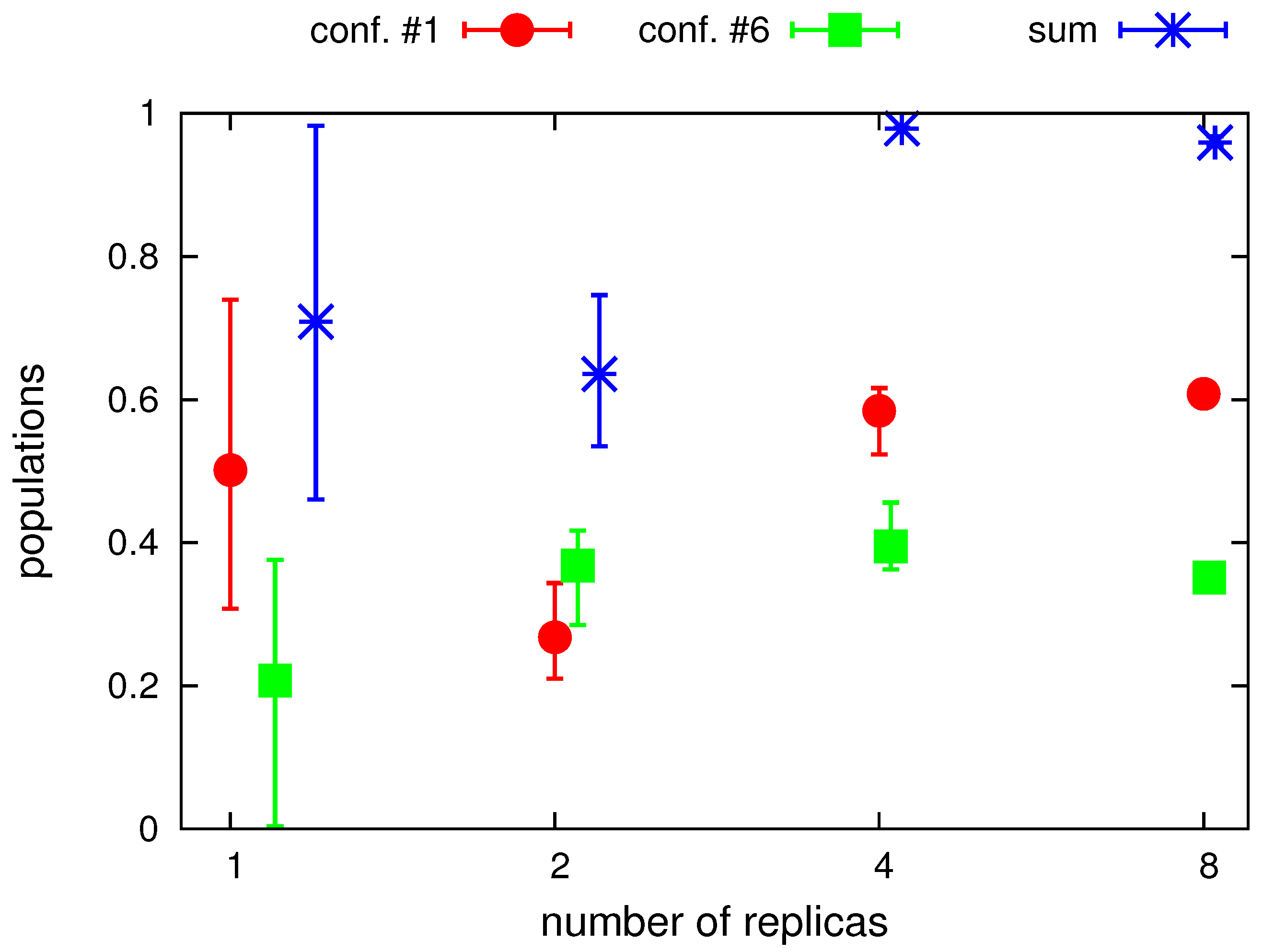
2.2. Tests with Synthetic Restraints
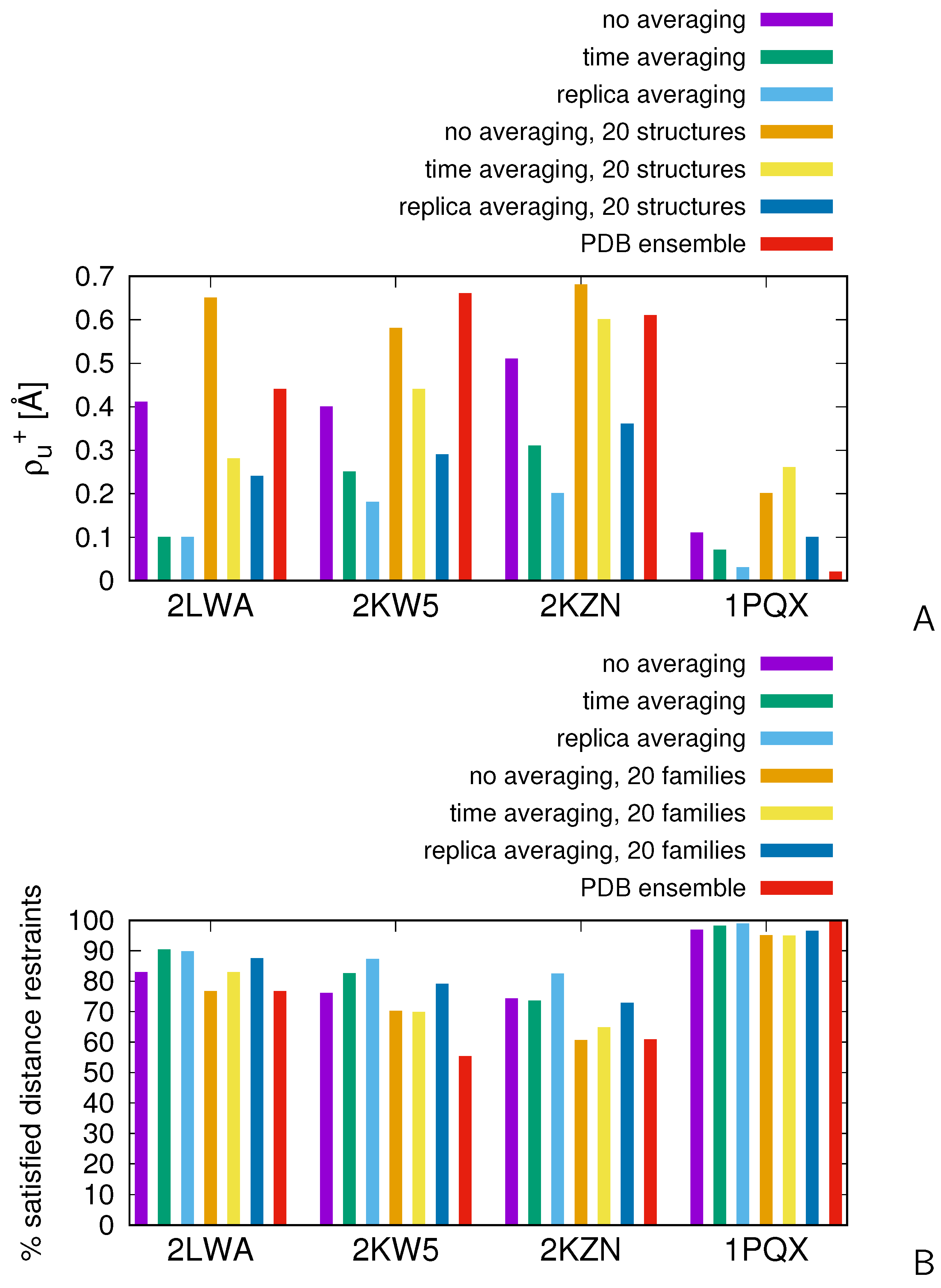
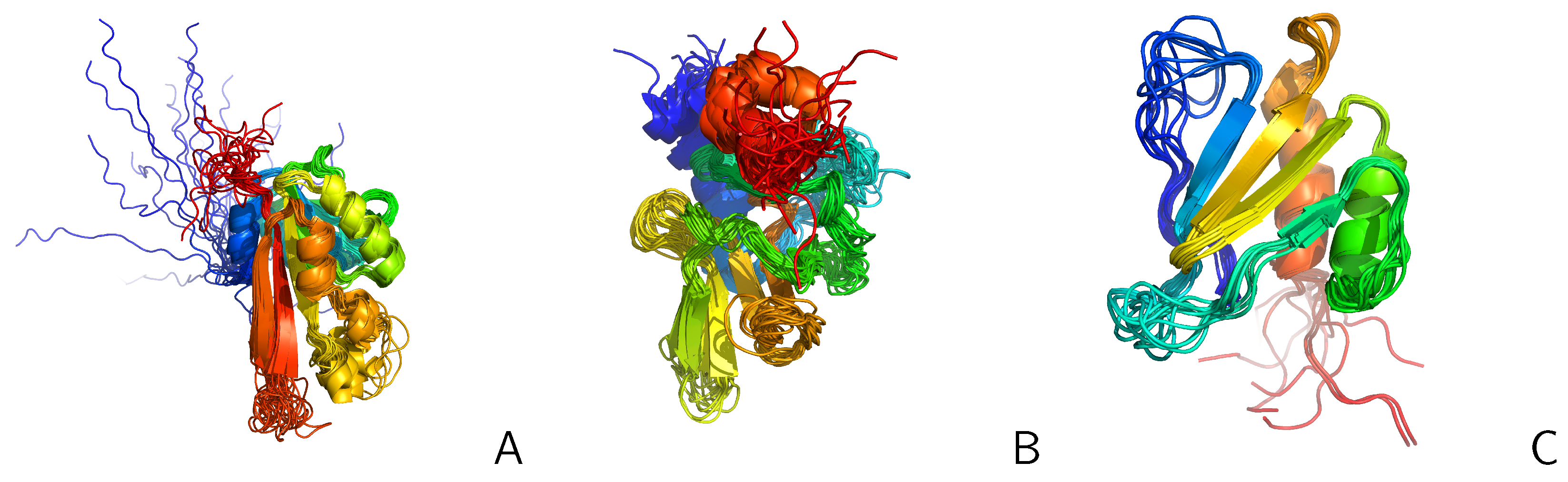
2.3. Tests with Experimental Restraints
3. Methods
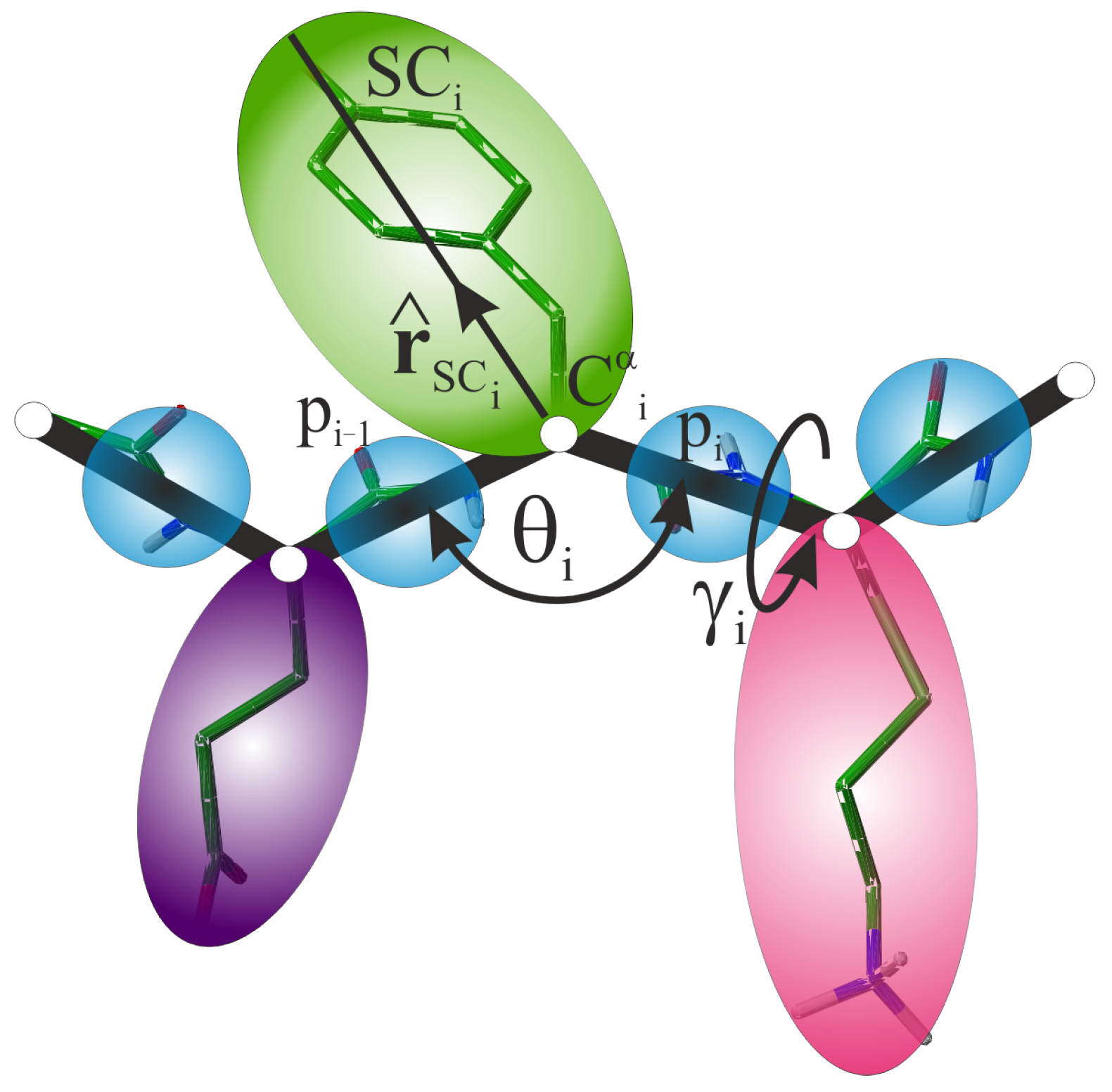
3.1. UNRES Model of Polypeptide Chains
3.2. Conformational Search with UNRES
3.3. Restraints from NMR with UNRES
3.4. Replica-Averaged Restraints
3.5. Systems Studied and Restraints
3.6. Calculation Procedure
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Senior, A.W.; Evans, R.; Jumper, J.; Kirkpatrick, J.; Sifre, L.; Green, T.; Qin, C.; Žídek, A.; Nelson, A.W.R.; Bridgland, A.; et al. Improved protein structure prediction using potentials from deep learning. Nature 2020, 577, 706–710. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- McCammon, J.A.; Gelin, B.R.; Karplus, M. Dynamics of folded proteins. Nature 1977, 267, 585–590. [Google Scholar] [CrossRef]
- Henzler-Wildman, K.; Kern, D. Dynamic personalities of proteins. Nature 2007, 450, 964–972. [Google Scholar] [CrossRef]
- Vendruscolo, M.; Dobson, C.M. Protein dynamics: Moore’s law in molecular biology. Curr. Biol. 2011, 21, R68–R70. [Google Scholar] [CrossRef]
- Dunker, A.K.; Lawson, J.D.; Brown, C.J.; Williams, R.M.; Romero, P.; Oh, J.S.; Oldfield, C.J.; Campen, A.M.; Ratliff, C.M.; Hipps, K.W.; et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001, 19, 26–59. [Google Scholar] [CrossRef]
- van der Lee, R.; Buljan, M.; Lang, B.; Weatheritt, R.J.; Daughdrill, G.W.; Dunker, A.K.; Fuxreiter, M.; Gough, J.; Gsponer, J.; Jones, D.T.; et al. Classification of intrinsically disordered regions and proteins. Chem. Rev. 2014, 114, 6589–6631. [Google Scholar] [CrossRef]
- Uversky, V.N. Intrinsically Disordered Proteins; SpringerBriefs in Molecular Science; Springer International Publishing: Cham, Switzerland, 2014. [Google Scholar] [CrossRef]
- Amaral, M.; Kokh, D.B.; Bomke, J.; Wegener, A.; Buchstaller, H.P.; Eggenweiler, H.M.; Matias, P.; Sirrenberg, C.; Wade, R.C.; Frech, M. Protein conformational flexibility modulates kinetics and thermodynamics of drug binding. Nat. Commun. 2017, 8, 2276. [Google Scholar] [CrossRef] [PubMed]
- Boehr, D.D.; Nussinov, R.; Wright, P.E. The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 2009, 5, 789–796. [Google Scholar] [CrossRef] [PubMed]
- Boura, E.; Rózycki, B.; Herrick, D.Z.; Chung, H.S.; Vecer, J.; Eaton, W.A.; Cafiso, D.S.; Hummer, G.; Hurley, J.H. Solution structure of the ESCRT-I complex by small-angle X-ray scattering, EPR, and FRET spectroscopy. Proc. Natl. Acad. Sci. USA 2011, 108, 9437–9442. [Google Scholar] [CrossRef] [PubMed]
- Ward, A.B.; Sali, A.; Wilson, I.A. Integrative structural biology. Science 2013, 339, 913–915. [Google Scholar] [CrossRef]
- Lu, X.; Chen, J.; Huang, J. The continuous evolution of biomolecular force fields. Structure 2025, 33, 1138–1149. [Google Scholar] [CrossRef] [PubMed]
- Bonomi, M.; Heller, G.T.; Camilloni, C.; Vendruscolo, M. Principles of protein structural ensemble determination. Curr. Opin. Struct. Biol. 2017, 42, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Gomes, G.N.W.; Krzeminski, M.; Namini, A.; Martin, E.W.; Mittag, T.; Head-Gordon, T.; Forman-Kay, J.D.; Gradinaru, C.C. Conformational ensembles of an intrinsically disordered protein consistent with NMR, SAXS, and single-molecule FRET. J. Am. Chem. Soc. 2020, 142, 15697–15710. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Brüschweiler, R. Quantitative prediction of ensemble dynamics, shapes and contact propensities of intrinsically disordered proteins. PLoS Comput. Biol. 2022, 18, e1010036. [Google Scholar] [CrossRef]
- Mittermaier, A.; Kay, L.E. New tools provide new insights in NMR studies of protein dynamics. Science 2006, 312, 224–228. [Google Scholar] [CrossRef]
- Salmon, L.; Nodet, G.; Ozenne, V.; Yin, G.; Jensen, M.R.; Zweckstetter, M.; Blackledge, M. NMR Characterization of long-range order in intrinsically disordered proteins. J. Am. Chem. Soc. 2010, 132, 8407–8418. [Google Scholar] [CrossRef]
- Konrat, R. NMR contributions to structural dynamics studies of intrinsically disordered proteins. J. Magn. Reson. 2014, 241, 74–85. [Google Scholar] [CrossRef]
- Adamski, W.; Salvi, N.; Maurin, D.; Magnat, J.; Milles, S.; Jensen, M.R.; Abyzov, A.; Moreau, C.J.; Blackledge, M.A. A unified description of intrinsically disordered protein dynamics under physiological conditions using NMR spectroscopy. J. Am. Chem. Soc. 2019, 141, 17817–17829. [Google Scholar] [CrossRef]
- Konarev, P.V.; Volkov, V.V.; Sokolova, A.V.; Koch, M.H.J.; Svergun, D.I. PRIMUS: A Windows PC-based system for small-angle scattering data analysis. J. Appl. Cryst. 2003, 36, 1277–1282. [Google Scholar] [CrossRef]
- Aznauryan, M.; Delgado, L.; Soranno, A.; Nettels, D.; Huang, J.; Labhardt, A.M.; Grzesiek, S.; Schuler, B. Comprehensive structural and dynamical view of an unfolded protein from the combination of single-molecule FRET, NMR, and SAXS. Proc. Natl. Acad. Sci. USA 2016, 113, E5389–E5398. [Google Scholar] [CrossRef] [PubMed]
- Schuler, B. Single-molecule FRET of protein structure and dynamics—A primer. J. Nanobiotechnol. 2013, 11, S2. [Google Scholar] [CrossRef] [PubMed]
- Bonomi, M.; Vendruscolo, M. Determination of protein structural ensembles using cryo-electron microscopy. Curr. Opin. Struct. Biol. 2019, 56, 37–45. [Google Scholar] [CrossRef]
- Fraser, J.S.; van den Bedem, H.; Samelson, A.J.; Lang, P.T.; Holton, J.M.; Echols, N.; Alber, T. Accessing protein conformational ensembles using room-temperature X-ray crystallography. Proc. Natl. Acad. Sci. USA 2011, 108, 16247–16252. [Google Scholar] [CrossRef]
- Rappsilber, J. The beginning of a beautiful friendship: Cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes. J. Struct. Biol. 2011, 173, 530–540. [Google Scholar] [CrossRef]
- Leitner, A.; Joachimiak, L.A.; Unverdorben, P.; Walzthoeni, T.; Frydman, J.; Förster, F.; Aebersold, R. Chemical cross-linking/mass spectrometry targeting acidic residues in proteins and protein complexes. Proc. Natl. Acad. Sci. USA 2014, 111, 9455–9460. [Google Scholar] [CrossRef]
- Piersimoni, L.; Kastritis, P.L.; Arlt, C.; Sinz, A. Cross-linking mass spectrometry for investigating protein conformations and protein-protein interactions—A method for all seasons. Chem. Rev. 2022, 122, 7500–7531. [Google Scholar] [CrossRef]
- Huang, Y.J.; Brock, K.P.; Ishida, Y.; Swapna, G.V.; Inouye, M.; Marks, D.S.; Sander, C.; Montelione, G.T. Chapter Thirteen—Combining Evolutionary Covariance and NMR Data for Protein Structure Determination. In Biological NMR Part A; Wand, A.J., Ed.; Methods in Enzymology; Academic Press: Cambridge, MA, USA, 2019; Volume 614, pp. 363–392. [Google Scholar] [CrossRef]
- Orioli, S.; Larsen, A.H.; Bottaro, S.; Lindorff-Larsen, K. How to learn from inconsistencies: Integrating molecular simulations with experimental data. Prog. Mol. Biol. Transl. Sci. 2020, 170, 123–176. [Google Scholar] [CrossRef]
- Shaw, D.E.; Deneroff, M.M.; Dror, R.O.; Kuskin, J.S.; Larson, R.H.; Salmon, J.K.; Young, C.; Batson, B.; Bowers, K.J.; Chao, J.C.; et al. Anton, a special-purpose machine for molecular dynamics simulation. Commun. ACM 2008, 51, 91–97. [Google Scholar] [CrossRef]
- Tozzini, V. Minimalist models for proteins: A comparative analysis. Q. Rev. Biophys. 2010, 43, 333–371. [Google Scholar] [CrossRef] [PubMed]
- Kmiecik, S.; Gront, D.; Kolinski, M.; Wieteska, L.; Dawid, A.E.; Kolinski, A. Coarse-grained protein models and their applications. Chem. Rev. 2016, 116, 7898–7936. [Google Scholar] [CrossRef] [PubMed]
- Noid, W.G. Perspective: Advances, challenges, and insight for predictive coarse-grained models. J. Phys. Chem. B 2023, 127, 4174–4207. [Google Scholar] [CrossRef] [PubMed]
- Borges-Araujo, L.; Patmanidis, I.; Singh, A.P.; Santos, L.H.S.; Sieradzan, A.K.; Vanni, S.; Czaplewski, C.; Pantano, S.; Shinoda, W.; Monticelli, L.; et al. Pragmatic coarse-graining of proteins: Models and applications. J. Chem. Theory Comput. 2023, 19, 7112–7135. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Liwo, A.; Sieradzan, A.K.; Lipska, A.G.; Czaplewski, C.; Joung, I.; Żmudzińska, W.; Hałabis, A.; Ołdziej, S. A general method for the derivation of the functional forms of the effective energy terms in coarse-grained energy functions of polymers. III. Determination of scale-consistent backbone-local and correlation potentials in the UNRES force field and force-field calibration and validation. J. Chem. Phys. 2019, 150, 155104. [Google Scholar] [CrossRef]
- Sieradzan, A.K.; Czaplewski, C.; Krupa, P.; Mozolewska, M.A.; Karczyńska, A.S.; Lipska, A.G.; Lubecka, E.A.; Gołaś, E.; Wirecki, T.; Makowski, M.; et al. Modeling the structure, dynamics, and transformations of proteins with the UNRES force field. In Protein Folding: Methods and Protocols; Muñoz, V., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2022; Volume 2376, Chapter 23; pp. 399–416. [Google Scholar] [CrossRef]
- Sieradzan, A.K.; Sans-Dueño, J.; Lubecka, E.A.; Czaplewski, C.; Lipska, A.G.; Leszczyński, H.; Ocetkiewicz, K.M.; Proficz, J.; Czarnul, P.; Krawczyk, H.; et al. Optimization of parallel implementation of UNRES package for coarse-grained simulations to treat large proteins. J. Comput. Chem. 2023, 44, 602–625. [Google Scholar] [CrossRef]
- Wüthrich, K. NMR of Proteins and Nucleic Acids; Wiley: New York, NY, USA, 1986. [Google Scholar]
- Schwieters, C.D.; Kuszewski, J.; Clore, G.M. Using Xplor-NIH for NMR molecular structure determination. Prog. Nucl. Magn. Reson. Spectrosc. 2006, 48, 47–62. [Google Scholar] [CrossRef]
- Torda, A.E.; Scheek, R.M.; van Gunsteren, W.F. Time-dependent distance restraints in molecular dynamics simulations. Chem. Phys. Lett. 1989, 157, 289–294. [Google Scholar] [CrossRef]
- Torda, A.E.; Brunne, R.M.; Huber, T.; Kessler, H.; van Gunsteren, W.F. Structure refinement using time-averaged J-coupling constant restraints. J. Biomol. NMR 1993, 3, 55–66. [Google Scholar] [CrossRef]
- Bonvin, A.M.J.J.; Boelens, R.; Kaptein, R. Time- and ensemble-averaged direct NOE restraints. J. Biomol. NMR 1994, 4, 143–149. [Google Scholar] [CrossRef]
- Hansen, N.; Heller, F.; Schmid, N.; van Gunsteren, W.F. Time-averaged order parameter restraints in molecular dynamics simulations. J. Biomol. NMR 2014, 60, 169–187. [Google Scholar] [CrossRef]
- Pearlman, D.A.; Case, D.A.; Caldwell, J.W.; Ross, W.S.; Cheatham, T.E.; DeBolt, S.; Ferguson, D.; Seibel, G.; Kollman, P. AMBER: A package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput. Phys. Commun. 1995, 91, 1–41. [Google Scholar] [CrossRef]
- Co, N.T.; Czaplewski, C.; Lubecka, E.A.; Liwo, A. Implementation of time-averaged restraints with UNRES coarse-grained model of polypeptide chains. J. Chem. Theory Comput. 2025, 21, 1476–1493. [Google Scholar] [CrossRef]
- Lubecka, E.A.; Liwo, A. ESCASA: Analytical estimation of atomic coordinates from coarse-grained geometry for nuclear-magnetic-resonance-assisted protein structure modeling. I. Backbone and Hβ protons. J. Comput. Chem. 2021, 42, 1579–1589. [Google Scholar] [CrossRef]
- Nikiforovich, G.V.; Vesterman, B.; Betins, J.; Podins, L. The space structure of a conformationally labile oligopeptide in solution: Angiotensin. J. Biomol. Struct. Dyn. 1987, 4, 1119–1135. [Google Scholar] [CrossRef]
- Bonomi, M.; Camilloni, C.; Cavalli, A.; Vendruscolo, M. Metainference: A Bayesian inference method for heterogeneous systems. Sci. Adv. 2016, 2, e1501177. [Google Scholar] [CrossRef] [PubMed]
- Schwieters, C.D.; Bermejo, G.A.; Clore, G.M. Xplor-NIH for molecular structure determination from NMR and other data sources. Protein Sci. 2018, 27, 26–40. [Google Scholar] [CrossRef] [PubMed]
- Medeiros Selegato, D.; Bracco, C.; Giannelli, C.; Parigi, G.; Luchinat, C.; Sgheri, L.; Ravera, E. Comparison of different reweighting approaches for the calculation of conformational variability of macromolecules from molecular simulations. ChemPhysChem 2020, 22, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Pitera, J.W.; Chodera, J.D. On the use of experimental observations to bias simulated ensembles. J. Chem. Theory Comput. 2012, 8, 3445–3451. [Google Scholar] [CrossRef]
- Roux, B.; Weare, J. On the statistical equivalence of restrained-ensemble simulations with the maximum entropy method. J. Chem. Phys. 2013, 138, 085102. [Google Scholar] [CrossRef]
- Hummer, G.; Köfinger, J. Bayesian ensemble refinement by replica simulations and reweighting. J. Chem. Phys. 2015, 143, 243150. [Google Scholar] [CrossRef]
- Olsson, S.; Cavalli, A. Quantification of entropy-loss in replica-averaged modeling. J. Chem. Theory Comput. 2015, 11, 3973–3977. [Google Scholar] [CrossRef]
- Cavalli, A.; Camilloni, C.; Vendruscolo, M. Molecular dynamics simulations with replica-averaged structural restraints generate structural ensembles according to the maximum entropy principle. J. Chem. Phys. 2013, 138, 094112. [Google Scholar] [CrossRef]
- Camilloni, C.; Cavalli, A.; Vendruscolo, M. Replica-averaged metadynamics. J. Chem. Theory Comput. 2013, 9, 5610–5617. [Google Scholar] [CrossRef]
- Camilloni, C.; Vendruscolo, M. Statistical mechanics of the denatured state of a protein using replica-averaged metadynamics. J. Am. Chem. Soc. 2014, 136, 8982–8991. [Google Scholar] [CrossRef] [PubMed]
- Raddi, R.M.; Marshall, T.; Ge, Y.; Voelz, V.A. Model selection using replica averaging with Bayesian inference of conformational populations. J. Chem. Theory Comput. 2025, 21, 5880–5889. [Google Scholar] [CrossRef] [PubMed]
- Birkhoff, G.D. Proof of the ergodic theorem. Proc. Natl. Acad. Sci. USA 1931, 17, 656–660. [Google Scholar] [CrossRef] [PubMed]
- von Neumann, J.V. Proof of the quasi-ergodic hypothesis. Proc. Natl. Acad. Sci. USA 1932, 18, 70–82. [Google Scholar] [CrossRef]
- Rhee, Y.M.; Pande, V.S. Multiplexed-replica exchange molecular dynamics method for protein folding simulation. Biophys. J. 2003, 84, 775–786. [Google Scholar] [CrossRef]
- Czaplewski, C.; Kalinowski, S.; Liwo, A.; Scheraga, H.A. Application of multiplexed replica exchange molecular dynamics to the UNRES force field: Tests with α and α+β proteins. J. Chem. Theory Comput. 2009, 5, 627–640. [Google Scholar] [CrossRef]
- Schrödinger, LLC. The PyMOL Molecular Graphics System, Version 3.1.6.1; Schrödinger, LLC: New York, NY, USA, 2025.
- Williams, T.; Kelley, C.; Bröker, H.-B.; Campbell, J.; Cunningham, R.; Denholm, D.; Elber, G.; Fearick, R.; Grammes, C.; Hart, L.; et al. Gnuplot: An Interactive Plotting Program, Version 6.0.3. 2025. Available online: https://gnuplot.sourceforge.net/ (accessed on 7 June 2025).
- Köfinger, J.; Hummer, G. Encoding prior knowledge in ensemble refinement. J. Chem. Phys. 2024, 160, 114111. [Google Scholar] [CrossRef] [PubMed]
- Kelley, D.; Galbraith, P. GRI: Scientific Graphics Language, Version 2.12.23. 2023. Available online: http://gri.sourceforge.net/ (accessed on 5 November 2024).
- Lorieau, J.L.; Louis, J.M.; Schwieters, C.D.; Bax, A. pH-triggered, activated-state conformations of the influenza hemagglutinin fusion peptide revealed by NMR. Proc. Natl. Acad. Sci. USA 2012, 109, 19994–19999. [Google Scholar] [CrossRef]
- Güntert, P.; Buchner, L. Combined automated NOE assignment and structure calculation with CYANA. J. Biomol. NMR 2015, 62, 453–471. [Google Scholar] [CrossRef]
- Zaborowski, B.; Jagieła, D.; Czaplewski, C.; Hałabis, A.; Lewandowska, A.; Żmudzińska, W.; Ołdziej, S.; Karczyńska, A.; Omieczynski, C.; Wirecki, T.; et al. A maximum-likelihood approach to force-field calibration. J. Chem. Inf. Model. 2015, 55, 2050–2070. [Google Scholar] [CrossRef]
- Sieradzan, A.K.; Makowski, M.; Augustynowicz, A.; Liwo, A. A general method for the derivation of the functional forms of the effective energy terms in coarse-grained energy functions of polymers. I. Backbone potentials of coarse-grained polypeptide chains. J. Chem. Phys. 2017, 146, 124106. [Google Scholar] [CrossRef]
- Liwo, A.; Khalili, M.; Czaplewski, C.; Kalinowski, S.; Ołdziej, S.; Wachucik, K.; Scheraga, H.A. Modification and optimization of the United-Residue (UNRES) potential energy function for canonical simulations. I. Temperature dependence of the effective energy function and tests of the optimization method with single training proteins. J. Phys. Chem. B 2007, 111, 260–285. [Google Scholar] [CrossRef]
- Khalili, M.; Liwo, A.; Rakowski, F.; Grochowski, P.; Scheraga, H.A. Molecular dynamics with the united-residue model of polypeptide chains. I. Lagrange equations of motion and tests of numerical stability in the microcanonical mode. J. Phys. Chem. B 2005, 109, 13785–13797. [Google Scholar] [CrossRef] [PubMed]
- Khalili, M.; Liwo, A.; Jagielska, A.; Scheraga, H.A. Molecular dynamics with the united-residue model of polypeptide chains. II. Langevin and Berendsen-bath dynamics and tests on model α-helical systems. J. Phys. Chem. B 2005, 109, 13798–13810. [Google Scholar] [CrossRef]
- Swope, W.C.; Andersen, H.C.; Berens, P.H.; Wilson, K.R. A computer simulation method for the calculation of equilibrium constants for the formation of physical clusters of molecules: Application to small water clusters. J. Chem. Phys. 1982, 76, 637–649. [Google Scholar] [CrossRef]
- Hansmann, U.H.E. Parallel tempering algorithm for conformational studies of biological molecules. Chem. Phys. Lett. 1997, 281, 140–150. [Google Scholar] [CrossRef]
- Trebst, S.; Troyer, M.; Hansmann, U.H.E. Optimized parallel tempering simulations of proteins. J. Chem. Phys. 2006, 124, 174903. [Google Scholar] [CrossRef]
- Kumar, S.; Rosenberg, J.M.; Bouzida, D.; Swendsen, R.H.; Kollman, P.A. The weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J. Comput. Chem. 1992, 13, 1011–1021. [Google Scholar] [CrossRef]
- Nishikawa, K.; Momany, F.A.; Scheraga, H.A. Low-energy structures of two dipeptides and their relationship to bend conformations. Macromolecules 1974, 7, 797–806. [Google Scholar] [CrossRef] [PubMed]
- Lubecka, E.A.; Liwo, A. A coarse-grained approach to NMR-data-assisted modeling of protein structures. J. Comput. Chem. 2022, 43, 2047–2059. [Google Scholar] [CrossRef]
- Lange, O.F.; Rossi, P.; Sgourakis, N.G.; Song, Y.; Lee, H.W.; Aramini, J.M.; Ertekin, A.; Xiao, R.; Acton, T.B.; Montelione, G.T.; et al. Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples. Proc. Natl. Acad. Sci. USA 2012, 109, 10873–10878. [Google Scholar] [CrossRef]
- Everett, J.K.; Tejero, R.; Murthy, S.B.K.; Acton, T.B.; Aramini, J.M.; Baran, M.C.; Benach, J.; Cort, J.R.; Eletsky, A.; Forouhar, F.; et al. A community resource of experimental data for NMR / X-Ray crystal structure pairs. Protein Sci. 2016, 25, 30–45. [Google Scholar] [CrossRef]
- Lee, J.; Liwo, A.; Scheraga, H.A. Energy-based de novo protein folding by conformational space annealing and an off-lattice united-residue force field: Application to the 10–55 fragment of staphylococcal protein A and to apo calbindin D9K. Proc. Natl. Acad. Sci. USA 1999, 96, 2025–2030. [Google Scholar] [CrossRef]
- Heo, L.; Feig, M. One Bead Per Residue Can Describe All-Atom Protein Structures. cg2all Version v1.3.1. 2023. Available online: https://github.com/huhlim/cg2all (accessed on 5 November 2024).
- Heo, L.; Feig, M. One bead per residue can describe all-atom protein structures. Structure 2024, 32, 97–111.e6. [Google Scholar] [CrossRef]
- Murtagh, F.; Heck, A. Multivariate Data Analysis; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1987. [Google Scholar] [CrossRef]
- Gorba, C.; Miyashita, O.; Tama, F. Normal-mode flexible fitting of high-resolution structure of biological molecules toward one-dimensional low-resolution data. Biophys. J. 2008, 94, 1589–1599. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Roux, B. EROS: Better than SAXS! Structure 2011, 19, 3–4. [Google Scholar] [CrossRef]
- Kimanius, D.; Pettersson, I.; Schluckebier, G.; Lindahl, E.; Andersson, M. SAXS-guided metadynamics. J. Chem. Theory Comput. 2015, 11, 3491–3498. [Google Scholar] [CrossRef] [PubMed]
- Leśniewski, M.; Pyrka, M.; Czaplewski, C.; Co, N.T.; Jiang, Y.; Gong, Z.; Tang, C.; Liwo, A. Assessment of two restraint potentials for coarse-grained chemical-cross-link-assisted modeling of protein structures. J. Chem. Inf. Model. 2024, 64, 1377–1393. [Google Scholar] [CrossRef] [PubMed]
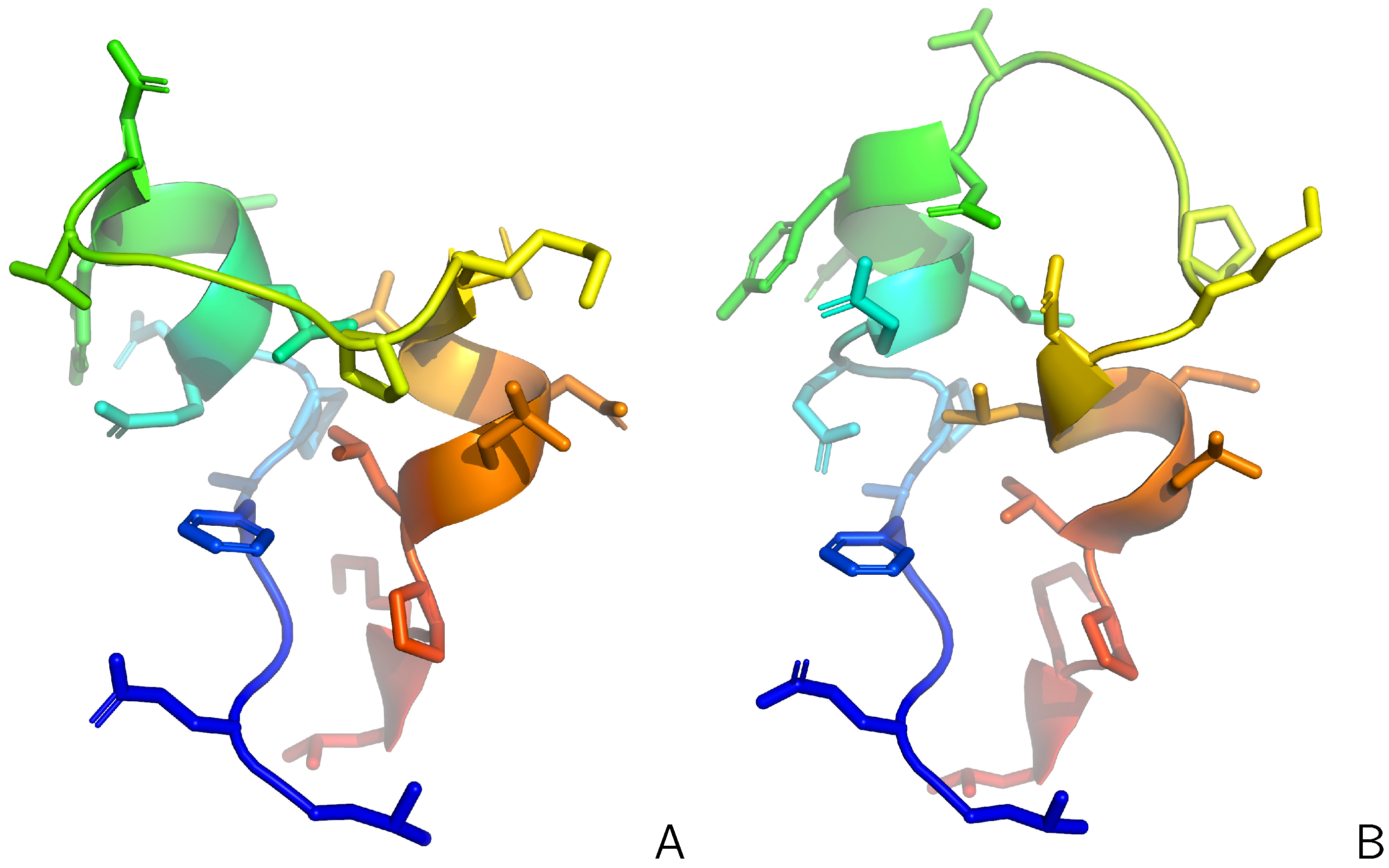

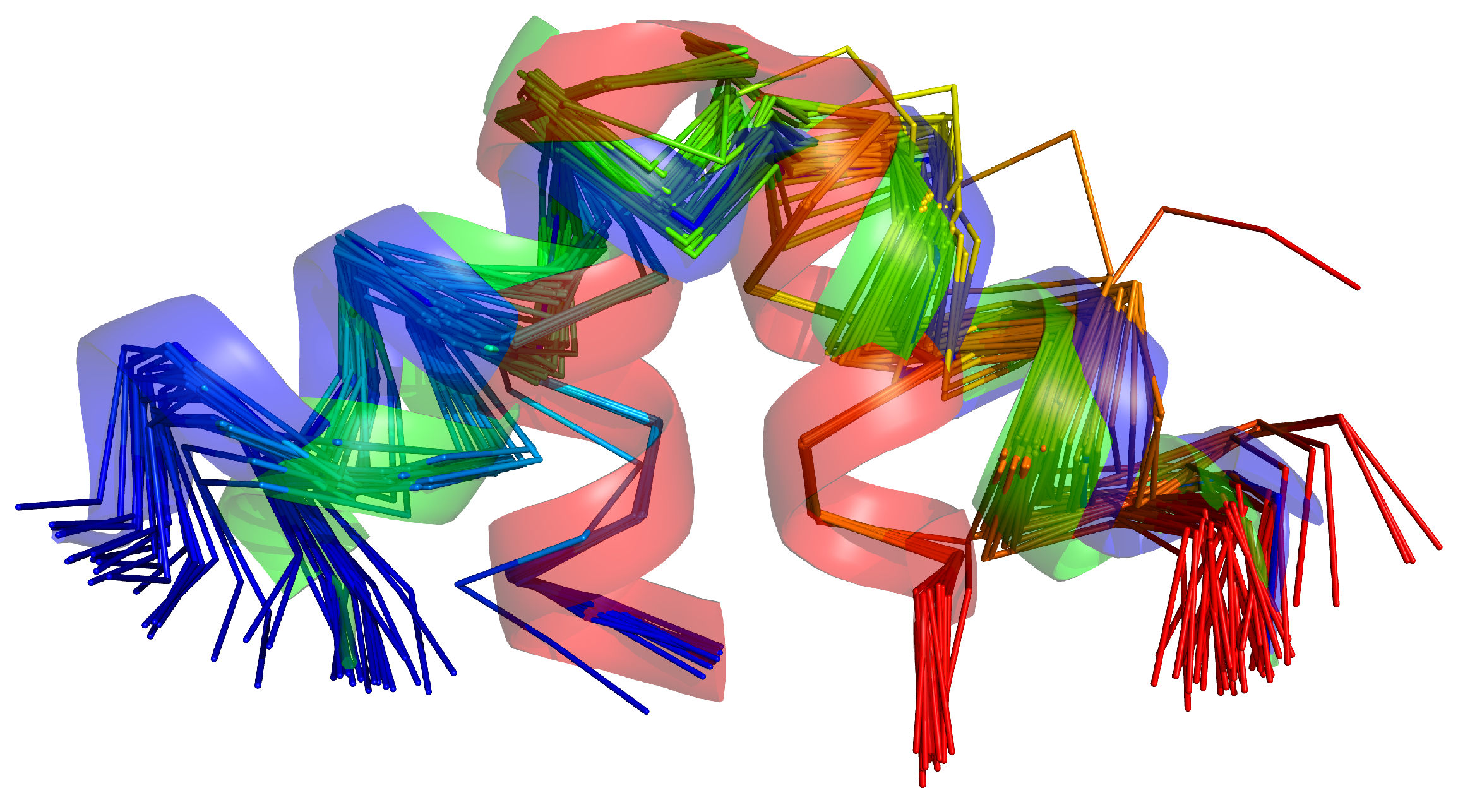
| Protein | # res. | Restraint Type | ||
|---|---|---|---|---|
| 2LWA | 25 | 175 | 18 | 20 |
| 2KW5 | 202 | 947 | 134 | 147 |
| 2KZN | 147 | 578 | 104 | 121 |
| 1PQX | 91 | 1015 | 64 | 74 |
| Set | Temperatures [K] | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 262 | 264 | 267 | 270 | 274 | 277 | 279 | 282 | 285 | 288 | 290 | 292 | |
| 295 | 298 | 301 | 305 | 308 | 315 | 327 | 333 | 340 | 355 | 362 | 370 | |
| 262 | 267 | 274 | 279 | 285 | 290 | 295 | 301 | 308 | 333 | 355 | 370 | |
| 274 | 279 | 285 | 290 | 295 | 301 | 308 | 333 | |||||
| 279 | 285 | 290 | 295 | 301 | 308 | |||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shirkov, L.; Czaplewski, C.; Liwo, A. Implementation of Replica-Averaged Restraints from Nuclear Magnetic Resonance Measurement with UNRES Coarse Grained Model of Polypeptide Chains. Molecules 2025, 30, 4354. https://doi.org/10.3390/molecules30224354
Shirkov L, Czaplewski C, Liwo A. Implementation of Replica-Averaged Restraints from Nuclear Magnetic Resonance Measurement with UNRES Coarse Grained Model of Polypeptide Chains. Molecules. 2025; 30(22):4354. https://doi.org/10.3390/molecules30224354
Chicago/Turabian StyleShirkov, Leonid, Cezary Czaplewski, and Adam Liwo. 2025. "Implementation of Replica-Averaged Restraints from Nuclear Magnetic Resonance Measurement with UNRES Coarse Grained Model of Polypeptide Chains" Molecules 30, no. 22: 4354. https://doi.org/10.3390/molecules30224354
APA StyleShirkov, L., Czaplewski, C., & Liwo, A. (2025). Implementation of Replica-Averaged Restraints from Nuclear Magnetic Resonance Measurement with UNRES Coarse Grained Model of Polypeptide Chains. Molecules, 30(22), 4354. https://doi.org/10.3390/molecules30224354







