How Aqueous Solvation Impacts the Frequencies and Intensities of Infrared Absorption Bands in Flavin: The Quest for a Suitable Solvent Model
Abstract
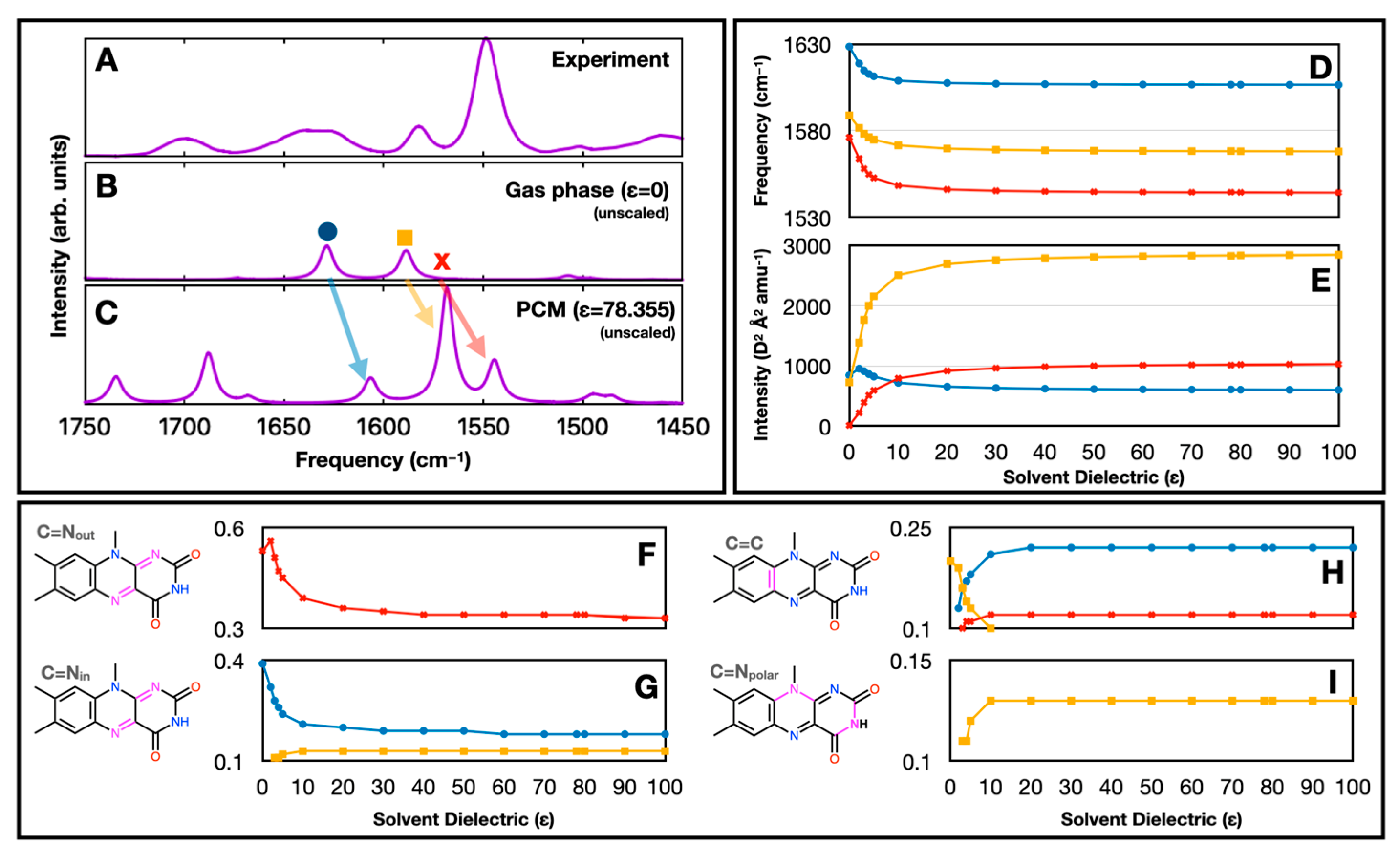
1. Introduction
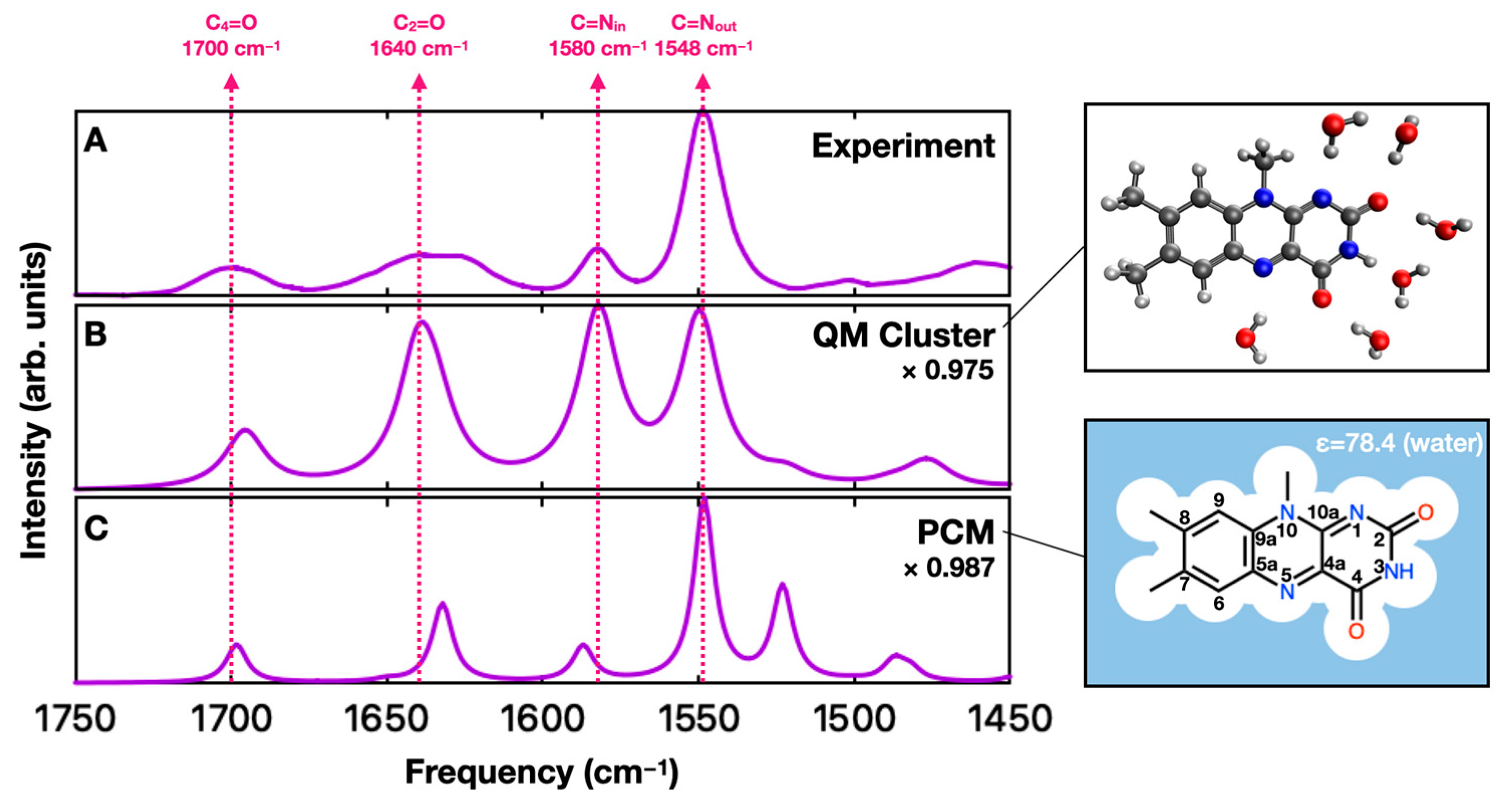
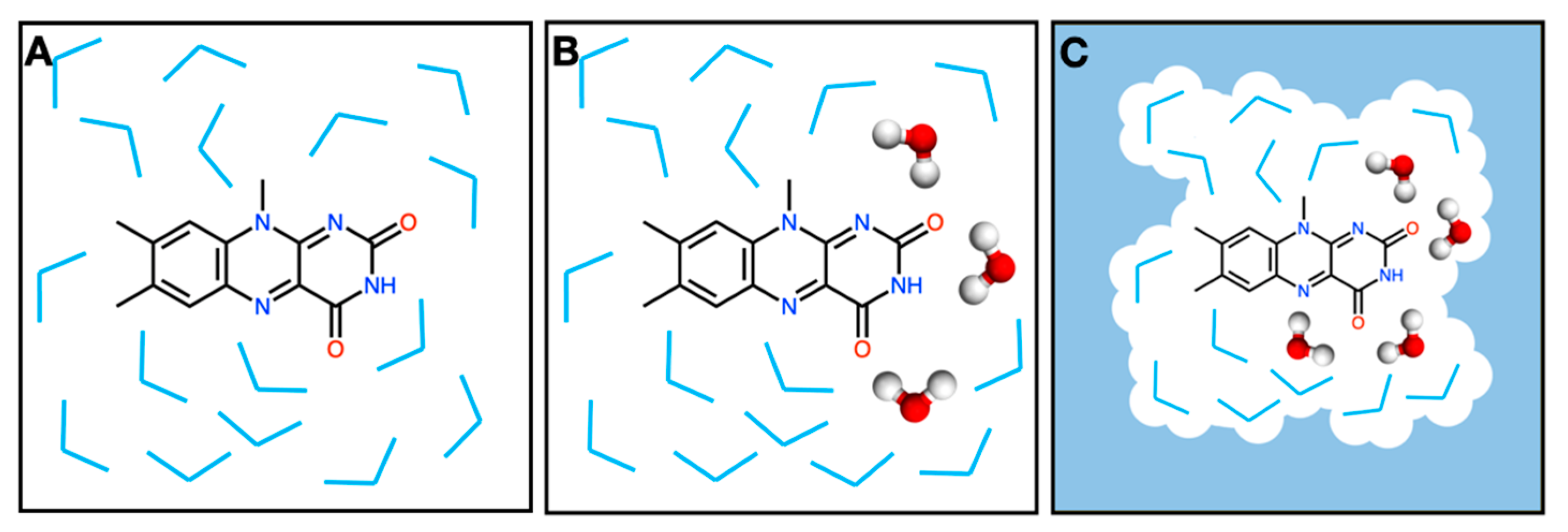
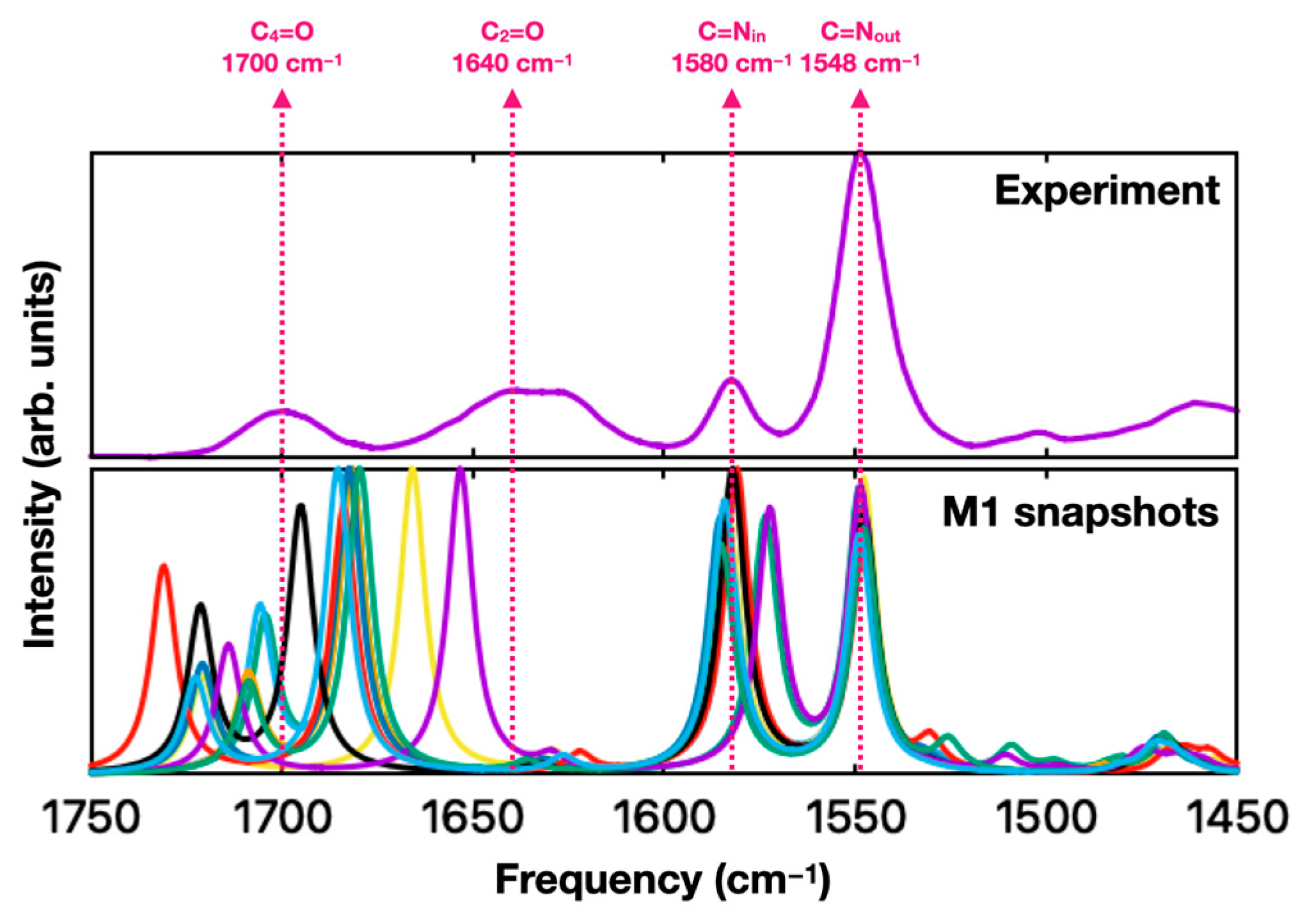
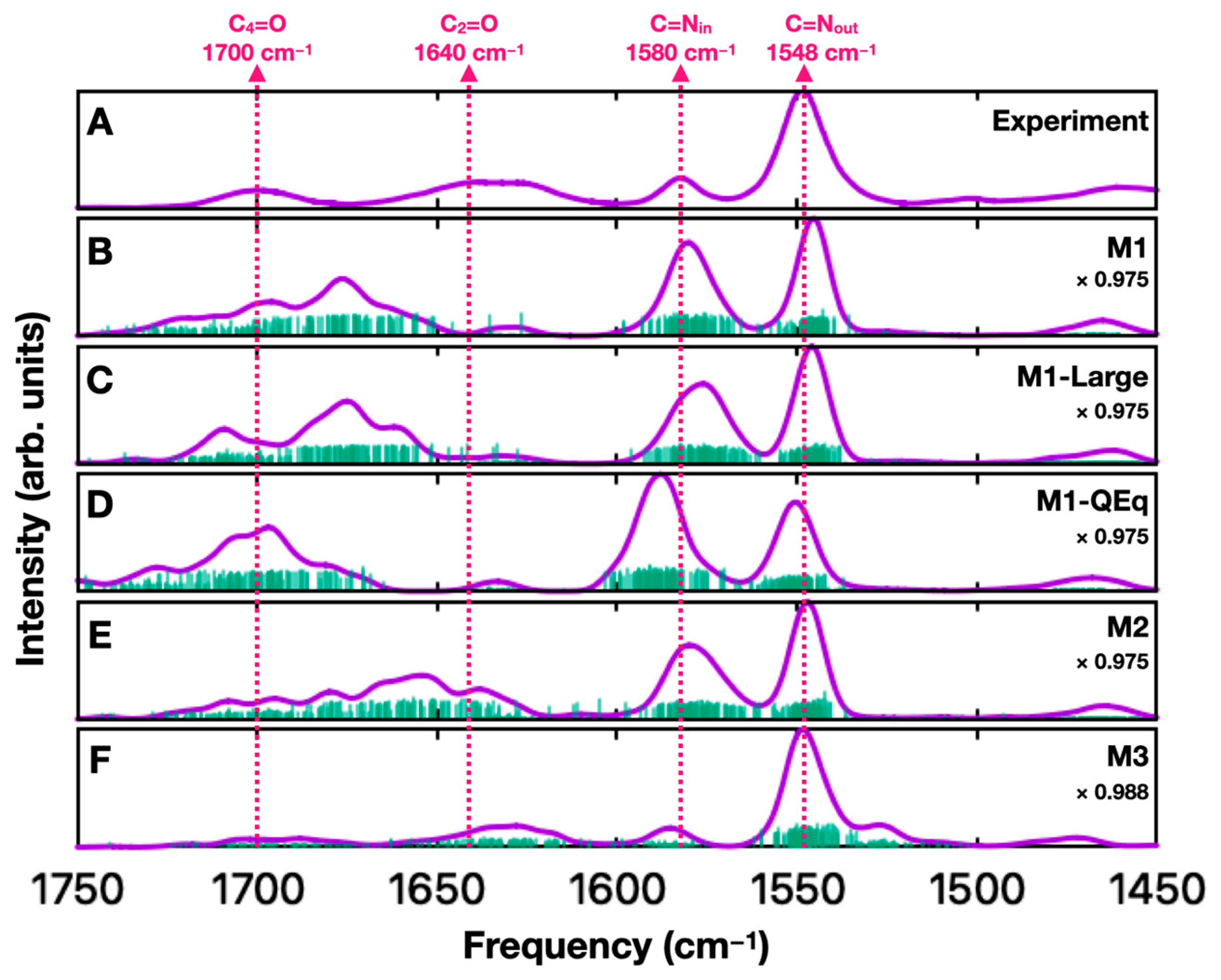
2. Results and Discussion
3. Materials and Methods
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lorenz-Fonfria, V.A. Infrared Difference Spectroscopy of Proteins: From Bands to Bonds. Chem. Rev. 2020, 120, 3466–3576. [Google Scholar] [CrossRef] [PubMed]
- Mäntele, W. Reaction-induced infrared difference spectroscopy for the study of protein function and reaction mechanisms. Trends Biochem. Sci. 1993, 18, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Thompson, L.M.; Lasoroski, A.; Champion, P.M.; Sage, J.T.; Frisch, M.J.; van Thor, J.J.; Bearpark, M.J. Analytical Harmonic Vibrational Frequencies for the Green Fluorescent Protein Computed with ONIOM: Chromophore Mode Character and Its Response to Environment. J. Chem. Theory Comput. 2014, 10, 751–766. [Google Scholar] [CrossRef] [PubMed]
- Huix-Rotllant, M.; Schwinn, K.; Ferré, N. Infrared spectroscopy from electrostatic embedding QM/MM: Local normal mode analysis of infrared spectra of arabidopsis thaliana plant cryptochrome. Phys. Chem. Chem. Phys. 2021, 23, 1666–1674. [Google Scholar] [CrossRef] [PubMed]
- Yamada, D.; Kandori, H. FTIR spectroscopy of flavin-binding photoreceptors. Methods Mol. Biol. 2014, 1146, 361–376. [Google Scholar] [CrossRef]
- Nishina, Y.; Sato, K.; Setoyama, C.; Tamaoki, H.; Miura, R.; Shiga, K. Intramolecular and intermolecular perturbation on electronic state of FAD free in solution and bound to flavoproteins: FTIR spectroscopic study by using the C=O stretching vibrations as probes. J. Biochem. 2007, 142, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Thöing, C.; Pfeifer, A.; Kakorin, S.; Kottke, T. Protonated triplet-excited flavin resolved by step-scan FTIR spectroscopy: Implications for photosensory LOV domains. Phys. Chem. Chem. Phys. 2013, 15, 5916–5926. [Google Scholar] [CrossRef]
- Kottke, T.; Batschauer, A.; Ahmad, M.; Heberle, J. Blue-Light-Induced Changes in Arabidopsis Cryptochrome 1 Probed by FTIR Difference Spectroscopy. Biochemistry 2006, 45, 2472–2479. [Google Scholar] [CrossRef][Green Version]
- Ihalainen, J.A.; Gustavsson, E.; Schroeder, L.; Donnini, S.; Lehtivuori, H.; Isaksson, L.; Thöing, C.; Modi, V.; Berntsson, O.; Stucki-Buchli, B.; et al. Chromophore–Protein Interplay during the Phytochrome Photocycle Revealed by Step-Scan FTIR Spectroscopy. J. Am. Chem. Soc. 2018, 140, 12396–12404. [Google Scholar] [CrossRef]
- Unno, M.; Sano, R.; Masuda, S.; Ono, T.A.; Yamauchi, S. Light-induced structural changes in the active site of the BLUF domain in AppA by Raman spectroscopy. J. Phys. Chem. B 2005, 109, 12620–12626. [Google Scholar] [CrossRef]
- Takahashi, R.; Okajima, K.; Suzuki, H.; Nakamura, H.; Ikeuchi, M.; Noguchi, T. FTIR study on the hydrogen bond structure of a key tyrosine residue in the flavin-binding blue light sensor TePixD from Thermosynechococcus elongatus. Biochemistry 2007, 46, 6459–6467. [Google Scholar] [CrossRef] [PubMed]
- Mehlhorn, J.; Steinocher, H.; Beck, S.; Kennis, J.T.; Hegemann, P.; Mathes, T. A set of engineered Escherichia coli expression strains for selective isotope and reactivity labeling of amino acid side chains and flavin cofactors. PLoS ONE 2013, 8, e79006. [Google Scholar] [CrossRef] [PubMed]
- Hontani, Y.; Mehlhorn, J.; Domratcheva, T.; Beck, S.; Kloz, M.; Hegemann, P.; Mathes, T.; Kennis, J.T.M. Spectroscopic and Computational Observation of Glutamine Tautomerization in the Blue Light Sensing Using Flavin Domain Photoreaction. J. Am. Chem. Soc. 2023, 145, 1040–1052. [Google Scholar] [CrossRef]
- Bonetti, C.; Mathes, T.; Van Stokkum, I.H.; Mullen, K.M.; Groot, M.-L.; Van Grondelle, R.; Hegemann, P.; Kennis, J.T. Hydrogen bond switching among flavin and amino acid side chains in the BLUF photoreceptor observed by ultrafast infrared spectroscopy. Biophys. J. 2008, 95, 4790–4802. [Google Scholar] [CrossRef] [PubMed]
- Alexandre, M.T.; van Grondelle, R.; Hellingwerf, K.J.; Kennis, J.T. Conformational heterogeneity and propagation of structural changes in the LOV2/Jα domain from Avena sativa phototropin 1 as recorded by temperature-dependent FTIR spectroscopy. Biophys. J. 2009, 97, 238–247. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, A.; Iwata, T.; Sato, Y.; Matsuoka, D.; Tokutomi, S.; Kandori, H. Light signal transduction pathway from flavin chromophore to the Jα helix of Arabidopsis phototropin1. Biophys. J. 2009, 96, 2771–2778. [Google Scholar] [CrossRef] [PubMed]
- Konold, P.E.; Mathes, T.; Weibetaenborn, J.; Groot, M.L.; Hegemann, P.; Kennis, J.T. Unfolding of the C-Terminal Jalpha Helix in the LOV2 Photoreceptor Domain Observed by Time-Resolved Vibrational Spectroscopy. J. Phys. Chem. Lett. 2016, 7, 3472–3476. [Google Scholar] [CrossRef] [PubMed]
- Iwata, T.; Nagai, T.; Ito, S.; Osoegawa, S.; Iseki, M.; Watanabe, M.; Unno, M.; Kitagawa, S.; Kandori, H. Hydrogen Bonding Environments in the Photocycle Process around the Flavin Chromophore of the AppA-BLUF domain. J. Am. Chem. Soc. 2018, 140, 11982–11991. [Google Scholar] [CrossRef]
- Domratcheva, T.; Grigorenko, B.L.; Schlichting, I.; Nemukhin, A.V. Molecular models predict light-induced glutamine tautomerization in BLUF photoreceptors. Biophys. J. 2008, 94, 3872–3879. [Google Scholar] [CrossRef][Green Version]
- Goings, J.J.; Li, P.F.; Zhu, Q.W.; Hammes-Schiffer, S. Formation of an unusual glutamine tautomer in a blue light using flavin photocycle characterizes the light-adapted state. Proc. Natl. Acad. Sci. USA 2020, 117, 26626–26632. [Google Scholar] [CrossRef]
- Rieff, B.; Bauer, S.; Mathias, G.; Tavan, P. DFT/MM description of flavin IR spectra in BLUF domains. J. Phys. Chem. B 2011, 115, 11239–11253. [Google Scholar] [CrossRef] [PubMed]
- Wille, G.; Ritter, M.; Friedemann, R.; Mantele, W.; Hubner, G. Redox-triggered FTIR difference spectra of FAD in aqueous solution and bound to flavoproteins. Biochemistry 2003, 42, 14814–14821. [Google Scholar] [CrossRef] [PubMed]
- Birss, V.I.; Hinman, A.S.; Mcgarvey, C.E.; Segal, J. In-Situ Ftir Thin-Layer Reflectance Spectroscopy of Flavin Adenine-Dinucleotide at a Mercury Gold Electrode. Electrochim. Acta 1994, 39, 2449–2454. [Google Scholar] [CrossRef]
- Iuliano, J.N.; French, J.B.; Tonge, P.J. Vibrational spectroscopy of flavoproteins. Methods Enzymol. 2019, 620, 189–214. [Google Scholar] [CrossRef]
- El Khoury, Y.; Van Wilderen, L.J.; Bredenbeck, J. Ultrafast 2D-IR spectroelectrochemistry of flavin mononucleotide. J. Chem. Phys. 2015, 142, 212416. [Google Scholar] [CrossRef]
- Zhao, R. Photochemistry, Photophysics and Spectroscopy of Redox States of Flavins Relevant to Photoactive Flavoproteins. Ph.D. Thesis, University of East Anglia, Norwich, UK, 2012. [Google Scholar]
- Spexard, M.; Immeln, D.; Thoing, C.; Kottke, T. Infrared spectrum and absorption coefficient of the cofactor flavin in water. Vib. Spectrosc. 2011, 57, 282–287. [Google Scholar] [CrossRef]
- Langer, J.; Gunther, A.; Seidenbecher, S.; Berden, G.; Oomens, J.; Dopfer, O. Probing protonation sites of isolated flavins using IR spectroscopy: From lumichrome to the cofactor flavin mononucleotide. Chemphyschem 2014, 15, 2550–2562. [Google Scholar] [CrossRef]
- Wolf, M.M.; Schumann, C.; Gross, R.; Domratcheva, T.; Diller, R. Ultrafast infrared spectroscopy of riboflavin: Dynamics, electronic structure, and vibrational mode analysis. J. Phys. Chem. B 2008, 112, 13424–13432. [Google Scholar] [CrossRef]
- Kar, R.K.; Borin, V.A.; Ding, Y.; Matysik, J.; Schapiro, I. Spectroscopic Properties of Lumiflavin: A Quantum Chemical Study. Photochem. Photobiol. 2019, 95, 662–674. [Google Scholar] [CrossRef]
- Rieff, B.; Mathias, G.; Bauer, S.; Tavan, P. Density functional theory combined with molecular mechanics: The infrared spectra of flavin in solution. Photochem. Photobiol. 2011, 87, 511–523. [Google Scholar] [CrossRef]
- Klaumunzer, B.; Kroner, D.; Saalfrank, P. (TD-)DFT calculation of vibrational and vibronic spectra of riboflavin in solution. J. Phys. Chem. B 2010, 114, 10826–10834. [Google Scholar] [CrossRef]
- Rieff, B.; Bauer, S.; Mathias, G.; Tavan, P. IR spectra of flavins in solution: DFT/MM description of redox effects. J. Phys. Chem. B 2011, 115, 2117–2123. [Google Scholar] [CrossRef]
- Kabir, M.P.; Orozco-Gonzalez, Y.; Hastings, G.; Gozem, S. The effect of hydrogen-bonding on flavin’s infrared absorption spectrum. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 262, 120110. [Google Scholar] [CrossRef]
- Kabir, M.P.; Orozco-Gonzalez, Y.; Gozem, S. Electronic spectra of flavin in different redox and protonation states: A computational perspective on the effect of the electrostatic environment. Phys. Chem. Chem. Phys. 2019, 21, 16526–16537. [Google Scholar] [CrossRef]
- Gozem, S.; Krylov, A.I. The ezSpectra suite: An easy-to-use toolkit for spectroscopy modeling. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2022, 12, e1546. [Google Scholar] [CrossRef]
- Wu, M.; Eriksson, L.A. Absorption Spectra of Riboflavin—A Difficult Case for Computational Chemistry. J. Phys. Chem. A 2010, 114, 10234–10242. [Google Scholar] [CrossRef]
- Karasulu, B.; Götze, J.P.; Thiel, W. Assessment of Franck–Condon methods for computing vibrationally broadened UV–Vis absorption spectra of flavin derivatives: Riboflavin, roseoflavin, and 5-thioflavin. J. Chem. Theory Comput. 2014, 10, 5549–5566. [Google Scholar] [CrossRef]
- Davari, M.D.; Kopka, B.; Wingen, M.; Bocola, M.; Drepper, T.; Jaeger, K.-E.; Schwaneberg, U.; Krauss, U. Photophysics of the LOV-based fluorescent protein variant iLOV-Q489K determined by simulation and experiment. J. Phys. Chem. B 2016, 120, 3344–3352. [Google Scholar] [CrossRef]
- Climent, T.; González-Luque, R.; Merchán, M.; Serrano-Andrés, L. Theoretical insight into the spectroscopy and photochemistry of isoalloxazine, the flavin core ring. J. Phys. Chem. A 2006, 110, 13584–13590. [Google Scholar] [CrossRef]
- Thomas, M.; Brehm, M.; Fligg, R.; Vöhringer, P.; Kirchner, B. Computing vibrational spectra from ab initio molecular dynamics. Phys. Chem. Chem. Phys. 2013, 15, 6608–6622. [Google Scholar] [CrossRef] [PubMed]
- Nonella, M.; Mathias, G.; Tavan, P. Infrared spectrum of p-benzoquinone in water obtained from a QM/MM hybrid molecular dynamics simulation. J. Phys. Chem. A 2003, 107, 8638–8647. [Google Scholar] [CrossRef]
- Schwinn, K.; Ferré, N.; Huix-Rotllant, M. Efficient analytic second derivative of electrostatic embedding QM/MM energy: Normal mode analysis of plant cryptochrome. J. Chem. Theory Comput. 2020, 16, 3816–3824. [Google Scholar] [CrossRef] [PubMed]
- Groenhof, G. Introduction to QM/MM simulations. Methods Mol. Biol. 2013, 924, 43–66. [Google Scholar] [CrossRef]
- Senn, H.M.; Thiel, W. QM/MM methods for biomolecular systems. Angew. Chem. Int. Ed. 2009, 48, 1198–1229. [Google Scholar] [CrossRef] [PubMed]
- Warshel, A.; Levitt, M. Theoretical studies of enzymic reactions: Dielectric, electrostatic and steric stabilization of the carbonium ion in the reaction of lysozyme. J. Mol. Biol. 1976, 103, 227–249. [Google Scholar] [CrossRef]
- Dapprich, S.; Komáromi, I.; Byun, K.S.; Morokuma, K.; Frisch, M.J. A new ONIOM implementation in Gaussian98. Part I. The calculation of energies, gradients, vibrational frequencies and electric field derivatives1Dedicated to Professor Keiji Morokuma in celebration of his 65th birthday.1. J. Mol. Struct. Theochem. 1999, 461–462, 1–21. [Google Scholar] [CrossRef]
- Chung, L.W.; Sameera, W.; Ramozzi, R.; Page, A.J.; Hatanaka, M.; Petrova, G.P.; Harris, T.V.; Li, X.; Ke, Z.; Liu, F. The ONIOM method and its applications. Chem. Rev. 2015, 115, 5678–5796. [Google Scholar] [CrossRef] [PubMed]
- Palivec, V.; Kopecky, V., Jr.; Jungwirth, P.; Bour, P.; Kaminsky, J.; Martinez-Seara, H. Simulation of Raman and Raman optical activity of saccharides in solution. Phys. Chem. Chem. Phys. 2020, 22, 1983–1993. [Google Scholar] [CrossRef]
- Rappé, A.K.; Bormann-Rochotte, L.M.; Wiser, D.C.; Hart, J.R.; Pietsch, M.A.; Casewit, C.J.; Skiff, W.M. APT a next generation QM-based reactive force field model. Mol. Phys. 2007, 105, 301–324. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Vreven, T.; Mennucci, B.; da Silva, C.O.; Morokuma, K.; Tomasi, J. The ONIOM-PCM method: Combining the hybrid molecular orbital method and the polarizable continuum model for solvation. Application to the geometry and properties of a merocyanine in solution. J. Chem. Phys. 2001, 115, 62–72. [Google Scholar] [CrossRef]
- Herbert, J.M. Dielectric continuum methods for quantum chemistry. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2021, 11, e1519. [Google Scholar] [CrossRef]
- Jamróz, M.H. Vibrational Energy Distribution Analysis (VEDA): Scopes and limitations. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2013, 114, 220–230. [Google Scholar] [CrossRef]
- Abe, M.; Kyogoku, Y.; Kitagawa, T.; Kawano, K.; Ohishi, N.; Takai-Suzuki, A.; Yagi, K. lnfrared spectra and molecular association of lumiflavin and riboflavin derivatives. Spectrochim. Acta Part A Mol. Spectroscopy 1986, 42, 1059–1068. [Google Scholar] [CrossRef]
- Martin, C.B.; Tsao, M.-L.; Hadad, C.M.; Platz, M.S. The Reaction of Triplet Flavin with Indole. A Study of the Cascade of Reactive Intermediates Using Density Functional Theory and Time Resolved Infrared Spectroscopy. J. Am. Chem. Soc. 2002, 124, 7226–7234. [Google Scholar] [CrossRef]
- Salomon-Ferrer, R.; Case, D.A.; Walker, R.C. An overview of the Amber biomolecular simulation package. WIREs Comput. Mol. Sci. 2013, 3, 198–210. [Google Scholar] [CrossRef]
- Case, D.A.; Cheatham III, T.E.; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M., Jr.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Wang, J.; Wang, W.; Kollman, P.A.; Case, D.A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006, 25, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Jakalian, A.; Jack, D.B.; Bayly, C.I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 2002, 23, 1623–1641. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16 Rev. C.01; Wallingford, CT, USA, 2016. [Google Scholar]
- Demichelis, R.; Civalleri, B.; Ferrabone, M.; Dovesi, R. On the performance of eleven DFT functionals in the description of the vibrational properties of aluminosilicates. Int. J. Quantum Chem. 2010, 110, 406–415. [Google Scholar] [CrossRef]
- Neugebauer, J.; Reiher, M.; Hess, B.A. Coupled-cluster Raman intensities: Assessment and comparison with multiconfiguration and density functional methods. J. Chem. Phys. 2002, 117, 8623–8633. [Google Scholar] [CrossRef]
- Halls, M.D.; Velkovski, J.; Schlegel, H.B. Harmonic frequency scaling factors for Hartree-Fock, S-VWN, B-LYP, B3-LYP, B3-PW91 and MP2 with the Sadlej pVTZ electric property basis set. Theor. Chem. Acc. 2001, 105, 413–421. [Google Scholar] [CrossRef]
- Jimenez-Hoyos, C.A.; Janesko, B.G.; Scuseria, G.E. Evaluation of range-separated hybrid density functionals for the prediction of vibrational frequencies, infrared intensities, and Raman activities. Phys. Chem. Chem. Phys. 2008, 10, 6621–6629. [Google Scholar] [CrossRef] [PubMed]
- Zvereva, E.E.; Shagidullin, A.R.; Katsyuba, S.A. Ab initio and DFT predictions of infrared intensities and Raman activities. J. Phys. Chem. A 2011, 115, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Tazhigulov, R.N.; Gurunathan, P.K.; Kim, Y.; Slipchenko, L.V.; Bravaya, K.B. Polarizable embedding for simulating redox potentials of biomolecules. Phys. Chem. Chem. Phys. 2019, 21, 11642–11650. [Google Scholar] [CrossRef]
- Karnaukh, E.A.; Bravaya, K.B. The redox potential of a heme cofactor in Nitrosomonas europaea cytochrome c peroxidase: A polarizable QM/MM study. Phys. Chem. Chem. Phys. 2021, 23, 16506–16515. [Google Scholar] [CrossRef]
- Tazhigulov, R.N.; Bravaya, K.B. Free energies of redox half-reactions from first-principles calculations. J. Phys. Chem. Lett. 2016, 7, 2490–2495. [Google Scholar] [CrossRef]
- Provorse Long, M.R.; Isborn, C.M. Combining explicit quantum solvent with a polarizable continuum model. J. Phys. Chem. B 2017, 121, 10105–10117. [Google Scholar] [CrossRef]
- Ikonnikov, E.; Paolino, M.; Garcia-Alvarez, J.; Orozco-Gonzalez, Y.; Granados, C.; Röder, A.; Léonard, J.; Olivucci, M.; Haacke, S.; Kornilov, O. Photoelectron Spectroscopy of Oppositely Charged Molecular Switches in the Aqueous Phase: Theory and Experiment. J. Phys. Chem. Lett. 2023, 14, 6061–6070. [Google Scholar] [CrossRef] [PubMed]
- Dratch, B.D.; Orozco-Gonzalez, Y.; Gadda, G.; Gozem, S. Ionic atmosphere effect on the absorption spectrum of a flavoprotein: A reminder to consider solution ions. J. Phys. Chem. Lett. 2021, 12, 8384–8396. [Google Scholar] [CrossRef] [PubMed]
- Barrozo, A.; Xu, B.; Gunina, A.O.; Jacobs, M.I.; Wilson, K.; Kostko, O.; Ahmed, M.; Krylov, A.I. To be or not to be a molecular ion: The role of the solvent in photoionization of arginine. J. Phys. Chem. Lett. 2019, 10, 1860–1865. [Google Scholar] [CrossRef] [PubMed]
- Makita, H.; Rohani, L.; Zhao, N.; Hastings, G. Quinones in the A1 binding site in photosystem I studied using time-resolved FTIR difference spectroscopy. Biochim. Biophys. Acta (BBA)-Bioenerg. 2017, 1858, 804–813. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le, D.P.N.; Hastings, G.; Gozem, S. How Aqueous Solvation Impacts the Frequencies and Intensities of Infrared Absorption Bands in Flavin: The Quest for a Suitable Solvent Model. Molecules 2024, 29, 520. https://doi.org/10.3390/molecules29020520
Le DPN, Hastings G, Gozem S. How Aqueous Solvation Impacts the Frequencies and Intensities of Infrared Absorption Bands in Flavin: The Quest for a Suitable Solvent Model. Molecules. 2024; 29(2):520. https://doi.org/10.3390/molecules29020520
Chicago/Turabian StyleLe, D. P. Ngan, Gary Hastings, and Samer Gozem. 2024. "How Aqueous Solvation Impacts the Frequencies and Intensities of Infrared Absorption Bands in Flavin: The Quest for a Suitable Solvent Model" Molecules 29, no. 2: 520. https://doi.org/10.3390/molecules29020520
APA StyleLe, D. P. N., Hastings, G., & Gozem, S. (2024). How Aqueous Solvation Impacts the Frequencies and Intensities of Infrared Absorption Bands in Flavin: The Quest for a Suitable Solvent Model. Molecules, 29(2), 520. https://doi.org/10.3390/molecules29020520





