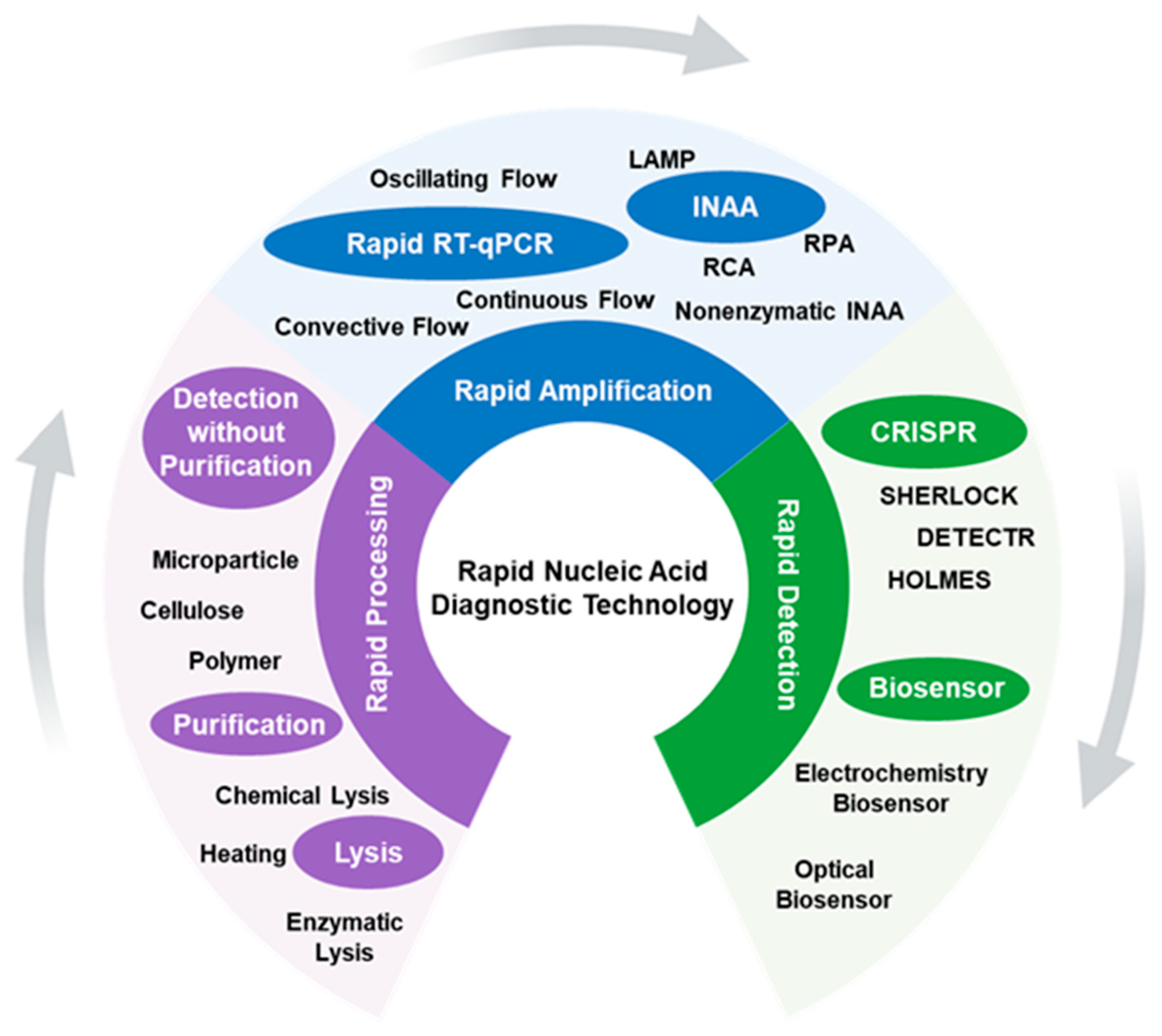
Rapid Nucleic Acid Diagnostic Technology for Pandemic Diseases
Abstract
1. Introduction
2. Rapid Processing
3. Rapid Amplification
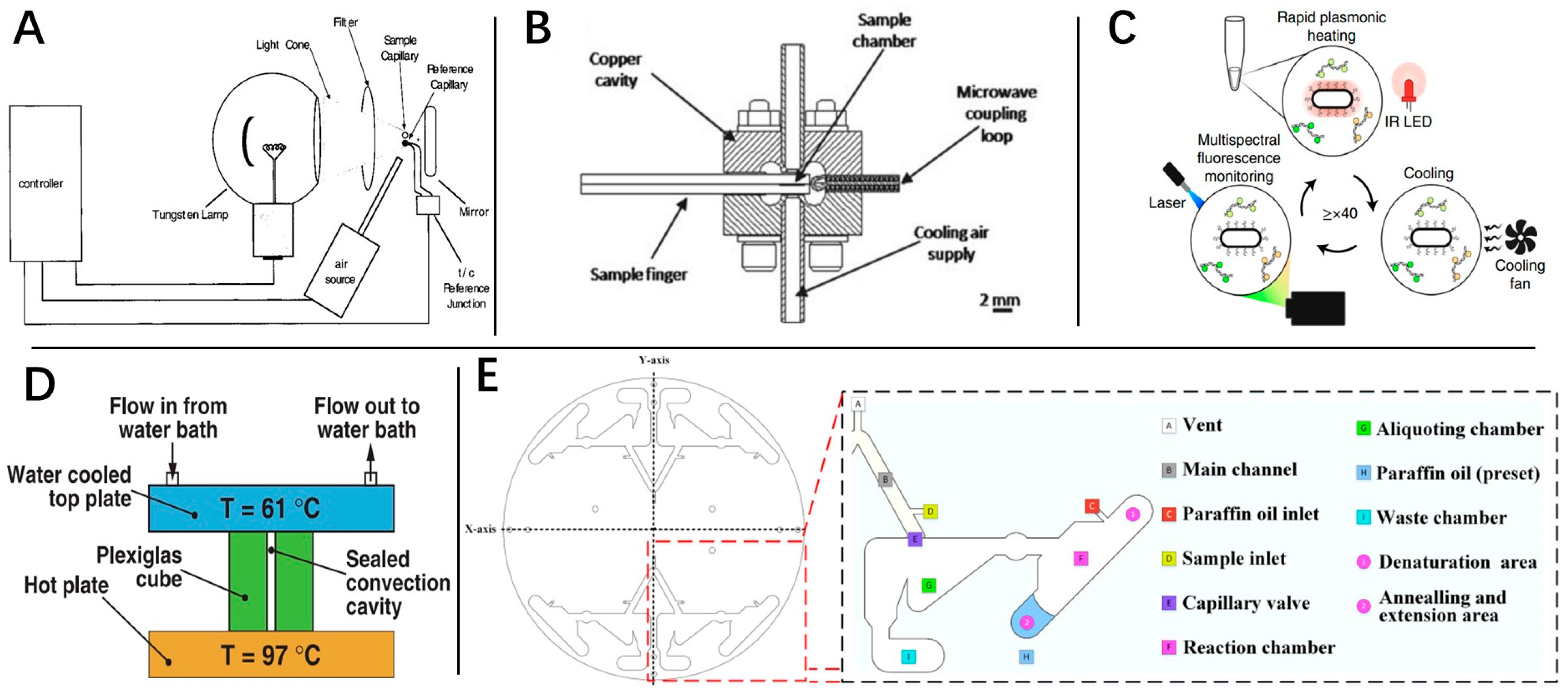
3.1. Rapid RT-qPCR
3.2. Rapid Isothermal Amplification
3.2.1. Loop-Mediated Isothermal Amplification
3.2.2. Rolling Circle Amplification
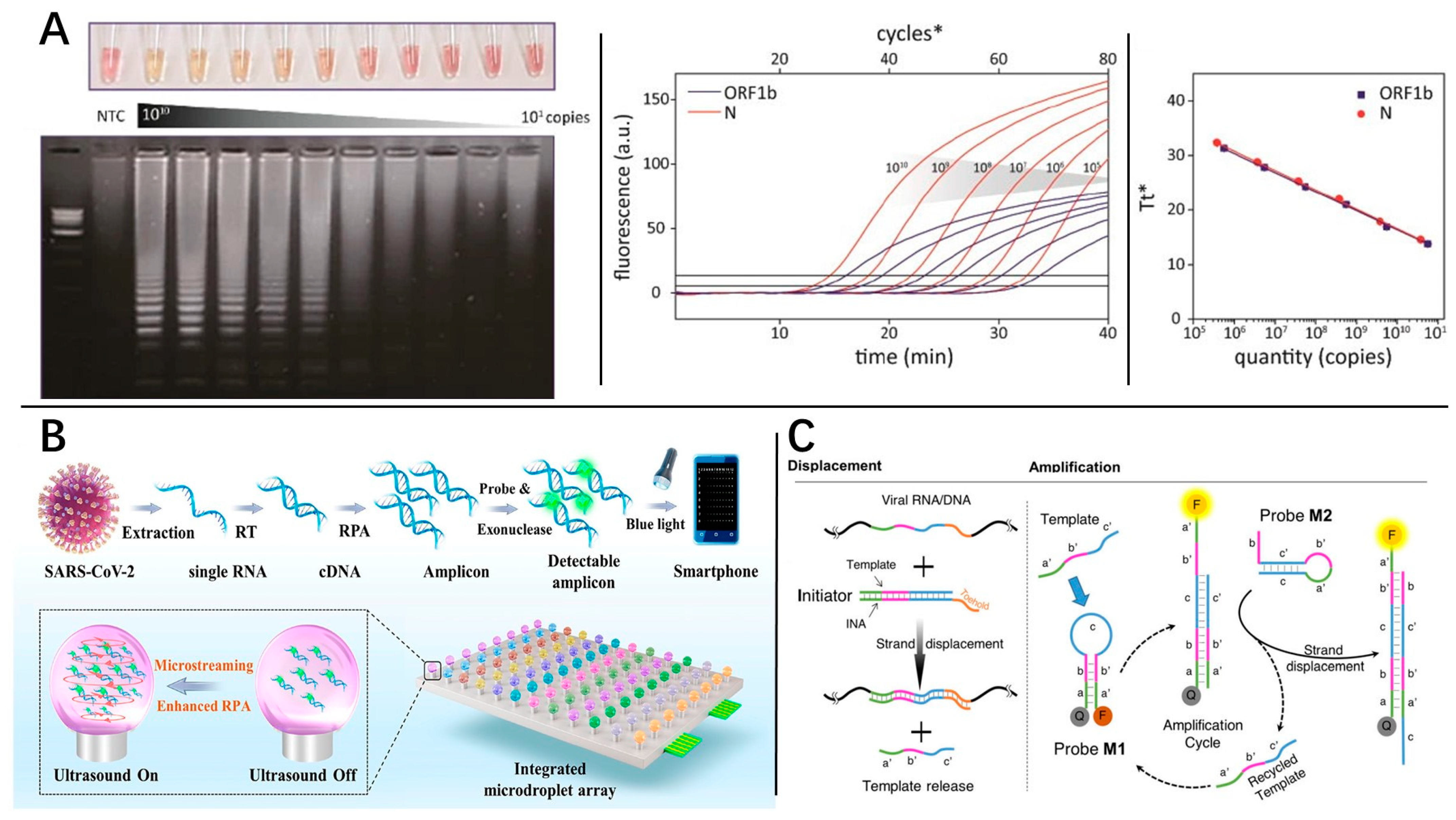
3.2.3. Recombinase Polymerase Amplification
3.2.4. Nonenzymatic INAA
4. Rapid Detection
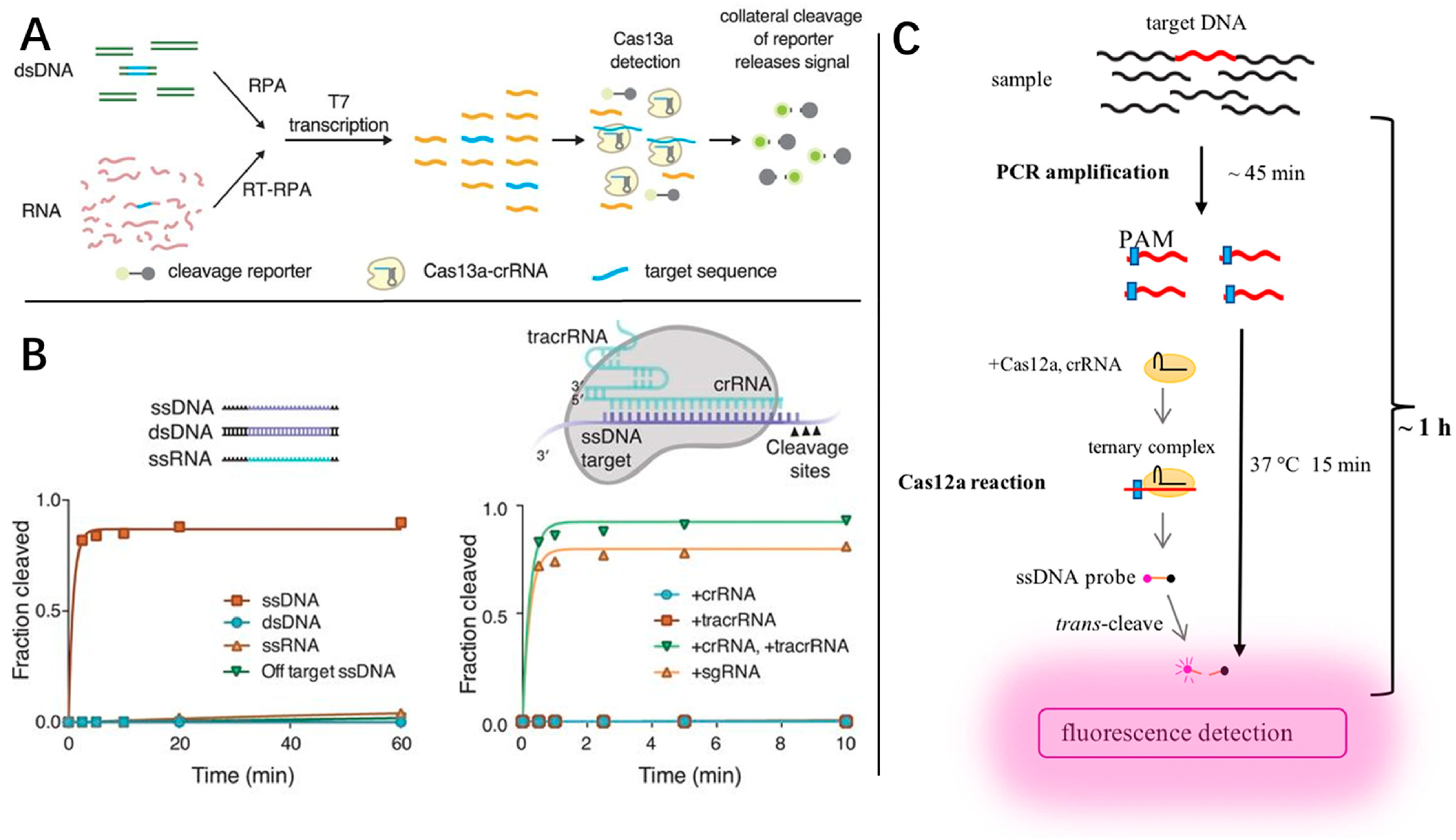
4.1. CRISPR
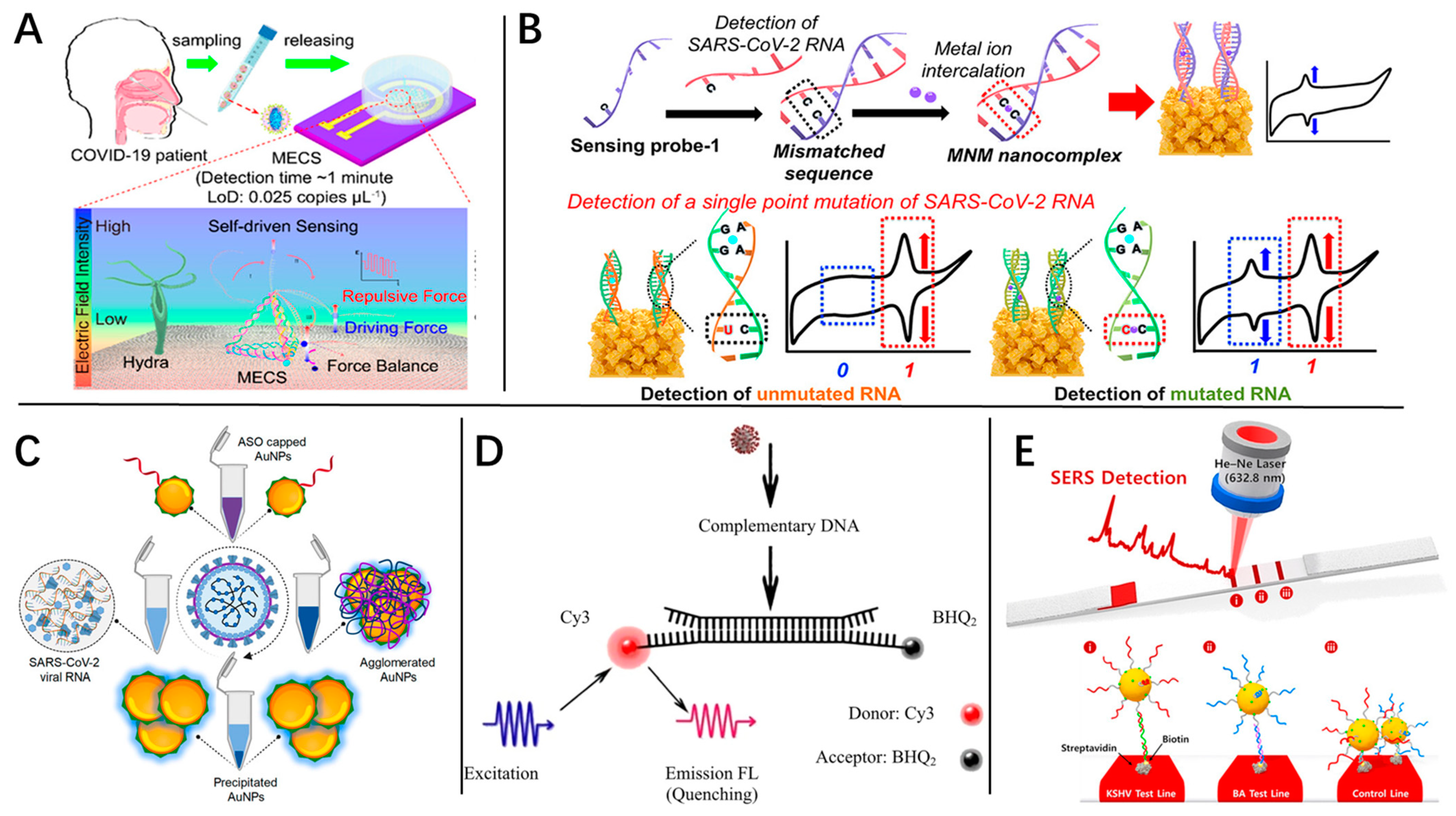
4.2. Physical and Chemical Biosensor
4.2.1. Electrochemistry Biosensor
4.2.2. Optical Biosensor
5. Conclusions and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19—11 March 2020. Available online: https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 (accessed on 15 December 2023).
- Yan, C.; Cui, J.; Huang, L.; Du, B.; Chen, L.; Xue, G.; Li, S.; Zhang, W.; Zhao, L.; Sun, Y.; et al. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin. Microbiol. Infect. 2020, 26, 773–779. [Google Scholar] [CrossRef]
- Pokhrel, P.; Hu, C.; Mao, H. Detecting the Coronavirus (COVID-19). ACS Sens. 2020, 5, 2283–2296. [Google Scholar] [CrossRef] [PubMed]
- Visseaux, B.; Collin, G.; Houhou-Fidouh, N.; Le Hingrat, Q.; Ferre, V.M.; Damond, F.; Ichou, H.; Descamps, D.; Charpentier, C. Evaluation of three extraction-free SARS-CoV-2 RT-PCR assays: A feasible alternative approach with low technical requirements. J. Virol. Methods 2021, 291, 114086. [Google Scholar] [CrossRef]
- Blumenfeld, N.R.; Bolene, M.A.E.; Jaspan, M.; Ayers, A.G.; Zarrandikoetxea, S.; Freudman, J.; Shah, N.; Tolwani, A.M.; Hu, Y.; Chern, T.L.; et al. Multiplexed reverse-transcriptase quantitative polymerase chain reaction using plasmonic nanoparticles for point-of-care COVID-19 diagnosis. Nat. Nanotechnol. 2022, 17, 984–992. [Google Scholar] [CrossRef]
- Lalli, M.A.; Langmade, S.J.; Chen, X.; Fronick, C.C.; Sawyer, C.S.; Burcea, L.C.; Wilkinson, M.N.; Fulton, R.S.; Heinz, M.; Buchser, W.J.; et al. Rapid and extraction-free detection of SARS-CoV-2 from saliva with colorimetric LAMP. medRxiv 2020, 67, 415–424. [Google Scholar] [CrossRef]
- Hwang, O.J.; Back, K. Simultaneous suppression of two distinct serotonin N-acetyltransferase isogenes by RNA interference leads to severe decreases in melatonin and accelerated seed deterioration in rice. Biomolecules 2020, 10, 141. [Google Scholar] [CrossRef] [PubMed]
- Nolan, T.; Bustin, S.A.; Burroughs, N.; Karteris, E. Ultra-High-Speed PCR Instrument Development. In PCR Technology: Current Innovations; Nolan, T., Bustin, S.A., Eds.; CRC Press: Boca Raton, FL, USA, 2013. [Google Scholar]
- Struijk, R.; van den Ouden, A.; Louwerse, J.; Curova, K.; Burggrave, R.; McNally, B.; de Groot, T.; Mulder, B.; de Vos, G. Ultrafast RNA extraction-free SARS-CoV-2 detection by direct RT-PCR using a rapid thermal cycling approach. Diagn. Microbiol. Infect. Dis. 2023, 107, 115975. [Google Scholar] [CrossRef] [PubMed]
- Farfour, E.; Asso-Bonnet, M.; Vasse, M. The ID NOW COVID-19, a high-speed high-performance assay. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 2041–2045. [Google Scholar] [CrossRef]
- Zahavi, M.; Rohana, H.; Azrad, M.; Shinberg, B.; Peretz, A. Rapid SARS-CoV-2 Detection Using the Lucira™ Check It COVID-19 Test Kit. Diagnostics 2022, 12, 1877. [Google Scholar] [CrossRef]
- Ullerich, L.; Campbell, S.; Krieg-Schneider, F.; Bürsgens, F.; Stehr, J. Ultra-fast PCR technologies for point-of-care testing. Laboratoriumsmedizin 2017, 41, 239–244. [Google Scholar] [CrossRef]
- Wawina-Bokalanga, T.; Sklenovska, N.; Vanmechelen, B.; Bloemen, M.; Vergote, V.; Laenen, L.; André, E.; Van Ranst, M.; Muyembe-Tamfum, J.-J.; Maes, P. An accurate and rapid Real-time PCR approach for human Monkeypox virus diagnosis. medRxiv 2022. [Google Scholar] [CrossRef]
- Coronavirus (COVID-19) Update: FDA Issues First Emergency Use Authorization for Point of Care Diagnostic. Available online: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-issues-first-emergency-use-authorization-point-care-diagnostic (accessed on 15 December 2023).
- Hansen, G.; Marino, J.; Wang, Z.-X.; Beavis, K.G.; Rodrigo, J.; Labog, K.; Westblade, L.F.; Jin, R.; Love, N.; Ding, K.; et al. Clinical Performance of the Point-of-Care cobas Liat for Detection of SARS-CoV-2 in 20 Minutes: A Multicenter Study. J. Clin. Microbiol. 2021, 59, e02811-20. [Google Scholar] [CrossRef]
- Diagnostics, R. Roche’s cobas SARS-CoV-2 Test to Detect Novel Coronavirus Receives FDA Emergency Use Authorization and Is Available in Markets Accepting the CE Mark. Available online: https://diagnostics.roche.com/global/en/news-listing/2020/roche-receives-fda-emergency-use-authorization-for-cobas-sars-co.html (accessed on 15 December 2023).
- Lownik, J.C.; Way, G.W.; Farrar, J.S.; Martin, R.K. Extraction-Free Rapid Cycle Quantitative RT-PCR and Extreme RT-PCR for SARS-CoV-2 Virus Detection. J. Mol. Diagn. 2021, 23, 1671–1679. [Google Scholar] [CrossRef]
- Nagura-Ikeda, M.; Imai, K.; Tabata, S.; Miyoshi, K.; Murahara, N.; Mizuno, T.; Horiuchi, M.; Kato, K.; Imoto, Y.; Iwata, M. Clinical evaluation of self-collected saliva by quantitative reverse transcription-PCR (RT-qPCR), direct RT-qPCR, reverse transcription–loop-mediated isothermal amplification, and a rapid antigen test to diagnose COVID-19. J. Clin. Microbiol. 2020, 58, e01438-20. [Google Scholar] [CrossRef] [PubMed]
- Hassan, F.; Hays, L.M.; Bonner, A.; Bradford, B.J.; Franklin, R.; Hendry, P.; Kaminetsky, J.; Vaughn, M.; Cieslak, K.; Moffatt, M.E.; et al. Multicenter Clinical Evaluation of the Alere i Respiratory Syncytial Virus Isothermal Nucleic Acid Amplification Assay. J. Clin. Microbiol. 2018, 56, e01777-17. [Google Scholar] [CrossRef]
- Katzman, B.M.; Wockenfus, A.M.; Kelley, B.R.; Karon, B.S.; Donato, L.J. Evaluation of the Visby medical COVID-19 point of care nucleic acid amplification test. Clin. Biochem. 2023, 117, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Kostyusheva, A.; Brezgin, S.; Babin, Y.; Vasilyeva, I.; Glebe, D.; Kostyushev, D.; Chulanov, V. CRISPR-Cas systems for diagnosing infectious diseases. Methods 2022, 203, 431–446. [Google Scholar] [CrossRef] [PubMed]
- Lino, C.; Barrias, S.; Chaves, R.; Adega, F.; Martins-Lopes, P.; Fernandes, J.R. Biosensors as diagnostic tools in clinical applications. Biochim. Biophys. Acta Rev. Cancer 2022, 1877, 188726. [Google Scholar] [CrossRef]
- Boehringer, H.R.; O’Farrell, B.J. Lateral Flow Assays in Infectious Disease Diagnosis. Clin. Chem. 2021, 68, 52–58. [Google Scholar] [CrossRef]
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA 2020, 323, 1843–1844. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Interim Guidelines for Clinical Specimens for COVID-19. Available online: https://www.cdc.gov/coronavirus/2019-ncov/index.html (accessed on 15 December 2023).
- European Centre for Disease Prevention and Control. Testing Strategies for SARS-CoV-2. Available online: https://www.ecdc.europa.eu/en/covid-19/surveillance/testing-strategies (accessed on 15 December 2023).
- Ott, I.M.; Strine, M.S.; Watkins, A.E.; Boot, M.; Kalinich, C.C.; Harden, C.A.; Vogels, C.B.F.; Casanovas-Massana, A.; Moore, A.J.; Muenker, M.C.; et al. Simply saliva: Stability of SARS-CoV-2 detection negates the need for expensive collection devices. medRxiv 2020. [Google Scholar] [CrossRef]
- Watkins, A.E.; Fenichel, E.P.; Weinberger, D.M.; Vogels, C.B.F.; Brackney, D.E.; Casanovas-Massana, A.; Campbell, M.; Fournier, J.; Bermejo, S.; Datta, R.; et al. Pooling saliva to increase SARS-CoV-2 testing capacity. MedRxiv 2020. [Google Scholar] [CrossRef]
- Ibrahimi, N.; Delaunay-Moisan, A.; Hill, C.; Le Teuff, G.; Rupprecht, J.-F.; Thuret, J.-Y.; Chaltiel, D.; Potier, M.-C. Screening for SARS-CoV-2 by RT-PCR: Saliva or nasopharyngeal swab? Rapid review and meta-analysis. PLoS ONE 2021, 16, e0253007. [Google Scholar] [CrossRef] [PubMed]
- Esbin, M.N.; Whitney, O.N.; Chong, S.; Maurer, A.; Darzacq, X.; Tjian, R. Overcoming the bottleneck to widespread testing: A rapid review of nucleic acid testing approaches for COVID-19 detection. RNA 2020, 26, 771–783. [Google Scholar] [CrossRef]
- Smyrlaki, I.; Ekman, M.; Lentini, A.; Rufino de Sousa, N.; Papanicolaou, N.; Vondracek, M.; Aarum, J.; Safari, H.; Muradrasoli, S.; Rothfuchs, A.G.; et al. Massive and rapid COVID-19 testing is feasible by extraction-free SARS-CoV-2 RT-PCR. Nat. Commun. 2020, 11, 4812. [Google Scholar] [CrossRef] [PubMed]
- Srivatsan, S.; Heidl, S.; Pfau, B.; Martin, B.K.; Han, P.D.; Zhong, W.; van Raay, K.; McDermot, E.; Opsahl, J.; Gamboa, L.; et al. SwabExpress: An End-to-End Protocol for Extraction-Free COVID-19 Testing. Clin. Chem. 2021, 68, 143–152. [Google Scholar] [CrossRef]
- Mason, M.G.; Botella, J.R. Rapid (30-second), equipment-free purification of nucleic acids using easy-to-make dipsticks. Nat. Protoc. 2020, 15, 3663–3677. [Google Scholar] [CrossRef]
- Zou, Y.; Mason, M.G.; Wang, Y.; Wee, E.; Turni, C.; Blackall, P.J.; Trau, M.; Botella, J.R. Nucleic acid purification from plants, animals and microbes in under 30 seconds. PLoS Biol. 2017, 15, e2003916. [Google Scholar] [CrossRef]
- Qian, S.; Chen, Y.; Peng, C.; Wang, X.; Wu, H.; Che, Y.; Wang, H.; Xu, J.; Wu, J. Dipstick-based rapid nucleic acids purification and CRISPR/Cas12a-mediated isothermal amplification for visual detection of African swine fever virus. Talanta 2022, 242, 123294. [Google Scholar] [CrossRef]
- Li, P.; Li, M.; Yuan, Z.; Jiang, X.; Yue, D.; Ye, B.; Zhao, Z.; Jiang, J.; Fan, Q.; Zhou, Z.; et al. 3D printed integrated separator with hybrid micro-structures for high throughput and magnetic-free nucleic acid separation from organism samples. Sep. Purif. Technol. 2021, 271, 118881. [Google Scholar] [CrossRef]
- Song, M.; Hong, S.; Lee, L.P. Multiplexed Ultrasensitive Sample-to-Answer RT-LAMP Chip for the Identification of SARS-CoV-2 and Influenza Viruses. Adv. Mater. 2023, 35, e2207138. [Google Scholar] [CrossRef]
- Lin, X.; Fang, M.; Yi, C.; Jiang, Y.; Zhang, C.; Pan, X.; Luo, Z. Functional hydrogel for fast, precise and inhibition-free point-of-care bacteria analysis in crude food samples. Biomaterials 2022, 280, 121278. [Google Scholar] [CrossRef]
- Yi, C.; Luo, Z.; Lu, Y.; Belwal, T.; Pan, X.; Lin, X. Nanoporous hydrogel for direct digital nucleic acid amplification in untreated complex matrices for single bacteria counting. Biosens. Bioelectron. 2021, 184, 113199. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zhang, S.; Chen, Y.; Qian, C.; Liu, Y.; Shen, H.; Wang, Z.; Ping, J.; Wu, J.; Zhang, Y.; et al. Progress in molecular detection with high-speed nucleic acids thermocyclers. J. Pharm. Biomed. Anal. 2020, 190, 113489. [Google Scholar] [CrossRef] [PubMed]
- Millington, A.L.; Houskeeper, J.A.; Quackenbush, J.F.; Trauba, J.M.; Wittwer, C.T. The kinetic requirements of extreme qPCR. Biomol. Detect. Quantif. 2019, 17, 100081. [Google Scholar] [CrossRef]
- Rejali, N.A.; Ye, F.D.; Zuiter, A.M.; Keller, C.C.; Wittwer, C.T. Nearest-neighbour transition-state analysis for nucleic acid kinetics. Nucleic Acids Res. 2021, 49, 4574–4585. [Google Scholar] [CrossRef] [PubMed]
- Crews, N.; Wittwer, C.; Gale, B. Continuous-flow thermal gradient PCR. Biomed. Microdevices 2008, 10, 187–195. [Google Scholar] [CrossRef]
- Chen, R.; Lu, X.; Li, M.; Chen, G.; Deng, Y.; Du, F.; Dong, J.; Huang, X.; Cui, X.; Tang, Z. Polymerase Chain Reaction using “V” Shape Thermal Cycling Program. Theranostics 2019, 9, 1572–1579. [Google Scholar] [CrossRef]
- Wittwer, C.T. Rapid Cycle and Extreme Polymerase Chain Reaction. Methods Mol. Biol. 2023, 2621, 257–266. [Google Scholar] [CrossRef]
- Wittwer, C.T.; Reed, G.B.; Ririe, K.M. Rapid cycle DNA amplification. In The Polymerase Chain Reaction; Springer: Berlin/Heidelberg, Germany, 1994; pp. 174–181. [Google Scholar]
- Wittwer, C.; Ririe, K.; Andrew, R.; David, D.; Gundry, R.; Balis, U. The LightCyclerTM: A microvolume multisample fluorimeter with rapid temperature control. Biotechniques 1997, 22, 176–181. [Google Scholar] [CrossRef]
- Wittwer, C.T.; Herrmann, M.G.; Moss, A.A.; Rasmussen, R.P. Continuous fluorescence monitoring of rapid cycle DNA amplification. Biotechniques 1997, 22, 130–138. [Google Scholar] [CrossRef]
- Roper, M.G.; Easley, C.J.; Landers, J.P. Advances in polymerase chain reaction on microfluidic chips. Anal. Chem. 2005, 77, 3887–3894. [Google Scholar] [CrossRef]
- Zhang, C.; Xing, D. Miniaturized PCR chips for nucleic acid amplification and analysis: Latest advances and future trends. Nucleic Acids Res. 2007, 35, 4223–4237. [Google Scholar] [CrossRef]
- Tong, R.; Zhang, L.; Song, Q.; Hu, C.; Chen, X.; Lou, K.; Gong, X.; Gao, Y.; Wen, W. A fully portable microchip real-time polymerase chain reaction for rapid detection of pathogen. Electrophoresis 2019, 40, 1699–1707. [Google Scholar] [CrossRef]
- Yin, B.; Wan, X.; Sohan, A.; Lin, X. Microfluidics-Based POCT for SARS-CoV-2 Diagnostics. Micromachines 2022, 13, 1238. [Google Scholar] [CrossRef]
- Ding, Y.; Howes, P.D.; de Mello, A.J. Recent advances in droplet microfluidics. Anal. Chem. 2019, 92, 132–149. [Google Scholar] [CrossRef]
- Sohrabi, S.; Moraveji, M.K. Droplet microfluidics: Fundamentals and its advanced applications. RSC Adv. 2020, 10, 27560. [Google Scholar] [CrossRef] [PubMed]
- Ji, M.; Xia, Y.; Loo, J.F.-C.; Li, L.; Ho, H.-P.; He, J.; Gu, D. Automated multiplex nucleic acid tests for rapid detection of SARS-CoV-2, influenza A and B infection with direct reverse-transcription quantitative PCR (dirRT-qPCR) assay in a centrifugal microfluidic platform. RSC Adv. 2020, 10, 34088–34098. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Liu, L.; Tu, Y.; Zhang, J.; Miao, G.; Zhang, L.; Ge, S.; Xia, N.; Yu, D.; Qiu, X. Rapid PCR powered by microfluidics: A quick review under the background of COVID-19 pandemic. Trends Anal. Chem. 2021, 143, 116377. [Google Scholar] [CrossRef] [PubMed]
- Easley, C.J.; Humphrey, J.A.C.; Landers, J.P. Thermal isolation of microchip reaction chambers for rapid non-contact DNA amplification. J. Micromech. Microeng. 2007, 17, 1758. [Google Scholar] [CrossRef]
- Ko, J.; Yoo, J.C. Non-Contact Temperature Control System Applicable to Polymerase Chain Reaction on a Lab-on-a-Disc. Sensors 2019, 19, 2621. [Google Scholar] [CrossRef] [PubMed]
- Hühmer, A.F.R.; Landers, J.P. Noncontact Infrared-Mediated Thermocycling for Effective Polymerase Chain Reaction Amplification of DNA in Nanoliter Volumes. Anal. Chem. 2000, 72, 5507–5512. [Google Scholar] [CrossRef] [PubMed]
- Markovic, T.; Nauwelaers, B. Analysis of Microwave Heating Devices for Microfluidics. In Proceedings of the 2022 IEEE MTT-S International Microwave Biomedical Conference (IMBioC), Suzhou, China, 16–18 May 2022; pp. 28–30. [Google Scholar]
- Shaw, K.J.; Docker, P.T.; Yelland, J.V.; Dyer, C.E.; Greenman, J.; Greenway, G.M.; Haswell, S.J. Rapid PCR amplification using a microfluidic device with integrated microwave heating and air impingement cooling. Lab. Chip 2010, 10, 1725–1728. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Vanloon, J.; Yan, H. Influence of microwave irradiation on DNA hybridization and polymerase reactions. Tetrahedron Lett. 2019, 60, 151060. [Google Scholar] [CrossRef]
- You, M.; Li, Z.; Feng, S.; Gao, B.; Yao, C.; Hu, J.; Xu, F. Ultrafast Photonic PCR Based on Photothermal Nanomaterials. Trends Biotechnol. 2020, 38, 637–649. [Google Scholar] [CrossRef]
- Krishnan, M.; Ugaz, V.M.; Burns, M.A. PCR in a Rayleigh-Benard convection cell. Science 2002, 298, 793. [Google Scholar] [CrossRef]
- Chou, W.P.; Chen, P.H.; Miao, M., Jr.; Kuo, L.S.; Yeh, S.H.; Chen, P.J. Rapid DNA amplification in a capillary tube by natural convection with a single isothermal heater. Biotechniques 2011, 50, 52–57. [Google Scholar] [CrossRef]
- Miao, G.; Zhang, L.; Zhang, J.; Ge, S.; Xia, N.; Qian, S.; Yu, D.; Qiu, X. Free convective PCR: From principle study to commercial applications-A critical review. Anal. Chim. Acta 2020, 1108, 177–197. [Google Scholar] [CrossRef]
- Qiu, X.; Zhang, S.; Xiang, F.; Wu, D.; Guo, M.; Ge, S.; Li, K.; Ye, X.; Xia, N.; Qian, S. Instrument-free point-of-care molecular diagnosis of H1N1 based on microfluidic convective PCR. Sens. Actuators B Chem. 2017, 243, 738–744. [Google Scholar] [CrossRef]
- Shu, B.; Zhang, C.; Xing, D. A sample-to-answer, real-time convective polymerase chain reaction system for point-of-care diagnostics. Biosens. Bioelectron. 2017, 97, 360–368. [Google Scholar] [CrossRef]
- Qiu, X.; Zhang, S.; Mei, L.; Wu, D.; Guo, Q.; Li, K.; Ge, S.; Ye, X.; Xia, N.; Mauk, M.G. Characterization and analysis of real-time capillary convective PCR toward commercialization. Biomicrofluidics 2017, 11, 024103. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Ge, S.; Gao, P.; Li, K.; Yang, Y.; Zhang, S.; Ye, X.; Xia, N.; Qian, S. A Low-Cost and Fast Real-Time PCR System Based on Capillary Convection. SLAS Technol. 2017, 22, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Khodakov, D.; Li, J.; Zhang, J.X.; Zhang, D.Y. Highly multiplexed rapid DNA detection with single-nucleotide specificity via convective PCR in a portable device. Nat. Biomed. Eng. 2021, 5, 702–712. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Dai, R.; Lu, S.; Liu, X.; Zhou, T.; Yang, C.; Hu, X.; Lv, X.; Li, X. Oscillatory-Flow PCR Microfluidic Chip Driven by Low Speed Biaxial Centrifugation. Biosensors 2023, 13, 555. [Google Scholar] [CrossRef] [PubMed]
- Farrar, J.S.; Wittwer, C.T. Extreme PCR: Efficient and specific DNA amplification in 15-60 seconds. Clin. Chem. 2015, 61, 145–153. [Google Scholar] [CrossRef]
- Rejali, N.A.; Zuiter, A.M.; Quackenbush, J.F.; Wittwer, C.T. Reverse transcriptase kinetics for one-step RT-PCR. Anal. Biochem. 2020, 601, 113768. [Google Scholar] [CrossRef]
- Chen, J.J.; Lin, Z.H. Fabrication of an Oscillating Thermocycler to Analyze the Canine Distemper Virus by Utilizing Reverse Transcription Polymerase Chain Reaction. Micromachines 2022, 13, 600. [Google Scholar] [CrossRef]
- Nguyen, P.Q.M.; Wang, M.; Ann Maria, N.; Li, A.Y.; Tan, H.Y.; Xiong, G.M.; Tan, M.M.; Bhagat, A.A.S.; Ong, C.W.M.; Lim, C.T. Modular micro-PCR system for the onsite rapid diagnosis of COVID-19. Microsyst. Nanoeng. 2022, 8, 82. [Google Scholar] [CrossRef]
- Kopp, M.U.; Mello, A.J.d.; Manz, A. Chemical Amplification: Continuous-Flow PCR on a Chip. Science 1998, 280, 1046–1048. [Google Scholar] [CrossRef]
- Yang, B.; Wang, P.; Li, Z.; You, Q.; Sekine, S.; Ma, J.; Zhuang, S.; Zhang, D.; Yamaguchi, Y. Simultaneous amplification of DNA in a multiplex circular array shaped continuous flow PCR microfluidic chip for on-site detection of bacterial. Lab. Chip 2023, 23, 2633–2639. [Google Scholar] [CrossRef]
- Huergo, M.A.C.; Thanh, N.T.K. Current advances in the detection of COVID-19 and evaluation of the humoral response. Analyst 2021, 146, 382–402. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, R.S.; de Oliveira Silva, J.; Gomes, K.B.; Azevedo, R.B.; Townsend, D.M.; de Paula Sabino, A.; Branco de Barros, A.L. Recent advances in point of care testing for COVID-19 detection. Biomed. Pharmacother. 2022, 153, 113538. [Google Scholar] [CrossRef]
- Zhou, H.; Xu, Z.; He, L.; Wang, Z.; Zhang, T.; Hu, T.; Huang, F.; Chen, D.; Li, Y.; Yang, Y.; et al. Coupling CRISPR/Cas12a and Recombinase Polymerase Amplification on a Stand-Alone Microfluidics Platform for Fast and Parallel Nucleic Acid Detection. Anal. Chem. 2023, 95, 3379–3389. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Fan, C.; Wang, L.; Xu, T.; Zhang, X. Enhanced Isothermal Amplification for Ultrafast Sensing of SARS-CoV-2 in Microdroplets. Anal. Chem. 2022, 94, 4135–4140. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Dong, Q.-Y.; Ding, Q.; Yang, X.; Yuan, R.; Yuan, Y.-L. Photoactive conjugated microporous polymer/carbon nanotube coupled with T-junction recycling dual-strand displacement amplification for sensing N-Gene of COVID-19. Sens. Actuators B Chem. 2023, 376, 132974. [Google Scholar] [CrossRef]
- Rubel, M.S.; Shkodenko, L.A.; Gorbenko, D.A.; Solyanikova, V.V.; Maltzeva, Y.I.; Rubel, A.A.; Koshel, E.I.; Kolpashchikov, D.M. Detection of Multiplex NASBA RNA Products Using Colorimetric Split G Quadruplex Probes. In RNA Nanostructures: Design, Characterization, and Applications; Afonin, K.A., Ed.; Springer: New York, NY, USA, 2023; pp. 287–298. [Google Scholar]
- Carter, J.G.; Orueta Iturbe, L.; Duprey, J.H.A.; Carter, I.R.; Southern, C.D.; Rana, M.; Whalley, C.M.; Bosworth, A.; Beggs, A.D.; Hicks, M.R.; et al. Ultrarapid detection of SARS-CoV-2 RNA using a reverse transcription-free exponential amplification reaction, RTF-EXPAR. Proc. Natl. Acad. Sci. USA 2021, 118, e2100347118. [Google Scholar] [CrossRef]
- Klein, S.; Muller, T.G.; Khalid, D.; Sonntag-Buck, V.; Heuser, A.M.; Glass, B.; Meurer, M.; Morales, I.; Schillak, A.; Freistaedter, A.; et al. SARS-CoV-2 RNA Extraction Using Magnetic Beads for Rapid Large-Scale Testing by RT-qPCR and RT-LAMP. Viruses 2020, 12, 863. [Google Scholar] [CrossRef]
- Dao Thi, V.L.; Herbst, K.; Boerner, K.; Meurer, M.; Kremer, L.P.; Kirrmaier, D.; Freistaedter, A.; Papagiannidis, D.; Galmozzi, C.; Stanifer, M.L.; et al. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020, 12, eabc7075. [Google Scholar] [CrossRef]
- De Felice, M.; De Falco, M.; Zappi, D.; Antonacci, A.; Scognamiglio, V. Isothermal amplification-assisted diagnostics for COVID-19. Biosens. Bioelectron. 2022, 205, 114101. [Google Scholar] [CrossRef]
- Kim, J.H.; Kang, M.; Park, E.; Chung, D.R.; Kim, J.; Hwang, E.S. A Simple and Multiplex Loop-Mediated Isothermal Amplification (LAMP) Assay for Rapid Detection of SARS-CoV. BioChip J. 2019, 13, 341–351. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.Q. Rolling replication of short DNA circle. Proc. Natl. Acad. Sci. USA 1995, 92, 4641–4645. [Google Scholar] [CrossRef] [PubMed]
- Dahl, F.; Banér, J.; Gullberg, M.; Mendel-Hartvig, M.; Landegren, U.; Nilsson, M. Circle-to-circle amplification for precise and sensitive DNA analysis. Proc. Natl. Acad. Sci. USA 2004, 101, 4548–4553. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Lou, Z. Ultrasensitive Detection of Nasopharyngeal Carcinoma-Related MiRNA through Garland Rolling Circle Amplification Integrated Catalytic Hairpin Assembly. ACS Omega 2021, 6, 6460–6465. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Xu, Y.; Mao, X.; Wei, Y.; Song, H.; Chen, N.; Huang, Q.; Fan, C.; Li, D. DNAzyme-Based Rolling-Circle Amplification DNA Machine for Ultrasensitive Analysis of MicroRNA in Drosophila Larva. Anal. Chem. 2012, 84, 7664–7669. [Google Scholar] [CrossRef] [PubMed]
- Khoothiam, K.; Treerattrakoon, K.; Iempridee, T.; Luksirikul, P.; Dharakul, T.; Japrung, D. Ultrasensitive detection of lung cancer-associated miRNAs by multiple primer-mediated rolling circle amplification coupled with a graphene oxide fluorescence-based (MPRCA-GO) sensor. Analyst 2019, 144, 4180–4187. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Qian, J. Performance analysis of and test research on decommissioned milling machine spindle after repair. Ferroelectrics 2021, 581, 32–39. [Google Scholar] [CrossRef]
- Qing, M.; Chen, S.L.; Sun, Z.; Fan, Y.; Luo, H.Q.; Li, N.B. Universal and Programmable Rolling Circle Amplification-CRISPR/Cas12a-Mediated Immobilization-Free Electrochemical Biosensor. Anal. Chem. 2021, 93, 7499–7507. [Google Scholar] [CrossRef]
- Piepenburg, O.; Williams, C.H.; Stemple, D.L.; Armes, N.A. DNA detection using recombination proteins. PLoS Biol. 2006, 4, e204. [Google Scholar] [CrossRef]
- Mohammadniaei, M.; Zhang, M.; Ashley, J.; Christensen, U.B.; Friis-Hansen, L.J.; Gregersen, R.; Lisby, J.G.; Benfield, T.L.; Nielsen, F.E.; Henning Rasmussen, J.; et al. A non-enzymatic, isothermal strand displacement and amplification assay for rapid detection of SARS-CoV-2 RNA. Nat. Commun. 2021, 12, 5089. [Google Scholar] [CrossRef]
- Yurke, B.; Turberfield, A.J.; Mills, A.P., Jr.; Simmel, F.C.; Neumann, J.L. A DNA-fuelled molecular machine made of DNA. Nature 2000, 406, 605–608. [Google Scholar] [CrossRef]
- Oliveira, B.B.; Veigas, B.; Baptista, P.V. Isothermal Amplification of Nucleic Acids: The Race for the Next “Gold Standard”. Front. Sens. 2021, 2, 752600. [Google Scholar] [CrossRef]
- Huang, T.; Zhang, R.; Li, J. CRISPR-Cas-based techniques for pathogen detection: Retrospect, recent advances, and future perspectives. J. Adv. Res. 2023, 50, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef] [PubMed]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef] [PubMed]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef]
- Harrington, L.B.; Burstein, D.; Chen, J.S.; Paez-Espino, D.; Ma, E.; Witte, I.P.; Cofsky, J.C.; Kyrpides, N.C.; Banfield, J.F.; Doudna, J.A. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science 2018, 362, 839–842. [Google Scholar] [CrossRef]
- Li, S.Y.; Cheng, Q.X.; Wang, J.M.; Li, X.Y.; Zhang, Z.L.; Gao, S.; Cao, R.B.; Zhao, G.P.; Wang, J. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 2018, 4, 20. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, R.; Li, J. CRISPR/cas systems redefine nucleic acid detection: Principles and methods. Biosens. Bioelectron. 2020, 165, 112430. [Google Scholar] [CrossRef]
- Wu, H.; Chen, X.; Zhang, M.; Wang, X.; Chen, Y.; Qian, C.; Wu, J.; Xu, J. Versatile detection with CRISPR/Cas system from applications to challenges. TrAC Trends Anal. Chem. 2021, 135, 116150. [Google Scholar] [CrossRef]
- Murugan, K.; Seetharam, A.S.; Severin, A.J.; Sashital, D.G. CRISPR-Cas12a has widespread off-target and dsDNA-nicking effects. J. Biol. Chem. 2020, 295, 5538–5553. [Google Scholar] [CrossRef]
- Gleditzsch, D.; Pausch, P.; Muller-Esparza, H.; Ozcan, A.; Guo, X.; Bange, G.; Randau, L. PAM identification by CRISPR-Cas effector complexes: Diversified mechanisms and structures. RNA Biol. 2019, 16, 504–517. [Google Scholar] [CrossRef]
- Wang, X.; Li, Y.; Zhao, M.; Wang, H.; Wan, Q.; Shi, C.; Ma, C. An ultrafast ratiometric electrochemical biosensor based on potential-assisted hybridization for nucleic acids detection. Anal. Chim. Acta 2022, 1211, 339915. [Google Scholar] [CrossRef]
- Kumar, N.; Shetti, N.P.; Jagannath, S.; Aminabhavi, T.M. Electrochemical sensors for the detection of SARS-CoV-2 virus. Chem. Eng. J. 2022, 430, 132966. [Google Scholar] [CrossRef] [PubMed]
- Yakoh, A.; Pimpitak, U.; Rengpipat, S.; Hirankarn, N.; Chailapakul, O.; Chaiyo, S. Paper-based electrochemical biosensor for diagnosing COVID-19: Detection of SARS-CoV-2 antibodies and antigen. Biosens. Bioelectron. 2021, 176, 112912. [Google Scholar] [CrossRef]
- Yao, B.; Zhang, J.; Fan, Z.; Ding, Y.; Zhou, B.; Yang, R.; Zhao, J.; Zhang, K. Rational Engineering of the DNA Walker Amplification Strategy by Using a Au@Ti3C2@PEI-Ru(dcbpy)32+ Nanocomposite Biosensor for Detection of the SARS-CoV-2 RdRp Gene. ACS Appl. Mater. Interfaces 2021, 13, 19816–19824. [Google Scholar] [CrossRef] [PubMed]
- Ramanujam, A.; Almodovar, S.; Botte, G.G. Ultra-Fast Electrochemical Sensor for Point-of-Care COVID-19 Diagnosis Using Non-Invasive Saliva Sampling. Processes 2021, 9, 1236. [Google Scholar] [CrossRef]
- Xu, M.; Li, Y.; Lin, C.; Peng, Y.; Zhao, S.; Yang, X.; Yang, Y. Recent Advances of Representative Optical Biosensors for Rapid and Sensitive Diagnostics of SARS-CoV-2. Biosensors 2022, 12, 862. [Google Scholar] [CrossRef]
- Lin, C.; Li, Y.; Peng, Y.; Zhao, S.; Xu, M.; Zhang, L.; Huang, Z.; Shi, J.; Yang, Y. Recent development of surface-enhanced Raman scattering for biosensing. J. Nanobiotechnol. 2023, 21, 149. [Google Scholar] [CrossRef]
- Park, S.; Su Jeon, C.; Choi, N.; Moon, J.I.; Min Lee, K.; Hyun Pyun, S.; Kang, T.; Choo, J. Sensitive and reproducible detection of SARS-CoV-2 using SERS-based microdroplet sensor. Chem. Eng. J. 2022, 446, 137085. [Google Scholar] [CrossRef] [PubMed]
- Edman, C.F.; Raymond, D.E.; Wu, D.J.; Tu, E.; Sosnowski, R.G.; Butler, W.F.; Nerenberg, M.; Heller, M.J. Electric Field Directed Nucleic Acid Hybridization on Microchips. Nucleic Acids Res. 1997, 25, 4907–4914. [Google Scholar] [CrossRef] [PubMed]
- Erickson, D.; Liu, X.; Krull, U.; Li, D. Electrokinetically Controlled DNA Hybridization Microfluidic Chip Enabling Rapid Target Analysis. Anal. Chem. 2004, 76, 7269–7277. [Google Scholar] [CrossRef]
- Wei, F.; Lin, C.-C.; Joon, A.; Feng, Z.; Troche, G.; Lira, M.E.; Chia, D.; Mao, M.; Ho, C.-L.; Su, W.-C.; et al. Noninvasive Saliva-based EGFR Gene Mutation Detection in Patients with Lung Cancer. Am. J. Respir. Crit. Care Med. 2014, 190, 1117–1126. [Google Scholar] [CrossRef] [PubMed]
- Uzunoglu, A.; Gunes Altuntas, E.; Huseyin Ipekci, H.; Ozoglu, O. Two-Dimensional (2D) materials in the detection of SARS-CoV-2. Microchem. J. 2023, 193, 108970. [Google Scholar] [CrossRef] [PubMed]
- Ji, D.; Guo, M.; Wu, Y.; Liu, W.; Luo, S.; Wang, X.; Kang, H.; Chen, Y.; Dai, C.; Kong, D.; et al. Electrochemical Detection of a Few Copies of Unamplified SARS-CoV-2 Nucleic Acids by a Self-Actuated Molecular System. J. Am. Chem. Soc. 2022, 144, 13526–13537. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, I.; Iwamoto, K.; Watanabe, Y.; Miyake, Y.; Ono, A. Metal-Ion Selectivity of Chemically Modified Uracil Pairs in DNA Duplexes. Angew. Chem. Int. Ed. 2009, 48, 1648–1651. [Google Scholar] [CrossRef] [PubMed]
- Alafeef, M.; Dighe, K.; Moitra, P.; Pan, D. Rapid, Ultrasensitive, and Quantitative Detection of SARS-CoV-2 Using Antisense Oligonucleotides Directed Electrochemical Biosensor Chip. ACS Nano 2020, 14, 17028–17045. [Google Scholar] [CrossRef]
- Yoon, J.; Conley, B.M.; Shin, M.; Choi, J.H.; Bektas, C.K.; Choi, J.W.; Lee, K.B. Ultrasensitive Electrochemical Detection of Mutated Viral RNAs with Single-Nucleotide Resolution Using a Nanoporous Electrode Array (NPEA). ACS Nano 2022, 16, 5764–5777. [Google Scholar] [CrossRef]
- Fan, Z.; Yao, B.; Ding, Y.; Zhao, J.; Xie, M.; Zhang, K. Entropy-driven amplified electrochemiluminescence biosensor for RdRp gene of SARS-CoV-2 detection with self-assembled DNA tetrahedron scaffolds. Biosens. Bioelectron. 2021, 178, 113015. [Google Scholar] [CrossRef]
- Moitra, P.; Alafeef, M.; Dighe, K.; Frieman, M.B.; Pan, D. Selective Naked-Eye Detection of SARS-CoV-2 Mediated by N Gene Targeted Antisense Oligonucleotide Capped Plasmonic Nanoparticles. ACS Nano 2020, 14, 7617–7627. [Google Scholar] [CrossRef]
- Teng, X.; Sun, X.; Pan, W.; Song, Z.; Wang, J. Carbon Dots Confined in Silica Nanoparticles for Triplet-to-Singlet Föster Resonance Energy-Transfer-Induced Delayed Fluorescence. ACS Appl. Nano Mater. 2022, 5, 5168–5175. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, J.; Li, Y.; Tan, G.; Sun, M.; Shan, Y.; Zhang, Y.; Wang, X.; Song, K.; Shi, R.; et al. SARS-CoV-2 detection using quantum dot fluorescence immunochromatography combined with isothermal amplification and CRISPR/Cas13a. Biosens. Bioelectron. 2022, 202, 113978. [Google Scholar] [CrossRef] [PubMed]
- Forster, T. Energy migration and fluorescence. J. Biomed. Opt. 2012, 17, 011002. [Google Scholar] [CrossRef] [PubMed]
- Bardajee, G.R.; Zamani, M.; Sharifi, M.; Rezanejad, H.; Motallebi, M. Rapid and Highly Sensitive Detection of Target DNA Related to COVID-19 Virus with a Fluorescent Bio-conjugated Probe via a FRET Mechanism. J. Fluoresc. 2022, 32, 1959–1967. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Deng, Y.; Zhao, D.; Fang, J. Plasmonic Silver Supercrystals with Ultrasmall Nanogaps for Ultrasensitive SERS-Based Molecule Detection. Adv. Opt. Mater. 2015, 3, 404–411. [Google Scholar] [CrossRef]
- Muhammad, M.; Huang, Q. A review of aptamer-based SERS biosensors: Design strategies and applications. Talanta 2021, 227, 122188. [Google Scholar] [CrossRef]
- Shan, B.; Pu, Y.; Chen, Y.; Liao, M.; Li, M. Novel SERS labels: Rational design, functional integration and biomedical applications. Coord. Chem. Rev. 2018, 371, 11–37. [Google Scholar] [CrossRef]
- Liang, X.; Li, N.; Zhang, R.; Yin, P.; Zhang, C.; Yang, N.; Liang, K.; Kong, B. Carbon-based SERS biosensor: From substrate design to sensing and bioapplication. NPG Asia Mater. 2021, 13, 8. [Google Scholar] [CrossRef]
- Zheng, X.S.; Jahn, I.J.; Weber, K.; Cialla-May, D.; Popp, J. Label-free SERS in biological and biomedical applications: Recent progress, current challenges and opportunities. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 197, 56–77. [Google Scholar] [CrossRef]
- Su, X.; Liu, X.; Xie, Y.; Chen, M.; Zhong, H.; Li, M. Quantitative Label-Free SERS Detection of Trace Fentanyl in Biofluids with a Freestanding Hydrophobic Plasmonic Paper Biosensor. Anal. Chem. 2023, 95, 3821–3829. [Google Scholar] [CrossRef]
- Pan, X.; Li, L.; Lin, H.; Tan, J.; Wang, H.; Liao, M.; Chen, C.; Shan, B.; Chen, Y.; Li, M. A graphene oxide-gold nanostar hybrid based-paper biosensor for label-free SERS detection of serum bilirubin for diagnosis of jaundice. Biosens. Bioelectron. 2019, 145, 111713. [Google Scholar] [CrossRef]
- Wang, X.; Choi, N.; Cheng, Z.; Ko, J.; Chen, L.; Choo, J. Simultaneous Detection of Dual Nucleic Acids Using a SERS-Based Lateral Flow Assay Biosensor. Anal. Chem. 2017, 89, 1163–1169. [Google Scholar] [CrossRef]
- Ravindran, N.; Kumar, S.; Yashini, M.; Rajeshwari, S.; Mamathi, C.A.; Nirmal Thirunavookarasu, S.; Sunil, C.K. Recent advances in Surface Plasmon Resonance (SPR) biosensors for food analysis: A review. Crit. Rev. Food Sci. Nutr. 2023, 63, 1055–1077. [Google Scholar] [CrossRef]
- Zhao, Y.; Tong, R.-J.; Xia, F.; Peng, Y. Current status of optical fiber biosensor based on surface plasmon resonance. Biosens. Bioelectron. 2019, 142, 111505. [Google Scholar] [CrossRef]
- Akib, T.B.A.; Mou, S.F.; Rahman, M.M.; Rana, M.M.; Islam, M.R.; Mehedi, I.M.; Mahmud, M.A.P.; Kouzani, A.Z. Design and Numerical Analysis of a Graphene-Coated SPR Biosensor for Rapid Detection of the Novel Coronavirus. Sensors 2021, 21, 3491. [Google Scholar] [CrossRef]
- Kettler, H.; White, K.; Hawkes, S.J. Mapping the Landscape of Diagnostics for Sexually Transmitted Infections: Key Findings and Recommendations; World Health Organization: Geneva, Switzerland, 2004. [Google Scholar]
- Wittwer, C.T.; Farrar, S.J. Methods for Fast Nucleic Acid Amplification. U.S. Patent No. 9,932,634, 3 April 2018. [Google Scholar]


| Machine Name | Fundamental Principle | Detection Time | LOD | Company | Reference |
|---|---|---|---|---|---|
| Mic qPCR Cycler | Magnetic induction of a more lightweight thermal block | 25 min | - | Bio Molecular Systems (Queensland, Australia) | [13] |
| αAmp® Cycler | Infrared light directly heating, and airflow cooling | 20 min | - | AlphaHelix Technologies AB (Stockholms Lan, Sweden) | [12] |
| NextGene-PCR | Pushing customized PCR plates with thin walls to different pre-heated temperature blocks | 27 min | 1000 copies/mL | Molecular Biology System B. V. (Goose, Zeeland) | [9] |
| GeneXpert® System | Covering the reaction liquid using thin foils to facilitate efficient heat transfer | 45 min | 100 copies/mL | Cepheid Inc (Sunnyvale, CA, USA) | [14] |
| Cobas® Liat System | Pressing the reaction sample forth and back with two differently pre-heated stamps | 20 min | 100–200 copies/mL | Roche Diagnostic International AG (Basel, Switzerland) | [15,16] |
| Light Cycler | Using thin capillary tubes | 20 min | 2000 copies/mL | [17] | |
| ID Now COVID-19 | LAMP | 5 min (positive) 13 min (negative) | 20,000 cpoies/mL | Abbott (Chicago, IL, USA) | [18] |
| BG-Nova-X8 | Compatible with RAP CRISPR and magnetic bead extraction | 30 min | - | Biogerm (Shanghai, China) | [12] |
| Alere i and q | Nicking enzyme amplification reaction | 15 min | - | Alere (Newport Beach, CA, USA) | [19] |
| Lucira COVID-19 | LAMP | 11 min (positive) 30 min (negative) | - | Lucira Health (Emeryville, CA, USA) | [11] |
| Visby Medical COVID-19 | Continuous flow system for PCR | 30 min | 500 copies/mL | Visby Medical (San Jose, CA, USA) | [20] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lei, Y.; Xu, D. Rapid Nucleic Acid Diagnostic Technology for Pandemic Diseases. Molecules 2024, 29, 1527. https://doi.org/10.3390/molecules29071527
Lei Y, Xu D. Rapid Nucleic Acid Diagnostic Technology for Pandemic Diseases. Molecules. 2024; 29(7):1527. https://doi.org/10.3390/molecules29071527
Chicago/Turabian StyleLei, Yu, and Dawei Xu. 2024. "Rapid Nucleic Acid Diagnostic Technology for Pandemic Diseases" Molecules 29, no. 7: 1527. https://doi.org/10.3390/molecules29071527
APA StyleLei, Y., & Xu, D. (2024). Rapid Nucleic Acid Diagnostic Technology for Pandemic Diseases. Molecules, 29(7), 1527. https://doi.org/10.3390/molecules29071527





