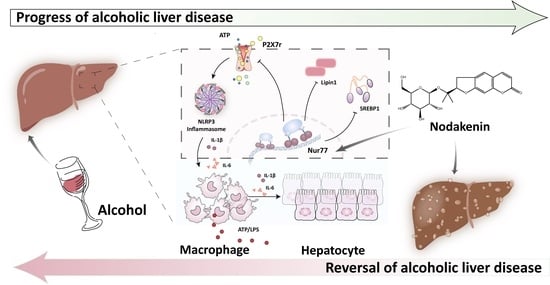
Regulation of the Nur77-P2X7r Signaling Pathway by Nodakenin: A Potential Protective Function against Alcoholic Liver Disease
Abstract
1. Introduction
2. Result
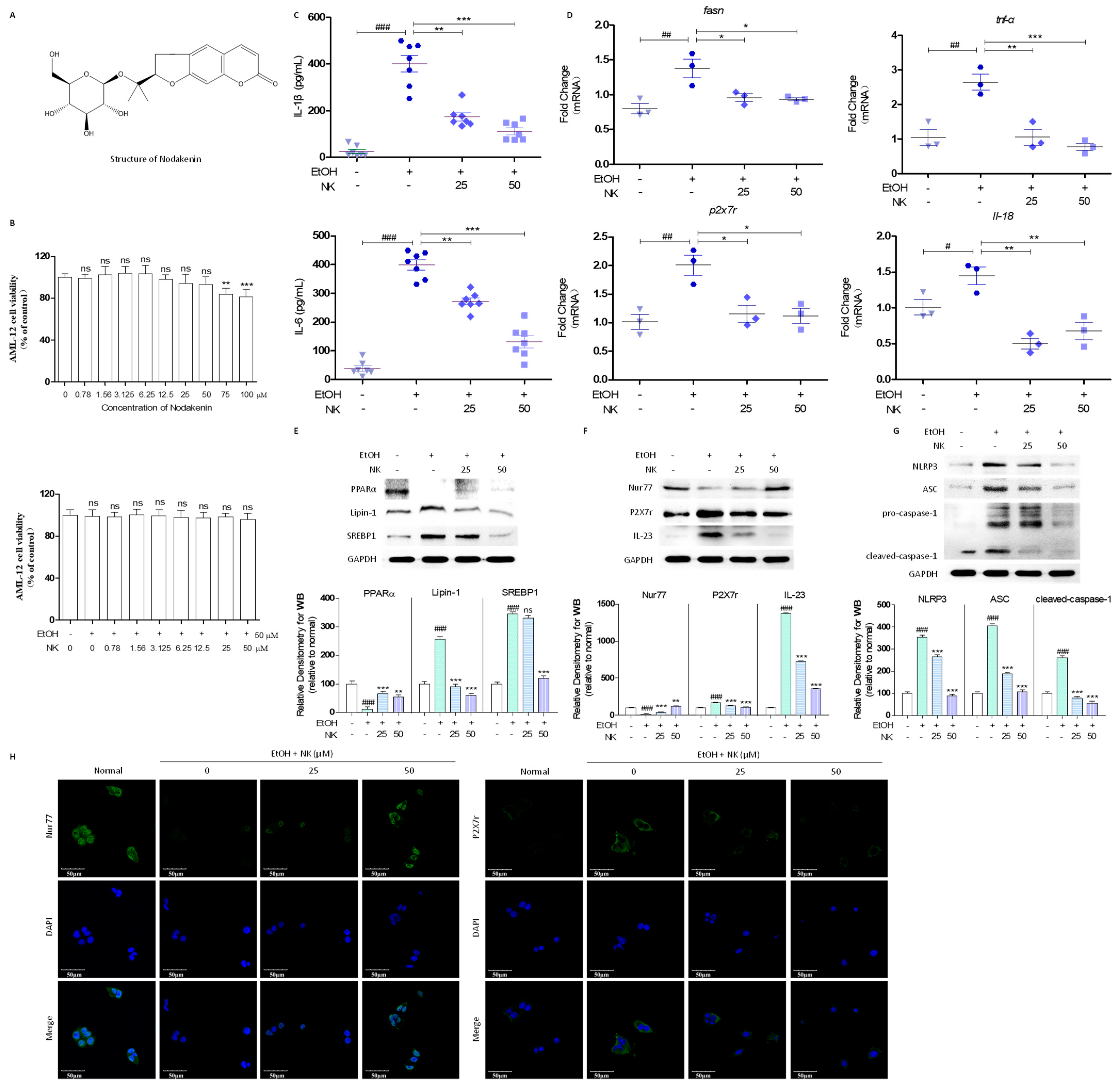
2.1. NK Regulates Lipid Deposition and Inflammatory Response in EtOH-Stimulated AML-12 Cells
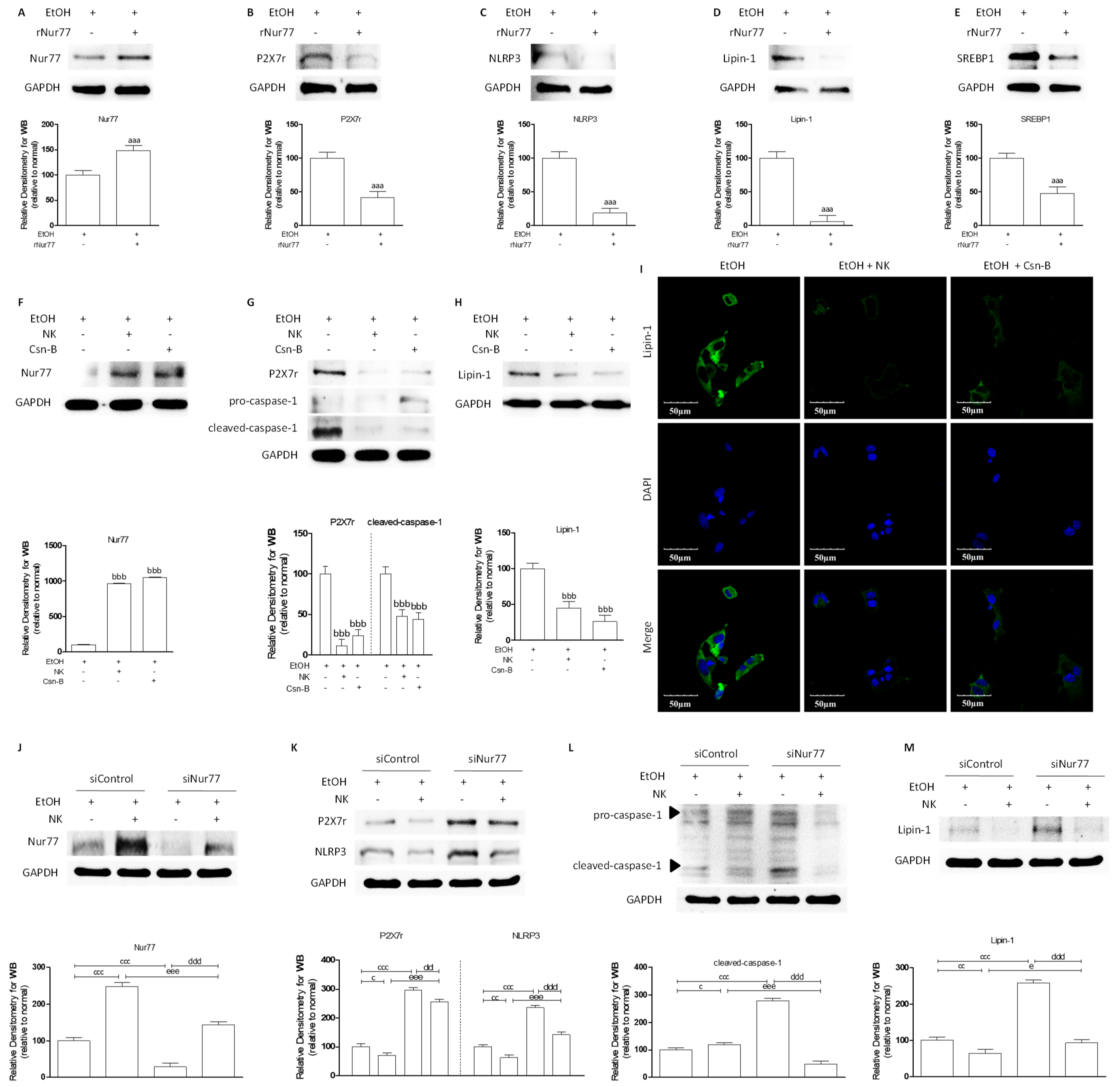
2.2. NK Ameliorated ALD via Enhanced Nur77-Mediated P2X7r Signaling and Lipid Accumulation
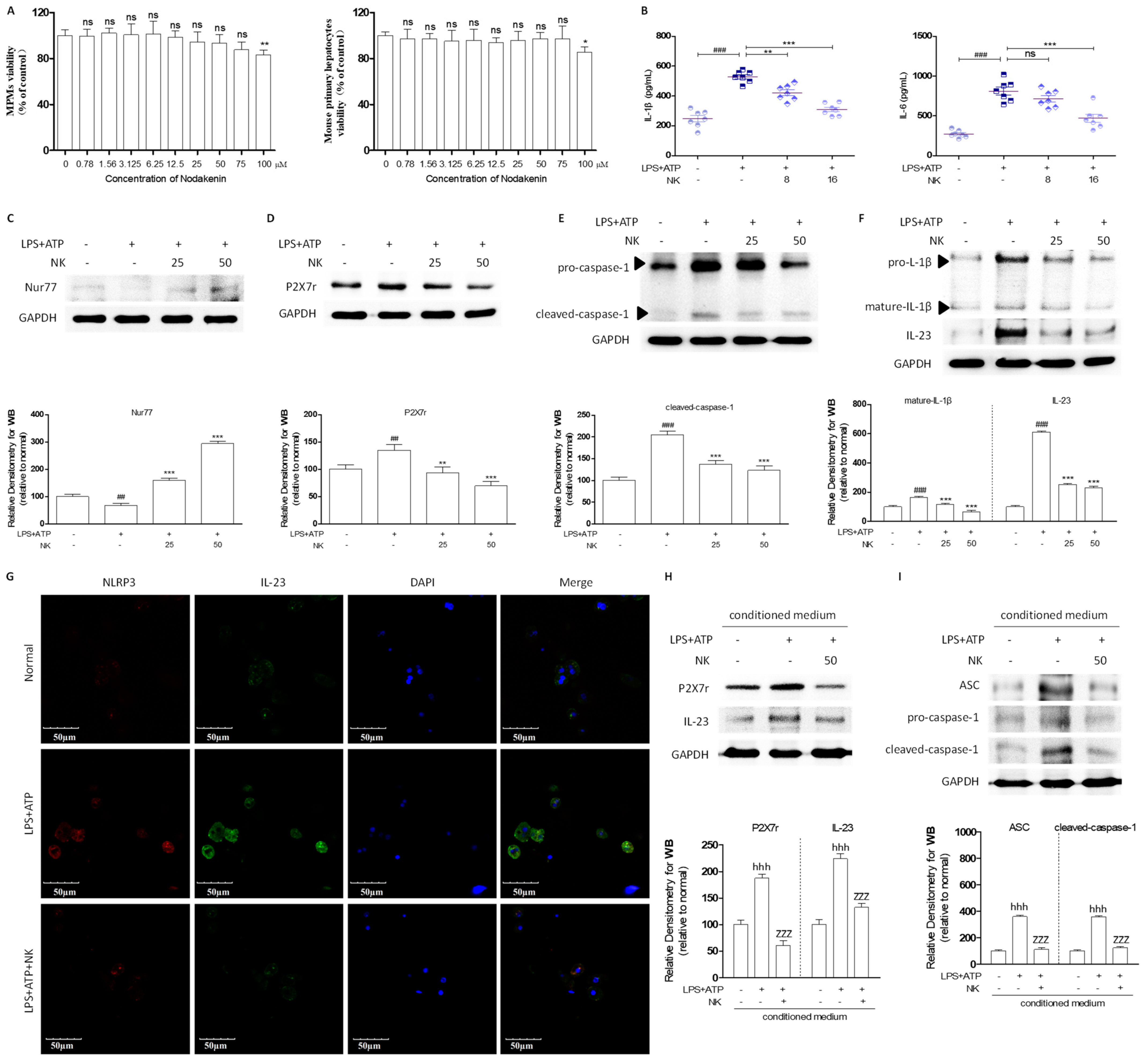
2.3. NK Attenuates the Inflammatory Response to Macrophage-to-Hepatocyte Communication
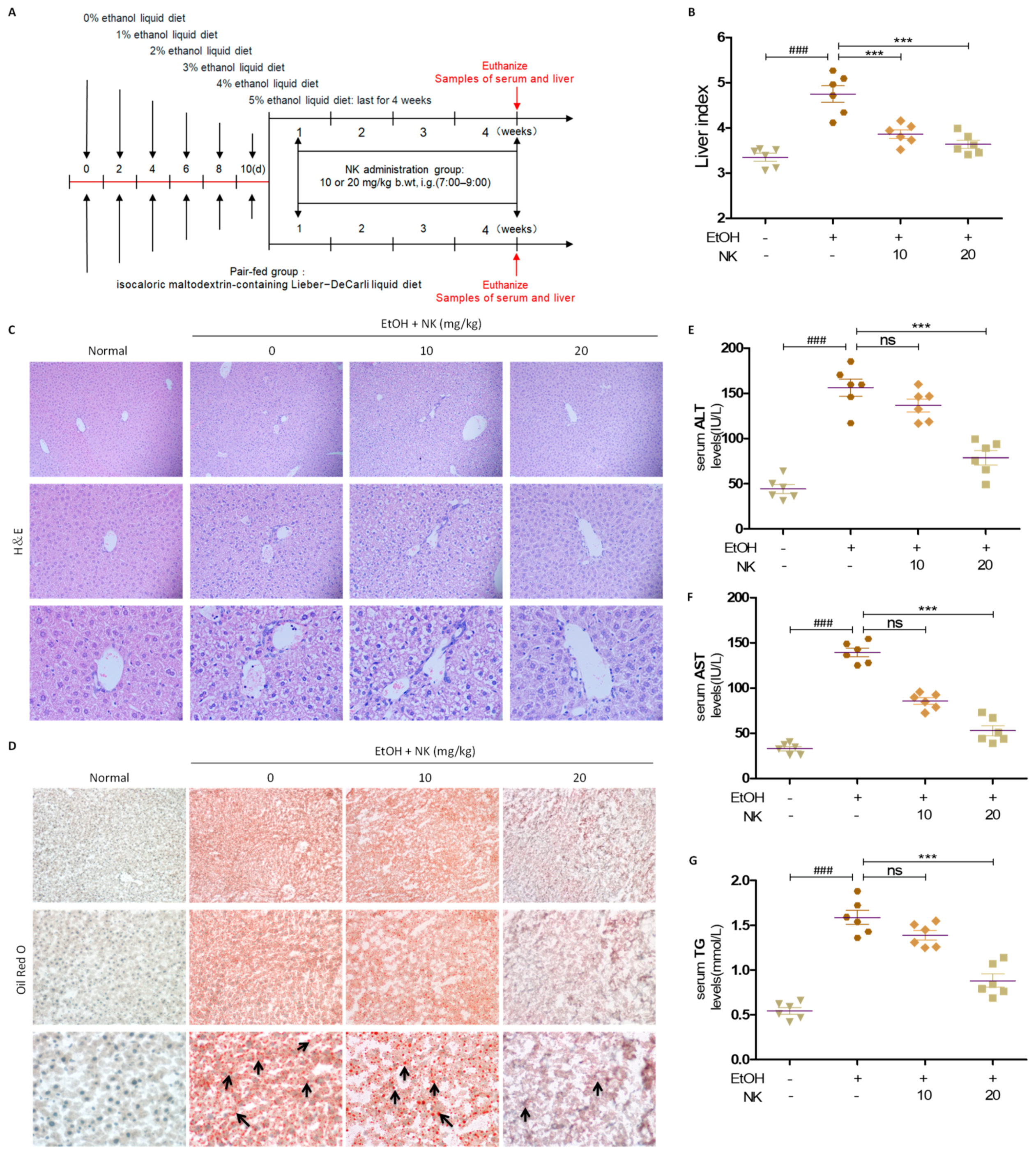
2.4. NK Improves Liver Injury in ALD Mice
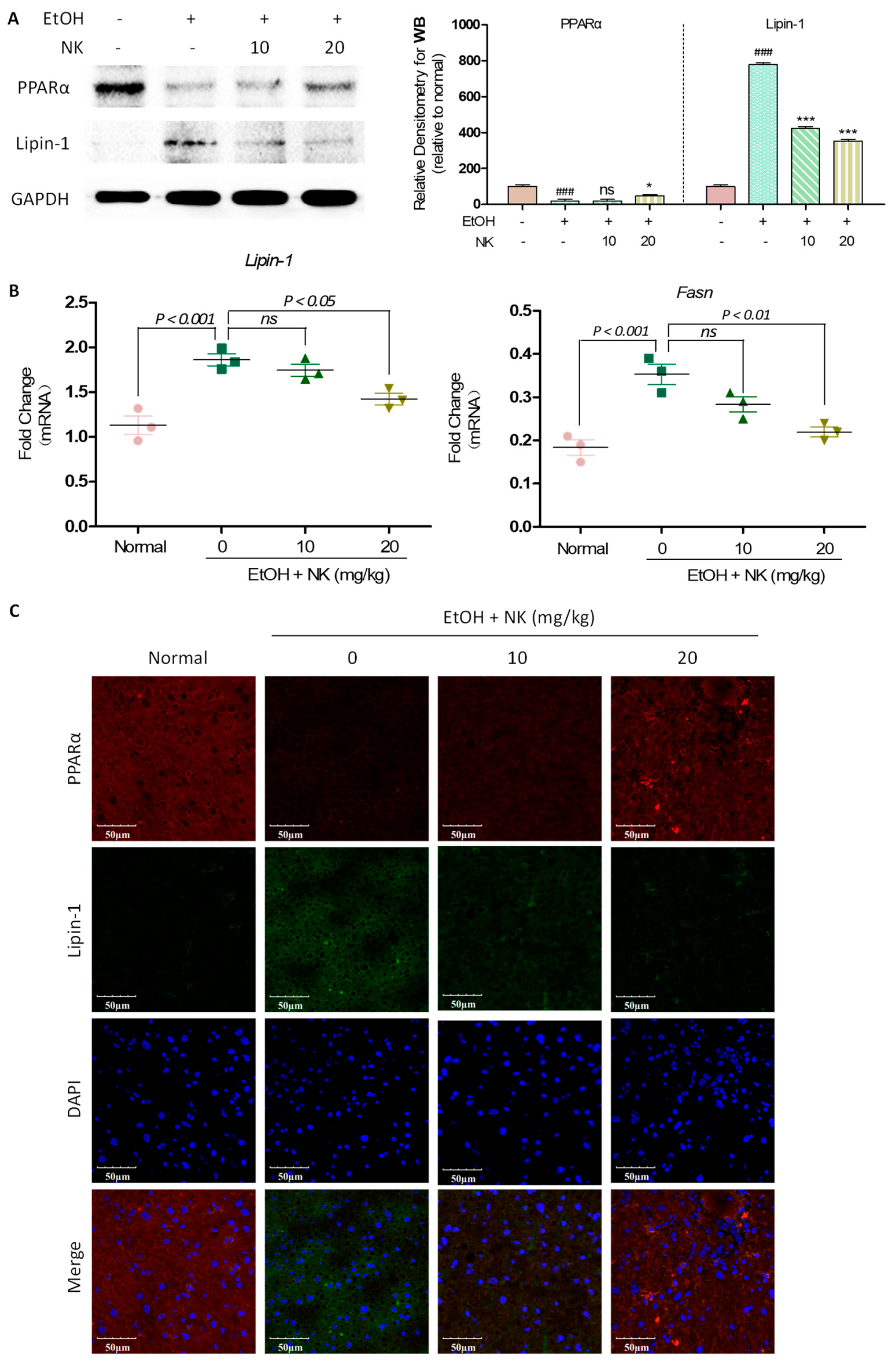
2.5. NK Regulates Lipid Accumulation in ALD Mice
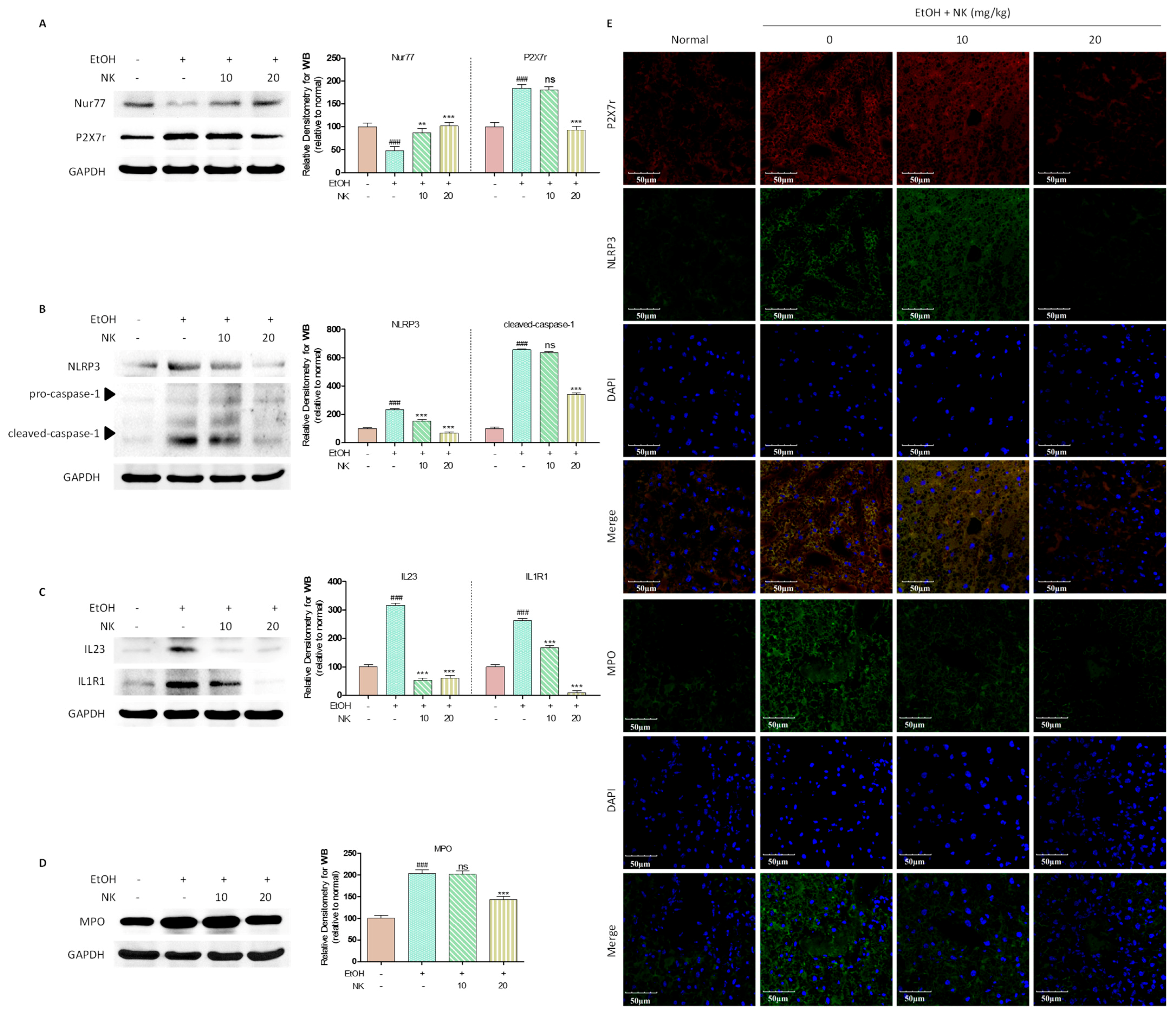
2.6. Nur77 May Be Involved in the Regulatory Effects of NK against the P2X7r-Mediated Inflammatory Response in ALD Mouse Livers
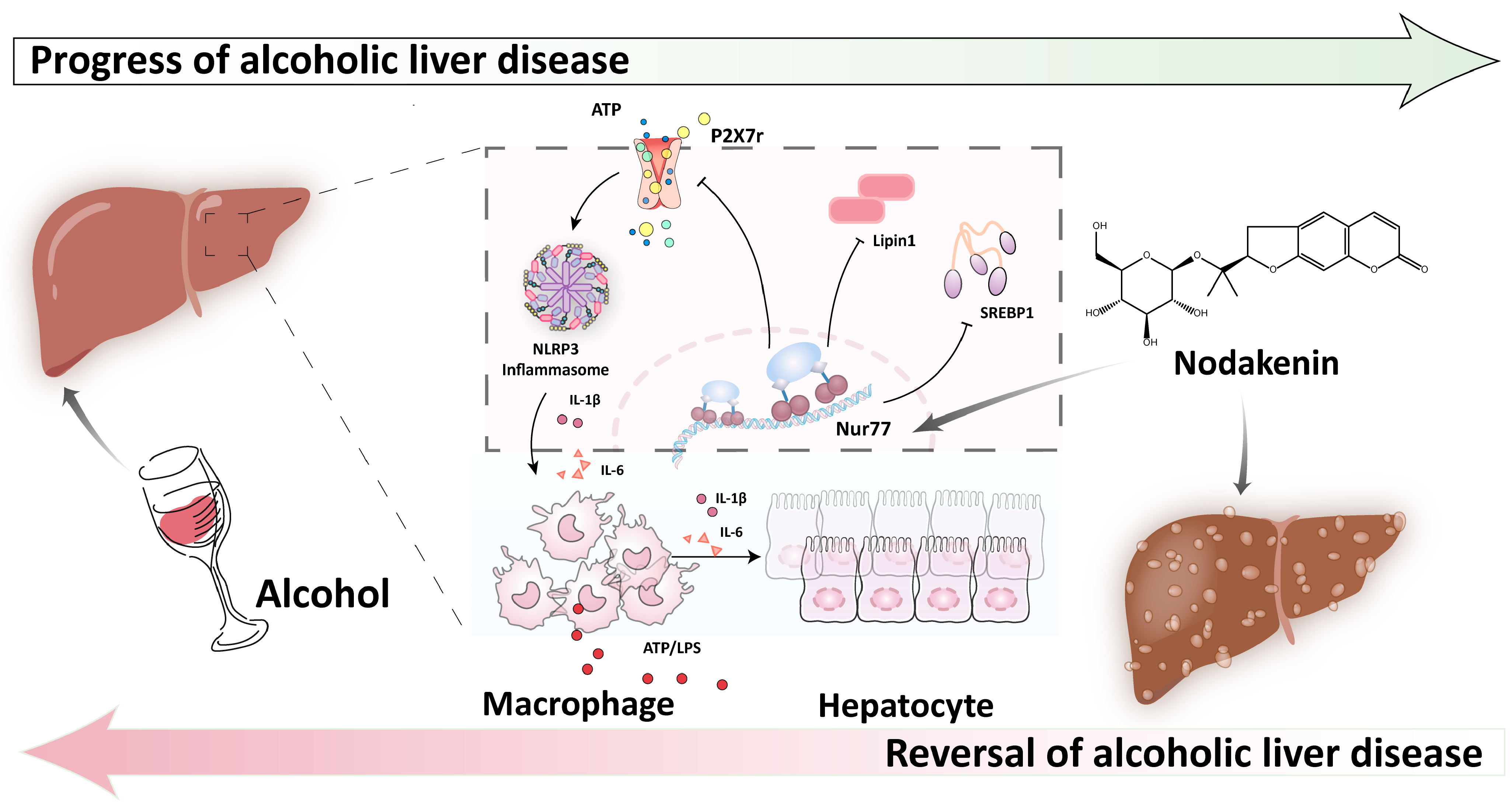
3. Discussion
4. Materials and Methods
4.1. Materials and Reagents
4.2. Animals
4.3. Isolation of Mouse Peritoneal Macrophages (MPMs) and Mouse Primary Hepatocytes
4.4. Cell Culture and Treatment
4.5. Cell Viability Assay
4.6. RT-qPCR
4.7. Knockdown of Nur77 by siRNA in Murine
4.8. Cell Immunofluorescence
4.9. Western Blotting
4.10. ELISA
4.11. Serum Aminotransferase Assays
4.12. Histopathological Analysis
4.13. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Chen, M.; Zhong, W.; Xu, W. Alcohol and the mechanisms of liver disease. J. Gastroenterol. Hepatol. 2023, 38, 1233–1240. [Google Scholar] [CrossRef]
- Lu, C.; Ge, T.; Shao, Y.; Cui, W.; Li, Z.; Xu, W.; Bao, X. ZNF281 drives hepatocyte senescence in alcoholic liver disease by reducing HK2-stabilized PINK1/Parkin-mediated mitophagy. Cell Prolif. 2023, 56, e13378. [Google Scholar] [CrossRef]
- Gopal, T.; Ai, W.; Casey, C.A.; Donohue, T.M.; Saraswathi, V. A review of the role of ethanol-induced adipose tissue dysfunction in alcohol-associated liver disease. Alcohol. Clin. Exp. Res. 2021, 45, 1927–1939. [Google Scholar] [CrossRef] [PubMed]
- Hyun, J.; Han, J.; Lee, C.; Yoon, M.; Jung, Y. Pathophysiological Aspects of Alcohol Metabolism in the Liver. Int. J. Mol. Sci. 2021, 22, 5717. [Google Scholar] [CrossRef] [PubMed]
- Osna, N.A.; Donohue, T.M.; Kharbanda, K.K. Alcoholic Liver Disease: Pathogenesis and Current Management. Alcohol. Res. 2017, 38, 147–161. [Google Scholar]
- Duddempudi, A.T. Immunology in alcoholic liver disease. Clin. Liver Dis. 2012, 16, 687–698. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.Z.; Chen, X.H.; Chen, S.J.; Zhang, J.; Wang, C.Y.; Liu, W.R.; Zhang, D.; Su, Y.; Zhang, X.K. Phase separation of Nur77 mediates celastrol-induced mitophagy by promoting the liquidity of p62/SQSTM1 condensates. Nat. Commun. 2021, 12, 5989. [Google Scholar] [CrossRef]
- Xu, Y.; Tian, J.; Kang, Q.; Yuan, H.; Liu, C.; Li, Z.; Liu, J.; Li, M. Knockout of Nur77 Leads to Amino Acid, Lipid, and Glucose Metabolism Disorders in Zebrafish. Front. Endocrinol. 2022, 3, 864631. [Google Scholar] [CrossRef]
- Jung, Y.S.; Lee, H.S.; Cho, H.R.; Kim, K.J.; Kim, J.H.; Safe, S.; Lee, S.O. Dual targeting of Nur77 and AMPKα by isoalantolactone inhibits adipogenesis in vitro and decreases body fat mass in vivo. Int. J. Obes. 2019, 43, 952–962. [Google Scholar] [CrossRef]
- Pei, L.; Waki, H.; Vaitheesvaran, B.; Wilpitz, D.C.; Kurland, I.J.; Tontonoz, P. NR4A Orphan Nuclear Receptors Are Transcriptional Regulators of Hepatic Glucose Metabolism. Nat. Med. 2006, 12, 1048–1055. [Google Scholar] [CrossRef]
- Miao, L.; Yang, Y.; Liu, Y.; Lai, L.; Wang, L.; Zhan, Y.; Yin, R.; Yu, M.; Li, C.; Yang, X.; et al. Glycerol kinase interacts with nuclear receptor NR4A1 and regulates glucose metabolism in the liver. FASEB J. 2019, 33, 6736–6747. [Google Scholar] [CrossRef]
- Fuchs, C.D.; Claudel, T.; Mlitz, V.; Riva, A.; Menz, M.; Brusilovskaya, K.; Haller, F.; Baumgartner, M.; Königshofer, P.; Unger, L.W.; et al. GLP-2 Improves Hepatic Inflammation and Fibrosis in Mdr2−/− Mice Via Activation of NR4a1/Nur77 in Hepatic Stellate Cells and Intestinal FXR Signaling. Cell Mol. Gastroenterol. Hepatol. 2023, 16, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Vuerich, M. P2X7R: Pivotal player in sepsis-induced liver damage. Purinergic Signal. 2020, 16, 473–474. [Google Scholar] [CrossRef]
- Wang, Y.; Dong, L.; Zhang, Y.; Zhang, Y.; Qin, G.; Zhang, D.; Chen, L.; He, W.; Zhou, J. Activation of the microglial P2X7R/NLRP3 inflammasome mediates central sensitization in a mouse model of medication overuse headache. Front. Mol. Neurosci. 2023, 16, 1177171. [Google Scholar] [CrossRef]
- Xia, G.Q.; Fang, Q.; Cai, J.N.; Li, Z.X.; Zhang, F.Z.; Lv, X.W. P2X7 Receptor in Alcoholic Steatohepatitis and Alcoholic Liver Fibrosis. J. Clin. Transl. Hepatol. 2022, 10, 1205–1212. [Google Scholar] [CrossRef]
- Zuo, R.M.; Jiao, J.Y.; Chen, N.; Jiang, X.L.; Wu, Y.L.; Nan, J.X.; Lian, L.H. Carnosic acid suppressed the formation of NETs in alcoholic hepatosteatosis based on P2X7R-NLRP3 axis. Phytomedicine 2023, 110, 154599. [Google Scholar] [CrossRef]
- Shang, Y.; Yang, H.X.; Li, X.; Zhang, Y.; Chen, N.; Jiang, X.L.; Zhang, Z.H.; Zuo, R.M.; Wang, H.; Lan, X.Q.; et al. Modulation of interleukin-36 based inflammatory feedback loop through the hepatocyte-derived IL-36R-P2X7R axis improves steatosis in alcoholic steatohepatitis. Br. J. Pharmacol. 2022, 179, 4378–4399. [Google Scholar] [CrossRef] [PubMed]
- Ok, S.; Oh, S.R.; Jung, T.S.; Jeon, S.O.; Jung, J.W.; Ryu, D.S. Effects of angelica gigas nakai as an anti-inflammatory agent in in vitro and in vivo atopic dermatitis models. Evid. Based Complement. Alternat Med. 2018, 2018, 2450712. [Google Scholar] [CrossRef] [PubMed]
- Bae, U.J.; Choi, E.K.; Oh, M.R.; Jung, S.J.; Park, J.; Jung, T.S.; Park, T.S.; Chae, S.W.; Park, B.H. Angelica gigas Ameliorates Hyperglycemia and Hepatic Steatosis in C57BL/KsJ-db/db Mice via Activation of AMP-Activated Protein Kinase Signaling Pathway. Am. J. Chin. Med. 2016, 44, 1627–1638. [Google Scholar] [CrossRef]
- Cho, J.H.; Kwon, J.E.; Cho, Y.; Kim, I.; Kang, S.C. Anti-inflammatory effect of angelica gigas via heme oxygenase (HO)-1 expression. Nutrients 2015, 7, 4862–4874. [Google Scholar] [CrossRef] [PubMed]
- Jin, B.R.; Lee, M.; An, H.J. Nodakenin represses obesity and its complications via the inhibition of the VLDLR signalling pathway in vivo and in vitro. Cell Prolif. 2021, 54, e13083. [Google Scholar] [CrossRef]
- Lim, J.Y.; Lee, J.H.; Yun, D.H.; Lee, Y.M.; Kim, D.K. Inhibitory effects of nodakenin on inflammation and cell death in lipopolysaccharide-induced liver injury mice. Phytomedicine 2021, 81, 153411. [Google Scholar] [CrossRef]
- Dong, H.; Zhong, W.; Zhang, W.; Hao, L.; Guo, W.; Yue, R.; Sun, X.; Sun, Z.; Bataller, R.; Zhou, Z. Loss of long-chain acyl-CoA synthetase 1 promotes hepatocyte death in alcohol-induced steatohepatitis. Metabolism 2023, 138, 155334. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Li, S.; Song, Z.; Luo, Q.; Zhang, Y.; Wang, H. The Regulatory Roles of Polysaccharides and Ferroptosis-Related Phytochemicals in Liver Diseases. Nutrients 2022, 14, 2303. [Google Scholar] [CrossRef] [PubMed]
- Ambade, A.; Lowe, P.; Kodys, K.; Catalano, D.; Gyongyosi, B.; Cho, Y.; Iracheta-Vellve, A.; Adejumo, A.; Saha, B.; Calenda, C.; et al. Pharmacological Inhibition of CCR2/5 Signaling Prevents and Reverses Alcohol-Induced Liver Damage, Steatosis, and Inflammation in Mice. Hepatology 2019, 69, 1105–1121. [Google Scholar] [CrossRef]
- Louvet, A.; Mathurin, P. Alcoholic liver disease: Mechanisms of injury and targeted treatment. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 231–242. [Google Scholar] [CrossRef]
- Wang, H.; Mehal, W.; Nagy, L.E.; Rotman, Y. Immunological mechanisms and therapeutic targets of fatty liver diseases. Cell Mol. Immunol. 2021, 18, 73–91. [Google Scholar] [CrossRef]
- Han, J.; Li, E.; Chen, L.; Zhang, Y.; Wei, F.; Liu, J.; Deng, H.; Wang, Y. The CREB coactivator CRTC2 controls hepatic lipid metabolism by regulating SREBP1. Nature 2015, 524, 243–246. [Google Scholar] [CrossRef]
- Tahri-Joutey, M.; Andreoletti, P.; Surapureddi, S.; Nasser, B.; Cherkaoui-Malki, M.; Latruffe, N. Mechanisms Mediating the Regulation of Peroxisomal Fatty Acid Beta-Oxidation by PPARα. Int. J. Mol. Sci. 2021, 22, 8969. [Google Scholar] [CrossRef] [PubMed]
- Piper, C.; Drobyski, W.R. Inflammatory Cytokine Networks in Gastrointestinal Tract Graft vs. Host Disease. Front. Immunol. 2019, 10, 163. [Google Scholar] [CrossRef]
- Rodríguez-González, R.; Baluja, A.; Veiras Del Río, S.; Rodríguez, A.; Rodríguez, J.; Taboada, M.; Brea, D.; Álvarez, J. Effects of sevoflurane postconditioning on cell death, inflammation and TLR expression in human endothelial cells exposed to LPS. J. Transl. Med. 2013, 11, 87. [Google Scholar] [CrossRef]
- Quagliariello, V.; Masarone, M.; Armenia, E.; Giudice, A.; Barbarisi, M.; Caraglia, M.; Barbarisi, A.; Persico, M. Chitosan-coated liposomes loaded with butyric acid demonstrate anticancer and anti-inflammatory activity in human hepatoma HepG2 cells. Oncol. Rep. 2019, 41, 1476–1486. [Google Scholar] [CrossRef]
- Hu, Y.W.; Zhang, P.; Yang, J.Y.; Huang, J.L.; Ma, X.; Li, S.F.; Zhao, J.Y.; Hu, Y.R.; Wang, Y.C.; Gao, J.J.; et al. Nur77 decreases atherosclerosis progression in apoE−/− mice fed a high-fat/high-cholesterol diet. PLoS ONE 2014, 9, e87313. [Google Scholar] [CrossRef]
- Fang, H.; Cao, Y.; Zhang, J.; Wang, X.; Li, M.; Hong, Z.; Wu, Z.; Fang, M. Lipidome remodeling activities of DPA-EA in palmitic acid-stimulated HepG2 cells and the in vivo anti-obesity effect of the DPA-EA and DHA-EA mixture prepared from algae oil. Front. Pharmacol. 2023, 14, 1146276. [Google Scholar] [CrossRef]
- Zhao, Y.; Bruemmer, D. NR4A orphan nuclear receptors: Transcriptional regulators of gene expression in metabolism and vascular biology. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 1535–1541. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, S.R.; Paul-Heng, M.; Krycer, J.R.; Fazakerley, D.J.; Sharland, A.F.; Hoy, A.J. Lipid and glucose metabolism in hepatocyte cell lines and primary mouse hepatocytes: A comprehensive resource for in vitro studies of hepatic metabolism. Am. J. Physiol. Endocrinol. Metab. 2019, 316, E578–E589. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Du, X.; Chen, H.; Liu, J.; Zhao, B.; Huang, D.; Li, G.; Xu, Q.; Zhang, M.; Weimer, B.C.; et al. Cytosporone B is an agonist for nuclear orphan receptor Nur77. Nat. Chem. Biol. 2008, 4, 548–556. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Tan, Y.J.; Xu, S.Q.; Qin, B.F.; Xiu, M.X.; Zhang, X.; Shi, L.Q.; Sun, H.M.; Song, J. Ginsenoside Rd, a natural production for attenuating fibrogenesis and inflammation in hepatic fibrosis by regulating the ERRα-mediated P2X7r pathway. Food. Funct. 2023, 14, 5606–5619. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, J.; Qin, B.-F.; Zhang, J.-J.; Feng, Q.-Y.; Liu, G.-C.; Zhao, G.-Y.; Sun, H.-M. Regulation of the Nur77-P2X7r Signaling Pathway by Nodakenin: A Potential Protective Function against Alcoholic Liver Disease. Molecules 2024, 29, 1078. https://doi.org/10.3390/molecules29051078
Song J, Qin B-F, Zhang J-J, Feng Q-Y, Liu G-C, Zhao G-Y, Sun H-M. Regulation of the Nur77-P2X7r Signaling Pathway by Nodakenin: A Potential Protective Function against Alcoholic Liver Disease. Molecules. 2024; 29(5):1078. https://doi.org/10.3390/molecules29051078
Chicago/Turabian StyleSong, Jian, Bo-Feng Qin, Jin-Jin Zhang, Qi-Yuan Feng, Guan-Cheng Liu, Gui-Yun Zhao, and Hai-Ming Sun. 2024. "Regulation of the Nur77-P2X7r Signaling Pathway by Nodakenin: A Potential Protective Function against Alcoholic Liver Disease" Molecules 29, no. 5: 1078. https://doi.org/10.3390/molecules29051078
APA StyleSong, J., Qin, B.-F., Zhang, J.-J., Feng, Q.-Y., Liu, G.-C., Zhao, G.-Y., & Sun, H.-M. (2024). Regulation of the Nur77-P2X7r Signaling Pathway by Nodakenin: A Potential Protective Function against Alcoholic Liver Disease. Molecules, 29(5), 1078. https://doi.org/10.3390/molecules29051078