Efficient Escorting Strategy for Aggregation-Prone Notch EGF Repeats with Sparcl1
Abstract
1. Introduction
2. Results
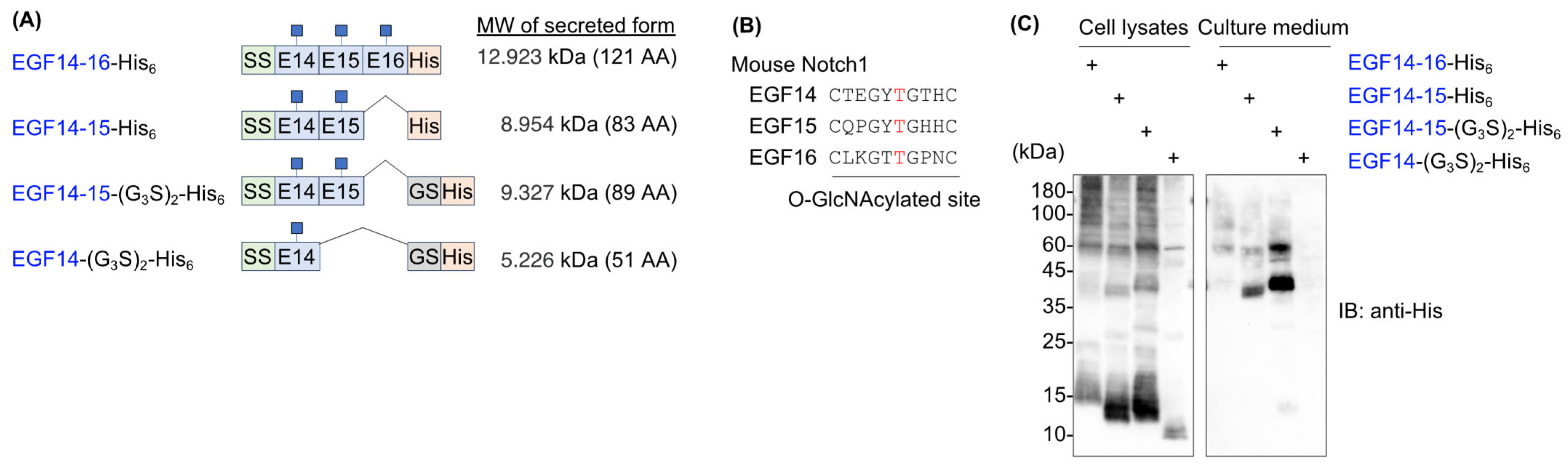
2.1. Mouse Notch1 Fragments Containing EGF14-16 Tended to Aggregate
2.2. Chemical Chaperone 4-PBA Does Not Enhance the Secretion of EGF14-15-(G3S)2-His6
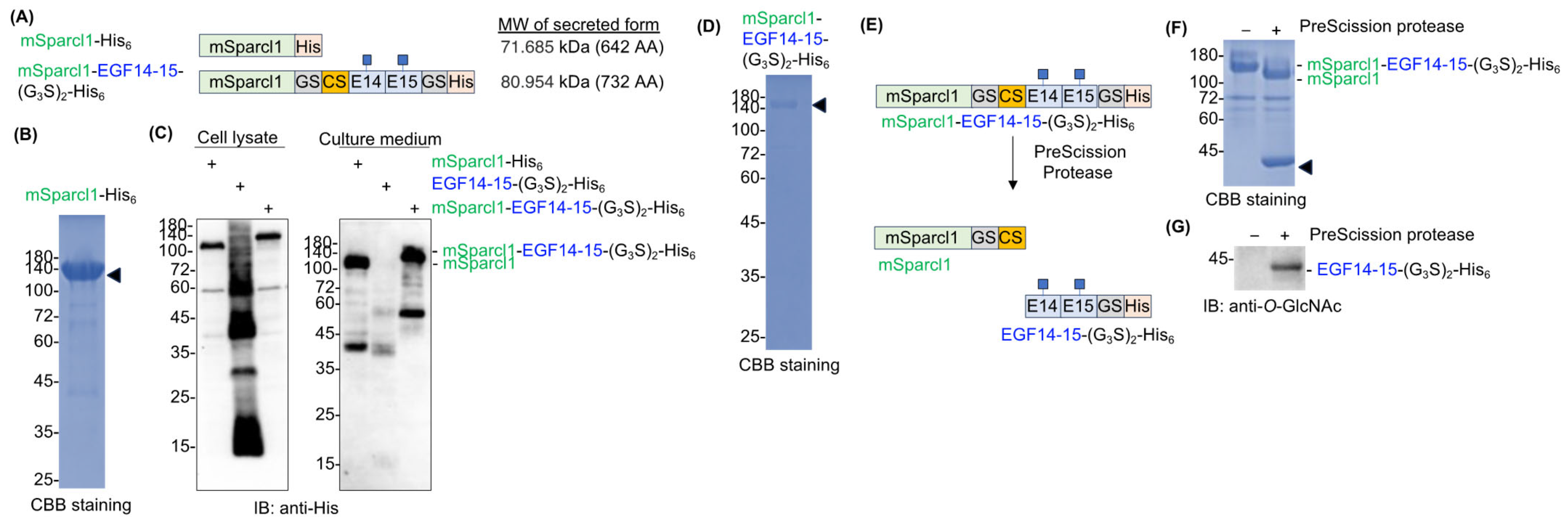
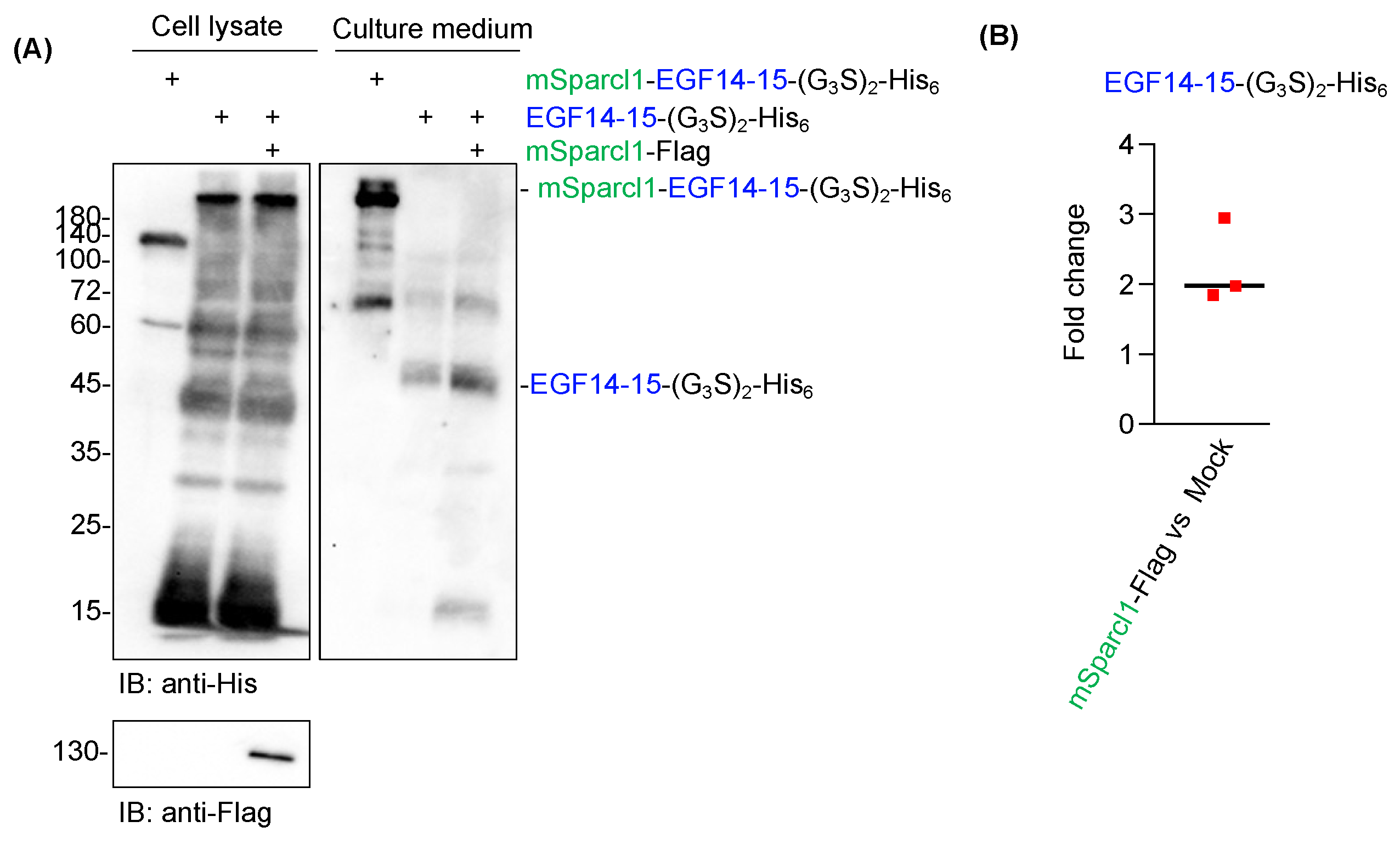
2.3. Sparcl1 Fusion Facilitates Extracellular Secretion of EGF14-15-(G3S)2-His6
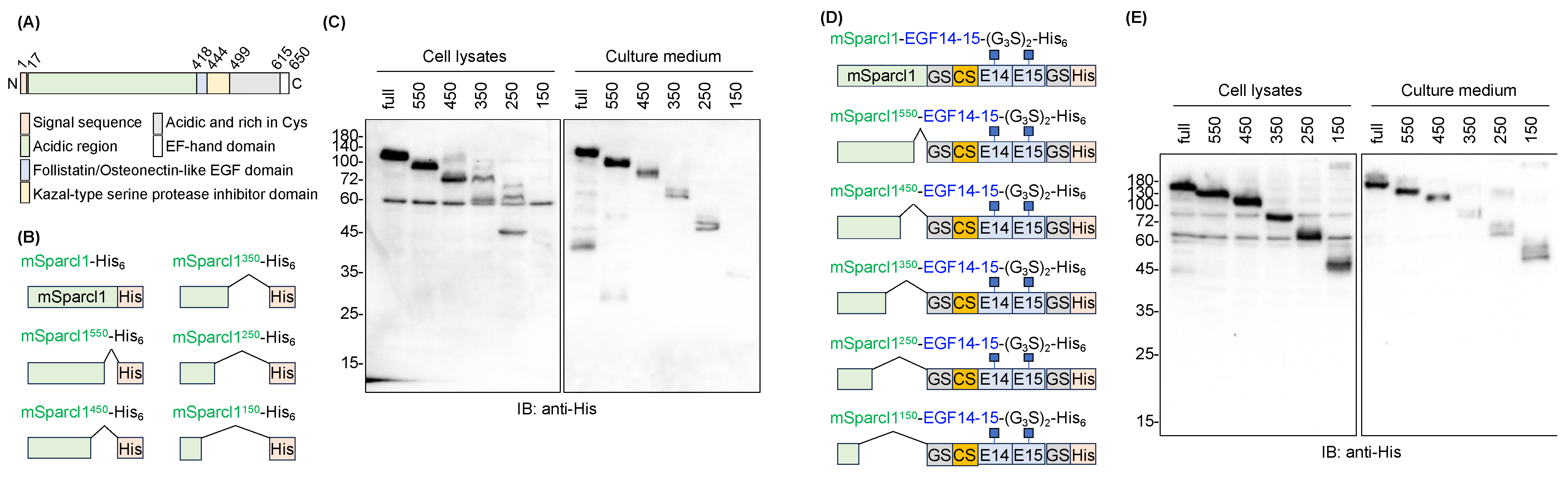
2.4. Deletion Mutants of mSparcl1 and Their Effects on the Secretion of EGF14-15-(G3S)2-His6
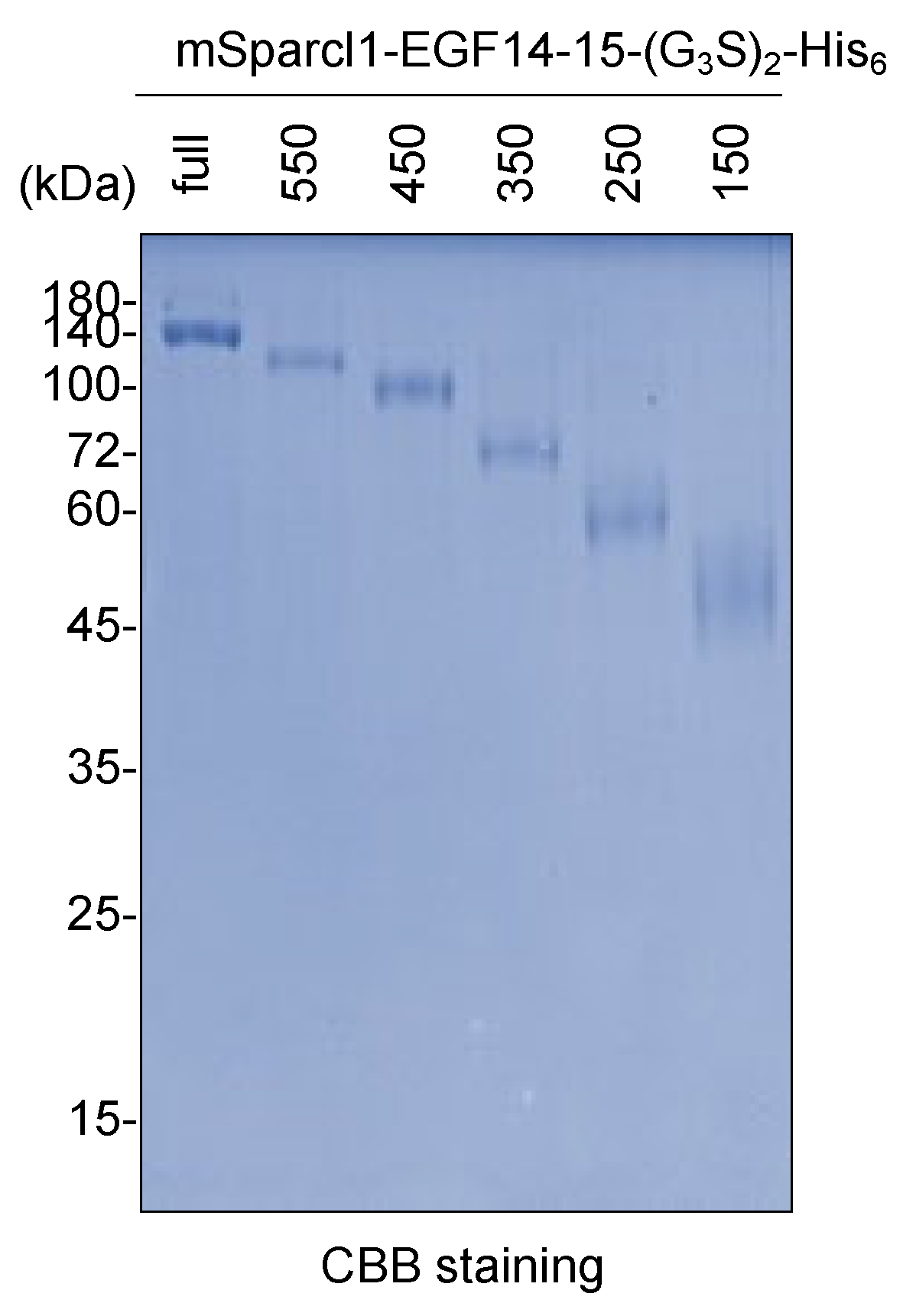
2.5. Purification of Deletion Mutants of mSparcl1-EGF14-15-(G3S)2-His6
2.6. Co-Transfection of mSparcl1 Slightly Enhances Secretion of EGF14-15-(G3S)2-His6
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Plasmids
4.3. SDS-PAGE, Immunoblotting, and CBB Staining
4.4. Recombinant Protein Expression and Purification
4.5. Immunofluorescence and Confocal Microscopy
4.6. Aggresome Formation Assay
4.7. Native PAGE
4.8. Data Analysis and Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Saiki, W.; Ma, C.; Okajima, T.; Takeuchi, H. Current Views on the Roles of O-Glycosylation in Controlling Notch-Ligand Interactions. Biomolecules 2021, 11, 309. [Google Scholar] [CrossRef]
- Takeuchi, H.; Haltiwanger, R.S. Significance of glycosylation in Notch signaling. Biochem. Biophys. Res. Commun. 2014, 453, 235–242. [Google Scholar] [CrossRef]
- Takeuchi, H.; Yu, H.; Hao, H.; Takeuchi, M.; Ito, A.; Li, H.; Haltiwanger, R.S. O-Glycosylation modulates the stability of epidermal growth factor-like repeats and thereby regulates Notch trafficking. J. Biol. Chem. 2017, 292, 15964–15973. [Google Scholar] [CrossRef]
- Tsukamoto, Y.; Ogawa, M.; Yogi, K.; Tashima, Y.; Takeuchi, H.; Okajima, T. Glycoproteomics of NOTCH1 EGF repeat fragments overexpressed with different glycosyltransferases in HEK293T cells reveals insights into O-GlcNAcylation of NOTCH1. Glycobiology 2022, 32, 616–628. [Google Scholar] [CrossRef]
- Sakaidani, Y.; Ichiyanagi, N.; Saito, C.; Nomura, T.; Ito, M.; Nishio, Y.; Nadano, D.; Matsuda, T.; Furukawa, K.; Okajima, T. O-linked-N-acetylglucosamine modification of mammalian Notch receptors by an atypical O-GlcNAc transferase Eogt1. Biochem. Biophys. Res. Commun. 2012, 419, 14–19. [Google Scholar] [CrossRef]
- Shi, S.; Stanley, P. Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proc. Natl. Acad. Sci. USA 2003, 100, 5234–5239. [Google Scholar] [CrossRef]
- Fernandez-Valdivia, R.; Takeuchi, H.; Samarghandi, A.; Lopez, M.; Leonardi, J.; Haltiwanger, R.S.; Jafar-Nejad, H. Regulation of mammalian Notch signaling and embryonic development by the protein O-glucosyltransferase Rumi. Development 2011, 138, 1925–1934. [Google Scholar] [CrossRef]
- Sawaguchi, S.; Varshney, S.; Ogawa, M.; Sakaidani, Y.; Yagi, H.; Takeshita, K.; Murohara, T.; Kato, K.; Sundaram, S.; Stanley, P.; et al. O-GlcNAc on NOTCH1 EGF repeats regulates ligand-induced Notch signaling and vascular development in mammals. eLife 2017, 6, e24419. [Google Scholar] [CrossRef]
- Tashima, Y.; Okajima, T. Congenital diseases caused by defective O-glycosylation of Notch receptors. Nagoya J. Med. Sci. 2018, 80, 299–307. [Google Scholar]
- Cortez, L.; Sim, V. The therapeutic potential of chemical chaperones in protein folding diseases. Prion 2014, 8, 197–202. [Google Scholar] [CrossRef]
- Delic, M.; Gongrich, R.; Mattanovich, D.; Gasser, B. Engineering of protein folding and secretion-strategies to overcome bottlenecks for efficient production of recombinant proteins. Antioxid. Redox Signal. 2014, 21, 414–437. [Google Scholar] [CrossRef]
- Andersen, E.; Chollet, M.E.; Baroni, M.; Pinotti, M.; Bernardi, F.; Skarpen, E.; Sandset, P.M.; Skretting, G. The effect of the chemical chaperone 4-phenylbutyrate on secretion and activity of the p.Q160R missense variant of coagulation factor FVII. Cell Biosci. 2019, 9, 69. [Google Scholar] [CrossRef]
- Besio, R.; Iula, G.; Garibaldi, N.; Cipolla, L.; Sabbioneda, S.; Biggiogera, M.; Marini, J.C.; Rossi, A.; Forlino, A. 4-PBA ameliorates cellular homeostasis in fibroblasts from osteogenesis imperfecta patients by enhancing autophagy and stimulating protein secretion. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864 Pt A, 1642–1652. [Google Scholar] [CrossRef]
- Iwayama, T.; Iwashita, M.; Miyashita, K.; Sakashita, H.; Matsumoto, S.; Tomita, K.; Bhongsatiern, P.; Kitayama, T.; Ikegami, K.; Shimbo, T.; et al. Plap-1 lineage tracing and single-cell transcriptomics reveal cellular dynamics in the periodontal ligament. Development 2022, 149, dev201203. [Google Scholar] [CrossRef]
- Klingler, A.; Regensburger, D.; Tenkerian, C.; Britzen-Laurent, N.; Hartmann, A.; Sturzl, M.; Naschberger, E. Species-, organ- and cell-type-dependent expression of SPARCL1 in human and mouse tissues. PLoS ONE 2020, 15, e0233422. [Google Scholar] [CrossRef]
- Marchi, S.; Corricelli, M.; Trapani, E.; Bravi, L.; Pittaro, A.; Delle Monache, S.; Ferroni, L.; Patergnani, S.; Missiroli, S.; Goitre, L.; et al. Defective autophagy is a key feature of cerebral cavernous malformations. EMBO Mol. Med. 2015, 7, 1403–1417. [Google Scholar] [CrossRef]
- Shen, D.; Coleman, J.; Chan, E.; Nicholson, T.P.; Dai, L.; Sheppard, P.W.; Patton, W.F. Novel cell- and tissue-based assays for detecting misfolded and aggregated protein accumulation within aggresomes and inclusion bodies. Cell Biochem. Biophys. 2011, 60, 173–185. [Google Scholar] [CrossRef]
- Viloria, K.; Munasinghe, A.; Asher, S.; Bogyere, R.; Jones, L.; Hill, N.J. A holistic approach to dissecting SPARC family protein complexity reveals FSTL-1 as an inhibitor of pancreatic cancer cell growth. Sci. Rep. 2016, 6, 37839. [Google Scholar] [CrossRef]
- Bradshaw, A.D. Diverse biological functions of the SPARC family of proteins. Int. J. Biochem. Cell Biol. 2012, 44, 480–488. [Google Scholar] [CrossRef]
- Fan, S.; Gangwar, S.P.; Machius, M.; Rudenko, G. Interplay between hevin, SPARC, and MDGAs: Modulators of neurexin-neuroligin transsynaptic bridges. Structure 2021, 29, 664–678.e6. [Google Scholar] [CrossRef]
- Naschberger, E.; Liebl, A.; Schellerer, V.S.; Schutz, M.; Britzen-Laurent, N.; Kolbel, P.; Schaal, U.; Haep, L.; Regensburger, D.; Wittmann, T.; et al. Matricellular protein SPARCL1 regulates tumor microenvironment-dependent endothelial cell heterogeneity in colorectal carcinoma. J. Clin. Investig. 2016, 126, 4187–4204. [Google Scholar] [CrossRef]
- Kalucka, J.; de Rooij, L.; Goveia, J.; Rohlenova, K.; Dumas, S.J.; Meta, E.; Conchinha, N.V.; Taverna, F.; Teuwen, L.A.; Veys, K.; et al. Single-Cell Transcriptome Atlas of Murine Endothelial Cells. Cell 2020, 180, 764–779.e20. [Google Scholar] [CrossRef]
- Bae, J.H.; Sung, B.H.; Kim, H.J.; Park, S.H.; Lim, K.M.; Kim, M.J.; Lee, C.R.; Sohn, J.H. An Efficient Genome-Wide Fusion Partner Screening System for Secretion of Recombinant Proteins in Yeast. Sci. Rep. 2015, 5, 12229. [Google Scholar] [CrossRef]
- Li, Y.; Xiao, C.; Pan, Y.; Qin, L.; Zheng, L.; Zhao, M.; Huang, M. Optimization of Protein Folding for Improved Secretion of Human Serum Albumin Fusion Proteins in Saccharomyces cerevisiae. J. Agric. Food Chem. 2023, 71, 18414–18423. [Google Scholar] [CrossRef]
- Wang, Y.; Kim, K.A.; Kim, J.H.; Sul, H.S. Pref-1, a preadipocyte secreted factor that inhibits adipogenesis. J. Nutr. 2006, 136, 2953–2956. [Google Scholar] [CrossRef]
- Sul, H.S. Minireview: Pref-1: Role in adipogenesis and mesenchymal cell fate. Mol. Endocrinol. 2009, 23, 1717–1725. [Google Scholar] [CrossRef]
- Bruijn, L.I.; Houseweart, M.K.; Kato, S.; Anderson, K.L.; Anderson, S.D.; Ohama, E.; Reaume, A.G.; Scott, R.W.; Cleveland, D.W. Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1. Science 1998, 281, 1851–1854. [Google Scholar] [CrossRef]
- DiFiglia, M.; Sapp, E.; Chase, K.O.; Davies, S.W.; Bates, G.P.; Vonsattel, J.P.; Aronin, N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 1997, 277, 1990–1993. [Google Scholar] [CrossRef]
- Narhi, L.; Wood, S.J.; Steavenson, S.; Jiang, Y.; Wu, G.M.; Anafi, D.; Kaufman, S.A.; Martin, F.; Sitney, K.; Denis, P.; et al. Both familial Parkinson’s disease mutations accelerate alpha-synuclein aggregation. J. Biol. Chem. 1999, 274, 9843–9846. [Google Scholar] [CrossRef]
- Ross, C.A.; Poirier, M.A. Protein aggregation and neurodegenerative disease. Nat. Med. 2004, 10, S10–S17. [Google Scholar] [CrossRef]
- Risher, W.C.; Patel, S.; Kim, I.H.; Uezu, A.; Bhagat, S.; Wilton, D.K.; Pilaz, L.J.; Singh Alvarado, J.; Calhan, O.Y.; Silver, D.L.; et al. Astrocytes refine cortical connectivity at dendritic spines. eLife 2014, 3, e04047. [Google Scholar] [CrossRef] [PubMed]
- Scalabrini, D.; Fenoglio, C.; Scarpini, E.; De Riz, M.; Comi, C.; Venturelli, E.; Cortini, F.; Piola, M.; Villa, C.; Naldi, P.; et al. Candidate gene analysis of SPARCL1 gene in patients with multiple sclerosis. Neurosci. Lett. 2007, 425, 173–176. [Google Scholar] [CrossRef] [PubMed]
- Taketomi, T.; Yasuda, T.; Morita, R.; Kim, J.; Shigeta, Y.; Eroglu, C.; Harada, R.; Tsuruta, F. Autism-associated mutation in Hevin/Sparcl1 induces endoplasmic reticulum stress through structural instability. Sci. Rep. 2022, 12, 11891. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, S.R. One-Dimensional Electrophoresis Using Nondenaturing Conditions. Curr. Protoc. Protein Sci. 2018, 94, e73. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kondo, Y.; Li, Y.; Okajima, T. Efficient Escorting Strategy for Aggregation-Prone Notch EGF Repeats with Sparcl1. Molecules 2024, 29, 1031. https://doi.org/10.3390/molecules29051031
Kondo Y, Li Y, Okajima T. Efficient Escorting Strategy for Aggregation-Prone Notch EGF Repeats with Sparcl1. Molecules. 2024; 29(5):1031. https://doi.org/10.3390/molecules29051031
Chicago/Turabian StyleKondo, Yuji, Yuxin Li, and Tetsuya Okajima. 2024. "Efficient Escorting Strategy for Aggregation-Prone Notch EGF Repeats with Sparcl1" Molecules 29, no. 5: 1031. https://doi.org/10.3390/molecules29051031
APA StyleKondo, Y., Li, Y., & Okajima, T. (2024). Efficient Escorting Strategy for Aggregation-Prone Notch EGF Repeats with Sparcl1. Molecules, 29(5), 1031. https://doi.org/10.3390/molecules29051031






