Quorum Sensing and Mobility Inhibition of Pathogenic Bacteria by Fulvifomes mexicanus sp. nov.
Abstract
1. Introduction
2. Results
2.1. Taxonomy
Basidiomycota, Agaricomycetes, Hymenochaetales, Hymenochaetaceae
- Fulvifomes mexicanus Mart.-Pineda, R. Valenz. & Raymundo sp. nov.
- Figure 1: A–I
- Mycobank: 859269
- Type. Mexico. Sonora State, Alamos-Río Cuchujaqui Biosphere Reserve, Alamos Municipality, Promontorios, LN 26°59′55″, LW 109°03′21″, alt. 370 m, 26 October 2018, Tropical dry forest, T. Raymundo 8046 (Holotype ENCB).
- Etymology. mexicanus (Lat.): referring to country of the locality type.
- Description. Basidiomata 70–135 × 50−70 × 25–55 mm, perennial, pileate-sessile, aplanate with umbo, broadly attached to the substrate, consistency woody hard. Pileus brown (7D-E8) to reddish brown (8E7) with tones dark brown (8F8,4) and the old part of the pileus (center) greyish brown (8F3) to black, concentrically zonate to sulcate, with some lines brownish grey (7E2) to the margin, slightly velvety in younger zones, glabrous, to slightly rimose to cracked, with a distinct crust, up to 5 mm thick to the center. Margin obtuse, sterile to fertile, yellowish brown (5E8), brown (6E8) to dark brown (7F6). Hymenophore poroid, brown (7D7) to dark brown, pores circular, 5–7 per mm, dissepiments (70–) 85–90 (–100) µm thick, and lumen 140–160 µm diam. Tubes brown (7E8), woody hard, up to 45 mm thick, tube layers distinctly stratified with intermittent context layers, individual tube layer up to 3 mm thick. Context up to 10 mm thick, brown (6E8) to dark brown in the surface.
2.2. Phylogenetic Analysis of Fulvifomes mexicanus sp. nov.

2.3. Antimicrobial Activity and Inhibition of Swimming Motility
2.4. Inhibition of Violacein Production
2.5. Metabolomic Profiling and Molecular Networking
3. Discussion
4. Materials and Methods
4.1. Fungal Specimen
4.2. Genetic Identification
4.3. Extracts Preparation
4.4. Antimicrobial Activity and Minimum Inhibitory Concentration (MIC).
4.5. Mobility Inhibition Tests
4.6. Inhibition of Violacein Production.
4.7. LC-MS, Untargeted Metabolomic and Feature-Based Molecular Network Analysis
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| LC-HRESIMS | Liquid Chromatography–High-Resolution Electrospray Ionization Mass Spectrometry |
| QS | Quorum Sensing |
| nrITS | Internal Transcribed Spacer of the nuclear ribosomal DNA |
| MIC | Minimum Inhibitory Concentration |
| GNPS | Global Natural Product Social Molecular Networking |
| MS | Mass Spectrometry |
| MP | Maximum Parsimony |
| ML | Maximum Likelihood |
| BI | Bayesian Inference |
| MIC | Minimum Inhibitory Concentration |
| CLSI | Clinical and Laboratory Standards Institute |
| MH | Mueller–Hinton |
| LB | Luria Broth |
| C6-AHL | N-hexanoyl-L-homoserine lactone |
| FBMN | Feature-Based Molecular Networking |
References
- Tang, K.W.K.; Millar, B.C.; Moore, J.E. Antimicrobial Resistance (AMR). Br. J. Biomed. Sci. 2023, 80, 11387. [Google Scholar] [CrossRef] [PubMed]
- Djordjevic, S.P.; Jarocki, V.M.; Seemann, T.; Cummins, M.L.; Watt, A.E.; Drigo, B.; Wyrsch, E.R.; Reid, C.J.; Donner, E.; Howden, B.P. Genomic Surveillance for Antimicrobial Resistance—A One Health Perspective. Nat. Rev. Genet. 2024, 25, 142–157. [Google Scholar] [CrossRef] [PubMed]
- Ernst, M.; Kang, K.B.; Caraballo-Rodríguez, A.M.; Nothias, L.-F.; Wandy, J.; Chen, C.; Wang, M.; Rogers, S.; Medema, M.H.; Dorrestein, P.C.; et al. MolNetEnhancer: Enhanced Molecular Networks by Integrating Metabolome Mining and Annotation Tools. Metabolites 2019, 9, 144. [Google Scholar] [CrossRef]
- Escaich, S. Antivirulence as a New Antibacterial Approach for Chemotherapy. Curr. Opin. Chem. Biol. 2008, 12, 400–408. [Google Scholar] [CrossRef]
- Escaich, S. Novel Agents to Inhibit Microbial Virulence and Pathogenicity. Expert Opin. Ther. Pat. 2010, 20, 1401–1418. [Google Scholar] [CrossRef] [PubMed]
- Hashem, A.H.; Attia, M.S.; Kandil, E.K.; Fawzi, M.M.; Abdelrahman, A.S.; Khader, M.S.; Khodaira, M.A.; Emam, A.E.; Goma, M.A.; Abdelaziz, A.M. Bioactive Compounds and Biomedical Applications of Endophytic Fungi: A Recent Review. Microb. Cell Fact. 2023, 22, 107. [Google Scholar] [CrossRef]
- Liao, C.; Huang, X.; Wang, Q.; Yao, D.; Lu, W. Virulence Factors of Pseudomonas aeruginosa and Antivirulence Strategies to Combat Its Drug Resistance. Front. Cell. Infect. Microbiol. 2022, 12, 926758. [Google Scholar] [CrossRef]
- Wang, Y.; Bian, Z.; Wang, Y. Biofilm Formation and Inhibition Mediated by Bacterial Quorum Sensing. Appl. Microbiol. Biotechnol. 2022, 106, 6365–6381. [Google Scholar] [CrossRef]
- Fuentes-Gutiérrez, A.; Curiel-Quesada, E.; Correa-Basurto, J.; Martínez-Muñoz, A.; Reyes-Arellano, A. N-Heterocycles Scaffolds as Quorum Sensing Inhibitors. Design, Synthesis, Biological and Docking Studies. Int. J. Mol. Sci. 2020, 21, 9512. [Google Scholar] [CrossRef]
- Conrado, R.; Gomes, T.C.; Roque, G.S.C.; De Souza, A.O. Overview of Bioactive Fungal Secondary Metabolites: Cytotoxic and Antimicrobial Compounds. Antibiotics 2022, 11, 1604. [Google Scholar] [CrossRef]
- Buroni, S.; Chiarelli, L.R. Antivirulence Compounds: A Future Direction to Overcome Antibiotic Resistance? Future Microbiol. 2020, 15, 299–301. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.T.; Wencewicz, T.A. Prospects for New Antibiotics: A Molecule-Centered Perspective. J. Antibiot. 2014, 67, 7–22. [Google Scholar] [CrossRef] [PubMed]
- Du, P.; Cao, T.-X.; Wu, Y.-D.; Zhou, M.; Liu, Z.-B. Two New Species of Hymenochaetaceae on Dracaena cambodiana from Tropical China. MycoKeys 2021, 80, 1–17. [Google Scholar] [CrossRef]
- Isaka, M.; Palasarn, S.; Sakayaroj, J.; Srichomthong, K.; Nithithanasilp, S.; Sappan, M. Limonoids from Fruiting Bodies of the Wood-Rot Basidiomycete Fulvifomes xylocarpicola Associated with the Mangrove Tree Xylocarpus granatum. Phytochemistry 2021, 181, 112555. [Google Scholar] [CrossRef]
- Schwarze, F.W.M.R.; Engels, J.; Mattheck, C. Host-Fungus Interactions: Development and Prognosis of Wood Decay in the Sapwood. In Fungal Strategies of Wood Decay in Trees; Springer: Berlin/Heidelberg, Germany, 2000; pp. 139–167. ISBN 978-3-642-63133-7. [Google Scholar]
- Hattori, T.; Ota, Y.; Sotome, K. Two New Species of Fulvifomes (Basidiomycota, Hymenochaetaceae) on Threatened or near Threatened Tree Species in Japan. Mycoscience 2022, 63, 131–141. [Google Scholar] [CrossRef]
- Suh, H.; Cho, Y.; Seo, C.W.; Kim, D.; Lee, H.-S.; Lim, Y.W. A Taxonomic Study of Fulvifomes (Hymenochaetales, Basidiomycota) in the Federated States of Micronesia and Identification of Two New Species. Mycol. Prog. 2024, 23, 15. [Google Scholar] [CrossRef]
- Wu, F.; Zhou, L.-W.; Vlasák, J.; Dai, Y.-C. Global Diversity and Systematics of Hymenochaetaceae with Poroid Hymenophore. Fungal Divers. 2022, 113, 1–192. [Google Scholar] [CrossRef]
- Fernando, D.; Adhikari, A.; Nanayakkara, C.; De Silva, E.D.; Wijesundera, R.; Soysa, P. Cytotoxic Effects of Ergone, a Compound Isolated from Fulviformes fastuosus. BMC Complement. Altern. Med. 2016, 16, 484. [Google Scholar] [CrossRef] [PubMed]
- Govindharaj, M.; Arumugam, S.; Nirmala, G.; Bharadwaj, M.; Murugiyan, K. Effect of Marine Basidiomycetes Fulvifomes sp. Derived Ergosterol Peroxide on Cytotoxicity and Apoptosis Induction in MCF-7 Cell Line. J. Fungi 2019, 5, 16. [Google Scholar] [CrossRef]
- Aslam, B.; Khurshid, M.; Arshad, M.I.; Muzammil, S.; Rasool, M.; Yasmeen, N.; Shah, T.; Chaudhry, T.H.; Rasool, M.H.; Shahid, A.; et al. Antibiotic Resistance: One Health One World Outlook. Front. Cell. Infect. Microbiol. 2021, 11, 771510. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef] [PubMed]
- LaSarre, B.; Federle, M.J. Exploiting Quorum Sensing to Confuse Bacterial Pathogens. Microbiol. Mol. Biol. Rev. 2013, 77, 73–111. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wei, J.; Liu, Z.; Wang, Y.; Yuan, X.; Wang, D.; Niu, J.; Yang, Y.; Zhou, J. Comparative Genomic Analysis of Sanghuangporus sanghuang with Other Hymenochaetaceae Species. Braz. J. Microbiol. 2024, 55, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Miao, K.; Liu, Y.; Zhao, Y.; Zhang, M.; Pan, S.; Dai, Y. Chemical Diversity of Biologically Active Metabolites in the Sclerotia of Inonotus obliquus and Submerged Culture Strategies for Up-Regulating Their Production. Appl. Microbiol. Biotechnol. 2010, 87, 1237–1254. [Google Scholar] [CrossRef]
- Chaban, B.; Hughes, H.V.; Beeby, M. The Flagellum in Bacterial Pathogens: For Motility and a Whole Lot More. Semin. Cell Dev. Biol. 2015, 46, 91–103. [Google Scholar] [CrossRef]
- Conrad, J.C.; Gibiansky, M.L.; Jin, F.; Gordon, V.D.; Motto, D.A.; Mathewson, M.A.; Stopka, W.G.; Zelasko, D.C.; Shrout, J.D.; Wong, G.C.L. Flagella and Pili-Mediated Near-Surface Single-Cell Motility Mechanisms in P. aeruginosa. Biophys. J. 2011, 100, 1608–1616. [Google Scholar] [CrossRef]
- De La Fuente-Núñez, C.; Korolik, V.; Bains, M.; Nguyen, U.; Breidenstein, E.B.M.; Horsman, S.; Lewenza, S.; Burrows, L.; Hancock, R.E.W. Inhibition of Bacterial Biofilm Formation and Swarming Motility by a Small Synthetic Cationic Peptide. Antimicrob. Agents Chemother. 2012, 56, 2696–2704. [Google Scholar] [CrossRef]
- Jarrell, K.F.; McBride, M.J. The Surprisingly Diverse Ways That Prokaryotes Move. Nat. Rev. Microbiol. 2008, 6, 466–476. [Google Scholar] [CrossRef]
- Khan, F.; Pham, D.T.N.; Oloketuyi, S.F.; Kim, Y.-M. Regulation and Controlling the Motility Properties of Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 2020, 104, 33–49. [Google Scholar] [CrossRef]
- O’Toole, G.A.; Kolter, R. Flagellar and Twitching Motility Are Necessary for Pseudomonas aeruginosa Biofilm Development. Mol. Microbiol. 1998, 30, 295–304. [Google Scholar] [CrossRef]
- Yeung, A.T.Y.; Torfs, E.C.W.; Jamshidi, F.; Bains, M.; Wiegand, I.; Hancock, R.E.W.; Overhage, J. Swarming of Pseudomonas aeruginosa Is Controlled by a Broad Spectrum of Transcriptional Regulators, Including MetR. J. Bacteriol. 2009, 191, 5592–5602. [Google Scholar] [CrossRef] [PubMed]
- Di Pasqua, R.; Betts, G.; Hoskins, N.; Edwards, M.; Ercolini, D.; Mauriello, G. Membrane Toxicity of Antimicrobial Compounds from Essential Oils. J. Agric. Food Chem. 2007, 55, 4863–4870. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.; Ge, X.; Wang, J.; Wei, Z.; Feng, W.; Wang, J. Ergosterol Peroxide Exhibits Antiviral and Immunomodulatory Abilities against Porcine Deltacoronavirus (PDCoV) via Suppression of NF-κB and P38/MAPK Signaling Pathways in vitro. Int. Immunopharmacol. 2021, 93, 107317. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, X.; Wang, J.; Zhang, J.; Duan, C.; Wang, J. Ergosterol Peroxide Inhibits Porcine Epidemic Diarrhea Virus Infection in Vero Cells by Suppressing ROS Generation and P53 Activation. Viruses 2022, 14, 402. [Google Scholar] [CrossRef]
- Merdivan, S.; Lindequist, U. Ergosterol Peroxide: A Mushroom-Derived Compound with Promising Biological Activities-A Review. Int. J. Med. Mushrooms 2017, 19, 93–105. [Google Scholar] [CrossRef]
- Meza-Menchaca, T.; Ramos-Ligonio, A.; López-Monteon, A.; Vidal Limón, A.; Kaluzhskiy, L.A.; Shkel, T.V.; Strushkevich, N.V.; Jiménez-García, L.F.; Agredano Moreno, L.T.; Gallegos-García, V.; et al. Insights into Ergosterol Peroxide’s Trypanocidal Activity. Biomolecules 2019, 9, 484. [Google Scholar] [CrossRef]
- Wang, F.-W. Bioactive Metabolites from Guignardia sp., an Endophytic Fungus Residing in Undaria Pinnatifida. Chin. J. Nat. Med. 2012, 10, 72–76. [Google Scholar] [CrossRef]
- Zhou, B.; Liang, X.; Feng, Q.; Li, J.; Pan, X.; Xie, P.; Jiang, Z.; Yang, Z. Ergosterol Peroxide Suppresses Influenza A Virus-Induced pro-Inflammatory Response and Apoptosis by Blocking RIG-I Signaling. Eur. J. Pharmacol. 2019, 860, 172543. [Google Scholar] [CrossRef]
- Durán, N.; Justo, G.Z.; Durán, M.; Brocchi, M.; Cordi, L.; Tasic, L.; Castro, G.R.; Nakazato, G. Advances in Chromobacterium violaceum and Properties of Violacein-Its Main Secondary Metabolite: A Review. Biotechnol. Adv. 2016, 34, 1030–1045. [Google Scholar] [CrossRef]
- Koh, C.-L.; Sam, C.-K.; Yin, W.-F.; Tan, L.; Krishnan, T.; Chong, Y.; Chan, K.-G. Plant-Derived Natural Products as Sources of Anti-Quorum Sensing Compounds. Sensors 2013, 13, 6217–6228. [Google Scholar] [CrossRef]
- Zhu, H.; Wang, S.; Zhang, S.; Cao, C. Inhibiting Effect of Bioactive Metabolites Produced by Mushroom Cultivation on Bacterial Quorum Sensing-Regulated Behaviors. Chemotherapy 2011, 57, 292–297. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Liu, W.; Wang, S.; Tian, B.; Zhang, S. Evaluation of Anti-Quorum-Sensing Activity of Fermentation Metabolites from Different Strains of a Medicinal Mushroom, Phellinus igniarius. Chemotherapy 2012, 58, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Chaverra Daza, K.E.; Silva Gómez, E.; Moreno Murillo, B.D.; Mayorga Wandurraga, H. Natural and Enantiopure Alkylglycerols as Antibiofilms Against Clinical Bacterial Isolates and Quorum Sensing Inhibitors of Chromobacterium violaceum ATCC 12472. Antibiotics 2021, 10, 430. [Google Scholar] [CrossRef] [PubMed]
- Grabski, H.; Hunanyan, L.; Tiratsuyan, S.; Vardapetyan, H. Interaction of Quercetin with Transcriptional Regulator LasR of Pseudomonas aeruginosa: Mechanistic Insights of the Inhibition of Virulence through Quorum Sensing. bioRxiv 2017. [Google Scholar] [CrossRef]
- Hernando-Amado, S.; Alcalde-Rico, M.; Gil-Gil, T.; Valverde, J.R.; Martínez, J.L. Naringenin Inhibition of the Pseudomonas aeruginosa Quorum Sensing Response Is Based on Its Time-Dependent Competition With N-(3-Oxo-Dodecanoyl)-L-Homoserine Lactone for LasR Binding. Front. Mol. Biosci. 2020, 7, 25. [Google Scholar] [CrossRef]
- Manefield, M.; De Nys, R.; Naresh, K.; Roger, R.; Givskov, M.; Peter, S.; Kjelleberg, S. Evidence That Halogenated Furanones from Delisea Pulchra Inhibit Acylated Homoserine Lactone (AHL)-Mediated Gene Expression by Displacing the AHL Signal from Its Receptor Protein. Microbiology 1999, 145, 283–291. [Google Scholar] [CrossRef]
- Santos, C.A.; Lima, E.M.F.; Franco, B.D.G.D.M.; Pinto, U.M. Exploring Phenolic Compounds as Quorum Sensing Inhibitors in Foodborne Bacteria. Front. Microbiol. 2021, 12, 735931. [Google Scholar] [CrossRef]
- Shi, R.; Luo, Q.; Liu, Y.; Meng, G.; Chen, W.; Wang, C. Effect of γ-Butyrolactone, a Quorum Sensing Molecule, on Morphology and Secondary Metabolism in Monascus. LWT 2022, 172, 114225. [Google Scholar] [CrossRef]
- Wang, W.; Li, D.; Huang, X.; Yang, H.; Qiu, Z.; Zou, L.; Liang, Q.; Shi, Y.; Wu, Y.; Wu, S.; et al. Study on Antibacterial and Quorum-Sensing Inhibition Activities of Cinnamomum camphora Leaf Essential Oil. Molecules 2019, 24, 3792. [Google Scholar] [CrossRef]
- Wu, C.-S.; Lin, Z.-M.; Wang, L.-N.; Guo, D.-X.; Wang, S.-Q.; Liu, Y.-Q.; Yuan, H.-Q.; Lou, H.-X. Phenolic Compounds with NF-κB Inhibitory Effects from the Fungus Phellinus baumii. Bioorganic Med. Chem. Lett. 2011, 21, 3261–3267. [Google Scholar] [CrossRef]
- Kodiyalmath, J.K.; Krishnappa, M. Evaluation of Antimicrobial Activity of Phellinus linteus (Berk. & M.A Curtis.) with Their Wild Collections from Western Ghats of India. Trop. Plant Res. 2017, 4, 351–357. [Google Scholar] [CrossRef]
- Ayala-Zavala, J.F.; Silva-Espinoza, B.A.; Cruz-Valenzuela, M.R.; Villegas-Ochoa, M.A.; Esqueda, M.; González-Aguilar, G.A.; Calderón-López, Y. Antioxidant and Antifungal Potential of Methanol Extracts of Phellinus spp. from Sonora, Mexico. Rev. Iberoam. Micol. 2012, 29, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Yoon, K.-N.; Jang, H.S. Antioxidant and Antimicrobial Activities of Fruiting Bodies of Phellinus gilvus Collected in Korea. Korean J. Clin. Lab. Sci. 2016, 48, 355–364. [Google Scholar] [CrossRef]
- Contato, A.G.; De Araújo, C.A.V.; Zanzarin, D.M.; Aranha, G.M.; Sybuia, P.A.; Pilau, E.J.; Castoldi, R.; Peralta, R.M.; De Souza, C.G.M. Biological Characterization and Antimicrobial Bioactives of Mycelium Extracts from Medicinal Mushrooms Phellinus linteus and Pleurotus albidus (Agaricomycetes). Int. J. Med. Mushrooms 2022, 24, 47–55. [Google Scholar] [CrossRef]
- Dokhaharani, S.C.; Ghobad-Nejhad, M.; Moghimi, H.; Farazmand, A.; Rahmani, H. Biological Activities of Two Polypore Macrofungi (Basidiomycota) and Characterization of Their Compounds Using HPLC–DAD and LC–ESI–MS/MS. Folia Microbiol. 2021, 66, 775–786. [Google Scholar] [CrossRef] [PubMed]
- Smolskaitė, L.; Venskutonis, P.R.; Talou, T. Comprehensive Evaluation of Antioxidant and Antimicrobial Properties of Different Mushroom Species. LWT Food Sci. Technol. 2015, 60, 462–471. [Google Scholar] [CrossRef]
- Lee, I.-K.; Yun, B.-S. Hispidin Analogs from the Mushroom Inonotus xeranticus and Their Free Radical Scavenging Activity. Bioorganic Med. Chem. Lett. 2006, 16, 2376–2379. [Google Scholar] [CrossRef]
- Lee, I.-K.; Seok, S.-J.; Kim, W.-K.; Yun, B.-S. Hispidin Derivatives from the Mushroom Inonotus x Eranticus and Their Antioxidant Activity. J. Nat. Prod. 2006, 69, 299–301. [Google Scholar] [CrossRef]
- Min, B.-S.; Yun, B.-S.; Lee, H.-K.; Jung, H.-J.; Jung, H.-A.; Choi, J.-S. Two Novel Furan Derivatives from Phellinus linteus with Anti-Complement Activity. Bioorganic Med. Chem. Lett. 2006, 16, 3255–3257. [Google Scholar] [CrossRef]
- Goodrich-Tanrikulu, M.; Howe, K.; Stafford, A.; Nelson, M.A. Changes in Fatty Acid Composition of Neurospora crassa Accompany Sexual Development and Ascospore Germination. Microbiology 1998, 144, 1713–1720. [Google Scholar] [CrossRef]
- Liu, P.; Ivanova-Petropulos, V.; Duan, C.; Yan, G. Effect of Unsaturated Fatty Acids on Intra-Metabolites and Aroma Compounds of Saccharomyces cerevisiae in Wine Fermentation. Foods 2021, 10, 277. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.-C. Hymenochaetaceae (Basidiomycota) in China. Fungal Divers. 2010, 45, 131–343. [Google Scholar] [CrossRef]
- Gramss, G.R.L. Gilbertson and L. Ryvarden, North American Polypores. Volume 1: Abortiporus—Lindtneria. 433 S., 209 Abb. Oslo 1986. Fungiflora A/S. J. Basic Microbiol. 1987, 27, 282. [Google Scholar] [CrossRef]
- Kornerup, A.; Wanscher, J.H. Methuen Handbook of Colour, 3rd ed.; Eyre Methuen: London, UK, 1978. [Google Scholar]
- Zheng, H.-F.; Huang, F.-C.; Liu, B.; Shao, Y.-Y.; Qin, P.-S. Fulvifomes nonggangensis and F. tubogeneratus (Hymenochaetales, Basidiomycota): Two New Species from Southern China Based on Morphological and Molecular Evidences. Mycobiology 2021, 49, 213–222. [Google Scholar] [CrossRef] [PubMed]
- García-Jacobo, I.; Raymundo, T.; Martínez-González, C.R.; Martínez-Pineda, M.; Valenzuela, R. Phylogenetic and Morphological Analyses Reveal Twelve New Species of the Genus Patellaria (Dothideomycetes, Ascomycota) from Mexico. J. Fungi 2025, 11, 44. [Google Scholar] [CrossRef] [PubMed]
- Martínez-González, C.R.; Ramírez-Mendoza, R.; Jiménez-Ramírez, J.; Gallegos-Vázquez, C.; Luna-Vega, I. Improved Method for Genomic DNA Extraction for Opuntia Mill. (Cactaceae). Plant Methods 2017, 13, 82. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols; Elsevier: Amsterdam, The Netherlands, 1990; pp. 315–322. ISBN 978-0-12-372180-8. [Google Scholar]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A Greedy Algorithm for Aligning DNA Sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Müller, K.; Quandt, D.; Müller, J. PhyDE®-Phylogenetic Data Editor. Program Distributed by the Author 2010. Available online: http://www.phyde.de (accessed on 3 March 2025).
- Peng, J.; Swofford, D.L.; Kubatko, L. Estimation of Speciation Times under the Multispecies Coalescent. Bioinformatics 2022, 38, 5182–5190. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2016, 34, 772–773. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree—A Graphical Viewer of Phylogenetic Trees, v.1.4.4.; Institute of Evolutionary Biology, Univ of Edinburgh 2018. Available online: http://tree.bio.ed.ac.uk/software/figtree (accessed on 10 March 2025).
- Abdullah, S.; Jang, S.-E.; Kwak, M.-K.; Chong, K. Ganoderma boninense Mycelia for Phytochemicals and Secondary Metabolites with Antibacterial Activity. J. Microbiol. 2020, 58, 1054–1064. [Google Scholar] [CrossRef]
- Chakarwarti, J.; Anand, V.; Nayaka, S.; Srivastava, S. In Vitro Antibacterial Activity and Secondary Metabolite Profiling of Endolichenic Fungi Isolated from Genus Parmotrema. Curr. Microbiol. 2024, 81, 195. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, M.P.; Patel, J.B. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically: M07-A11, 11th ed.; Documents/Clinical and Laboratory Standards Institute; Committee for Clinical Laboratory Standards: Wayne, PA, USA, 2018; ISBN 978-1-56238-836-2. [Google Scholar]
- Ibarra, J.A.; Knodler, L.A.; Sturdevant, D.E.; Virtaneva, K.; Carmody, A.B.; Fischer, E.R.; Porcella, S.F.; Steele-Mortimer, O. Induction of Salmonella Pathogenicity Island 1 under Different Growth Conditions Can Affect Salmonella–Host Cell Interactions in Vitro. Microbiology 2010, 156, 1120–1133. [Google Scholar] [CrossRef]
- Choo, J.H.; Rukayadi, Y.; Hwang, J.-K. Inhibition of Bacterial Quorum Sensing by Vanilla Extract. Lett. Appl. Microbiol. 2006, 42, 637–641. [Google Scholar] [CrossRef]
- El-Elimat, T.; Figueroa, M.; Ehrmann, B.M.; Cech, N.B.; Pearce, C.J.; Oberlies, N.H. High-Resolution MS, MS/MS, and UV Database of Fungal Secondary Metabolites as a Dereplication Protocol for Bioactive Natural Products. J. Nat. Prod. 2013, 76, 1709–1716. [Google Scholar] [CrossRef]
- Mohimani, H.; Gurevich, A.; Shlemov, A.; Mikheenko, A.; Korobeynikov, A.; Cao, L.; Shcherbin, E.; Nothias, L.-F.; Dorrestein, P.C.; Pevzner, P.A. Dereplication of Microbial Metabolites through Database Search of Mass Spectra. Nat. Commun. 2018, 9, 4035. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and Community Curation of Mass Spectrometry Data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Schmid, R.; Heuckeroth, S.; Korf, A.; Smirnov, A.; Myers, O.; Dyrlund, T.S.; Bushuiev, R.; Murray, K.J.; Hoffmann, N.; Lu, M.; et al. Integrative Analysis of Multimodal Mass Spectrometry Data in MZmine 3. Nat. Biotechnol. 2023, 41, 447–449. [Google Scholar] [CrossRef] [PubMed]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. MassBank: A Public Repository for Sharing Mass Spectral Data for Life Sciences. J. Mass. Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Van Der Hooft, J.J.J.; Wandy, J.; Barrett, M.P.; Burgess, K.E.V.; Rogers, S. Topic Modeling for Untargeted Substructure Exploration in Metabolomics. Proc. Natl. Acad. Sci. USA 2016, 113, 13738–13743. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Schymanski, E.L.; Jeon, J.; Gulde, R.; Fenner, K.; Ruff, M.; Singer, H.P.; Hollender, J. Identifying Small Molecules via High Resolution Mass Spectrometry: Communicating Confidence. Environ. Sci. Technol. 2014, 48, 2097–2098. [Google Scholar] [CrossRef]
- Shun, Z.; Silverberg, A.; Chang, C.K.; Ouyang, P. Dunnett’s Many-to-One Test and Least Square Means. J. Biopharm. Stat. 2003, 13, 17–28. [Google Scholar] [CrossRef]


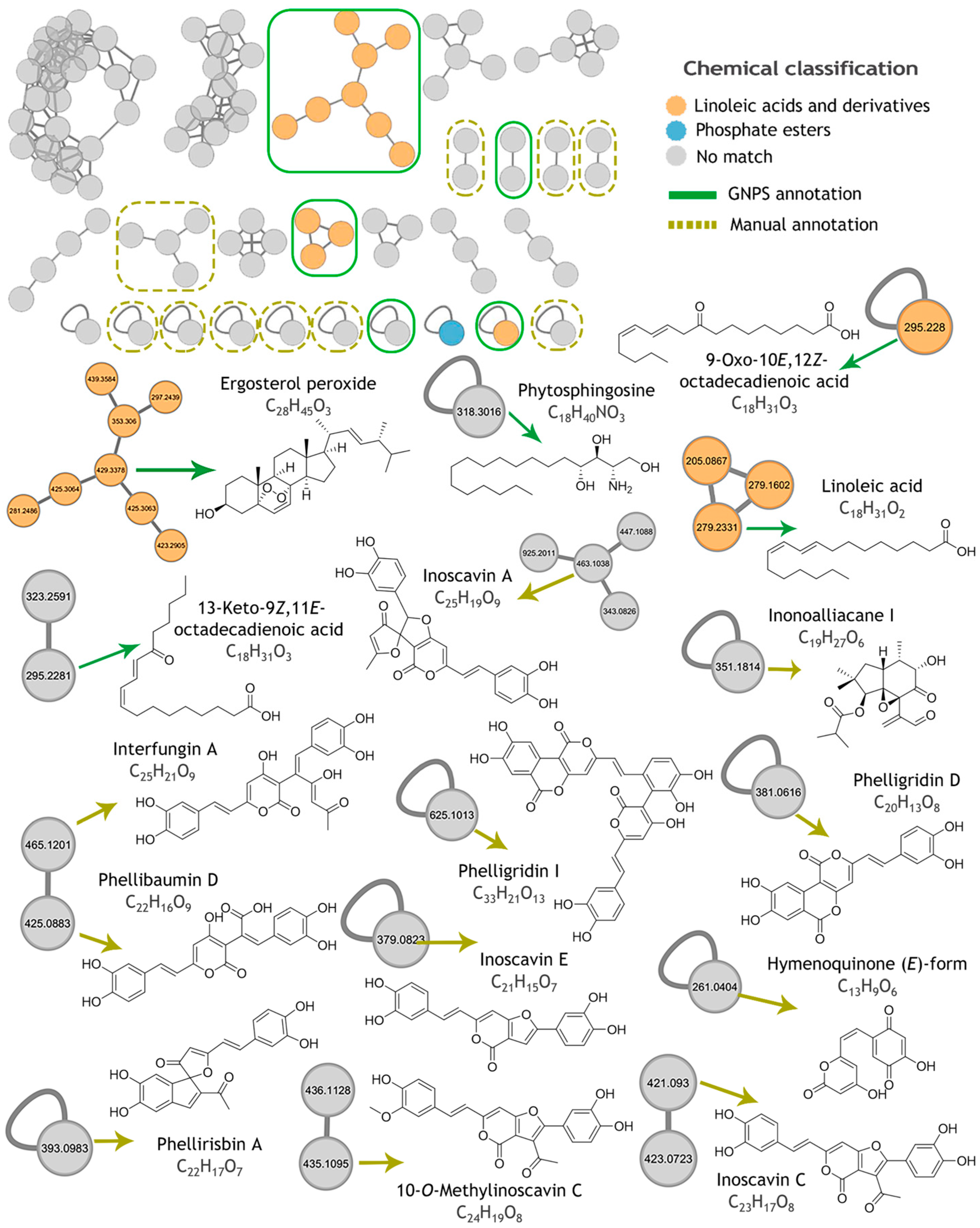
| Compound | Observed Ion (m/z) a | Adduct a | Molecular Formula (Adduct) | Exact Mass | Mass Accuracy (ppm) |
|---|---|---|---|---|---|
| 13-Keto-9Z,11E-octadecadienoic acid | 295.2281 | [M + H]+ | C18H31O3 | 295.2268 | +4.5 |
| 9-Oxo-10E,12Z-octadecadienoic acid | 295.228 | [M + H]+ | C18H31O3 | 295.2268 | +4.2 |
| Linoleic acid | 279.2331 | [M + H]+ | C18H31O2 | 279.2329 | +0.5 |
| Phytosphingosine | 318.3016 | [M + H]+ | C18H40NO3 | 318.3003 | +4.2 |
| Ergosterol peroxide | 429.3378 | [M + H]+ | C28H45O3 | 429.3363 | +3.4 |
| Compound | Observed Ion (m/z) | Adduct | Molecular Formula (Adduct) | Exact Mass | Mass Accuracy (ppm) a | Fungal Source |
|---|---|---|---|---|---|---|
| Interfungin A | 465.1201 | [M + H]+ | C25H21O9 | 465.1180 | +4.5 | Hymenochaete xerantica |
| Inoscavin A | 465.1201 | [M + H]+ | C25H19O9 | 463.1023 | +3.1 | Hymenochaete xerantica, Sanghuangporus baumii, Fulvifomes fastuosus. |
| Phelligridin I (syn Inonoblin A) | 625.1013 | [M + H]+ | C33H21O13 | 625.0977 | +5.8 | Inonotus obliquus, Phellinus igniarius |
| 10-O-Methylinoscavin C | 435.1095 | [M + H]+ | C24H19O8 | 435.1074 | +4.7 | Hymenochaete xerantica |
| Phelliribsin A | 393.0983 | [M + H]+ | C22H17O7 | 393.0969 | +3.6 | Phylloporia ribis |
| Hymenoquinone (E)-form | 261.0404 | [M + H]+ | C13H9O6 | 261.0394 | +4.0 | Hymenochaete mougeotii |
| Phellibaumin D | 425.0883 | [M + H]+ | C22H17O9 | 425.0867 | +3.7 | Sanghuangporus baumii |
| Inonoalliacane I | 351.1814 | [M + H]+ | C19H27O6 | 351.1802 | +3.4 | Inonotus sp. |
| Phelligridin D | 379.0475 | [M + H]+ | C20H13O8 | 381.0605 | +2.9 | Inonotus obliquus, Phellinus igniarius, Phellinus linteus |
| Inoscavin C | 421.0930 | [M + H]+ | C23H17O8 | 421.0918 | +2.9 | Phellinus ellipsoideus, Hymenochaete xerantica, Phellinus igniarius. |
| Inoscavin E | 379.0823 | [M + H]+ | C21H15O7 | 379.0812 | +2.8 | Phellinus linteus, Hymenochaete xerantica |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bolaños-Nuñez, A.; Martínez-Pineda, M.; Valenzuela, R.; Figueroa, M.; D. Patiño, A.; Curiel-Quesada, E.; Martínez-Gonzáles, C.R.; Villanueva-Silva, R.; Raymundo, T.; Pérez-Valdespino, A. Quorum Sensing and Mobility Inhibition of Pathogenic Bacteria by Fulvifomes mexicanus sp. nov. Molecules 2025, 30, 2278. https://doi.org/10.3390/molecules30112278
Bolaños-Nuñez A, Martínez-Pineda M, Valenzuela R, Figueroa M, D. Patiño A, Curiel-Quesada E, Martínez-Gonzáles CR, Villanueva-Silva R, Raymundo T, Pérez-Valdespino A. Quorum Sensing and Mobility Inhibition of Pathogenic Bacteria by Fulvifomes mexicanus sp. nov. Molecules. 2025; 30(11):2278. https://doi.org/10.3390/molecules30112278
Chicago/Turabian StyleBolaños-Nuñez, Angelica, Michelle Martínez-Pineda, Ricardo Valenzuela, Mario Figueroa, Albert D. Patiño, Everardo Curiel-Quesada, César Ramiro Martínez-Gonzáles, Rodrigo Villanueva-Silva, Tania Raymundo, and Abigail Pérez-Valdespino. 2025. "Quorum Sensing and Mobility Inhibition of Pathogenic Bacteria by Fulvifomes mexicanus sp. nov." Molecules 30, no. 11: 2278. https://doi.org/10.3390/molecules30112278
APA StyleBolaños-Nuñez, A., Martínez-Pineda, M., Valenzuela, R., Figueroa, M., D. Patiño, A., Curiel-Quesada, E., Martínez-Gonzáles, C. R., Villanueva-Silva, R., Raymundo, T., & Pérez-Valdespino, A. (2025). Quorum Sensing and Mobility Inhibition of Pathogenic Bacteria by Fulvifomes mexicanus sp. nov. Molecules, 30(11), 2278. https://doi.org/10.3390/molecules30112278








