Impact of Post-Harvest Apple Scab on Peel Microbiota, Fermentation Dynamics, and the Volatile/Non-Volatile Composition of Cider
Abstract
1. Introduction
2. Results
2.1. Physicochemical Analysis of Apples, Progress of Alcoholic Fermentation, and Chemical Features of Ciders
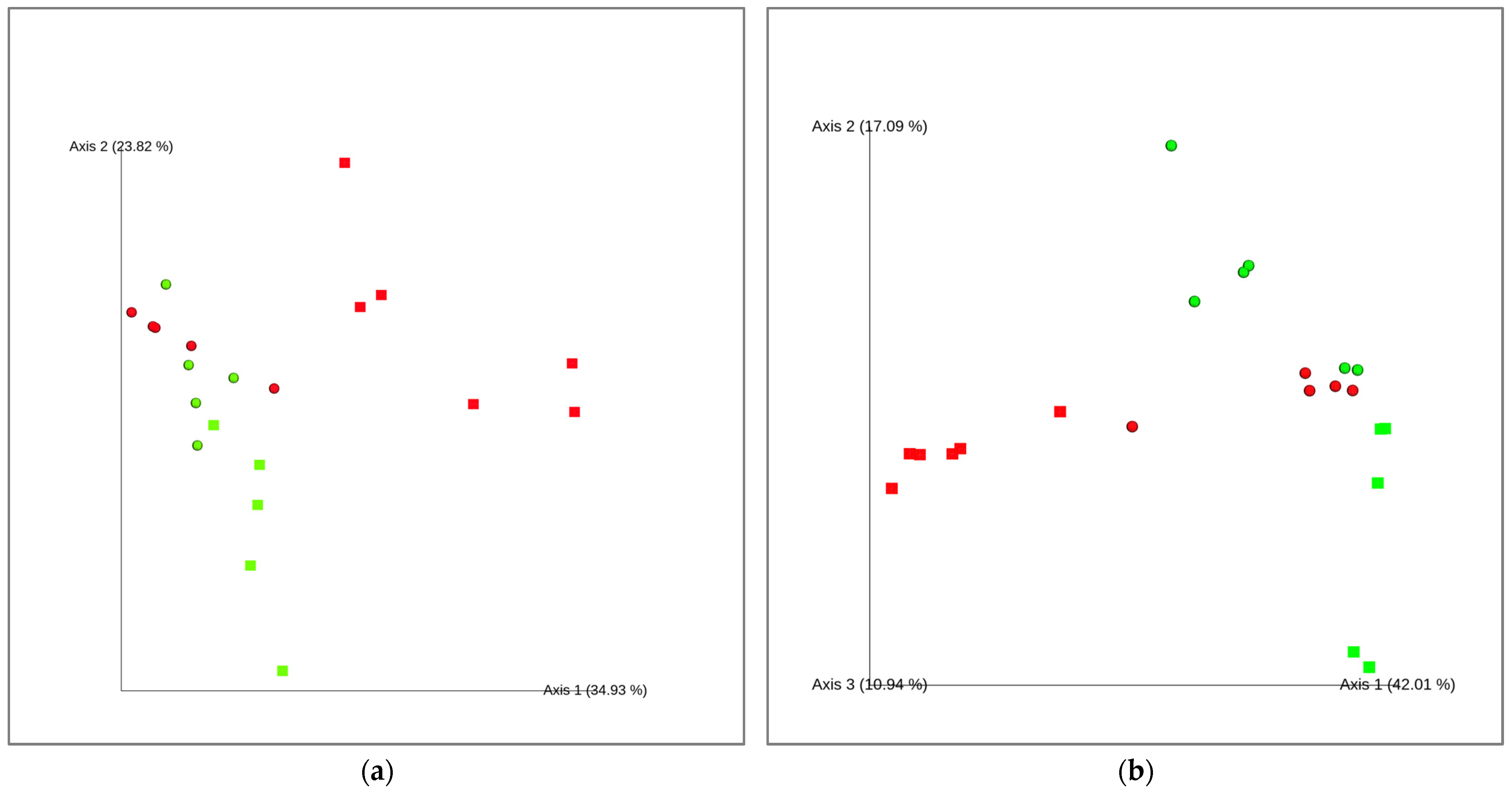
2.2. Analysis of the Apple Peel Microbiota in Healthy and Scab-Affected Fruit
3. Discussion
4. Materials and Methods
4.1. Sampling and Storage of Apples
4.2. Cider Production
4.3. Physicochemical Analysis of Apples and Ciders
4.4. Quantification of Culturable Microbiota on Apple Peel
4.5. Identification of the Microbiota on Apple Peel
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bowen, J.K.; Mesarich, C.H.; Bus, V.G.M.; Beresford, R.M.; Plummer, K.M.; Templeton, M.D. Venturia inaequalis: The causal agent of apple scab. Mol. Plant Pathol. 2011, 12, 105–122. [Google Scholar] [CrossRef] [PubMed]
- MacHardy, W.E. Apple Scab: Biology, Epidemiology, and Management; American Phytopathological Society: St. Paul, MN, USA, 1997; pp. xvi + 545. [Google Scholar]
- Belete, T.; Boyraz, N. Critical review on apple scab (Venturia inaequalis) biology, epidemiology, economic importance, management and defence mechanisms to the causal agent. J. Plant Physiol. 2017, 5, 2. [Google Scholar] [CrossRef]
- Florian, V.C.; Carmen, P.U.I.A.; Groza, R.; Suciu, L.A.; Florian, T. Study of the major pathogens that lead to apple fruit decay during storage. Not. Bot. Horti Agrobot. Cluj-Napoca 2018, 46, 538–545. [Google Scholar] [CrossRef]
- Ghiorghiu, A.V.; Chira, C.L.; Ion, L. Research on limiting the manifestation of fruit storage diseases through the use of treatments with biological products. Sci. Pap. Ser. B Hortic. 2021, 65, 76–81. [Google Scholar]
- Khajuria, Y.P.; Kaul, S.; Wani, A.A.; Dhar, M.J. Genetics of resistance in apple against Venturia inaequalis (Wint.) Cke. Tree Genet. Genomes 2018, 14, 16. [Google Scholar] [CrossRef]
- Yin, J.; Wang, J.N.; Zhao, L.; Cui, Z.L.; Yao, S.; Li, G.X.; Yuan, J. Compost tea: Preparation, utilization mechanisms, and agricultural applications potential—A comprehensive review. Environ. Technol. Innov. 2025, 38, 104137. [Google Scholar] [CrossRef]
- Praveen, M.; Brogi, S. Microbial fermentation in food and beverage industries: Innovations, challenges, and opportunities. Foods 2025, 14, 114. [Google Scholar] [CrossRef]
- Campbell-Platt, G. Fermented foods, a world perspective. Food Res. Int. 1994, 27, 253–257. [Google Scholar] [CrossRef]
- Soemarie, Y.B.; Milanda, T.; Barliana, M.I. Fermented foods as probiotics: A review. J. Adv. Pharm. Technol. Res. 2021, 12, 335. [Google Scholar] [CrossRef]
- Parvez, S.; Malik, K.A.; Ah Kang, S.; Kim, H.Y. Probiotics and their fermented food products are beneficial for health. J. Appl. Microbiol. 2006, 100, 1171–1185. [Google Scholar] [CrossRef]
- Dammak, I.; Alsaiari, N.S.; Fhoula, I.; Amari, A.; Hamdi, Z.; Hassouna, M.; Ben Rebah, F.; Mechichi, T.; Lasram, S. Comparative evaluation of the capacity of commercial and autochthonous Saccharomyces cerevisiae strains to remove ochratoxin a from natural and synthetic grape juices. Toxins 2022, 14, 465. [Google Scholar] [CrossRef] [PubMed]
- Massoud, R.; Hadiani, M.R.; Hamzehlou, P.; Khosravi-Darani, K. Bioremediation of heavy metals in food industry: Application of Saccharomyces cerevisiae. Electron. J. Biotechnol. 2019, 37, 56–60. [Google Scholar] [CrossRef]
- Zolfaghari, H.; Khezerlou, A.; Ehsani, A.; Khosroushahi, A.Y. Detoxification of aflatoxin B1 by probiotic yeasts and bacteria isolated from dairy products of Iran. Adv. Pharm. Bull. 2020, 10, 482. [Google Scholar] [CrossRef]
- Zoghi, A.; Massoud, R.; Todorov, S.D.; Chikindas, M.L.; Popov, I.; Smith, S.; Khosravi-Darani, K. Role of the lactobacilli in food bio-decontamination: Friends with benefits. Enzym. Microb. Technol. 2021, 150, 109861. [Google Scholar] [CrossRef]
- Yang, Y.; Pei, J. Isolation and characterization of an Enterococcus strain from Chinese sauerkraut with potential for lead removal. Eur. Food Res. Technol. 2020, 246, 2055–2064. [Google Scholar] [CrossRef]
- Sionek, B.; Szydłowska, A.; Küçükgöz, K.; Kołożyn-Krajewska, D. Traditional and new microorganisms in lactic acid fermentation of food. Fermentation 2023, 9, 1019. [Google Scholar] [CrossRef]
- Guzzon, R.; Nicolini, G.; Nardin, T.; Malacarne, M.; Larcher, R. Survey about the microbiological features, the oenological performance and the influence on the character of wine of active dry yeast employed as starters of wine fermentation. Int. J. Sci. Technol. 2014, 49, 2142–2148. [Google Scholar] [CrossRef]
- Golombek, P.; Wacker, M.; Buck, N.; Durner, D. Impact of UV-C treatment and thermal pasteurization of grape must on sensory characteristics and volatiles of must and resulting wines. Food Chem. 2021, 338, 128003. [Google Scholar] [CrossRef]
- Al Riachy, R.; Strub, C.; Durand, N.; Guibert, B.; Guichard, H.; Constancias, F.; Chochois, V.; Lopez-Lauri, F.; Fontana, A. Microbiome status of cider-apples, from orchard to processing, with a special focus on Penicillium expansum occurrence and patulin contamination. J. Fungi 2021, 7, 244. [Google Scholar] [CrossRef]
- Cabranes, C.; Moreno, J.; Mangas, J.J. Dynamics of yeast populations during cider fermentation in the Asturian region of Spain. Appl. Environ. Microbiol. 1990, 56, 881–3884. [Google Scholar] [CrossRef]
- Padilla, B.; Gil, J.V.; Manzanares, P. Past and future of non-saccharomyces yeasts: From spoilage microorganisms to biotechnological tools for improving wine aroma complexity. Front. Microbiol. 2016, 7, 411. [Google Scholar] [CrossRef] [PubMed]
- Saranraj, P.; Geetha, M. Microbial spoilage of bakery products and its control by preservatives. Int. J. Pharm. Biol. Sci. Arch. 2012, 3, 38–48. [Google Scholar]
- Suiker, I.M.; Wösten, H.A.B. Spoilage yeasts in beer and beer products. Curr. Opin. Food Sci. 2022, 44, 100815. [Google Scholar] [CrossRef]
- Barata, A.; González, S.; Malfeito-Ferreira, M.; Querol, A.; Loureiro, V. Sour rot-damaged grapes are sources of wine spoilage yeasts. FEMS Yeast Res. 2008, 8, 1008–1017. [Google Scholar] [CrossRef]
- Nogueira, A.; Wosiacki, G. Apple cider fermentation. Handb. Plant-Based Fermented Food Beverage Technol. 2012, 19, 209–235. [Google Scholar]
- Jan, I.; Rab, A. Influence of storage duration on physico-chemical changes in fruit of apple cultivars. J. Anim. Plant Sci. 2012, 22, 708–714. [Google Scholar]
- Gandasi, R.; Sahana, B.B.; Kadanthottu, S.J.; Manikantan, P.; Wen-Chao, L.; Arun, M.; Hesam, K.; Shreeshivadasan, C.; Biljo, V.J. A review on ethanol tolerance mechanisms in yeast: Current knowledge in biotechnological applications and future directions. Process Biochem. 2024, 138, 1–13. [Google Scholar] [CrossRef]
- Rico-Munoz, E.; Samson, R.A.; Houbraken, J. Mould spoilage of foods and beverages: Using the right methodology. Food Microbiol. 2019, 81, 51–62. [Google Scholar] [CrossRef]
- Maslanka, R.; Bednarska, S.; Zadrag-Tecza, R. Virtually identical does not mean exactly identical. Discrepancy in energy metabolism between glucose and fructose fermentation influences the reproductive potential of yeast cells. Arch. Biochem. Biophys. 2024, 756, 110021. [Google Scholar] [CrossRef]
- Endoh, R.; Horiyama, M.; Ohkuma, M. D-Fructose Assimilation and Fermentation by Yeasts Belonging to Saccharomycetes: Rediscovery of universal phenotypes and elucidation of fructophilic behaviors in Ambrosiozyma platypodis and Cyberlindnera americana. Microorganisms 2021, 9, 758. [Google Scholar] [CrossRef]
- Fairbairn, S.C.; Monforte, A.R.; Brand, J.; Silva Ferreira, A.C.; Bauer, F. Yeast metabolic activity is sufficient to create a wine like aromatic feature in a synthetic grape must—A sensory-driven approach. OENO One 2022, 56, 29–40. [Google Scholar] [CrossRef]
- Barba, F.J.; Koubaa, M.; do Prado-Silva, L.; Orlien, V.; de Souza Sant’Ana, A. Mild processing applied to the inactivation of the main foodborne bacterial pathogens: A review. Trends Food Sci Technol. 2017, 66, 20–35. [Google Scholar] [CrossRef]
- Kuchen, B.; Vazquez, F.; Maturano, Y.P.; Scaglia, G.J.E.; Pera, L.M.; Vallejo, M.D. Toward application of biocontrol to inhibit wine spoilage yeasts: The use of statistical designs for screening and optimisation. OENO One 2021, 55, 75–96. [Google Scholar] [CrossRef]
- Alegbeleye, O.; Odeyemi, O.A.; Strateva, M.; Stratev, D. Microbial spoilage of vegetables, fruits and cereals. Appl. Food Res. 2022, 2, 100122. [Google Scholar] [CrossRef]
- Howell, K. Spoilage: Yeast Spoilage of Food and Beverages. Encycl. Food Health 2016, 113–117. [Google Scholar] [CrossRef]
- Moss, M.O. Fungi, quality and safety issues in fresh fruits and vegetables. J. Appl. Microbiol. 2008, 104, 1239–1243. [Google Scholar] [CrossRef]
- Ðurović, G.; Maddalena, G.; Alawamleh, A.; Guzzon, R.; Mazzoni, V.; Dalton, D.T.; Walton, V.M.; Suckling, D.M.; Butler, R.C.; Angeli, S.; et al. Liquid baits with Oenococcus oeni increase captures of Drosophila suzukii. Insects 2021, 12, 66. [Google Scholar] [CrossRef]
- Wassermann, B.; Kusstatscher, P.; Berg, G. Microbiome response to hot water treatment and potential synergy with biological control on stored apples. Front. Microbiol. 2019, 10, 2502. [Google Scholar] [CrossRef]
- Angeli, D.; Razack Sare, A.; Haissam Jijakli, H.; Pertot, I.; Massart, S. Insights gained from metagenomic shotgun sequencing of apple fruit epiphytic microbiota. Postharvest Biol. Technol. 2019, 153, 96–106. [Google Scholar] [CrossRef]
- Abdelfattah, A.; Freilich, S.; Bartuv, R.; Zhimo, V.Y.; Kumar, A.; Biasi, A.; Salim, S.; Feygenberg, O.; Burchard, E.; Dardick, C.; et al. Global analysis of the apple fruit microbiome are all apples the same. Environ. Microbiol. 2021, 23, 6038–6055. [Google Scholar] [CrossRef]
- Shen, Y.; Nie, J.; Dong, Y.; Kuang, L.; Li, Y.; Zhang, J. Compositional shifts in the surface fungal communities of apple fruits during cold storage. Postharvest Biol. Technol. 2018, 144, 55–62. [Google Scholar] [CrossRef]
- Reddy, K.R.N.; Spadaro, D.; Lorè, A.; Gullino, M.L.; Garibaldi, A. Potential of patulin production by Penicillium expansum strains on various fruits. Mycotoxin Res. 2010, 26, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Wenneker, M.; Pham, K.T.; Lemmers, M.E.; de Boer, F.A.; van der Lans, A.M.; van Leeuwen, P.J.; Hollinger, T.C.; Thomma, B.P.H.J. First report of Fusarium avenaceum causing wet core rot of ‘Elstar’ apples in the Netherlands. Plant Dis. 2016, 100, 1501. [Google Scholar] [CrossRef]
- Videira, S.I.; Groenewald, J.Z.; Kolecka, A.; van Haren, L.; Boekhout, T.; Crous, P.W. Elucidating the Ramularia eucalypti species complex. Persoonia 2015, 34, 50–64. [Google Scholar] [CrossRef]
- Garello, M.; Piombo, E.; Prencipe, S.; Schiavon, G.; Berra, L.; Wisniewski, M.; Droby, S.; Spadaro, D. Fruit microbiome: A powerful tool to study the epidemiology of dry lenticel rot and white haze. Emerging post-harvest diseases of apple. Postharvest Biol. Technol. 2023, 196, 112163. [Google Scholar] [CrossRef]
- Liu, K.; Liang, Z.; Yang, A.; Yan, J.; Cong, P.; Han, X.; Zhang, C. Comparative transcriptome analysis of apple cultivars reveals key genes and pathways in response to Alternaria alternata apple pathotype infection. Hortic. Plant J. 2023, 10, 641–656. [Google Scholar] [CrossRef]
- Spadaro, D.; Torres, R.; Errampalli, D.; Everett, K.; Ramos, L.; Mari, M. Postharvest diseases of pome fruit. In Postharvest Pathology of Fresh Horticultural Produce; Palou, L., Smilanick, J.L., Eds.; CRC Press: Boca Raton, FL, USA, 2019; pp. 55–109. [Google Scholar]
- Boekhout, T.; Gildemacher, P.; Theelen, B.; Müller, W.H.; Heijne, B.; Lutz, M. Extensive colonization of apples by smut anamorphs causes a new post-harvest disorder. FEMS Yeast Res. 2006, 6, 63–76. [Google Scholar] [CrossRef]
- Baric, S.; Lindner, L.; Marschall, K.; Dalla Via, J. Haplotype diversity of Tilletiopsis spp. causing white haze in apple orchards in Northern Italy. Plant Pathol. 2010, 59, 535–541. [Google Scholar] [CrossRef]
- Guarnaccia, V.; Remolif, M.E.G.; Nari, L.; Gualandri, V.; Angeli, D.; Oettl, S.; Dijksterhuis, J.; Boekhout, T.; Spadaro, D. Characterization of fungal species involved in white haze disorder on apples in Northern Italy and description of Golubevia mali sp. nov. and Entyloma mali sp. nov. Postharvest Biol. Technol. 2024, 209, 112678. [Google Scholar] [CrossRef]
- Gillis, M.; De Ley, J. The genera Chromobacterium and Janthinobacterium. In The Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 737–746. [Google Scholar]
- Dharmarha, V.; Pulido, N.; Ponder, M.A. Effect of post-harvest interventions on surficial carrot bacterial community dynamics, pathogen survival, and antibiotic resistance. Int. J. Food Microbiol. 2019, 291, 24–34. [Google Scholar] [CrossRef]
- Legein, M.; Smets, W.; Vandenheuvel, D.; Eilers, T.; Muyshondt, B.; Prinsen, E.; Samson, R.; Lebeer, S. Modes of action of microbial biocontrol in the phyllosphere. Front. Microbiol. 2020, 11, 1619. [Google Scholar] [CrossRef] [PubMed]
- Lynch, K.M.; Zannini, E.; Wilkinson, S.; Daenen, L.; Arendt, E.K. Physiology of acetic acid bacteria and their role in vinegar and fermented beverages. Compr. Rev. Food Sci. Food Saf. 2019, 18, 587–625. [Google Scholar] [CrossRef] [PubMed]
- Innerebner, G.; Knief, C.; Vorholt, J.A. Protection of Arabidopsis thaliana against Leaf-Pathogenic Pseudomonas syringae by Sphingomonas Strains in a controlled model system. Appl. Environ. Microbiol. 2011, 77, 10. [Google Scholar] [CrossRef]
- Höfte, M. The Use of Pseudomonas spp. as Bacterial Biocontrol Agents to Control Plant Disease; Bioprotectants for Plant Disease Management; Burleigh Dodds Series in Agricultural Science; Dodds, B., Ed.; Ghent University: Gent, Belgium, 2021. [Google Scholar]
- Testai, E.; Buratti, F.M.; Funari, E.; Manganelli, M.; Vichi, S.; Arnich, N.; Biré, R.; Fessard, V.; Sialehaamoa, A. Review and analysis of occurrence, exposure and toxicity of cyanobacteria toxins in food. EFSA Support. Publ. 2016, 13, 998E. [Google Scholar] [CrossRef]
- Binati, R.L.; Salvetti, E.; Bzducha-Wróbel, A.; Bašinskienė, L.; Čižeikienė, D.; Bolzonella, D.; Felis, G.E. Non-conventional yeasts for food and additives production in a circular economy perspective. FEMS Yeast Res. 2021, 21, foab052. [Google Scholar] [CrossRef]
- Torres-Guardado, R.; Esteve-Zarzoso, B.; Reguant, C.; Bordons, A. Microbial interactions in alcoholic beverages. Int. Microbiol. 2022, 25, 1–15. [Google Scholar] [CrossRef]
- Havis, N.D.; Brown, J.K.M.; Clemente, G.; Frei, P.; Jedryczka, M.; Kaczmarek, J.; Kaczmarek, M.; Matusinsky, P.; McGrann, G.R.; Pereyra, S.; et al. Ramularia collo-cygni—An Emerging Pathogen of Barley Crops. Phytopathology 2015, 105, 895–904. [Google Scholar] [CrossRef]
- Nian, L.; Xie, Y.; Zhang, H.; Wang, M.; Yuan, B.; Shujie, C.; Cao, C. Vishniacozyma victoriae: An endophytic antagonist yeast of kiwifruit with biocontrol effect to Botrytis cinerea. Food Chem. 2023, 411, 135442. [Google Scholar] [CrossRef]
- Gallo, A.; Larcher, R.; Cappello, N.; Paolini, M.; Moser, S.; Carrau, F.; Schneider, R.; Nardin, T.; Roman, T. Insights into the grape must composition effect on Hanseniaspora vineae performance and metabolic aroma compounds in Chardonnay base wine for sparkling wine production. J. Food Compos. Anal. 2023, 123, 105514. [Google Scholar] [CrossRef]
- Nicolini, G.; Román, T.; Carlin, S.; Malacarne, M.; Nardin, T.; Bertoldi, D.; Larcher, R. Characterisation of single-variety still ciders produced with dessert apples in the Italian Alps. J. Inst. Brew. 2018, 124, 457–466. [Google Scholar] [CrossRef]
- OIV. Compendium of International Methods of Wine and Must Analysis; International Organization of Vine and Wine: Dijon, France, 2023. [Google Scholar]
- Paolini, M.; Tonidandel, L.; Larcher, R. Development of a fast gas chromatography tandem mass spectrometry method for volatile aromatic compound analysis in oenological products. J. Mass Spectrom. 2018, 53, 801–810. [Google Scholar] [CrossRef] [PubMed]
- ISO 7218:2024; Microbiology of the Food Chain—General Requirements and Guidance for Microbiological Examinations. ISO: Geneva, Switzerland, 2024.
- Baker, G.C.; Smith, J.J.; Cowan, D.A. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 2003, 55, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Claesson, M.J.; Wang, Q.; O’Sullivan, O.; Greene-Diniz, R.; Cole, J.R.; Ross, R.P.; O’Toole, P.W. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 2010, 38, e200. [Google Scholar] [CrossRef]
- Mbareche, H.; Veillette, M.; Bilodeau, G.; Duchaine, C. Comparison of the performance of ITS1 and ITS2 as barcodes in amplicon-based sequencing of bioaerosols. PeerJ 2020, 8, e8523. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. Fast tree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.S.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef]
- Vázquez-Baeza, Y.; Pirrung, M.; Gonzalez, A.; Knight, R. EMPeror: A tool for visualizing high-throughput microbial community data. Gigascience 2013, 2, 16. [Google Scholar] [CrossRef]
- Mandal, S.; Van Treuren, W.; White, R.A.; Eggesbø, M.; Knight, R.; Peddada, S.D. Analysis of composition of microbiomes: A novel method for studying microbial composition. Microb. Ecol. Health Dis. 2015, 26, 27663. [Google Scholar] [CrossRef]

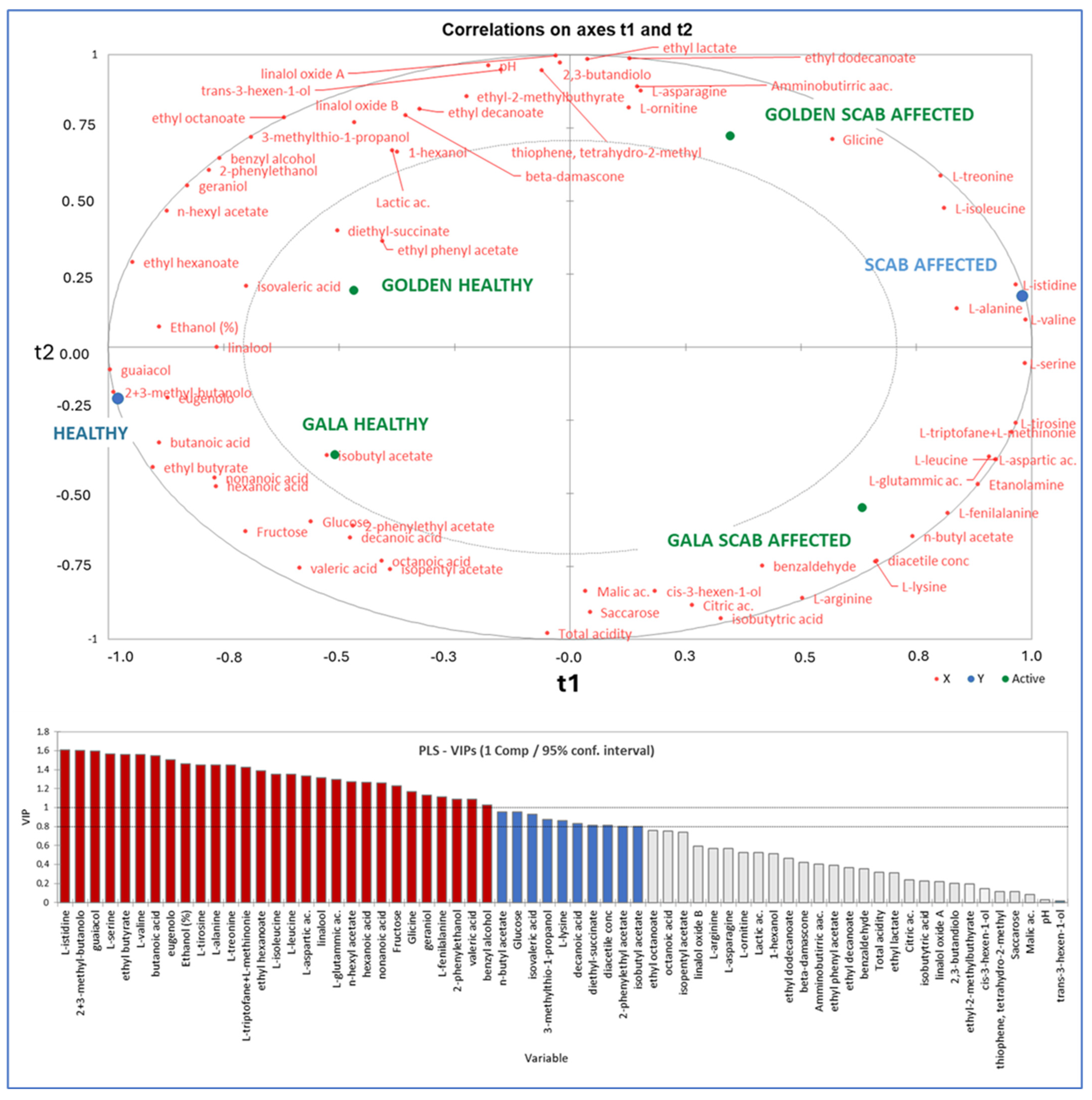
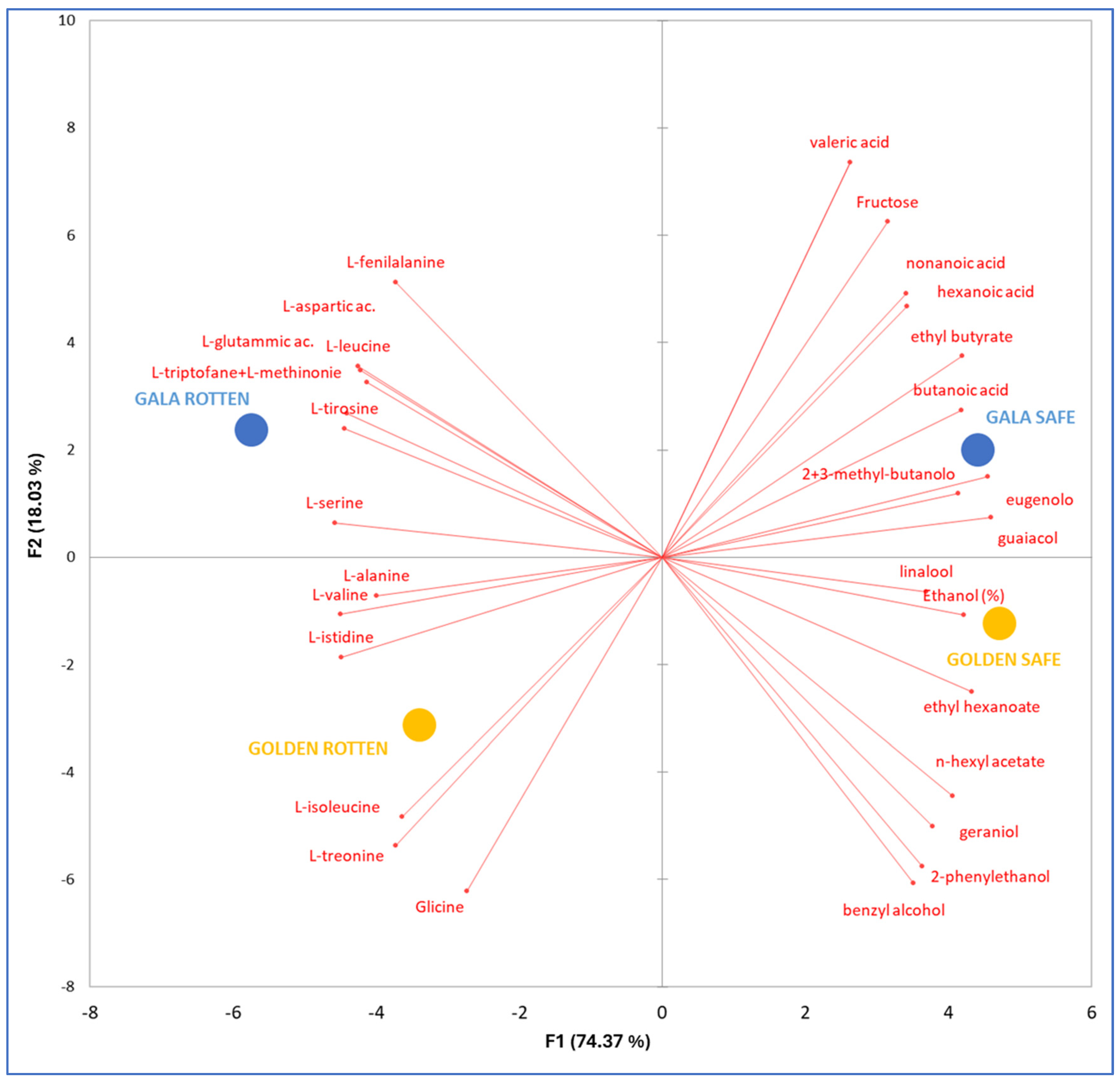

| Golden Healthy | Golden Scab-Affected | Gala Healthy | Gala Scab-Affected | |
|---|---|---|---|---|
| pH | 3.88 ± 0.10 a | 3.94 ± 0.13 a | 3.84 ± 0.01 a | 3.78 ± 0.02 b |
| Citric acid (g/L) | 0.1 ± 0.5 a | 0.1 ± 0.3 a | 0.1 ± 0.1 a | 0.1 ± 0.0 a |
| Malic acid (g/L) | 1.7 ± 2.1 a | 1.2 ± 1.9 a | 5.3 ± 0.6 a | 4.4 ± 0.1 b |
| Lactic acid (g/L) | 0.9 ± 1.4 a | 0.9 ± 0.7 a | 0.1 ± 0.1 a | 0.1 ± 0.3 a |
| Fructose (mg/L) | 36.5 ± 1.2 a | 29.2 ± 3.6 b | 52.7 ± 7.8 a | 36.2 ± 7.7 b |
| Glucose (mg/L) | 19.4 ± 3.5 a | 36.3 ± 1.2 b | 9.3 ± 5.6 a | 46.7 ± 7.7 b |
| Sucrose (mg/L) | 3.2 ± 2.1 a | 3.1 ± 2.9 a | 12.6 ± 6.4 a | 11.5 ± 5.6 a |
| Ethanol (%) | 7.5 ± 0.1 a | 7.1 ± 1.4 b | 7.3 ± 0.1 a | 7.1 ± 0.0 b |
| Sample | Gala Healthy | SD | Gala Scab-Affected | SD | Golden Healthy | SD | Golden Scab-Affected | SD | VIP |
|---|---|---|---|---|---|---|---|---|---|
| Volatile compounds (mg/L) | |||||||||
| Isobutyl Acetate | 0.007 | 0.001 | 0.004 | 0.001 | 0.004 | 0.001 | 0.004 | 0.001 | 0.802 |
| Ethyl Butyrate | 0.044 | 0.009 | 0.034 | 0.004 | 0.042 | 0.003 | 0.030 | 0.008 | 1.561 |
| Ethyl-2-methylbutyrate | 0.002 | 0.001 | 0.002 | 0.000 | 0.018 | 0.002 | 0.015 | 0.003 | 0.191 |
| n-Butyl Acetate | 1.326 | 0.124 | 1.658 | 0.021 | 1.293 | 0.075 | 1.318 | 0.038 | 0.957 |
| Isopentyl Acetate | 0.265 | 0.087 | 0.153 | 0.014 | 0.095 | 0.009 | 0.067 | 0.007 | 0.739 |
| Ethyl Hexanoate | 0.190 | 0.006 | 0.092 | 0.008 | 0.186 | 0.027 | 0.148 | 0.016 | 1.388 |
| n-Hexyl Acetate | 0.014 | 0.001 | 0.010 | 0.001 | 0.016 | 0.001 | 0.013 | 0.002 | 1.270 |
| Ethyl Lactate | 0.169 | 0.035 | 0.141 | 0.017 | 8.681 | 2.277 | 12.974 | 1.310 | 0.310 |
| 1-Hexanol | 1.407 | 0.040 | 1.339 | 0.050 | 4.515 | 0.400 | 2.923 | 1.054 | 0.513 |
| trans-3-Hexen-1-ol | 0.022 | 0.002 | 0.017 | 0.001 | 0.060 | 0.015 | 0.064 | 0.005 | 0.015 |
| cis-3-Hexen-1-ol | 0.026 | 0.002 | 0.025 | 0.003 | 0.004 | 0.001 | 0.009 | 0.004 | 0.146 |
| Ethyl Octanoate | 0.058 | 0.011 | 0.021 | 0.005 | 0.076 | 0.012 | 0.072 | 0.012 | 0.756 |
| Linalool Oxide A | 0.003 | 0.002 | 0.002 | 0.000 | 0.008 | 0.001 | 0.012 | 0.002 | 0.222 |
| Linalool Oxide B | 0.003 | 0.002 | 0.001 | 0.000 | 0.008 | 0.001 | 0.006 | 0.000 | 0.595 |
| Benzaldehyde | 0.016 | 0.000 | 0.029 | 0.002 | 0.019 | 0.001 | 0.012 | 0.002 | 0.356 |
| Isobutyric acid | 0.079 | 0.014 | 0.091 | 0.002 | 0.073 | 0.003 | 0.066 | 0.012 | 0.225 |
| Linalool | 0.024 | 0.001 | 0.021 | 0.001 | 0.032 | 0.003 | 0.019 | 0.002 | 1.317 |
| Butanoic acid | 0.351 | 0.003 | 0.292 | 0.078 | 0.368 | 0.019 | 0.263 | 0.091 | 1.549 |
| Ethyl Decanoate | 0.012 | 0.000 | 0.007 | 0.003 | 0.054 | 0.009 | 0.041 | 0.013 | 0.368 |
| Isovaleric Acid | 0.342 | 0.015 | 0.206 | 0.006 | 0.264 | 0.055 | 0.288 | 0.025 | 0.927 |
| Diethyl-Succinate | 0.004 | 0.000 | 0.003 | 0.000 | 0.050 | 0.004 | 0.012 | 0.004 | 0.815 |
| 3-Methylthio-1-Propanol | 1.202 | 0.024 | 0.149 | 0.085 | 1.510 | 0.150 | 1.392 | 0.030 | 0.875 |
| Valeric Acid | 0.025 | 0.001 | 0.021 | 0.004 | 0.021 | 0.001 | 0.018 | 0.002 | 1.086 |
| Ethyl Phenyl Acetate | 0.009 | 0.000 | 0.006 | 0.001 | 0.006 | 0.002 | 0.008 | 0.001 | 0.393 |
| 2-Phenylethyl Acetate | 0.038 | 0.008 | 0.016 | 0.002 | 0.010 | 0.004 | 0.009 | 0.001 | 0.802 |
| β-Damascone | 0.019 | 0.001 | 0.017 | 0.001 | 0.028 | 0.003 | 0.025 | 0.008 | 0.423 |
| Hexanoic Acid | 1.139 | 0.036 | 0.688 | 0.035 | 0.799 | 0.091 | 0.629 | 0.185 | 1.267 |
| Ethyl Dodecanoate | 0.102 | 0.023 | 0.104 | 0.007 | 0.144 | 0.023 | 0.177 | 0.008 | 0.467 |
| Geraniol | 0.318 | 0.036 | 0.188 | 0.068 | 0.337 | 0.021 | 0.303 | 0.014 | 1.134 |
| Guaiacol | 0.007 | 0.001 | 0.004 | 0.002 | 0.007 | 0.000 | 0.005 | 0.001 | 1.598 |
| Benzyl Alcohol | 0.218 | 0.008 | 0.098 | 0.009 | 0.275 | 0.031 | 0.228 | 0.046 | 1.028 |
| 2-Phenylethanol | 20.270 | 0.677 | 15.888 | 0.695 | 22.739 | 1.701 | 20.428 | 1.032 | 1.088 |
| Octanoic Acid | 1.707 | 0.214 | 1.150 | 0.123 | 0.893 | 0.155 | 0.780 | 0.170 | 0.754 |
| Nonanoic Acid | 0.008 | 0.002 | 0.003 | 0.001 | 0.005 | 0.001 | 0.003 | 0.000 | 1.260 |
| Decanoic Acid | 0.433 | 0.119 | 0.317 | 0.057 | 0.285 | 0.080 | 0.269 | 0.017 | 0.832 |
| Diacetyl | 6.227 | 2.014 | 7.653 | 0.196 | 5.975 | 0.257 | 5.964 | 0.175 | 0.815 |
| 3-Methyl-2-butanol | 60.365 | 1.573 | 52.434 | 2.848 | 59.376 | 2.449 | 53.082 | 4.611 | 1.600 |
| 2,3-Butanediol | 0.750 | 0.005 | 0.636 | 0.023 | 5.786 | 6.277 | 7.399 | 0.744 | 0.201 |
| Thiophene. Tetrahydro-2-Methyl | 1.532 | 0.390 | 1.544 | 0.033 | 2.339 | 0.126 | 2.451 | 0.035 | 0.115 |
| Eugenol | 0.544 | 0.045 | 0.222 | 0.006 | 0.794 | 0.093 | 0.102 | 0.004 | 1.502 |
| Non-volatile compounds (g/L) | |||||||||
| Aminobutyric acid | 0.333 | 0.122 | 0.433 | 0.117 | 0.633 | 0.221 | 0.675 | 0.121 | 0.405 |
| L-aspartic acid | 0.533 | 0.045 | 1.500 | 0.068 | 0.450 | 0.028 | 0.850 | 0.098 | 1.333 |
| L-glutamic acid | 0.500 | 0.023 | 1.933 | 0.021 | 0.575 | 0.036 | 0.975 | 0.078 | 1.294 |
| L-alanine | 1.067 | 0.087 | 1.300 | 0.045 | 0.800 | 0.021 | 1.400 | 0.121 | 1.450 |
| L-arginine | 0.867 | 0.122 | 1.500 | 0.138 | 0.300 | 0.184 | 0.350 | 0.089 | 0.568 |
| L-asparagine | 0.367 | 0.125 | 0.300 | 0.107 | 0.350 | 0.215 | 0.550 | 0.068 | 0.567 |
| Ethanolamine | 3.733 | 0.064 | 5.600 | 0.184 | 3.375 | 0.129 | 4.150 | 0.098 | 1.360 |
| L-phenylalanine | 0.333 | 0.022 | 0.767 | 0.092 | 0.275 | 0.027 | 0.375 | 0.111 | 1.111 |
| Glycine | 0.900 | 0.048 | 1.233 | 0.141 | 0.700 | 0.148 | 2.975 | 0.210 | 1.167 |
| L-isoleucine | 0.167 | 0.066 | 0.367 | 0.021 | 0.300 | 0.018 | 0.400 | 0.084 | 1.352 |
| L-histidine | 0.233 | 0.045 | 0.433 | 0.058 | 0.225 | 0.031 | 0.450 | 0.066 | 1.608 |
| L-leucine | 0.600 | 0.022 | 1.133 | 0.031 | 0.500 | 0.086 | 0.775 | 0.054 | 1.350 |
| L-lysine | 1.133 | 0.102 | 1.967 | 0.122 | 0.475 | 0.086 | 0.825 | 0.098 | 0.863 |
| L-ornithine | 0.600 | 0.148 | 0.467 | 0.089 | 0.525 | 0.128 | 0.850 | 0.036 | 0.528 |
| L-serine | 1.067 | 0.022 | 2.267 | 0.076 | 0.825 | 0.084 | 1.950 | 0.122 | 1.565 |
| L-tyrosine | 0.333 | 0.096 | 0.633 | 0.048 | 0.300 | 0.052 | 0.475 | 0.046 | 1.451 |
| L-threonine | 0.300 | 0.086 | 0.467 | 0.081 | 0.350 | 0.021 | 0.600 | 0.084 | 1.450 |
| L-tryptophan + L-methionine | 0.333 | 0.054 | 0.600 | 0.047 | 0.300 | 0.032 | 0.450 | 0.096 | 1.428 |
| L-valine | 0.233 | 0.048 | 0.533 | 0.032 | 0.300 | 0.048 | 0.475 | 0.021 | 1.557 |
| Apple Sample | Bacteria | Fungi |
|---|---|---|
| ×103 CFU/g | ||
| Golden healthy | 2.3 ± 1.2 a | 0.6 ± 0.4 a |
| Golden scab-affected | 6.1 ± 1.7 b | 2.8 ± 0.8 b |
| Gala healthy | 9.1 ± 1.4 a | 3.2 ± 1.2 a |
| Gala scab-affected | 10 ± 0.7 a | 4.5 ± 1.6 a |
| Bacteria (Relative Abundance) | ||||
|---|---|---|---|---|
| Gala Healthy | Gala Scab-Affected | Golden Healthy | Golden Scab-Affected | |
| Geodermatophilaceae | 0.395 | n.d. | 0.740 | 2.825 |
| Microbacteriaceae | 8.510 | 4.687 | 4.057 | 4.917 |
| Rhodococcus | 1.208 | 5.430 | n.d. | 1.262 |
| Nocardioidaceae | 0.334 | 1.162 | 0.183 | 1.840 |
| Williamsia | n.d. | 1.130 | n.d. | n.d. |
| Other Actinobacteria | 0.894 | 0.964 | 1.223 | 0.889 |
| Hymenobacter | 4.200 | 5.113 | 6.172 | 8.113 |
| Flavobacterium | 2.340 | 1.920 | 0.603 | 0.740 |
| Chryseobacterium | 1.722 | 0.213 | 0.393 | 0.390 |
| Sphingobacteriaceae | 3.152 | 0.920 | 1.298 | 0.553 |
| Other Bacteroidetes | 1.703 | 0.849 | 1.499 | 1.329 |
| Cyanobacteria | 1.911 | 1.304 | 3.434 | 1.803 |
| Aerococcus | 0.251 | n.d. | 2.519 | 0.258 |
| Other Firmicutes | 0.472 | 0.334 | 0.114 | 0.403 |
| Methylobacterium | 4.455 | 6.775 | 13.264 | 10.875 |
| Agrobacterium | 3.235 | 4.526 | 0.484 | 1.608 |
| Acetobacteraceae | 2.357 | 0.813 | 17.415 | 2.484 |
| Sphingomonas | 27.560 | 29.487 | 23.394 | 18.762 |
| Other Alphaproteobacteria | 1.665 | 0.406 | 1.628 | 0.354 |
| Comamonadaceae | 3.949 | 6.192 | 2.263 | 7.431 |
| Janthinobacterium | 16.103 | 10.259 | 3.083 | 7.073 |
| Enterobacteriaceae | 0.678 | 0.623 | 11.942 | 1.404 |
| Pseudomonas | 11.321 | 13.465 | 3.476 | 24.095 |
| Xanthomonadaceae | 0.260 | 3.407 | n.d. | 0.018 |
| Other Gammaproteobacteria | 0.756 | 0.020 | 0.307 | 0.142 |
| Fungi (relative abundance) | ||||
| Gala healthy | Gala scab-affected | Golden healthy | Golden scab-affected | |
| Ascomycota; Cladosporium | 12.430 | 9.285 | 12.263 | 15.358 |
| Ascomycota; Mycosphaerella | 3.701 | 3.397 | 8.204 | 9.243 |
| Ascomycota; Ramularia * | 32.646 | 2.355 | 6.503 | 3.376 |
| Ascomycota; Mycosphaerellaceae | 2.028 | 1.449 | 0.679 | 2.445 |
| Ascomycota; Aureobasidium | 1.975 | 4.438 | 2.647 | 5.285 |
| Ascomycota; Didymellaceae * | 1.831 | 26.948 | 21.655 | 15.838 |
| Ascomycota; Alternaria * | 1.004 | 7.848 | 4.295 | 8.084 |
| Ascomycota; Penicillium | 1.489 | 0.038 | 0.031 | 0.217 |
| Ascomycota; Saccharomycetales; | n.d. | 0.500 | 0.022 | n.d. |
| Ascomycota; Acremonium | 4.204 | 0.461 | 2.586 | 1.888 |
| Ascomycota; Nectriaceae * | 6.155 | 13.263 | 0.087 | 0.228 |
| Other Ascomycota | 3.575 | 3.247 | 6.781 | 5.525 |
| Basidiomycota; Cystobasidium * | 2.275 | 0.330 | 1.024 | 0.293 |
| Basidiomycota; Tilletiopsis | 2.647 | 1.532 | 0.722 | 0.661 |
| Basidiomycota; Cystofilobasidiales | 0.051 | 0.206 | 0.713 | 3.785 |
| Basidiomycota; Filobasidium * | 2.056 | 4.290 | 2.303 | 4.201 |
| Basidiomycota; Naganishia * | 0.135 | 1.784 | 0.011 | 0.015 |
| Basidiomycota; Vishniacozyma * | 15.765 | 5.090 | 24.183 | 17.060 |
| Other Basidiomycota | 4.189 | 8.889 | 4.811 | 5.549 |
| Sample | Observed OTUs | Shannon Index | Evenness Index |
|---|---|---|---|
| Bacteria | |||
| Golden healthy | 69 ± 6 a | 5.496 ± 0.378 ab | 0.890 ± 0.029 a |
| Golden scab-affected | 91 ± 25 b | 5.690 ± 0.520 b | 0.885 ± 0.026 a |
| Gala healthy | 58 ± 17 a | 5.411 ± 0.495 ab | 0.898 ± 0.027 a |
| Gala scab-affected | 155 ± 68 b | 5.362 ± 0.472 a | 0.893 ± 0.022 a |
| Fungi | |||
| Golden healthy | 74 ± 5 b | 4.502 ± 0.283 b | 0.707 ± 0.042 a |
| Golden scab-affected | 87 ± 14 c | 4.617 ± 0.232 c | 0.719 ± 0.025 b |
| Gala healthy | 61 ± 10 a | 4.391 ± 0.257 a | 0.711 ± 0.049 ab |
| Gala scab-affected | 74 ± 11 b | 4.462 ± 0.290 b | 0.708 ± 0.040 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gualandri, V.; Larcher, R.; Franciosi, E.; Paolini, M.; Nardin, T.; Pertot, I.; Guzzon, R. Impact of Post-Harvest Apple Scab on Peel Microbiota, Fermentation Dynamics, and the Volatile/Non-Volatile Composition of Cider. Molecules 2025, 30, 2322. https://doi.org/10.3390/molecules30112322
Gualandri V, Larcher R, Franciosi E, Paolini M, Nardin T, Pertot I, Guzzon R. Impact of Post-Harvest Apple Scab on Peel Microbiota, Fermentation Dynamics, and the Volatile/Non-Volatile Composition of Cider. Molecules. 2025; 30(11):2322. https://doi.org/10.3390/molecules30112322
Chicago/Turabian StyleGualandri, Valeria, Roberto Larcher, Elena Franciosi, Mauro Paolini, Tiziana Nardin, Ilaria Pertot, and Raffaele Guzzon. 2025. "Impact of Post-Harvest Apple Scab on Peel Microbiota, Fermentation Dynamics, and the Volatile/Non-Volatile Composition of Cider" Molecules 30, no. 11: 2322. https://doi.org/10.3390/molecules30112322
APA StyleGualandri, V., Larcher, R., Franciosi, E., Paolini, M., Nardin, T., Pertot, I., & Guzzon, R. (2025). Impact of Post-Harvest Apple Scab on Peel Microbiota, Fermentation Dynamics, and the Volatile/Non-Volatile Composition of Cider. Molecules, 30(11), 2322. https://doi.org/10.3390/molecules30112322










