Comparative Analysis of Metabolome and Transcriptome in Different Tissue Sites of Aquilaria sinensis (Lour.) Gilg
Abstract
1. Introduction
2. Results
2.1. Phenotypic Analysis of Aquilaria sinensis
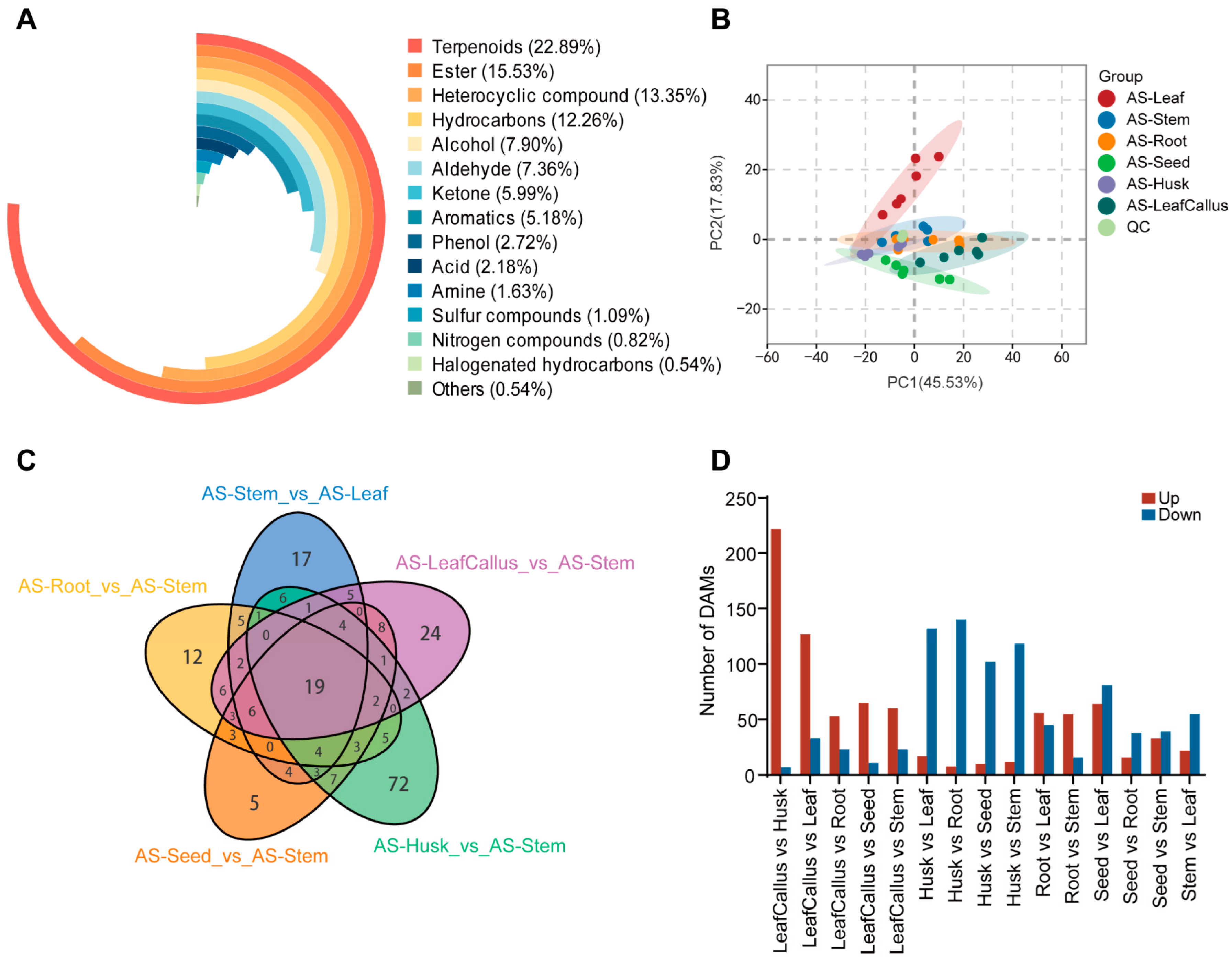
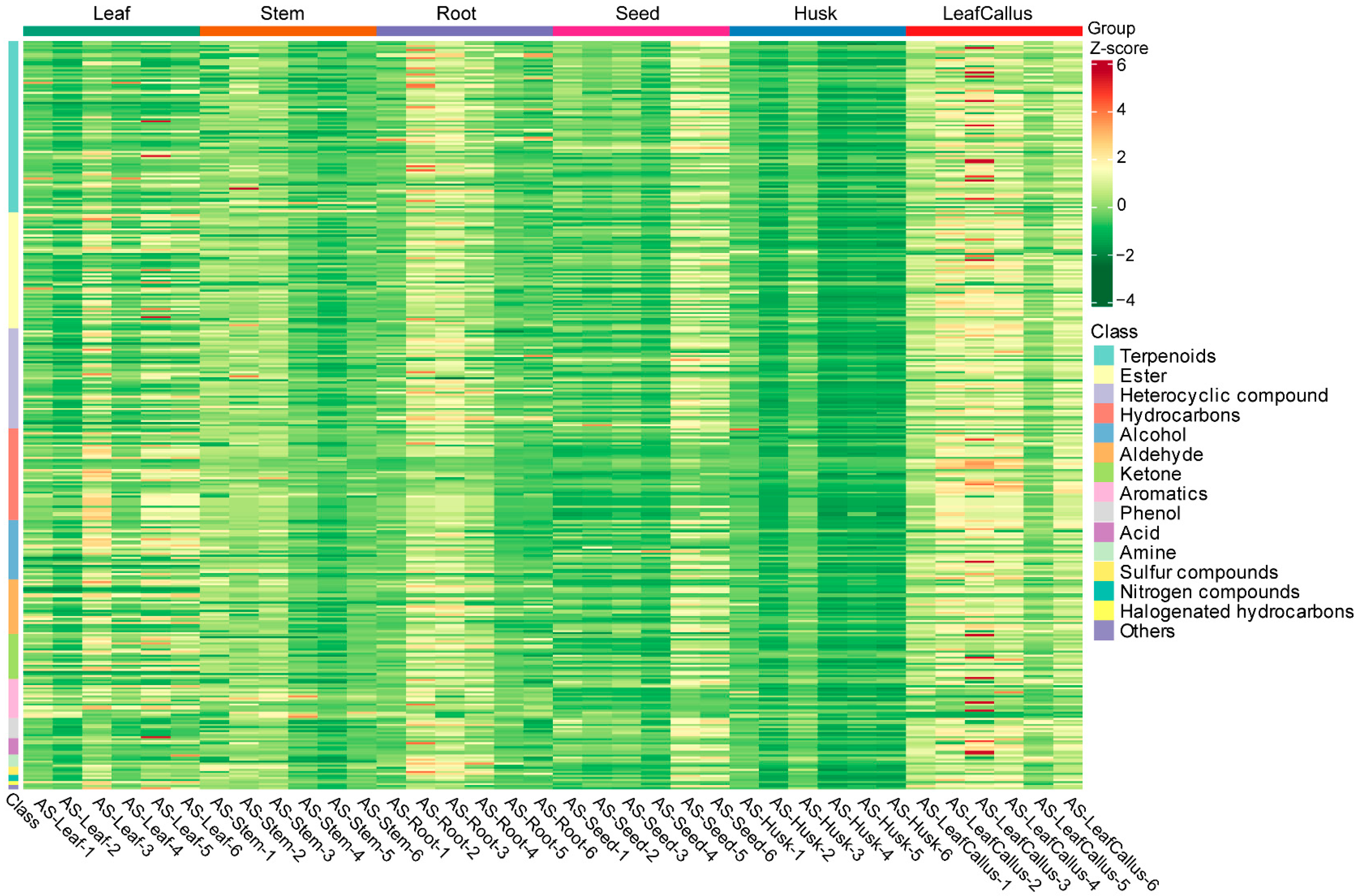
2.2. Metabolomi Analysis of Distinct Tissue Sites in Aquilaria sinensis
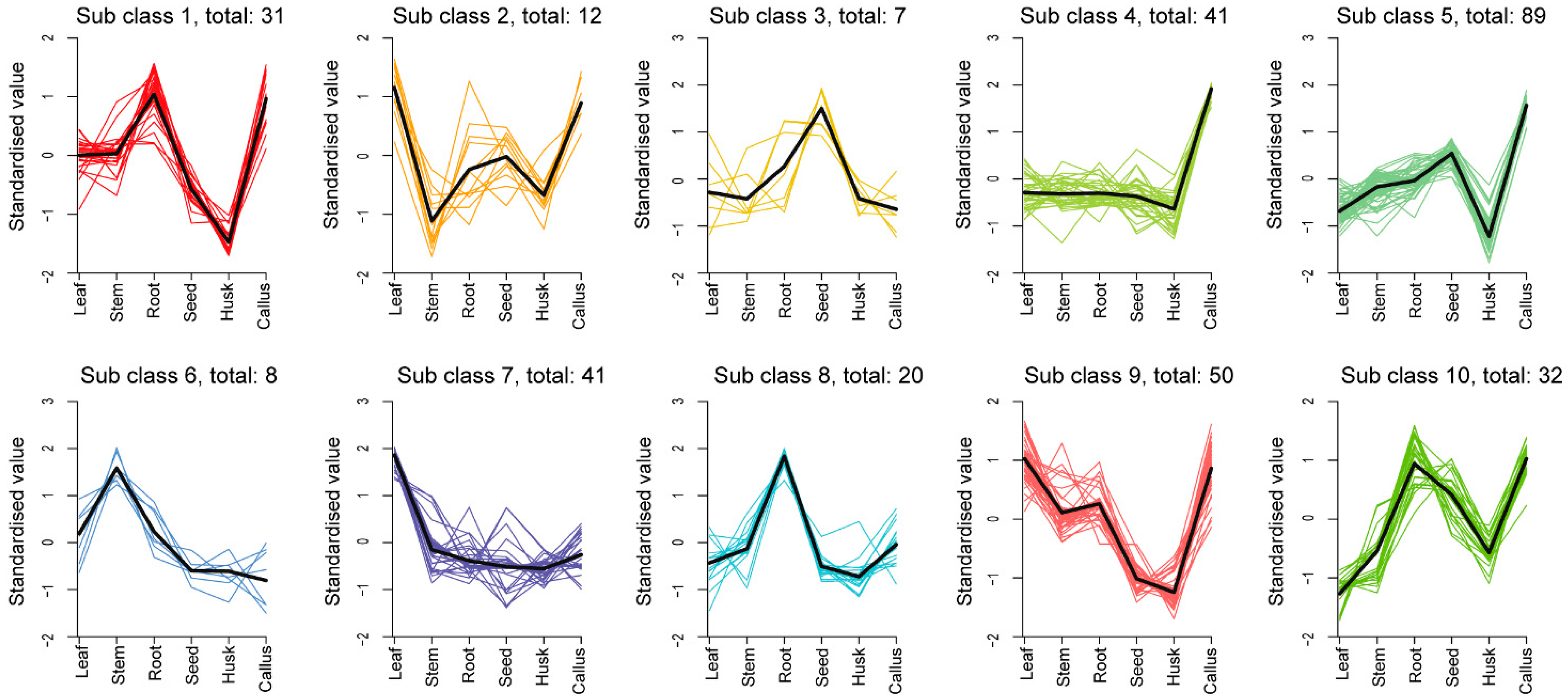
2.3. Identification and Classification of Differentially Accumulation Metabolites (DAMs)
2.4. Enrichment Analysis of the KEGG Metabolic Pathway for DAMs
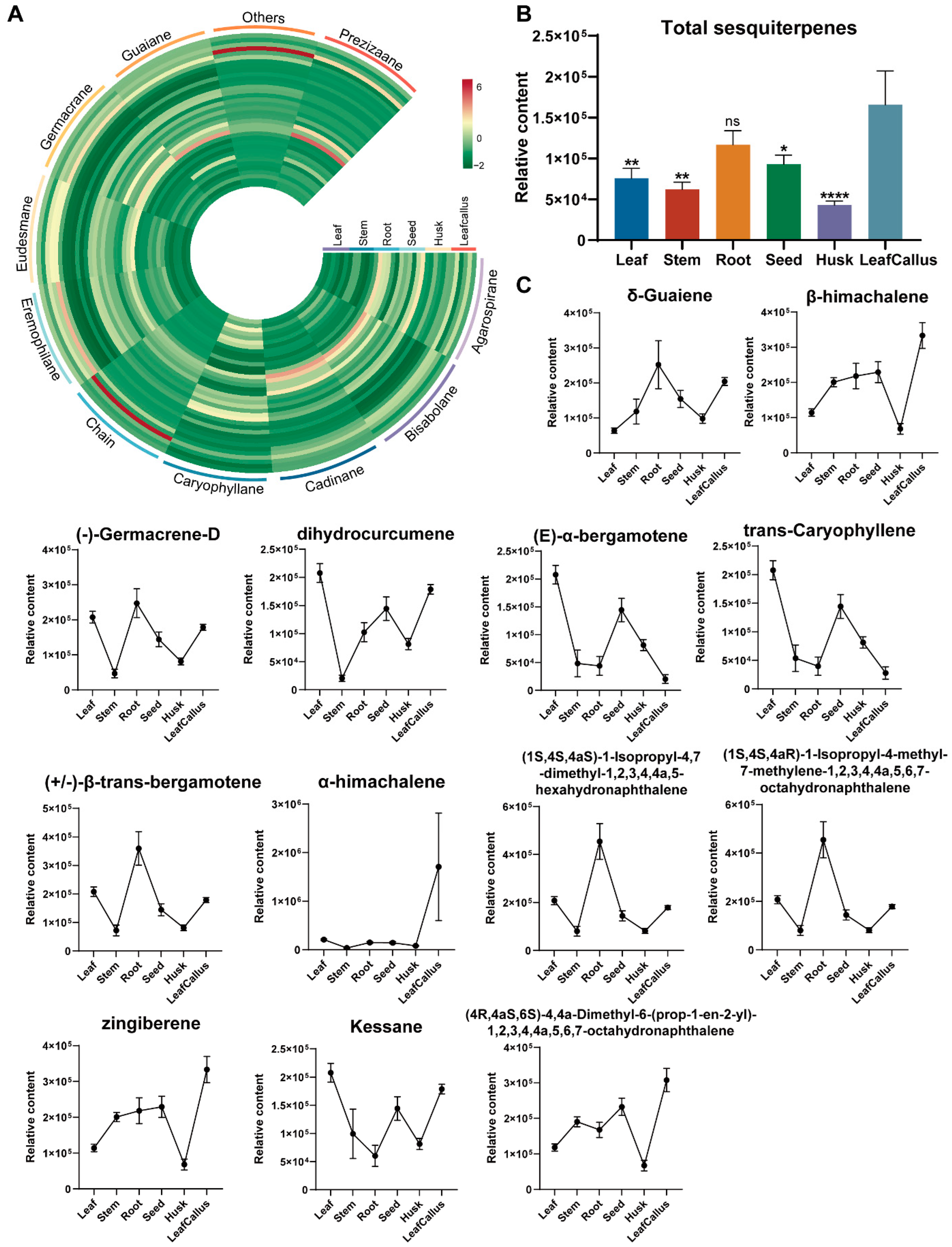
2.5. Analysis of Volatile Organic Compounds Related to Sesquiterpenes in A. sinensis
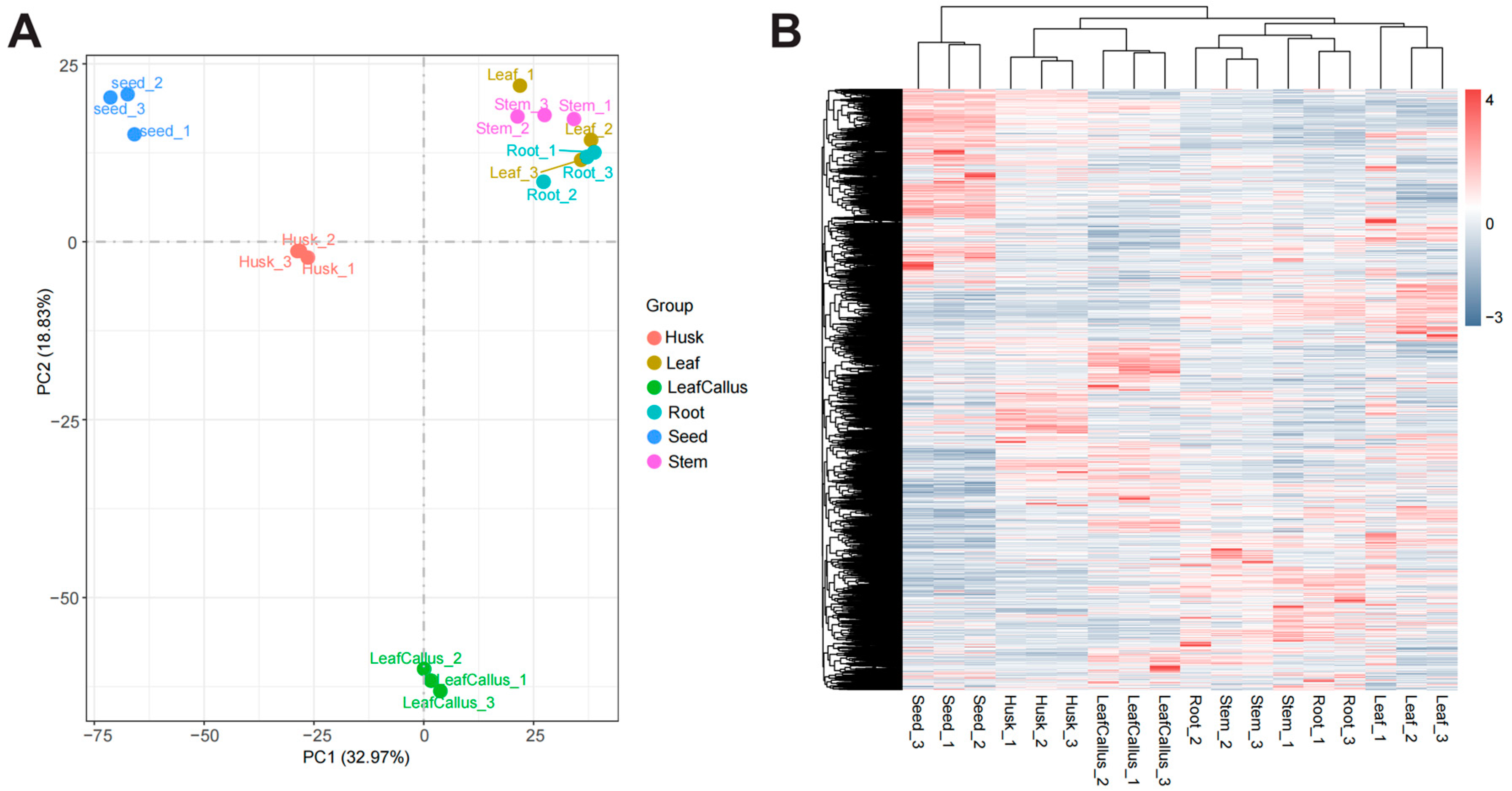
2.6. Transcriptome Analysis of Distinct Tissue Sites in Aquilaria sinensis
2.7. Identification and Classification of Differentially Expressed Genes (DEGs)
2.8. Functional Classification and Enrichment Analysis of DEGs
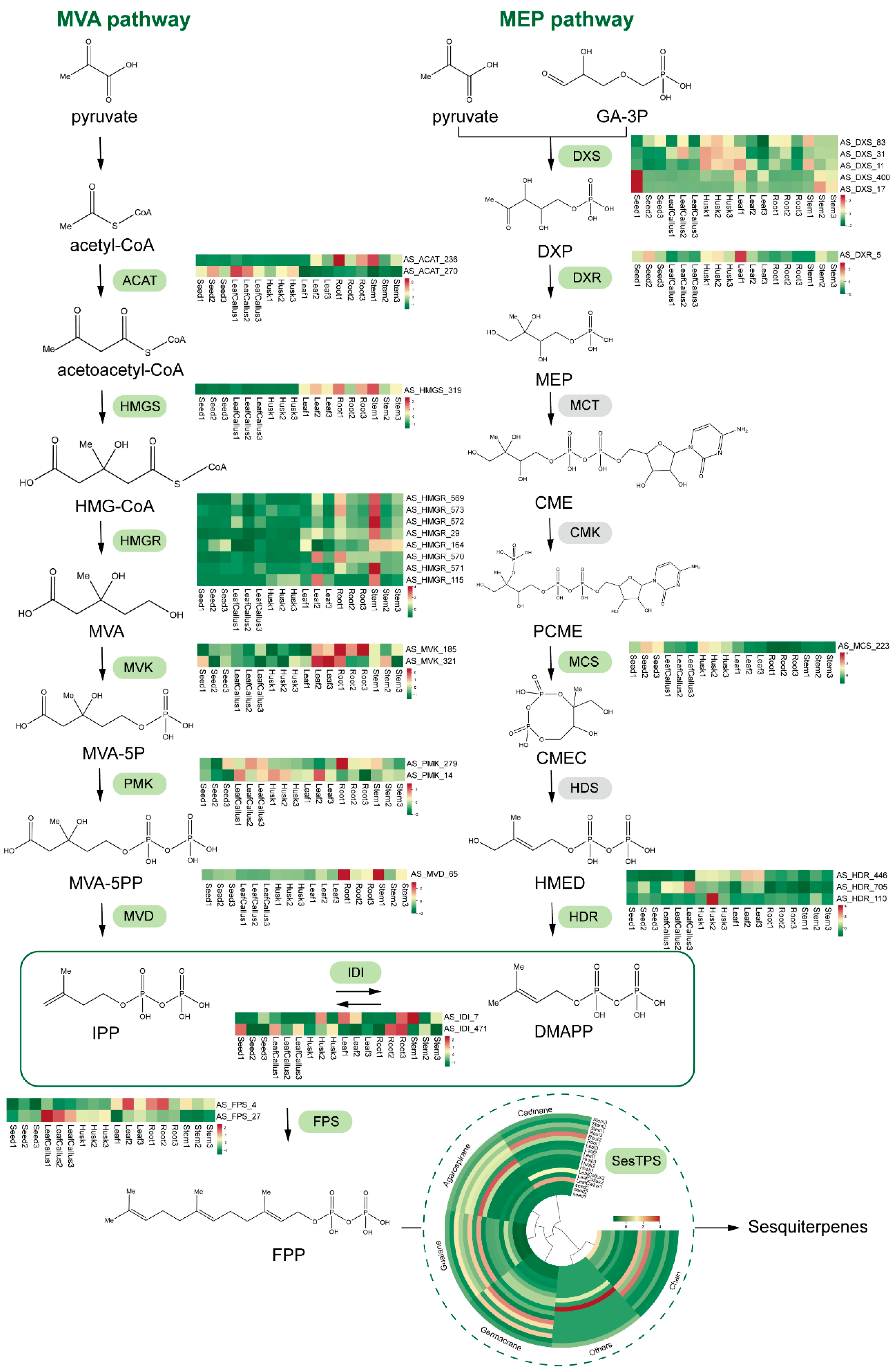
2.9. Analysis of Sesquiterpene Biosynthetic Genes in A. sinensis
2.10. Correlation Analysis of Transcriptome and Metabolome Expression Data
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Volatile Metabolomic Analysis of Sample Preparation and Extraction
4.3. Detection of Volatile Organic Compounds (VOCs)
4.4. RNA Extraction and Transcriptome Sequencing
4.5. Transcriptome and Metabolome Conjoint Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kalra, R.; Kaushik, N. A review of chemistry, quality and analysis of infected agarwood tree (Aquilaria sp.). Phytochem. Rev. 2017, 16, 1045–1079. [Google Scholar] [CrossRef]
- Gao, Z.H.; Wei, J.H.; Yang, Y.; Zhang, Z.; Zhao, W.T. Selection and validation of reference genes for studying stress-related agarwood formation of Aquilaria sinensis. Plant Cell Rep. 2012, 31, 1759–1768. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Yun, Y.; Jian-He, W.; Hui, M.; Chun, S.; Huai-Qiong, C. Advances in studies on mechanism of agarwood formation in Aquilaria sinensis and its hypothesis of agarwood formation induced by defense response. Chin. Tradit. Herb. Drugs 2010, 41, 156–159. [Google Scholar]
- Xu, Y.H.; Zhang, Z.; Wang, M.X.; Wei, J.H.; Chen, H.J.; Gao, Z.H.; Sui, C.; Luo, H.M.; Zhang, X.L.; Yang, Y.; et al. Identification of genes related to agarwood formation: Transcriptome analysis of healthy and wounded tissues of Aquilaria sinensis. BMC Genom. 2013, 14, 16. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, C.H.; Yu, Z.X.; Wu, C.M.; Peng, D.Q.; Liu, X.M.; Liu, Y.Y.; Yang, Y.; Guo, P.; Wei, J.H. Agarwood Essential Oil Ameliorates Restrain Stress-Induced Anxiety and Depression by Inhibiting HPA Axis Hyperactivity. Int. J. Mol. Sci. 2018, 19, 3468. [Google Scholar] [CrossRef] [PubMed]
- Azren, P.D.; Lee, S.Y.; Emang, D.; Mohamed, R. History and perspectives of induction technology for agarwood production from cultivated Aquilaria in Asia: A review. J. For. Res. 2019, 30, 1–11. [Google Scholar] [CrossRef]
- Li, W.; Chen, H.Q.; Wang, H.; Mei, W.L.; Dai, H.F. Natural products in agarwood and Aquilaria plants: Chemistry, biological activities and biosynthesis. Nat. Prod. Rep. 2021, 38, 528–565. [Google Scholar] [CrossRef]
- Hu, B.; Ling, S.-J.; Liu, X.; Huang, J.-B.; Cui, H.; Zhao, Z.-X. Two new 2-(2-phenylethyl)chromone derivatives and two sesquiterpenes from agarwood of Aquilaria sinensis with anti-inflammatory activity. Fitoterapia 2024, 173, 105824. [Google Scholar] [CrossRef]
- Yang, H.R.; Wang, P.; Liu, F.Z.; Yuan, J.Z.; Cai, C.H.; Wu, F.; Jiang, B.; Mei, W.L.; Dai, H.F. Dimeric 2-(2-phenethyl)chromones from agarwood of Aquilaria filaria. Fitoterapia 2023, 165, 7. [Google Scholar] [CrossRef]
- Chen, H.Q.; Wei, J.H.; Yang, J.S.; Zhang, Z.; Yang, Y.; Gao, Z.H.; Sui, C.; Gong, B. Chemical Constituents of Agarwood Originating from the Endemic Genus Aquilaria Plants. Chem. Biodivers. 2012, 9, 236–250. [Google Scholar] [CrossRef]
- Naef, R. The volatile and semi-volatile constituents of agarwood, the infected heartwood of Aquilaria species: A review. Flavour Frag. J. 2011, 26, 73–89. [Google Scholar] [CrossRef]
- Liang, J.J.; Lv, T.M.; Xu, Z.Y.; Huang, X.X.; Song, S.J. Aquilaria sinensis (Lour.) Spreng: Phytochemical review and Chemotaxonomic values. Biochem. Syst. Ecol. 2022, 102, 20. [Google Scholar] [CrossRef]
- Chen, D.; Bi, D.; Song, Y.L.; Tu, P.F. Flavanoids from the stems of Aquilaria sinensis. Chin. J. Nat. Med. 2012, 10, 287–291. [Google Scholar] [CrossRef]
- Yang, M.X.; Liang, Y.G.; Chen, H.R.; Huang, Y.F.; Gong, H.G.; Zhang, T.Y.; Ito, Y. Isolation of Flavonoids From Wild Aquilaria sinensis Leaves by an Improved Preparative High-Speed Counter-Current Chromatography Apparatus. J. Chromatogr. Sci. 2018, 56, 18–24. [Google Scholar] [CrossRef]
- Liu, C.M.; Perng, M.H.; Chen, C.Y. The antioxidation and antiproliferation activity of flavonoids from Aquilaria agallocha and Aquilaria sinensis. Biomed. Res. 2018, 29, 2191–2196. [Google Scholar]
- Qi, J.; Lu, J.-J.; Liu, J.-H.; Yu, B.-Y. Flavonoid and a rare benzophenone glycoside from the leaves of Aquilaria sinensis. Chem. Pharm. Bull. 2009, 57, 134–137. [Google Scholar] [CrossRef]
- Yuan, H.; Zhao, J.; Wang, M.; Khan, S.I.; Zhai, C.; Xu, Q.; Huang, J.; Peng, C.; Xiong, G.; Wang, W.; et al. Benzophenone glycosides from the flower buds of Aquilaria sinensis. Fitoterapia 2017, 121, 170–174. [Google Scholar] [CrossRef]
- Wu, Y.; Liu, C.; Li, H.F.; Sun, J.B.; Li, Y.Y.; Gu, W.; Wang, D.Y.; Liu, J.G.; Hu, Y.L. A novel neolignan glycoside from Aquilaria sinensis. Biochem. Syst. Ecol. 2014, 55, 41–45. [Google Scholar] [CrossRef]
- Hara, H.; Ise, Y.; Morimoto, N.; Shimazawa, M.; Ichihashi, K.; Ohyama, M.; Iinuma, M. Laxative effect of agarwood leaves and its mechanism. Biosci. Biotechnol. Biochem. 2008, 72, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.H.; Peng, K.; Tan, L.H.; Dai, H.F. Aquilarin A, a New Benzenoid Derivative from the Fresh Stem of Aquilaria sinensis. Molecules 2010, 15, 4011–4016. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Mei, W.-L.; Wang, H.; Zuo, W.-J.; Yang, D.-L.; Dai, H.-F. Chemical constituents from stems of Aquilaria sinensis. Zhongguo Zhong Yao Za Zhi 2013, 38, 2826–2831. [Google Scholar]
- Chen, C.T.; Yeh, Y.T.; Chao, D.; Chen, C.Y. Chemical constituents from the bark of Aquilaria sinensis. Chem. Nat. Compd. 2013, 48, 1074–1075. [Google Scholar] [CrossRef]
- Wang, S.L.; Hwang, T.L.; Chung, M.I.; Sung, P.J.; Shu, C.W.; Cheng, M.J.; Chen, J.J. New Flavones, a 2-(2-Phenylethyl)-4H-chromen-4-one Derivative, and Anti-Inflammatory Constituents from the Stem Barks of Aquilaria sinensis. Molecules 2015, 20, 20912–20925. [Google Scholar] [CrossRef]
- Wang, S.C.; Wang, F.; Yue, C.H. Chemical constituents from the petioles and leaves of Aquilaria sinensis. Biochem. Syst. Ecol. 2015, 61, 458–461. [Google Scholar] [CrossRef]
- Zhang, Z.X.; Wang, X.H.; Yang, W.Q.; Wang, J.; Su, C.; Liu, X.; Li, J.; Zhao, Y.F.; Shi, S.P.; Tu, P.F. Five 2-(2-Phenylethyl)chromones from Sodium Chloride-Elicited Aquilaria sinensis Cell Suspension Cultures. Molecules 2016, 21, 7. [Google Scholar] [CrossRef]
- Feng, J.; Yang, X. Liposolubility constituents from leaves of Aquilaria sinensis. Zhongguo Zhong Yao Za Zhi 2011, 36, 2092–2095. [Google Scholar]
- Wagh, V.D.; Korinek, M.; Lo, I.W.; Hsu, Y.-M.; Chen, S.-L.; Hsu, H.-Y.; Hwang, T.-L.; Wu, Y.-C.; Chen, B.-H.; Cheng, Y.-B.; et al. Inflammation Modulatory Phorbol Esters from the Seeds of Aquilaria malaccensis. J. Nat. Prod. 2017, 80, 1421–1427. [Google Scholar] [CrossRef]
- Korinek, M.; Wagh, V.D.; Lo, I.W.; Hsu, Y.-M.; Hsu, H.-Y.; Hwang, T.-L.; Wu, Y.-C.; Cheng, Y.-B.; Chen, B.-H.; Chang, F.-R. Antiallergic Phorbol Ester from the Seeds of Aquilaria malaccensis. Int. J. Mol. Sci. 2016, 17, 398. [Google Scholar] [CrossRef]
- Kang, Y.F.; Chien, S.L.; Wu, H.M.; Li, W.J.; Chen, C.T.; Li, H.T.; Chen, H.L.; Chao, D.; Chen, S.J.; Huang, C.T.; et al. Secondary Metabolites from the Leaves of Aquilaria sinensis. Chem. Nat. Compd. 2014, 50, 1110–1112. [Google Scholar] [CrossRef]
- Mei, W.L.; Lin, F.; Zuo, W.J.; Wang, H.; Dai, H.F. Cucurbitacins from fruits of Aquilaria sinensis. Chin. J. Nat. Med. 2012, 10, 234–237. [Google Scholar] [CrossRef]
- Bergman, M.E.; Davis, B.; Phillips, M.A. Medically Useful Plant Terpenoids: Biosynthesis, Occurrence, and Mechanism of Action. Molecules 2019, 24, 3961. [Google Scholar] [CrossRef]
- Lombard, J.; Moreira, D. Origins and early evolution of the mevalonate pathway of isoprenoid biosynthesis in the three domains of life. Mol. Biol. Evol. 2011, 28, 87–99. [Google Scholar] [CrossRef]
- Shang, C.-H.; Zhu, F.; Li, N.; Ou-Yang, X.; Shi, L.; Zhao, M.-W.; Li, Y.-X. Cloning and characterization of a gene encoding HMG-CoA reductase from Ganoderma lucidum and its functional identification in yeast. Biosci. Biotechnol. Biochem. 2008, 72, 1333–1339. [Google Scholar] [CrossRef][Green Version]
- Rohmer, M. The discovery of a mevalonate-independent pathway for isoprenoid biosynthesis in bacteria, algae and higher plants. Nat. Prod. Rep. 1999, 16, 565–574. [Google Scholar] [CrossRef]
- Kitaoka, N.; Lu, X.; Yang, B.; Peters, R.J. The application of synthetic biology to elucidation of plant mono-, sesqui-, and diterpenoid metabolism. Mol. Plant 2015, 8, 6–16. [Google Scholar] [CrossRef]
- Nagegowda, D.A. Plant volatile terpenoid metabolism: Biosynthetic genes, transcriptional regulation and subcellular compartmentation. FEBS Lett. 2010, 584, 2965–2973. [Google Scholar] [CrossRef]
- Vranová, E.; Coman, D.; Gruissem, W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 2013, 64, 665–700. [Google Scholar] [CrossRef]
- Maoz, I.; Lewinsohn, E.; Gonda, I. Amino acids metabolism as a source for aroma volatiles biosynthesis. Curr. Opin. Plant Biol. 2022, 67, 10. [Google Scholar] [CrossRef]
- Hunter, W.N. The non-mevalonate pathway of isoprenoid precursor biosynthesis. J. Biol. Chem. 2007, 282, 21573–21577. [Google Scholar] [CrossRef]
- Dubey, V.S.; Bhalla, R.; Luthra, R. An overview of the non-mevalonate pathway for terpenoid biosynthesis in plants. J. Biosci. 2003, 28, 637–646. [Google Scholar] [CrossRef]
- Cordoba, E.; Salmi, M.; León, P. Unravelling the regulatory mechanisms that modulate the MEP pathway in higher plants. J. Exp. Bot. 2009, 60, 2933–2943. [Google Scholar] [CrossRef]
- Park, J.; Pandya, V.R.; Ezekiel, S.J.; Berghuis, A.M. Phosphonate and Bisphosphonate Inhibitors of Farnesyl Pyrophosphate Synthases: A Structure-Guided Perspective. Front. Chem. 2020, 8, 612728. [Google Scholar] [CrossRef]
- Chen, F.; Tholl, D.; Bohlmann, J.; Pichersky, E. The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011, 66, 212–229. [Google Scholar] [CrossRef]
- Kumeta, Y.; Ito, M. Characterization of delta-guaiene synthases from cultured cells of Aquilaria, responsible for the formation of the sesquiterpenes in agarwood. Plant Physiol. 2010, 154, 1998–2007. [Google Scholar] [CrossRef]
- Siah, C.H.; Namasivayam, P.; Mohamed, R. Transcriptome reveals senescing callus tissue of Aquilaria malaccensis, an endangered tropical tree, triggers similar response as wounding with respect to terpenoid biosynthesis. Tree Genet. Genomes 2016, 12, 10. [Google Scholar] [CrossRef]
- Caser, M.; Chitarra, W.; D’Angiolillo, F.; Perrone, I.; Demasi, S.; Lovisolo, C.; Pistelli, L.; Pistelli, L.; Scariot, V. Drought stress adaptation modulates plant secondary metabolite production in Salvia dolomitica Codd. Ind. Crop. Prod. 2019, 129, 85–96. [Google Scholar] [CrossRef]
- Kumari, S.; Nazir, F.; Maheshwari, C.; Kaur, H.; Gupta, R.; Siddique, K.H.M.; Khan, M.I.R. Plant hormones and secondary metabolites under environmental stresses: Enlightening defense molecules. Plant Physiol. Biochem. 2024, 206, 108238. [Google Scholar] [CrossRef]
- Bhawana, M.; Prashant, S.P.; Deepak, P. Understanding the role of key metabolic genes, transcription factors, and trichome-related genes in-terms of temperature-stress management techniques in the rose-scented Geranium (Pelargonium graveolens) using transcriptomic analysis. Ind. Crop. Prod. 2023, 199, 14. [Google Scholar] [CrossRef]
- Zhang, Y.N.; Diao, S.; Ding, X.Y.; Sun, J.M.; Luan, Q.F.; Jiang, J.M. Transcriptional regulation modulates terpenoid biosynthesis of Pinus elliottii under drought stress. Ind. Crop. Prod. 2023, 202, 12. [Google Scholar] [CrossRef]
- Li, Y.; Fang, J.; Qi, X.; Lin, M.; Zhong, Y.; Sun, L.; Cui, W. Combined Analysis of the Fruit Metabolome and Transcriptome Reveals Candidate Genes Involved in Flavonoid Biosynthesis in Actinidia arguta. Int. J. Mol. Sci. 2018, 19, 1471. [Google Scholar] [CrossRef]
- Rattan, S.; Kumar, P.; Kaur, E.; Sood, A.; Acharya, V.; Warghat, A.R. Comparative transcriptome and tissue-specific expression analysis of genes reveal tissue-cultured plants as an alternative source for phenylethanoids and phenylpropanoids in Rhodiola imbricata (Edgew.). Gene 2022, 836, 146672. [Google Scholar] [CrossRef]
- Luo, J. Metabolite-based genome-wide association studies in plants. Curr. Opin. Plant Biol. 2015, 24, 31–38. [Google Scholar] [CrossRef]
- Luo, X.; Sun, D.; Wang, S.; Luo, S.; Fu, Y.; Niu, L.; Shi, Q.; Zhang, Y. Integrating full-length transcriptomics and metabolomics reveals the regulatory mechanisms underlying yellow pigmentation in tree peony (Paeonia suffruticosa Andr.) flowers. Hortic. Res. 2021, 8, 235. [Google Scholar] [CrossRef]
- Hirasawa, T.; Saito, M.; Yoshikawa, K.; Furusawa, C.; Shmizu, H. Integrated Analysis of the Transcriptome and Metabolome of Corynebacterium glutamicum during Penicillin-Induced Glutamic Acid Production. Biotechnol. J. 2018, 13, e1700612. [Google Scholar] [CrossRef]
- Mesnage, R.; Biserni, M.; Balu, S.; Frainay, C.; Poupin, N.; Jourdan, F.; Wozniak, E.; Xenakis, T.; Mein, C.A.; Antoniou, M.N. Integrated transcriptomics and metabolomics reveal signatures of lipid metabolism dysregulation in HepaRG liver cells exposed to PCB 126. Arch. Toxicol. 2018, 92, 2533–2547. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, R.; Song, Y.; He, J.; Sun, J.; Bai, J.; An, Z.; Dong, L.; Zhan, Q.; Abliz, Z. RRLC-MS/MS-based metabonomics combined with in-depth analysis of metabolic correlation network: Finding potential biomarkers for breast cancer. Analyst 2009, 134, 2003–2011. [Google Scholar] [CrossRef]
- Thévenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the Human Adult Urinary Metabolome Variations with Age, Body Mass Index, and Gender by Implementing a Comprehensive Workflow for Univariate and OPLS Statistical Analyses. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef]
- Okudera, Y.; Ito, M. Production of agarwood fragrant constituents in Aquilaria calli and cell suspension cultures. Plant Biotechnol. 2009, 26, 307–315. [Google Scholar] [CrossRef]
- Fujita, M.; Fujita, Y.; Noutoshi, Y.; Takahashi, F.; Narusaka, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Crosstalk between abiotic and biotic stress responses: A current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 2006, 9, 436–442. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, Y.; Wen, F.; Yao, D.; Wang, L.; Guo, J.; Ni, L.; Zhang, A.; Tan, M.; Jiang, M. A novel rice C2H2-type zinc finger protein, ZFP36, is a key player involved in abscisic acid-induced antioxidant defence and oxidative stress tolerance in rice. J. Exp. Bot. 2014, 65, 5795–5809. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, X.L.; Yang, Y.; Wei, J.H.; Meng, H.; Gao, Z.H.; Xu, Y.H. Hydrogen peroxide induces vessel occlusions and stimulates sesquiterpenes accumulation in stems of Aquilaria sinensis. Plant Growth Regul. 2014, 72, 81–87. [Google Scholar] [CrossRef]
- Xu, Y.H.; Liao, Y.C.; Zhang, Z.; Liu, J.; Sun, P.W.; Gao, Z.H.; Sui, C.; Wei, J.H. Jasmonic acid is a crucial signal transducer in heat shock induced sesquiterpene formation in Aquilaria sinensis. Sci. Rep. 2016, 6, 9. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xu, Y.H.; Zhang, Z.; Wei, J.H. Hydrogen peroxide promotes programmed cell death and salicylic acid accumulation during the induced production of sesquiterpenes in cultured cell suspensions of Aquilaria sinensis. Funct. Plant Biol. 2015, 42, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.Q.; Movahed, N.; Zheng, H.Q. LUNAPARK Is an E3 Ligase That Mediates Degradation of ROOT HAIR DEFECTIVE3 to Maintain a Tubular ER Network in Arabidopsis. Plant Cell 2020, 32, 2964–2978. [Google Scholar] [CrossRef] [PubMed]
- Kud, J.; Wang, W.J.; Yuan, Y.L.; Caplan, A.; Kuhl, J.C.; Dandurand, L.M.; Xiao, F.M. Functional Characterization of RING-Type E3 Ubiquitin Ligases In Vitro and In Planta. J. Vis. Exp. 2019, 7, e60533. [Google Scholar] [CrossRef]
- Bartholomew, D.M.; Van Dyk, D.E.; Lau, S.M.C.; O’Keefe, D.P.; Rea, P.A.; Viitanen, P.V. Alternate energy-dependent pathways for the vacuolar uptake of glucose and glutathione conjugates. Plant Physiol. 2002, 130, 1562–1572. [Google Scholar] [CrossRef] [PubMed]
- Jasinski, M.; Stukkens, Y.; Degand, H.; Purnelle, B.; Marchand-Brynaert, J.; Boutry, M. A plant plasma membrane ATP binding cassette-type transporter is involved in antifungal terpenoid secretion. Plant Cell 2001, 13, 1095–1107. [Google Scholar] [CrossRef][Green Version]
- Durán-Soria, S.; Pott, D.M.; Osorio, S.; Vallarino, J.G. Sugar Signaling during Fruit Ripening. Front. Plant Sci. 2020, 11, 564917. [Google Scholar] [CrossRef]
- Yang, Y.-Q.; Yin, H.-X.; Yuan, H.-B.; Jiang, Y.-W.; Dong, C.-W.; Deng, Y.-L. Characterization of the volatile components in green tea by IRAE-HS-SPME/GC-MS combined with multivariate analysis. PLoS ONE 2018, 13, e0193393. [Google Scholar] [CrossRef]
- Zhang, W.; Cao, J.; Li, Z.; Li, Q.; Lai, X.; Sun, L.; Chen, R.; Wen, S.; Sun, S.; Lai, Z. HS-SPME and GC/MS volatile component analysis of Yinghong No. 9 dark tea during the pile fermentation process. Food Chem. 2021, 357, 129654. [Google Scholar] [CrossRef]
- Yuan, H.; Cao, G.; Hou, X.; Huang, M.; Du, P.; Tan, T.; Zhang, Y.; Zhou, H.; Liu, X.; Liu, L.; et al. Development of a widely targeted volatilomics method for profiling volatilomes in plants. Mol. Plant 2022, 15, 189–202. [Google Scholar] [CrossRef] [PubMed]
- GB 23200.8-2016; Determination of 500 Pesticides and Related Chemicals Residues in Fruits and Vegetables Gas Chromatography-Mass Spectrometry. Standards Press of China: Beijing, China, 2016.
- Chong, J.; Xia, J. MetaboAnalystR: An R package for flexible and reproducible analysis of metabolomics data. Bioinformatics 2018, 34, 4313–4314. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, A.; Liu, J.; Huang, L. Comparative Analysis of Metabolome and Transcriptome in Different Tissue Sites of Aquilaria sinensis (Lour.) Gilg. Molecules 2024, 29, 1075. https://doi.org/10.3390/molecules29051075
Wang A, Liu J, Huang L. Comparative Analysis of Metabolome and Transcriptome in Different Tissue Sites of Aquilaria sinensis (Lour.) Gilg. Molecules. 2024; 29(5):1075. https://doi.org/10.3390/molecules29051075
Chicago/Turabian StyleWang, Anjun, Juan Liu, and Luqi Huang. 2024. "Comparative Analysis of Metabolome and Transcriptome in Different Tissue Sites of Aquilaria sinensis (Lour.) Gilg" Molecules 29, no. 5: 1075. https://doi.org/10.3390/molecules29051075
APA StyleWang, A., Liu, J., & Huang, L. (2024). Comparative Analysis of Metabolome and Transcriptome in Different Tissue Sites of Aquilaria sinensis (Lour.) Gilg. Molecules, 29(5), 1075. https://doi.org/10.3390/molecules29051075






