1. Introduction
Rubiae radix et rhizoma (RRR), known as “Qian Cao” in China, can cool blood, stop bleeding and remove blood stasis [
1]. It can be traced back to Shennong’s Classic of the Materia Medica and has been used for thousands of years [
2,
3]. As far as varietal origin is concerned, the 2020 edition of Ch.P stipulates that RRR is the dried root and rhizome of
Rubia cordifolia L, a Rubiaceae plant [
3]. Strictly speaking,
Rubia cordifolia L is the legal and only source of RRR, and the other varieties are fakes, of which
Rubia schumanniana E. Pritz. (RSP) and
Rubia magna P. G. Xiao (RMP) are the most common adulterants. However, to reduce costs and gain illegal benefits, unscrupulous traders often sell RSP and RMP as RRR or adulterate them into RRR, which seriously jeopardizes the interests of consumers and even endangers lives. Therefore, it is necessary to strengthen RRR identification evaluation.
To this end, Zhao et al. constructed the complete chloroplast genome of RRR [
4]. Based on morphological and microscopic characteristics, researchers identified RRR, RMP and RSP, and the results showed that RMP has few fine roots and the wood is lighter in colour, while the wood part of the section of RRR is mainly yellow-red in colour [
5,
6,
7]. Li et al. determined the contents of alizarin, lucidin, xanthopurpurin, purpurin, 6-hydroxyrubiadin, chrysophanol, and mollugin, and combined multivariate statistical analysis to construct a partial least squares discriminant analysis (PLS-DA) model, which can differentiate RRR, RMP, and RSP [
8]. All the above studies help improve RRR quality control and market regulation. However, these studies focus more on species differentiation, and it is not ideal to identify whether the RRR is adulterated with RMP or RSP. Therefore, to address the above issues, RRR adulterant evaluation was realized based on LC-MS and “ion identity”.
Due to the characteristics of high sensitivity and high accuracy, LC-MS technology has been widely used in the identification and analysis of traditional Chinese medicine (TCM) [
9,
10,
11]. Pu Y et al. used LC-MS technology to determine lucidin content and the results showed good linearity in the range of 0.005–2.64 ng and the average recovery rate was 97.17%; lucidin can be used for RRR identification and quality control [
12]. Zheng et al. developed an effective ultra-high-performance liquid chromatography coupled with quadrupole time-of-flight tandem mass spectrometry (UHPLC/Q-TOF/MS) method to analyze the chemical constituents in rat plasma and urine after the oral administration of RRR extract and the results showed alizarin-1-O-β-glucuronide and purpurin-3-O-β-glucuronide were identified in rat plasma, which is helpful for RRR metabolic analysis and mechanisms of action [
13]. Humbare et al. extracted RRR with methanol as the solvent, and identified more than 100 chemical constituents, such as 6-methoxygenic acid, Rubiprasin A, and 1-hydroxy-2-methyl anthraquinone, by UPLC-UV-MS, which is helpful for the identification and analysis of RRR [
14]. Hu et al. explored the UHPLC/Q-TOF/MS method to characterize 45 quinones and quantify 24 quinones from Rubia plants and used chemometric methods to evaluate the chemical relationship between Rubia samples according to the composition quinones, which is helpful for Rubia species quality control and identification [
15]. Zhang et al. established the UPLC-QQQ-MS/MS method, and qualitatively and quantitatively analyzed 14 kinds of cyclic peptide compounds in 20 Rubia plants; combined with chemometrics, a classification model of Rubia species was established, which provided a reference for the identification of RRR and its adulterants [
16]. On the other hand, “ion identity” has been established based on the ordered array of high-precision mass spectrometry data, which have better specificity than single ion equivalents and contributes to the identification evaluation of RRR [
17].
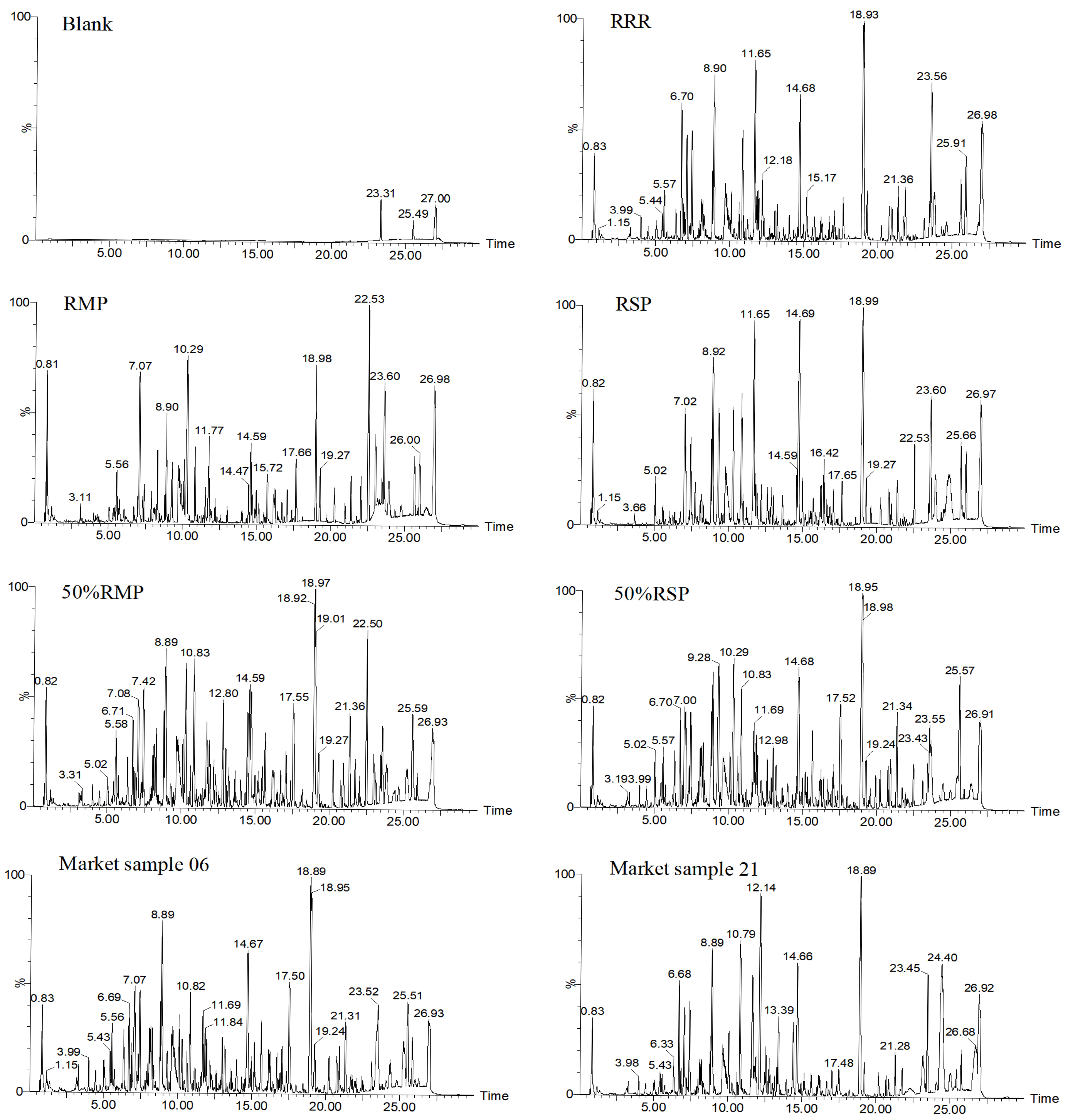
In this research, considering the high sensitivity and accuracy of LC-MS and the high-level specificity of “ion identity”, LC-MS was used to analyze RRR, RSP and RMP. After data conversion, the ion intersections were taken from different batches of RSP, RMP, and RRR. Then, the proprietary ions of three herbs were obtained by taking the de-intersection. Finally, the top n ions were treated as the “ion identity” of RSP, RMP, and RRR for matching test samples to obtain the recognition index (RI) and combined chemometrics for analysis.
3. Discussion
In this paper, the “ion identity” based on LC-MS was proposed to conduct RRR identification evaluation. Firstly, LC-MS was optimized to obtain MS data [
23,
24,
25,
26,
27]. We examined the ESI
+ and ESI
− mode. Samples have more data in the positive mode. The collision voltage was optimized for 10~40 V. In terms of extraction solvents, methanol had the best extraction results. Then, the deviations for
tR and
m/
z were optimized in the process of acquiring the “ion identity”. The
tR drifted but did not exceed 0.1 min. On the other hand, we comprehensively compare the “ion identity” cases when the Δ
m/
z ≤ 0, 0.01, and 0.05 Da. The results showed that when Δ
m/
z ≤ 0.01 Da, this ensures sufficient precision, as well as allowing us to screen further based on ionic strength. However, the shared ions cannot be acquired when Δ
m/
z ≤ 0.00 Da and if Δ
m/
z ≤ 0.05 Da, there are more ion data, but larger deviation will easily lead to false positive results. Finally, the ion number in the “ion identity” is equally important for RRR identification evaluation. Therefore, we examined the matching situation when 50, 100, and 150 ions were output as their respective “ion identities”. As shown in
Table 7, as the ion number in the “ion identity” increases, the RIs of the matches between RRR11, RMP06, and RSP06 and their own “ion identity” decreases, except for the RRR10 sample. Therefore, to take care of the matching between RMP, RSP and the “ion identity”, we finally chose to output the top 50 ions as their respective “ion identities”, which do not affect the matching of RRR simultaneously. If all ions are regarded as “ion identities” for matching identification, the matching effect becomes poor. Not all ions are valid data, some ions are invalid and jumbled, and the ion number in different herbs is different, which leads to the loss of comparative significance of RI. Reducing the number of ions in the “ion identity” to remove invalid data and focusing on high-abundance and highly specific ions can improve the matching accuracy.
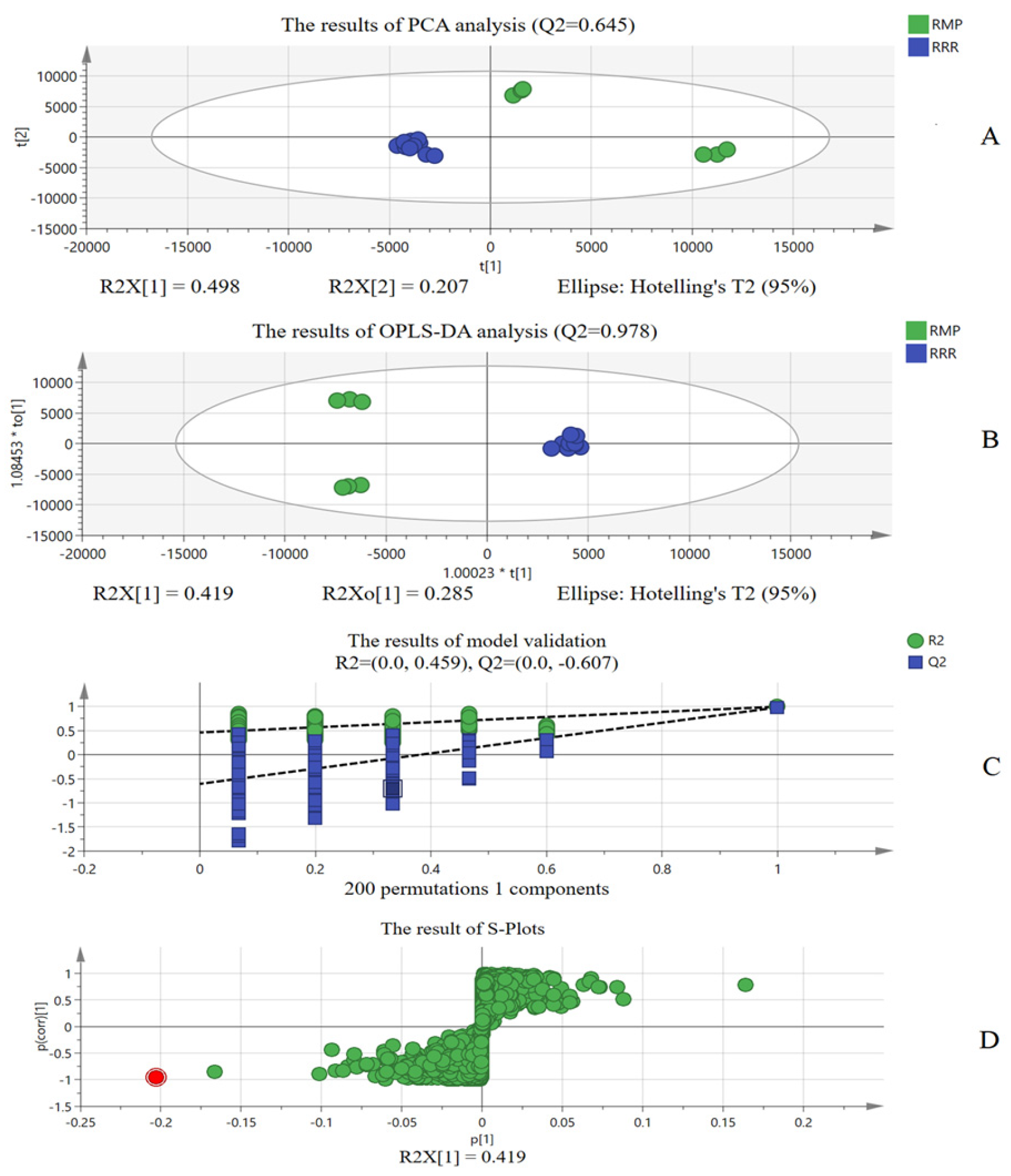
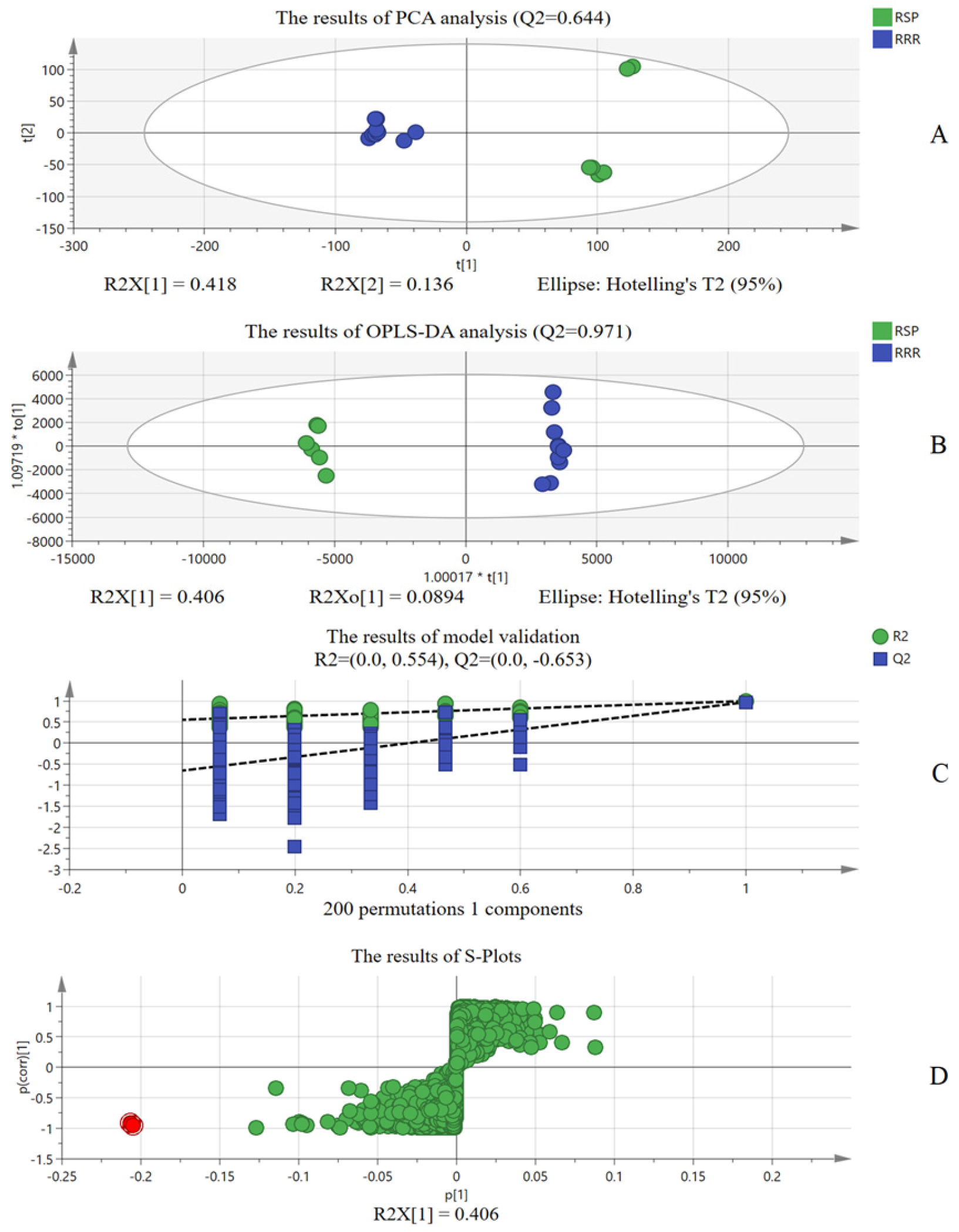
As for RRR market samples, identification evaluation based only on the “ion identity” of LC-MS is not sufficient. Therefore, combined with chemometric analysis, the differential ions were extracted for adulteration identification. “Ion identity” based on LC-MS and chemometrics based on LC-MS combine and confirm each other, which fully proves adulteration in the RRR market samples and makes the results more reliable. On the other hand, chemometrics also proved that identification evaluation based on the “ion identity” of LC-MS was reliable. Moreover, compared with previous studies [
17,
18], the present study focused on the adulteration of same-family herbs, which are more difficult to identify compared to herbs with great differences. The research also fully considered the differential chemical compositions of different herbs and used this to construct an ordered quantized matrix of chemical compositions according to the “ion identity”, which has more specificity than a single or a few chemical components. RRR identification evaluation based on “ion identity” uses more accurate high-resolution mass spectrometry data to realize information and automatic identification and analysis. Compared with traditional microscopic analysis and data modeling analysis, the analysis method based on “ion identity” is more accurate, fast and reliable.
As for the detection thresholds of RMP and RSP adulteration, these are different from ROC and confusion matrix evaluation based on data modeling and identification. The detection thresholds of RMP and RSP were based on the RI test result of 3% adulterated positive samples. Therefore, in each subsequent matching identification, it is necessary to prepare such a 3% positive adulterated sample as a reference to determine the RI detection threshold. For the samples whose matching results are very close to the detection threshold, they can be further extracted and verified by the differential ions analyzed by chemometrics.
In this study, RRR identification evaluation was conducted based on the “ion identity” of LC-MS and chemometrics. However, there are still some regrets: (1) Although the samples involve different producing areas and are collected in person, the sample size is indeed limited, which can only meet the requirements of a preliminary exploration. (2) Although it does not affect the final evaluation, and fully proves the rationality and reliability of analysis based on “ion identity”, chemometric analysis, based on a comparison between existing chemical substance standards and references, fails to clarify what compounds the different chemical components A and B are. Therefore, the sample size could be increased to carry out analysis and explore these proprietary chemical compositions.