Molecular Characterization of the Recombinant Ig1 Axl Receptor Domain: An Intriguing Bait for Screening in Drug Discovery
Abstract
1. Introduction
2. Results
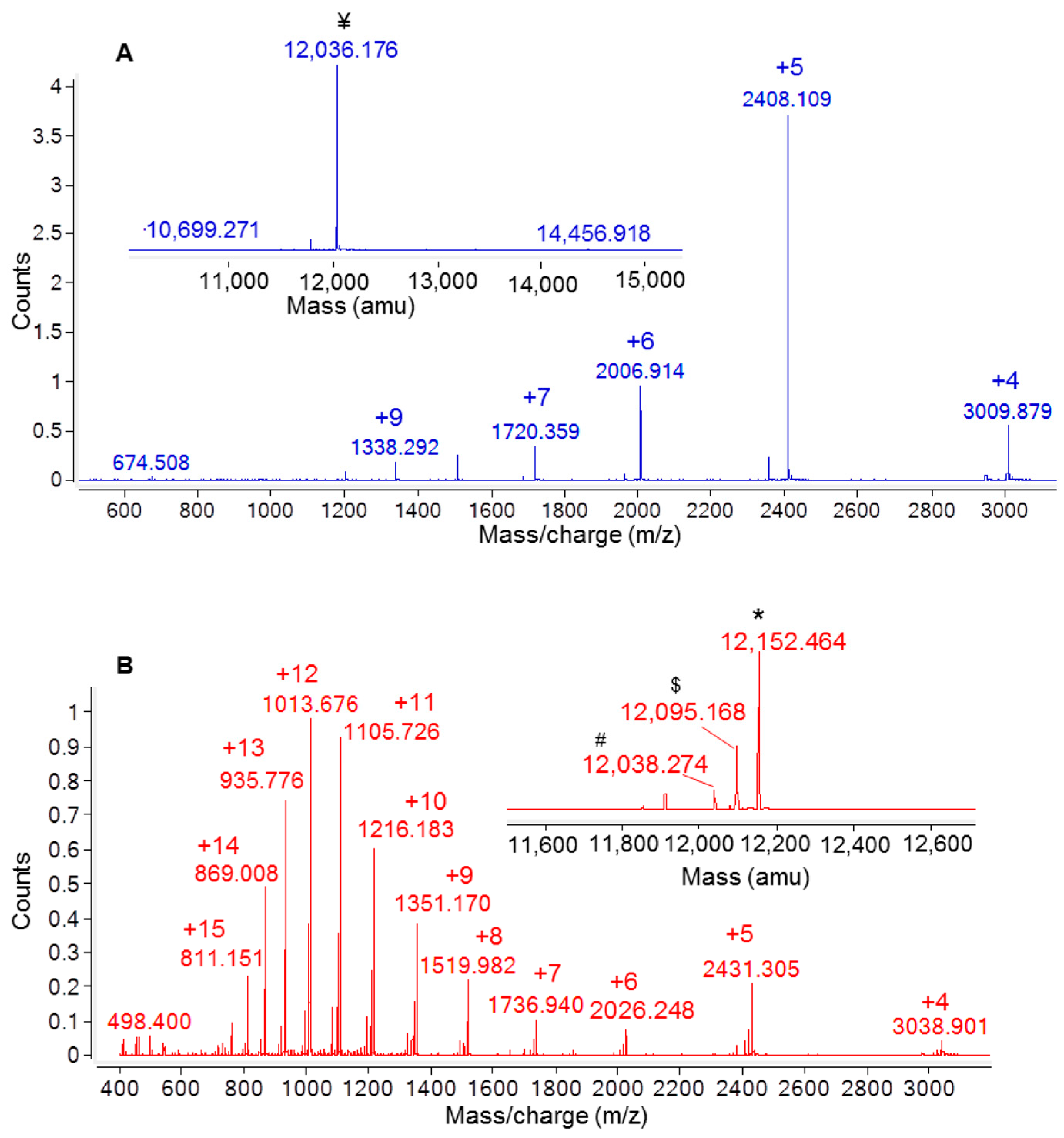
2.1. Expression and Purification of the His-Tagged Recombinant Ig1 Axl Protein
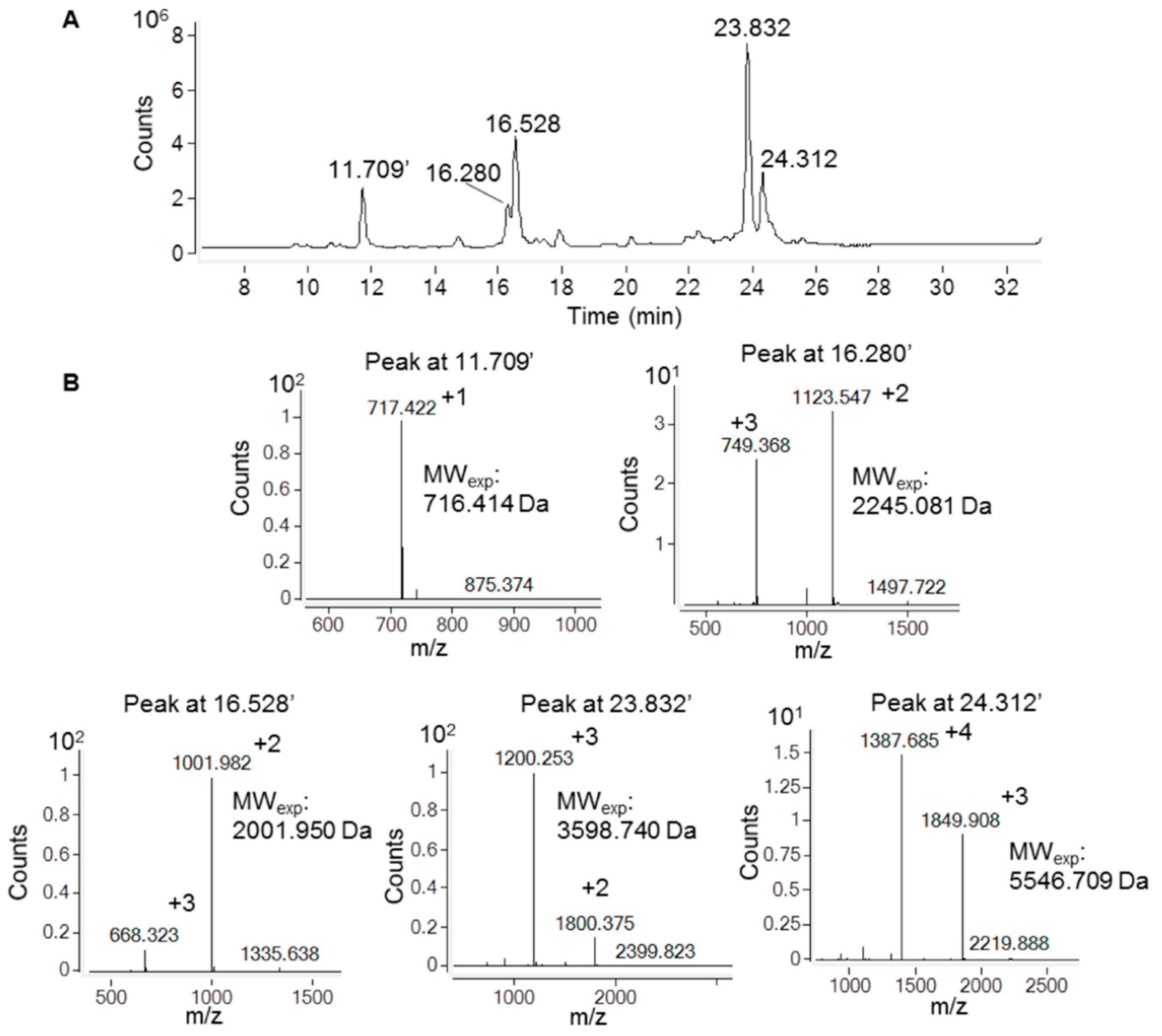
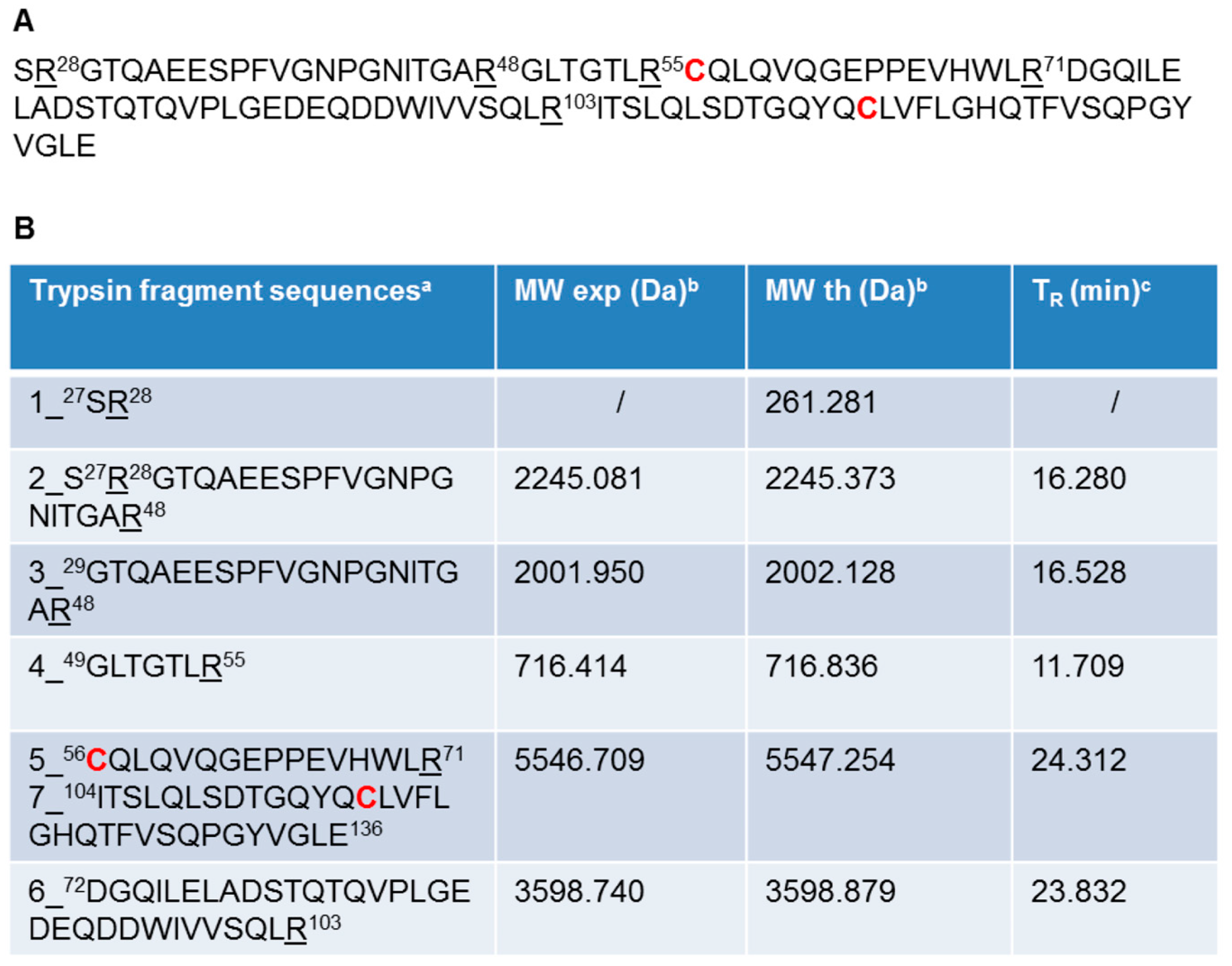
2.2. Recombinant Ig1 Axl Protein Disulfide Bond Analysis
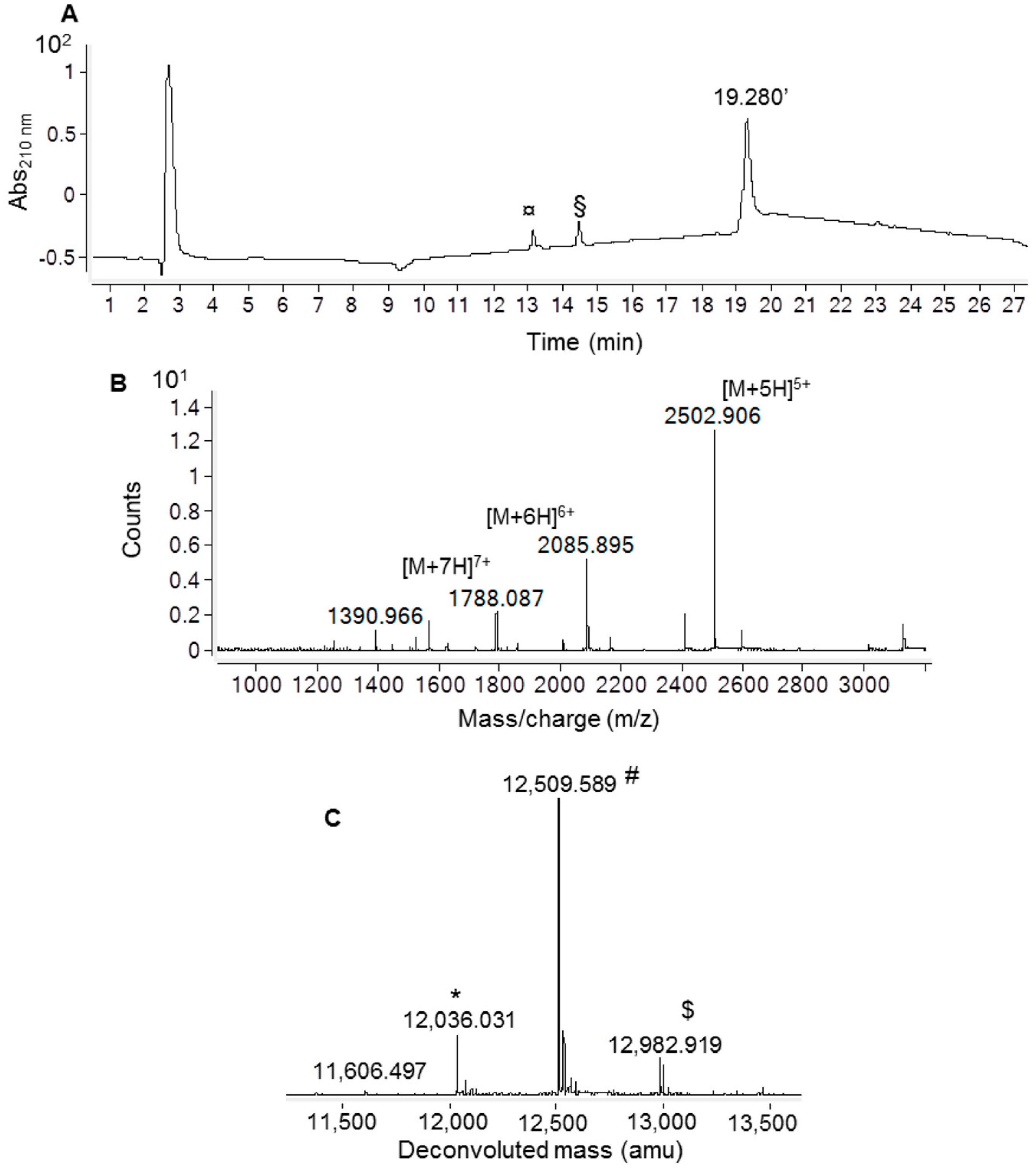
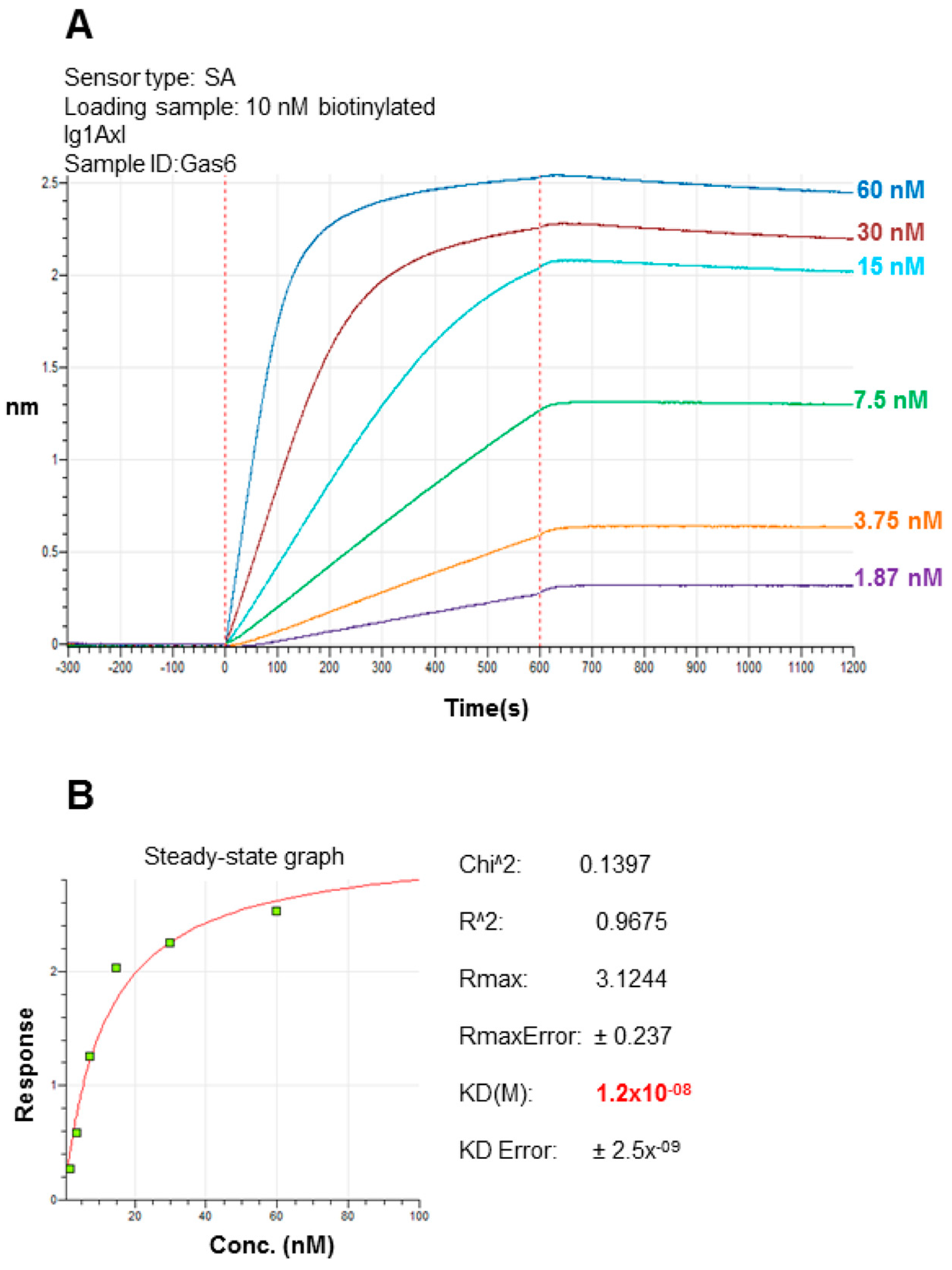
2.3. Recombinant Ig1 Axl-Gas6 Binding Analysis
2.4. Spectroscopic Characterization of the Recombinant Ig1 Axl Protein
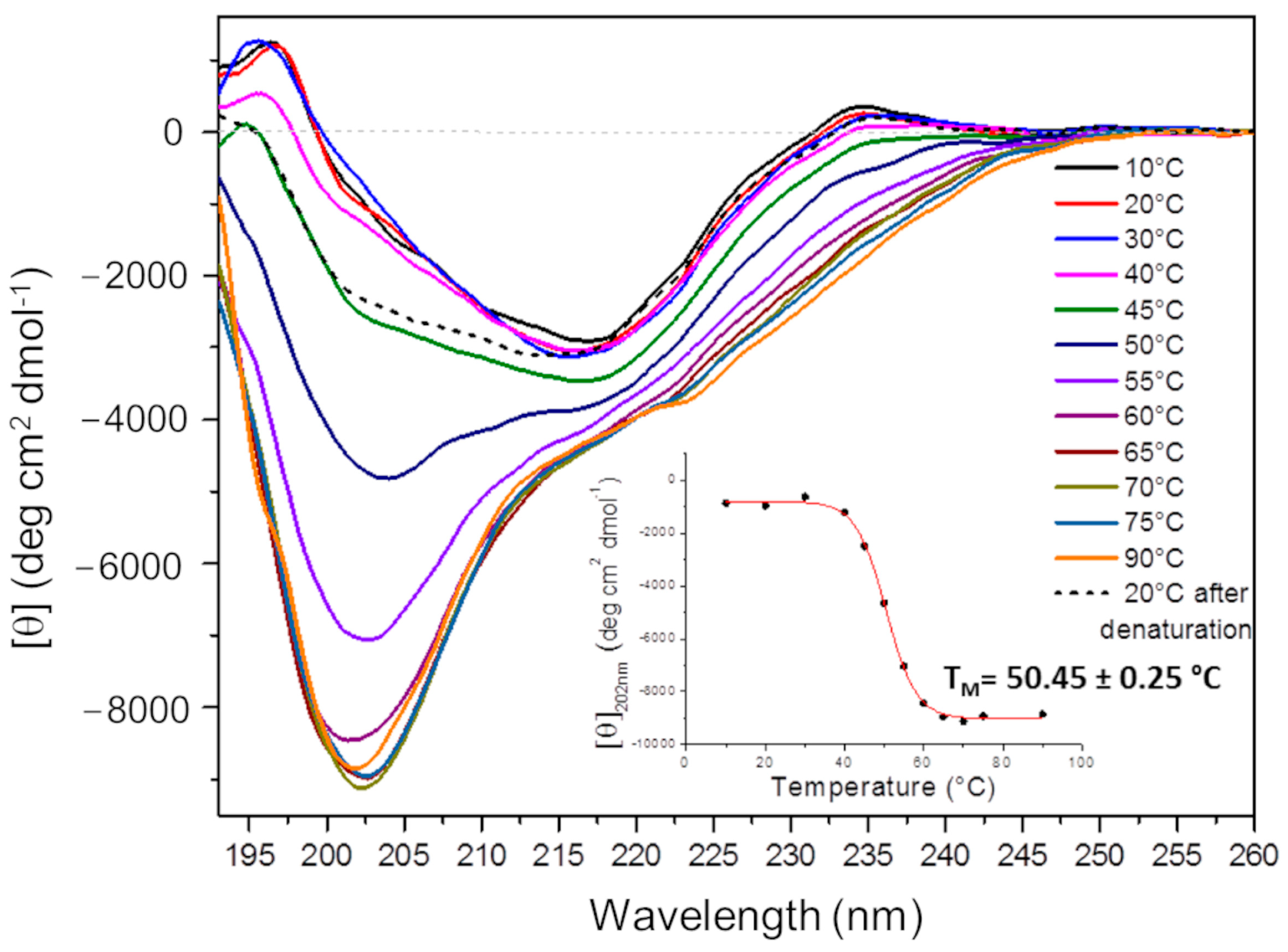
2.4.1. Circular Dichroism (CD) Analysis
2.4.2. Steady-State Fluorescence Studies
3. Discussion
4. Materials and Methods
4.1. Reagents, Strains, and Plasmids
4.2. Ig1 Axl Protein Expression, Refolding, and Purification
4.3. Ig1 Axl Iodoacetamide Alkylation and Trypsin Digestion
4.4. In Vitro Ig1 Axl/Gas6 Binding Assay using Biolayer Interferometry (BLI)
4.5. Recombinant Ig1 Axl Protein Circular Dichroism (CD) Analysis
4.6. Fluorescence Spectroscopy Analysis of the Recombinant Ig1 Axl Protein
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tsou, W.I.; Nguyen, K.Q.; Calarese, D.A.; Garforth, S.J.; Antes, A.L.; Smirnov, S.V.; Almo, S.C.; Birge, R.B.; Kotenko, S.V. Receptor tyrosine kinases, TYRO3, AXL, and MER, demonstrate distinct patterns and complex regulation of ligand-induced activation. J. Biol. Chem. 2014, 289, 25750–25763. [Google Scholar] [CrossRef] [PubMed]
- Lew, E.D.; Oh, J.; Burrola, P.G.; Lax, I.; Zagorska, A.; Traves, P.G.; Schlessinger, J.; Lemke, G. Differential TAM receptor-ligand-phospholipid interactions delimit differential TAM bioactivities. eLife 2014, 3, e03385. [Google Scholar] [CrossRef] [PubMed]
- Wium, M.; Paccez, J.D.; Zerbini, L.F. The Dual Role of TAM Receptors in Autoimmune Diseases and Cancer: An Overview. Cells 2018, 7, 166. [Google Scholar] [CrossRef] [PubMed]
- Bellan, M.; Pirisi, M.; Sainaghi, P.P. The Gas6/TAM System and Multiple Sclerosis. Int. J. Mol. Sci. 2016, 17, 1807. [Google Scholar] [CrossRef] [PubMed]
- Meertens, L.; Labeau, A.; Dejarnac, O.; Cipriani, S.; Sinigaglia, L.; Bonnet-Madin, L.; Le Charpentier, T.; Hafirassou, M.L.; Zamborlini, A.; Cao-Lormeau, V.M.; et al. Axl Mediates ZIKA Virus Entry in Human Glial Cells and Modulates Innate Immune Responses. Cell Rep. 2017, 18, 324–333. [Google Scholar] [CrossRef]
- Shimojima, M.; Ikeda, Y.; Kawaoka, Y. The mechanism of Axl-mediated Ebola virus infection. J. Infect. Dis. 2007, 196 (Suppl. 2), S259–S263. [Google Scholar] [CrossRef]
- Nakano, T.; Tani, M.; Ishibashi, Y.; Kimura, K.; Park, Y.B.; Imaizumi, N.; Tsuda, H.; Aoyagi, K.; Sasaki, H.; Ohwada, S.; et al. Biological properties and gene expression associated with metastatic potential of human osteosarcoma. Clin. Exp. Metastasis 2003, 20, 665–674. [Google Scholar] [CrossRef]
- Graham, D.K.; DeRyckere, D.; Davies, K.D.; Earp, H.S. The TAM family: Phosphatidylserine sensing receptor tyrosine kinases gone awry in cancer. Nat. Rev. Cancer 2014, 14, 769–785. [Google Scholar] [CrossRef]
- Gjerdrum, C.; Tiron, C.; Hoiby, T.; Stefansson, I.; Haugen, H.; Sandal, T.; Collett, K.; Li, S.; McCormack, E.; Gjertsen, B.T.; et al. Axl is an essential epithelial-to-mesenchymal transition-induced regulator of breast cancer metastasis and patient survival. Proc. Natl. Acad. Sci. USA 2010, 107, 1124–1129. [Google Scholar] [CrossRef]
- Yamagata, M.; Sanes, J.R.; Weiner, J.A. Synaptic adhesion molecules. Curr. Opin. Cell Biol. 2003, 15, 621–632. [Google Scholar] [CrossRef]
- Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 2000, 103, 211–225. [Google Scholar] [CrossRef] [PubMed]
- Mark, M.R.; Chen, J.; Hammonds, R.G.; Sadick, M.; Godowsk, P.J. Characterization of Gas6, a member of the superfamily of G domain-containing proteins, as a ligand for Rse and Axl. J. Biol. Chem. 1996, 271, 9785–9789. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, K.; Nagata, K.; Ohashi, K.; Nakano, T.; Arita, H.; Mizuno, K. Roles of gamma-carboxylation and a sex hormone-binding globulin-like domain in receptor-binding and in biological activities of Gas6. FEBS Lett. 1997, 408, 306–310. [Google Scholar] [CrossRef]
- Sasaki, T.; Knyazev, P.G.; Clout, N.J.; Cheburkin, Y.; Gohring, W.; Ullrich, A.; Timpl, R.; Hohenester, E. Structural basis for Gas6-Axl signalling. EMBO J. 2006, 25, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Di Stasi, R.; De Rosa, L.; D’Andrea, L.D. Therapeutic aspects of the Axl/Gas6 molecular system. Drug Discov. Today 2020, 25, 2130–2148. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Wei, Y.; Wei, X. AXL receptor tyrosine kinase as a promising anti-cancer approach: Functions, molecular mechanisms and clinical applications. Mol. Cancer 2019, 18, 153. [Google Scholar] [CrossRef] [PubMed]
- Bhalla, S.; Gerber, D.E. AXL Inhibitors: Status of Clinical Development. Curr. Oncol. Rep. 2023, 25, 521–529. [Google Scholar] [CrossRef]
- Kim, J.H.; Koh, B.; Ahn, D.G.; Lee, S.J.; Park, T.J.; Park, J.P. A screening study of high affinity peptide as molecular binder for AXL, tyrosine kinase receptor involving in Zika virus entry. Bioelectrochemistry 2021, 137, 107670. [Google Scholar] [CrossRef]
- Roskoski, R., Jr. Properties of FDA-approved small molecule protein kinase inhibitors: A 2020 update. Pharmacol. Res. 2020, 152, 104609. [Google Scholar] [CrossRef]
- Cruz, E.; Kayser, V. Monoclonal antibody therapy of solid tumors: Clinical limitations and novel strategies to enhance treatment efficacy. Biologics 2019, 13, 33–51. [Google Scholar] [CrossRef]
- Henninot, A.; Collins, J.C.; Nuss, J.M. The Current State of Peptide Drug Discovery: Back to the Future? J. Med. Chem. 2018, 61, 1382–1414. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, N.; Zhang, W.; Cheng, X.; Yan, Z.; Shao, G.; Wang, X.; Wang, R.; Fu, C. Therapeutic peptides: Current applications and future directions. Signal Transduct. Target. Ther. 2022, 7, 48. [Google Scholar] [CrossRef] [PubMed]
- Zaccai, N.; Jones, E.Y. Ig-Superfold and its Variable Uses in Molecular Recognition. In Handbook of Cell Signaling, 1st ed.; Bradshaw, R.A., Dennis, E.A., Eds.; Academic Press: Cambridge, MA, USA, 2003; Volume 1, pp. 57–62. [Google Scholar]
- De Rosa, L.; Di Stasi, R.; Romanelli, A.; D’Andrea, L.D. Exploiting Protein N-Terminus for Site-Specific Bioconjugation. Molecules 2021, 26, 3521. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef] [PubMed]
- Rucker, A.L.; Creamer, T.P. Polyproline II helical structure in protein unfolded states: Lysine peptides revisited. Protein Sci. 2002, 11, 980–985. [Google Scholar] [CrossRef] [PubMed]
- Provencher, S.W.; Glockner, J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry 1981, 20, 33–37. [Google Scholar] [CrossRef]
- Manavalan, P.; Johnson, W.C., Jr. Variable selection method improves the prediction of protein secondary structure from circular dichroism spectra. Anal. Biochem. 1987, 167, 76–85. [Google Scholar] [CrossRef]
- Sreerama, N.; Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000, 287, 252–260. [Google Scholar] [CrossRef]
- Gay, C.M.; Balaji, K.; Byers, L.A. Giving AXL the axe: Targeting AXL in human malignancy. Br. J. Cancer 2017, 116, 415–423. [Google Scholar] [CrossRef]
- Kariolis, M.S.; Miao, Y.R.; Jones, D.S., 2nd; Kapur, S.; Mathews, I.I.; Giaccia, A.J.; Cochran, J.R. An engineered Axl ‘decoy receptor’ effectively silences the Gas6-Axl signaling axis. Nat. Chem. Biol. 2014, 10, 977–983. [Google Scholar] [CrossRef]
- Di Stasi, R.; Diana, D.; Capasso, D.; Palumbo, R.; Romanelli, A.; Pedone, C.; Fattorusso, R.; D’Andrea, L.D. VEGFR1(D2) in drug discovery: Expression and molecular characterization. Biopolymers 2010, 94, 800–809. [Google Scholar] [CrossRef] [PubMed]
- Di Stasi, R.; Diana, D.; De Rosa, L.; Fattorusso, R.; D’Andrea, L.D. Biochemical and Conformational Characterization of Recombinant VEGFR2 Domain 7. Mol. Biotechnol. 2019, 61, 860–872. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat. Protoc. 2006, 1, 2527–2535. [Google Scholar] [CrossRef] [PubMed]


| Algorithm Used | % Helix | % Sheet | % Turn | % Unordered |
|---|---|---|---|---|
| Selcon3, 20 °C | 10.5 | 36 | 16 | 37.5 |
| Continll, 20 °C | 3.6 | 37.2 | 20.9 | 38.3 |
| CDSSTR, 20 °C | 3.5 | 38.3 | 19.9 | 38.3 |
| Selcon3, 90 °C | 10.2 | 19.6 | 13.6 | 56.6 |
| Continll, 90 °C | 7.5 | 15.8 | 12.8 | 63.9 |
| CDSSTR, 90 °C | 7.7 | 13.9 | 11.4 | 66 |
| Selcon3, 20 °C * | 3.1 | 28.1 | 12.1 | 56.7 |
| Continll, 20 °C * | 6.6 | 28.2 | 19.2 | 46 |
| CDSSTR, 20 °C * | 14 | 25.9 | 18.5 | 41.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Stasi, R.; De Rosa, L.; Izzi, G.; D’Andrea, L.D. Molecular Characterization of the Recombinant Ig1 Axl Receptor Domain: An Intriguing Bait for Screening in Drug Discovery. Molecules 2024, 29, 521. https://doi.org/10.3390/molecules29020521
Di Stasi R, De Rosa L, Izzi G, D’Andrea LD. Molecular Characterization of the Recombinant Ig1 Axl Receptor Domain: An Intriguing Bait for Screening in Drug Discovery. Molecules. 2024; 29(2):521. https://doi.org/10.3390/molecules29020521
Chicago/Turabian StyleDi Stasi, Rossella, Lucia De Rosa, Guido Izzi, and Luca Domenico D’Andrea. 2024. "Molecular Characterization of the Recombinant Ig1 Axl Receptor Domain: An Intriguing Bait for Screening in Drug Discovery" Molecules 29, no. 2: 521. https://doi.org/10.3390/molecules29020521
APA StyleDi Stasi, R., De Rosa, L., Izzi, G., & D’Andrea, L. D. (2024). Molecular Characterization of the Recombinant Ig1 Axl Receptor Domain: An Intriguing Bait for Screening in Drug Discovery. Molecules, 29(2), 521. https://doi.org/10.3390/molecules29020521






