Molecular Periphery Design Allows Control of the New Nitrofurans Antimicrobial Selectivity
Abstract
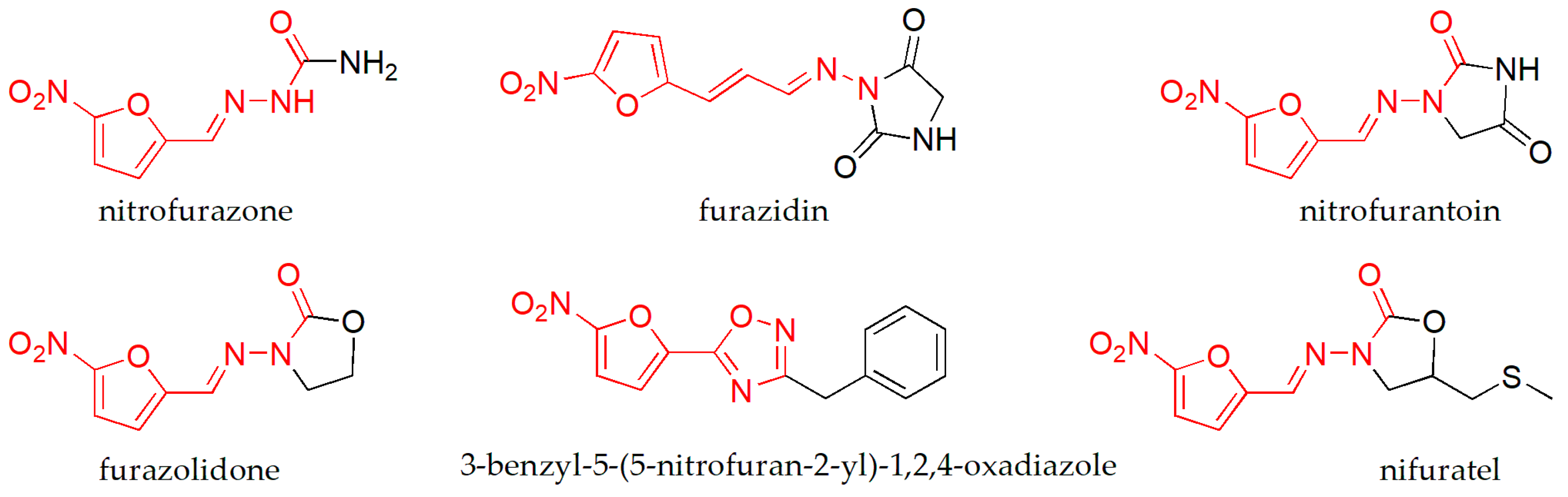
1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Antibacterial Activity
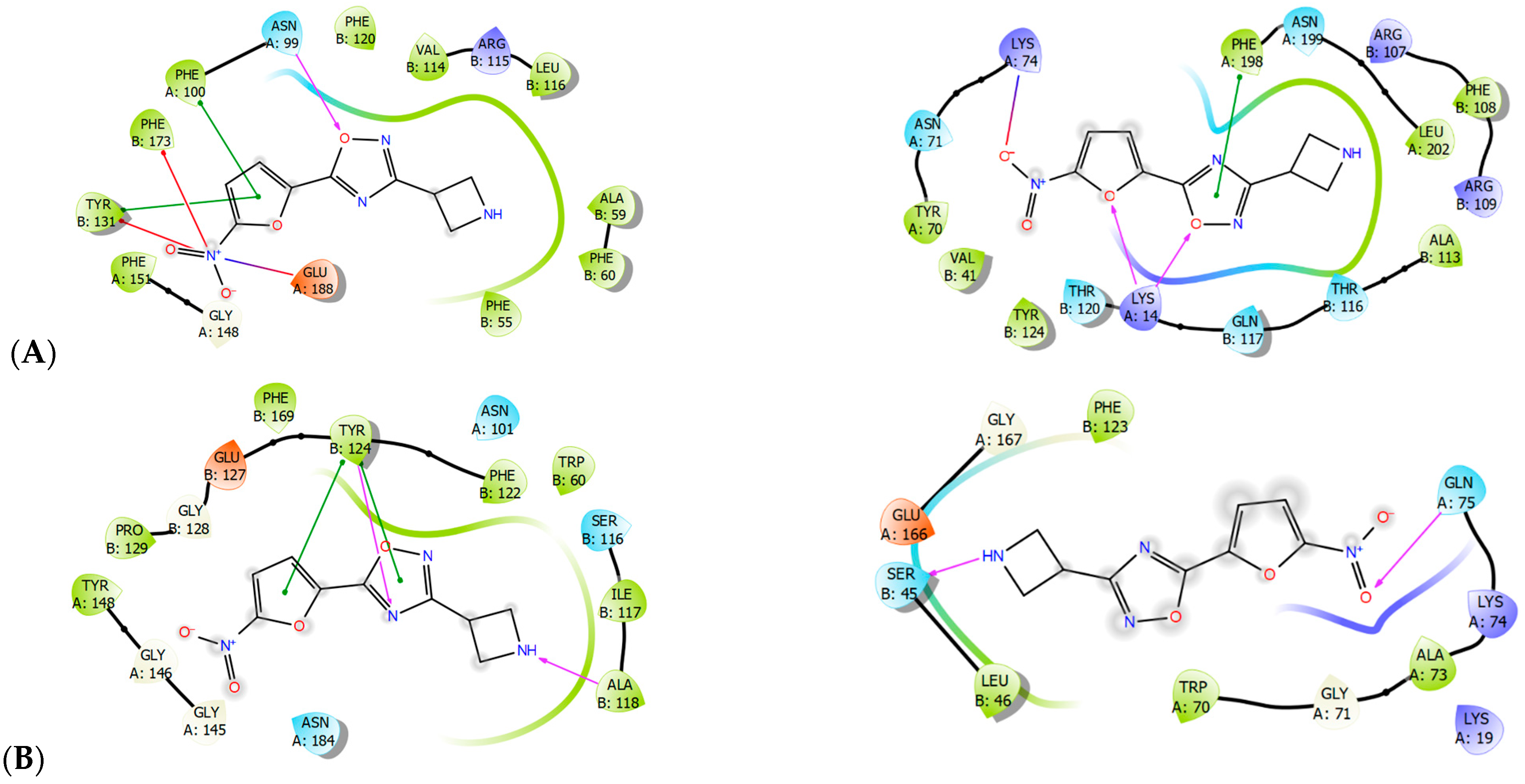
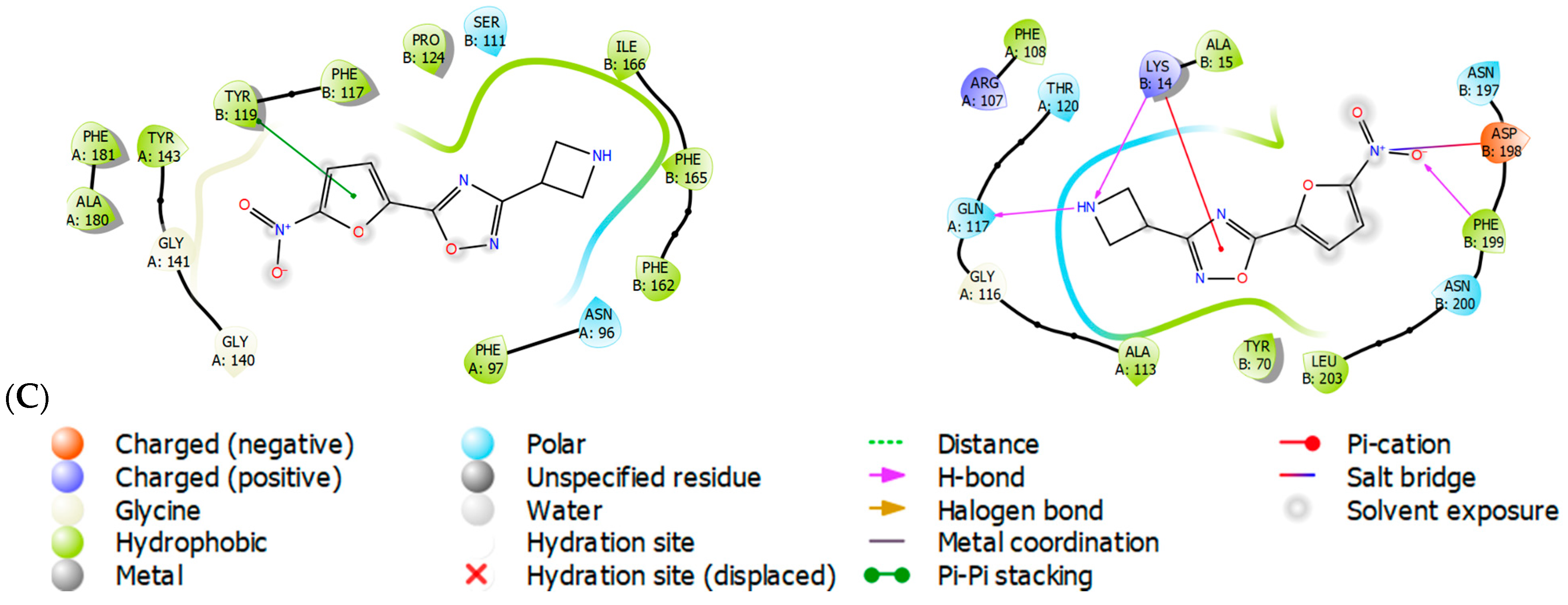
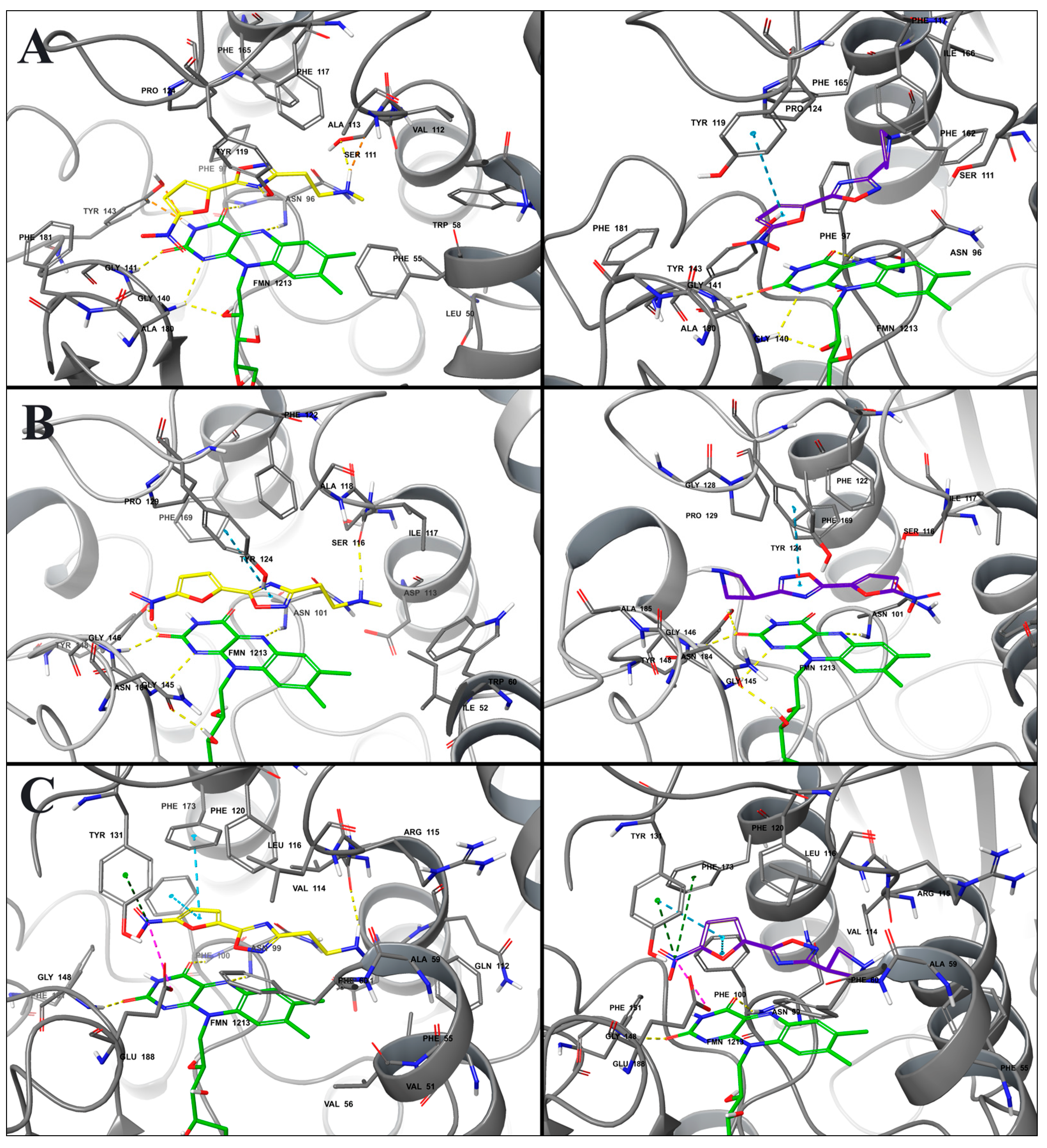
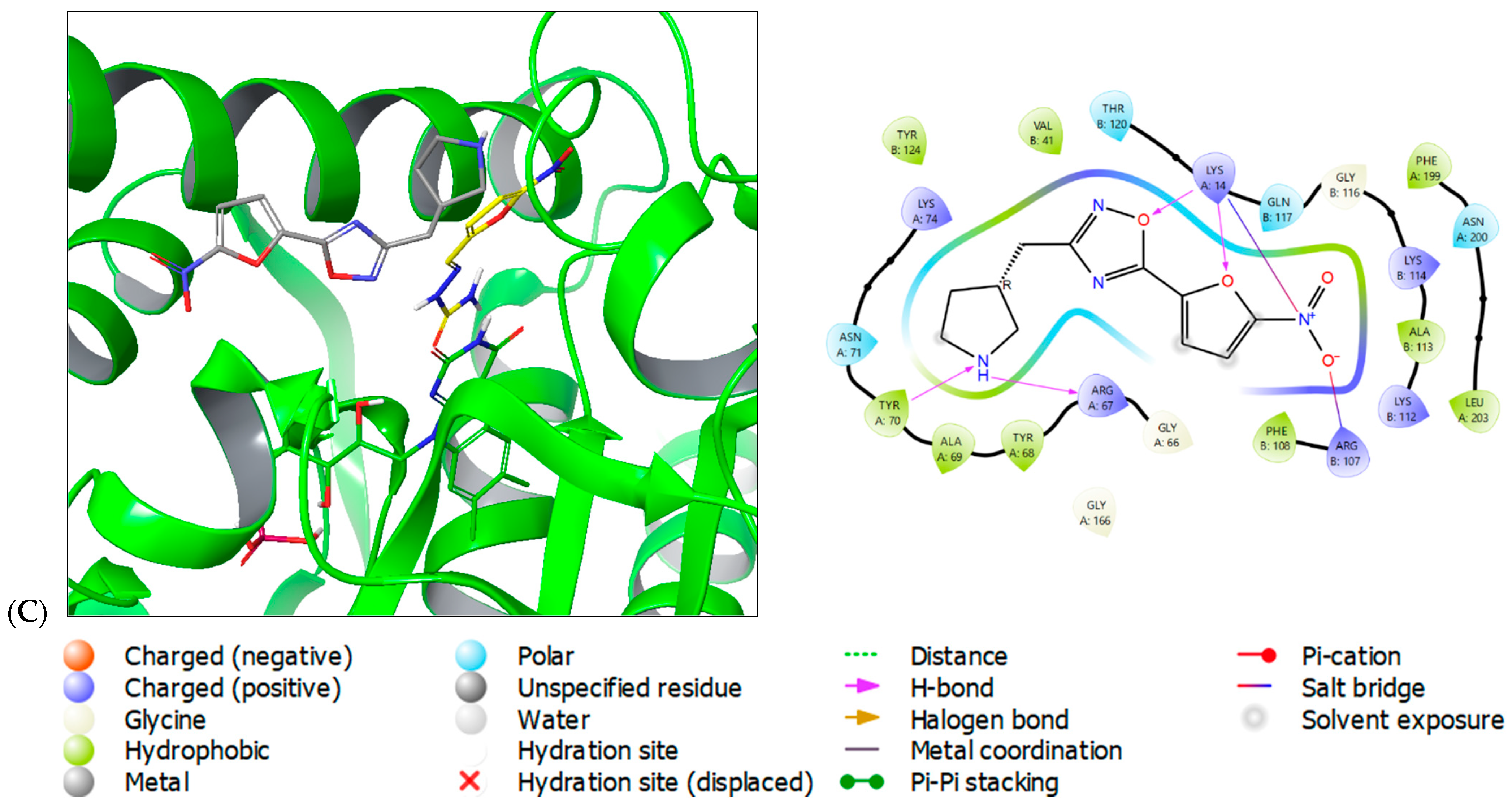
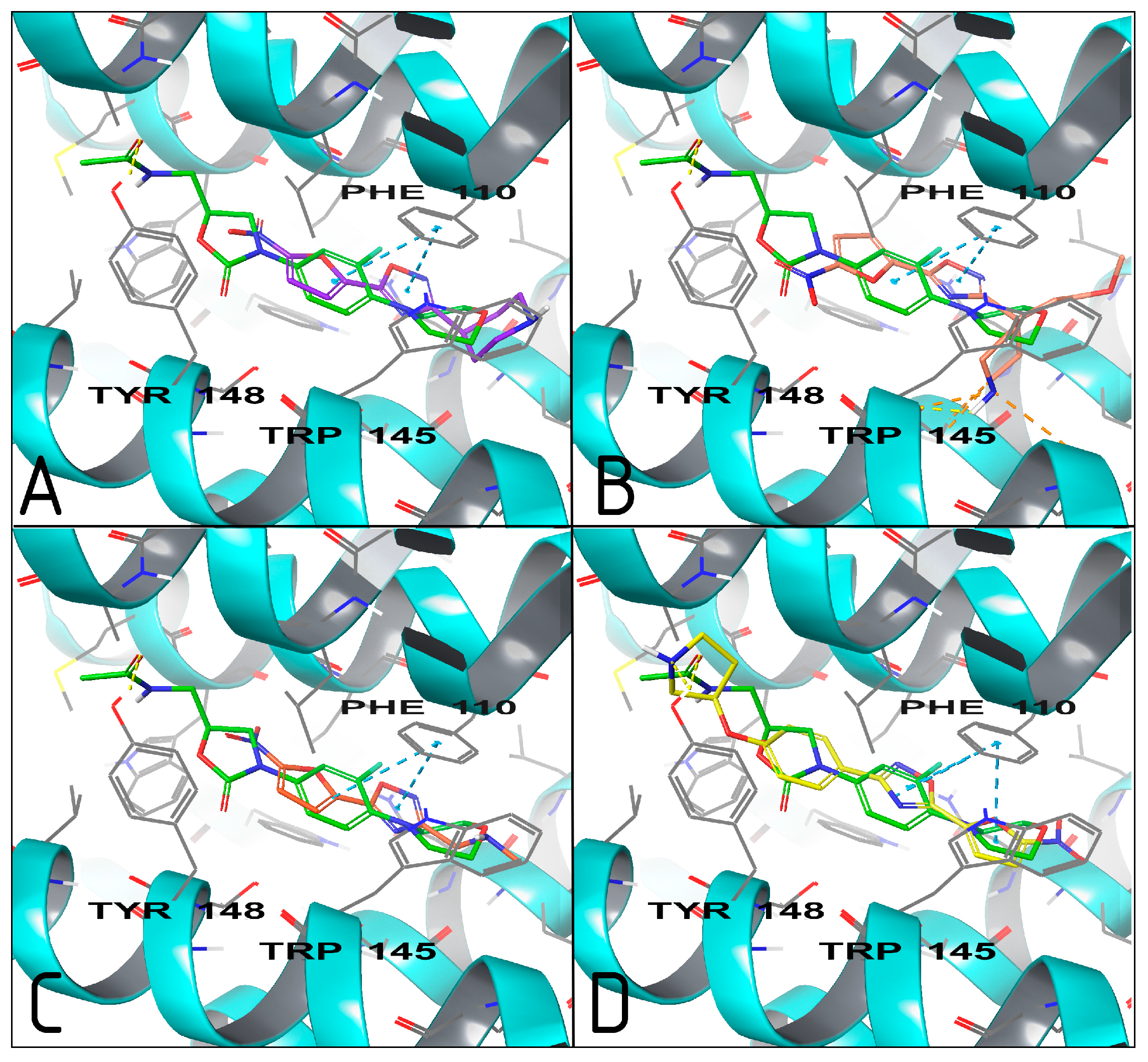
2.3. Molecular Modeling
- (1)
- The HTH-type transcriptional regulator EthR interacts with linezolid, as indicated by Pdb: 5NZ0. Inhibition of EthR has been shown to enhance the effectiveness of antibiotics and reduce drug resistance [29];
- (2)
- Arylamine N-acetyltransferase (TBNAT, Pdb: 4BGF) is responsible for the intracellular survival of M. tuberculosis within macrophages. It has been identified as a promising target [30,31]. In 2020, it was discovered that nitrofurans could inhibit it. Prior to this, there had been limited research on the subject [32].
- (3)
3. Materials and Methods
3.1. Chemistry
3.1.1. General Procedure for the Synthesis of Compounds 1a–m
- tert-butyl 4-[(hydroxyamino)(imino)methyl]piperidine-1-carboxylate (1a)
- tert-butyl 3-[(hydroxyamino)(imino)methyl]piperidine-1-carboxylate (1b)
- tert-butyl [3-(hydroxyamino)-3-iminopropyl]methylcarbamate (1c)
- tert-butyl [2-(hydroxyamino)-2-iminoethyl]methylcarbamate (1d)
- tert-butyl 3-{4-[(hydroxyamino)(imino)methyl]phenoxy}pyrrolidine-1-carboxylate (1e)
- tert-butyl 3-{2-[(hydroxyamino)(imino)methyl]phenoxy}pyrrolidine-1-carboxylate (1f)
- tert-butyl 4-[(hydroxyamino)(imino)methyl]-4-(2-methoxyethyl)piperidine-1-carboxylate (1g)
- tert-butyl 3-[(hydroxyamino)(imino)methyl]azetidine-1-carboxylate (1h)
- tert-butyl 2-[(hydroxyamino)(imino)methyl]piperidine-1-carboxylate (1i)
- tert-butyl 4-[(hydroxyamino)(imino)methyl]-4-methylpiperidine-1-carboxylate (1j)
- tert-butyl 3-[2-(hydroxyamino)-2-iminoethyl]pyrrolidine-1-carboxylate (1k)
- tert-butyl 3-[(hydroxyamino)(imino)methyl]pyrrolidine-1-carboxylate (1l)
- tert-butyl 3-[(hydroxyamino)(imino)methyl]-3-(2-methoxyethyl)piperidine-1-carboxylate (1m)
3.1.2. General Procedure for the Synthesis of Compounds 2a–m
- 4-[5-(5-nitro-2-furyl)-1,2,4-oxadiazol-3-yl]piperidine hydrochloride (2a)
- 3-[5-(5-nitro-2-furyl)-1,2,4-oxadiazol-3-yl]piperidine hydrochloride (2b)
- N-methyl-2-[5-(5-nitro-2-furyl)-1,2,4-oxadiazol-3-yl]ethanamine hydrochloride (2c)
- N-methyl-1-[5-(5-nitro-2-furyl)-1,2,4-oxadiazol-3-yl]methanamine hydrochloride (2d)
- 5-(5-nitro-2-furyl)-3-[4-(pyrrolidin-3-yloxy)phenyl]-1,2,4-oxadiazole hydrochloride (2e)
- 5-(5-nitro-2-furyl)-3-[2-(pyrrolidin-3-yloxy)phenyl]-1,2,4-oxadiazole hydrochloride (2f)
- 4-(2-methoxyethyl)-4-[5-(5-nitro-2-furyl)-1,2,4-oxadiazol-3-yl]piperidine hydrochloride (2g)
- 3-azetidin-3-yl-5-(5-nitro-2-furyl)-1,2,4-oxadiazole hydrochloride (2h)
- 2-[5-(5-nitro-2-furyl)-1,2,4-oxadiazol-3-yl]piperidine hydrochloride (2i)
- 4-methyl-4-[5-(5-nitro-2-furyl)-1,2,4-oxadiazol-3-yl]piperidine hydrochloride (2j)
- 5-(5-nitro-2-furyl)-3-(pyrrolidin-3-ylmethyl)-1,2,4-oxadiazole hydrochloride (2k)
- 5-(5-nitro-2-furyl)-3-pyrrolidin-3-yl-1,2,4-oxadiazole hydrochloride (2l)
- 3-(2-methoxyethyl)-4-[5-(5-nitro-2-furyl)-1,2,4-oxadiazol-3-yl]piperidine hydrochloride (2m)
3.2. Biological Activity Evaluation
3.3. In Silico Studies
3.3.1. Target Selection
3.3.2. Protein and Ligand Structure Preparation
3.3.3. Induced-Fit Docking of Molecules
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Akram, F.; Imtiaz, M.; Haq, I.U. Emergent crisis of antibiotic resistance: A silent pandemic threat to 21(st) century. Microb. Pathog. 2023, 174, 105923. [Google Scholar] [CrossRef] [PubMed]
- Miethke, M.; Pieroni, M.; Weber, T.; Brönstrup, M.; Hammann, P.; Halby, L.; Arimondo, P.B.; Glaser, P.; Aigle, B.; Bode, H.B.; et al. Towards the sustainable discovery and development of new antibiotics. Nat. Rev. Chem. 2021, 5, 726–749. [Google Scholar] [CrossRef] [PubMed]
- Paharik, A.E.; Schreiber, H.L.t.; Spaulding, C.N.; Dodson, K.W.; Hultgren, S.J. Narrowing the spectrum: The new frontier of precision antimicrobials. Genome Med. 2017, 9, 110. [Google Scholar] [CrossRef]
- Rogacheva, E.; Kraeva, L.; Lukin, A.; Vinogradova, L.; Komarova, K.; Chudinov, M.; Gureev, M.; Chupakhin, E. 5-Nitrofuran-Tagged Oxazolyl Pyrazolopiperidines: Synthesis and Activity against ESKAPE Pathogens. Molecules 2023, 28, 6491. [Google Scholar] [CrossRef] [PubMed]
- Krasavin, M.; Lukin, A.; Vedekhina, T.; Manicheva, O.; Dogonadze, M.; Vinogradova, T.; Zabolotnykh, N.; Rogacheva, E.; Kraeva, L.; Yablonsky, P. Conjugation of a 5-nitrofuran-2-oyl moiety to aminoalkylimidazoles produces non-toxic nitrofurans that are efficacious in vitro and in vivo against multidrug-resistant Mycobacterium tuberculosis. Eur. J. Med. Chem. 2018, 157, 1115–1126. [Google Scholar] [CrossRef]
- Vedekhina, T.S.; Chudinov, M.V.; Lukin, A.Y. Design and synthesis of 4-nitroimidazole derivatives with potential antitubercular activity. Fine Chem. Technol. 2023, 18, 219–229. [Google Scholar] [CrossRef]
- Ryan, A.; Kaplan, E.; Laurieri, N.; Lowe, E.; Sim, E. Activation of nitrofurazone by azoreductases: Multiple activities in one enzyme. Sci. Rep. 2011, 1, 63. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H. Remarkable diversification of bacterial azoreductases: Primary sequences, structures, substrates, physiological roles, and biotechnological applications. Appl. Microbiol. Biotechnol. 2019, 103, 3965–3978. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, P.S.; Sisson, G.; Croxen, M.A.; Welch, K.; Harman, W.D.; Cremades, N.; Morash, M.G. Antiparasitic drug nitazoxanide inhibits the pyruvate oxidoreductases of Helicobacter pylori, selected anaerobic bacteria and parasites, and Campylobacter jejuni. Antimicrob. Agents Chemother. 2007, 51, 868–876. [Google Scholar] [CrossRef]
- Munoz-Davila, M.J. Role of Old Antibiotics in the Era of Antibiotic Resistance. Highlighted Nitrofurantoin for the Treatment of Lower Urinary Tract Infections. Antibiotics 2014, 3, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Polycarpou-Schwarz, M.; Muller, K.; Denger, S.; Riddell, A.; Lewis, J.; Gannon, F.; Reid, G. Thanatop: A novel 5-nitrofuran that is a highly active, cell-permeable inhibitor of topoisomerase II. Cancer. Res. 2007, 67, 4451–4458. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Althagafy, H.S.; El-Aziz, M.K.A.; Ibrahim, I.M.; Abd-Alhameed, E.K.; Hassanein, E.H.M. Pharmacological updates of nifuroxazide: Promising preclinical effects and the underlying molecular mechanisms. Eur. J. Pharmacol. 2023, 951, 175776. [Google Scholar] [CrossRef]
- Bauer, A.W.; Kirby, W.M.M.; Sherris, J.C.; Turck, M. Antibiotic Susceptibility Testing by a Standardized Single Disk Method. Am. J. Clin. Pathol. 1966, 45, 493–496. [Google Scholar] [CrossRef] [PubMed]
- Zorzi, R.R.; Jorge, S.D.; Palace-Berl, F.; Pasqualoto, K.F.; Bortolozzo Lde, S.; de Castro Siqueira, A.M.; Tavares, L.C. Exploring 5-nitrofuran derivatives against nosocomial pathogens: Synthesis, antimicrobial activity and chemometric analysis. Bioorg. Med. Chem. 2014, 22, 2844–2854. [Google Scholar] [CrossRef] [PubMed]
- Krasavin, M.; Shetnev, A.; Panova, V.; Ivanovskyi, S.; Kalinin, S.; Vinogradova, T.; Sharoyko, V.; Yablonsky, P. Hetaryl- and heteroarylvinyl-substituted nitrofurans identified as non-cytotoxic selective antitubercular agents. Mendeleev. Commun. 2022, 32, 452–453. [Google Scholar] [CrossRef]
- Picconi, P.; Prabaharan, P.; Auer, J.L.; Sandiford, S.; Cascio, F.; Chowdhury, M.; Hind, C.; Wand, M.E.; Sutton, J.M.; Rahman, K.M. Novel pyridyl nitrofuranyl isoxazolines show antibacterial activity against multiple drug resistant Staphylococcus species. Bioorg. Med. Chem. 2017, 25, 3971–3979. [Google Scholar] [CrossRef] [PubMed]
- Ran, K.; Gao, C.; Deng, H.; Lei, Q.; You, X.; Wang, N.; Shi, Y.; Liu, Z.; Wei, W.; Peng, C.; et al. Identification of novel 2-aminothiazole conjugated nitrofuran as antitubercular and antibacterial agents. Bioorg. Med. Chem. Lett. 2016, 26, 3669–3674. [Google Scholar] [CrossRef] [PubMed]
- Pendleton, J.N.; Gorman, S.P.; Gilmore, B.F. Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti-Infect. Ther. 2013, 11, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Wolkenberg, S.E.; Zhao, Z.; Wisnoski, D.D.; Leister, W.H.; O’Brien, J.; Lemaire, W.; Williams, D.L., Jr.; Jacobson, M.A.; Sur, C.; Kinney, G.G.; et al. Discovery of GlyT1 inhibitors with improved pharmacokinetic properties. Bioorg. Med. Chem. Lett. 2009, 19, 1492–1495. [Google Scholar] [CrossRef] [PubMed]
- Chappie, T.A.; Humphrey, J.M.; Allen, M.P.; Estep, K.G.; Fox, C.B.; Lebel, L.A.; Liras, S.; Marr, E.S.; Menniti, F.S.; Pandit, J.; et al. Discovery of a series of 6,7-dimethoxy-4-pyrrolidylquinazoline PDE10A inhibitors. J. Med. Chem. 2007, 50, 182–185. [Google Scholar] [CrossRef] [PubMed]
- Falgueyret, J.P.; Oballa, R.M.; Okamoto, O.; Wesolowski, G.; Aubin, Y.; Rydzewski, R.M.; Prasit, P.; Riendeau, D.; Rodan, S.B.; Percival, M.D. Novel, nonpeptidic cyanamides as potent and reversible inhibitors of human cathepsins K and L. J. Med. Chem. 2001, 44, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Dhameliya, T.M.; Chudasma, S.J.; Patel, T.M.; Dave, B.P. A review on synthetic account of 1,2,4-oxadiazoles as anti-infective agents. Mol. Divers 2022, 26, 2967–2980. [Google Scholar] [CrossRef] [PubMed]
- Kayukova, L.A. Synthesis of 1,2,4-oxadiazoles (a review). Pharm. Chem. J. 2005, 39, 539–547. [Google Scholar] [CrossRef]
- Szulczyk, D.; Wozinski, M.; Kolinski, M.; Kmiecik, S.; Glogowska, A.; Augustynowicz-Kopec, E.; Dobrowolski, M.A.; Roszkowski, P.; Struga, M.; Ciura, K. Menthol- and thymol-based ciprofloxacin derivatives against Mycobacterium tuberculosis: In vitro activity, lipophilicity, and computational studies. Sci. Rep. 2023, 13, 16328. [Google Scholar] [CrossRef] [PubMed]
- McOsker, C.C.; Fitzpatrick, P.M. Nitrofurantoin: Mechanism of action and implications for resistance development in common uropathogens. J. Antimicrob. Chemother. 1994, 33 (Suppl. A), 23–30. [Google Scholar] [CrossRef] [PubMed]
- PDB Entry—6YJF. Available online: https://www.wwpdb.org/pdb?id=pdb_00006yjf (accessed on 20 March 2024).
- Baretta, K.; Garen, C.; Yin, J.; James, M.N. Expression, purification, crystallization and preliminary crystallographic analysis of the phosphoglycerate kinase from Acinetobacter baumannii. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2012, 68, 790–792. [Google Scholar] [CrossRef] [PubMed]
- Almasaudi, S.B. Acinetobacter spp. as nosocomial pathogens: Epidemiology and resistance features. Saudi J. Biol. Sci. 2018, 25, 586–596. [Google Scholar] [CrossRef] [PubMed]
- Tatum, N.J.; Duarte, F.; Kamerlin, S.C.L.; Pohl, E. Relative Binding Energies Predict Crystallographic Binding Modes of Ethionamide Booster Lead Compounds. J. Phys. Chem. Lett. 2019, 10, 2244–2249. [Google Scholar] [CrossRef] [PubMed]
- Abuhammad, A.; Fullam, E.; Lowe, E.D.; Staunton, D.; Kawamura, A.; Westwood, I.M.; Bhakta, S.; Garner, A.C.; Wilson, D.L.; Seden, P.T.; et al. Piperidinols that show anti-tubercular activity as inhibitors of arylamine N-acetyltransferase: An essential enzyme for mycobacterial survival inside macrophages. PLoS ONE 2012, 7, e52790. [Google Scholar] [CrossRef] [PubMed]
- Abuhammad, A.; Lowe, E.D.; McDonough, M.A.; Shaw Stewart, P.D.; Kolek, S.A.; Sim, E.; Garman, E.F. Structure of arylamine N-acetyltransferase from Mycobacterium tuberculosis determined by cross-seeding with the homologous protein from M. marinum: Triumph over adversity. Acta Crystallogr. D Biol. Crystallogr. 2013, 69, 1433–1446. [Google Scholar] [CrossRef] [PubMed]
- Agre, N.; Tawari, N.; Maitra, A.; Gupta, A.; Munshi, T.; Degani, M.; Bhakta, S. 3-(5-Nitrofuran-2-yl)prop-2-en-1-one Derivatives, with Potent Antituberculosis Activity, Inhibit A Novel Therapeutic Target, Arylamine N-acetyltransferase, in Mycobacteria. Antibiotics 2020, 9, 368. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Alian, A.; Stroud, R.; Ortiz de Montellano, P.R. Pyrrolidine carboxamides as a novel class of inhibitors of enoyl acyl carrier protein reductase from Mycobacterium tuberculosis. J. Med. Chem. 2006, 49, 6308–6323. [Google Scholar] [CrossRef] [PubMed]
- El Sawy, M.A.; Elshatanofy, M.M.; El Kilany, Y.; Kandeel, K.; Elwakil, B.H.; Hagar, M.; Aouad, M.R.; Albelwi, F.F.; Rezki, N.; Jaremko, M.; et al. Novel Hybrid 1,2,4- and 1,2,3-Triazoles Targeting Mycobacterium Tuberculosis Enoyl Acyl Carrier Protein Reductase (InhA): Design, Synthesis, and Molecular Docking. Int. J. Mol. Sci. 2022, 23, 4706. [Google Scholar] [CrossRef] [PubMed]
- Kahlmeter, G.; Brown, D.F.; Goldstein, F.W.; MacGowan, A.P.; Mouton, J.W.; Odenholt, I.; Rodloff, A.; Soussy, C.J.; Steinbakk, M.; Soriano, F.; et al. European Committee on Antimicrobial Susceptibility Testing (EUCAST) Technical Notes on antimicrobial susceptibility testing. Clin. Microbiol. Infect. 2006, 12, 501–503. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, I.; Hilpert, K.; Hancock, R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Chuprun, S.; Dar’in, D.; Rogacheva, E.; Kraeva, L.; Levin, O.; Manicheva, O.; Dogonadze, M.; Vinogradova, T.; Bakulina, O.; Krasavin, M. Mutually Isomeric 2- and 4-(3-nitro-1,2,4-triazol-1-yl)pyrimidines Inspired by an Antimycobacterial Screening Hit: Synthesis and Biological Activity against the ESKAPE Panel of Pathogens. Antibiotics 2020, 9, 666. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Wu, C.; Ghoreishi, D.; Chen, W.; Wang, L.; Damm, W.; Ross, G.A.; Dahlgren, M.K.; Russell, E.; Von Bargen, C.D.; et al. OPLS4: Improving Force Field Accuracy on Challenging Regimes of Chemical Space. J. Chem. Theory Comput. 2021, 17, 4291–4300. [Google Scholar] [CrossRef]
- O’Neill, A.G.; Beaupre, B.A.; Zheng, Y.; Liu, D.; Moran, G.R. NfoR: Chromate Reductase or Flavin Mononucleotide Reductase? Appl. Environ. Microbiol. 2020, 86, e01758-20. [Google Scholar] [CrossRef] [PubMed]
- Cellitti, S.E.; Shaffer, J.; Jones, D.H.; Mukherjee, T.; Gurumurthy, M.; Bursulaya, B.; Boshoff, H.I.; Choi, I.; Nayyar, A.; Lee, Y.S.; et al. Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824. Structure 2012, 20, 101–112. [Google Scholar] [CrossRef] [PubMed]



| ESKAPE Panel | Compound 2 | NFt | FZ | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a | b | c | d | e | f | g | h | i | j | k | l | m | |||
| E. faecium | 0 | 0 | 13 ± 1.6 | 0 | 7 ± 1.6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 15 ± 1.6 | 13 ± 1.0 |
| S. aureus | 21 ± 1.6 | 17 ± 1.0 | 21 ± 1.6 | 0 | 27 ± 0.4 | 21 ± 1.0 | 0 | 21 ± 1.6 | 21 | 25 ± 1.0 | 0 | 15 ± 0.3 | 17 ± 1.2 | 21 ± 0.3 | 23 ± 1.6 |
| K. pneumoniae | 0 | 0 | 25 ± 1.0 | 0 | 9 ± 1.6 | 0 | 0 | 17 ± 0.3 | 0 | 0 | 0 | 0 | 0 | 12 ± 1.0 | 14 ± 1.6 |
| A. baumannii | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 23 ± 1.0 | 0 | 0 | 13 ± 0.1 | 0 | 0 | 0 | 0 |
| P. aeruginosa | 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E. cloacae | 0 | 13 | 21 ± 1.6 | 0 | 0 | 0 | 0 | 19 ± 0.5 | 0 | 0 | 0 | 0 | 0 | 24 ± 0.4 | 21 ± 1.0 |
| Compounds | Pathogens | ||||||
|---|---|---|---|---|---|---|---|
| E. faecium | S. aureus | K. pneumoniae | A. baumannii | P. aeruginosa | E. cloacae | M. tuberculosis | |
| 2c | 4 ± 0.1 | 6 ± 0.6 | 0.5 ± 0.1 | 32 ± 0.1 | 4 ± 0.03 | 0.25 ± 0.01 | 100 ± 10.0 |
| 2h | 1 ± 0.03 | 0.8 ± 0.01 | 1.75 ± 0.5 | 0.4 ± 0.1 | 9 ± 0.6 | 3.5 ± 0.1 | 50 ± 3.0 |
| 2e | nt | nt | nt | nt | nt | nt | 6.2 ± 0.4 |
| 2d | nt | nt | nt | nt | nt | nt | 25 ± 1.8 |
| 2f,i,k | nt | nt | nt | nt | nt | nt | 50 ± 3.1 |
| 2a,b,g,j,l,m | nt | nt | nt | nt | nt | nt | ≥100 |
| nitrofurantoin | 8 ± 0.1 | 4 ± 0.1 | 64 ± 3.0 | nt | nt | 4 ± 0.1 | nt |
| furazidine | 32 ± 0.1 | 8 ± 0.1 | 32 ± 3.0 | nt | nt | 2 ± 0.1 | nt |
| ciprofloxacin | 1.25 ± 0.1 | 1.25 ± 0.1 | 0.6 ± 0.1 | 2.5 ± 0.1 | 0.6 ± 0.1 | 3 ± 0.1 | 0.5–0.25 [24] |
| Compounds | GlideScore (kcal/mol) and Binding Quality (Stars) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AzoR | NfsA | NfsB | Pgk | Ddn | |||||||||
| P. aeruginosa | S. aureus | A. baumannii | P. aeruginosa | S. aureus | A. baumannii | P. aeruginosa | S. aureus | A. baumannii | P. aeruginosa | S. aureus | A. baumannii | M. tuberculosis | |
| 2a | −7.51 (***) | −6.81 (**) | −8.44 (***) | −5.47 (***) | −7.18 (***) | −6.15 (***) | −5.16 (**) | −6.03 (***) | −5.41 (*) | −6.77 (***) | −6.50 (*) | −6.88 (***) | −6.70 (***) |
| 2c | −6.95 (***) | −7.53 (***) | −7.95 (***) | −5.14 (***) | −5.83 (***) | −6.15 (***) | −5.46 (*) | −5.75 (**) | −5.77 (**) | −8.24 (***) | −5.52 (*) | −8.40 (***) | n/a |
| 2d | −6.58 (*) | −6.81 (*) | −6.69 (*) | −6.25 (*) | −5.89 (*) | −6.09 (*) | −5.71 (*) | −7.20 (*) | −5.79 (*) | −6.98 (*) | −5.99 (*) | −6.18 (***) | n/a |
| 2e | −8.89 (***) | −8.28 (**) | −9.23 (*) | −4.77 (*) | −6.02 (*) | −5.40 (***) | −7.04 (*) | −6.63 (**) | −5.35 (*) | −8.38 (***) | −6.71 (*) | −7.57 (**) | −7.74 (***) |
| 2f | −6.51 (*) | −7.87 (***) | −8.24 (***) | −4.79 (*) | −6.60 (*) | −6.78 (***) | −6.44 (**) | −7.61 (***) | −5.24 (*) | −8.55 (***) | −7.32 (*) | −8.65 (***) | −6.01 (*) |
| 2g | −7.59 (*) | −8.64 (*) | −7.24 (*) | −5.63 (*) | −5.82 (*) | −7.05 (*) | −5.88 (*) | −6.16 (*) | −5.83 (*) | −7.00 (**) | −6.91 (*) | −6.45 (***) | n/a |
| 2h | −7.46 (***) | −7.11 (***) | −7.48 (***) | −5.13 (***) | −7.19 (***) | −6.94 (***) | −4.60 (**) | −5.98 (***) | −5.07 (*) | −6.60 (***) | −7.16 (*) | −7.02 (***) | n/a |
| 2i | −6.09 (***) | −7.01 (***) | −7.83 (***) | −4.77 (*) | −7.03 (***) | −6.35 (***) | −7.16 (**) | −7.43 (***) | −7.20 (*) | −6.98 (***) | −5.79 (*) | −8.13 (***) | −5.83 (***) |
| 2j | −6.37 (***) | −7.40 (**) | −7.89 (***) | −4.57 (*) | −6.62 (**) | −7.06 (**) | −5.63 (**) | −6.21 (***) | −6.11 (*) | −7.06 (***) | −6.28 (*) | −6.46 (***) | −6.54 (***) |
| 2k | −7.25 (***) | −7.55 (**) | −6.14 (***) | −5.32 (*) | −5.90 (*) | −4.66 (*) | −5.86 (*) | −6.67 (***) | −7.24 (*) | −7.65 (***) | −6.98 (*) | −8.14 (***) | −5.80 (***) |
| GlideScore (kcal/mol)/Quality of Binding Pose (Stars) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Protein | 2a | 2c | 2d | 2e | 2g | 2h | 2k | Control |
| EthR | −8.67 (*) | −7.80 (**) | −7.47 (**) | −10.32 (***) | −7.66 (*) | −8.05 (**) | −8.29 (**) | −10.80 (***) |
| TBNAT | −5.86 (*) | −4.91 (*) | −5.22 (**) | −5.85 (**) | −6.39 (*) | −4.70 (**) | −4.88 (*) | N/A |
| InhA | −7.23 (***) | −6.57 (***) | −5.57 (*) | −7.66 (***) | −6.23 (*) | −6.86 (***) | −7.20 (***) | −9.70 (***) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vinogradova, L.; Lukin, A.; Komarova, K.; Zhuravlev, M.; Fadeev, A.; Chudinov, M.; Rogacheva, E.; Kraeva, L.; Gureev, M.; Porozov, Y.; et al. Molecular Periphery Design Allows Control of the New Nitrofurans Antimicrobial Selectivity. Molecules 2024, 29, 3364. https://doi.org/10.3390/molecules29143364
Vinogradova L, Lukin A, Komarova K, Zhuravlev M, Fadeev A, Chudinov M, Rogacheva E, Kraeva L, Gureev M, Porozov Y, et al. Molecular Periphery Design Allows Control of the New Nitrofurans Antimicrobial Selectivity. Molecules. 2024; 29(14):3364. https://doi.org/10.3390/molecules29143364
Chicago/Turabian StyleVinogradova, Lyubov, Alexey Lukin, Kristina Komarova, Maxim Zhuravlev, Artem Fadeev, Mikhail Chudinov, Elizaveta Rogacheva, Lyudmila Kraeva, Maxim Gureev, Yuri Porozov, and et al. 2024. "Molecular Periphery Design Allows Control of the New Nitrofurans Antimicrobial Selectivity" Molecules 29, no. 14: 3364. https://doi.org/10.3390/molecules29143364
APA StyleVinogradova, L., Lukin, A., Komarova, K., Zhuravlev, M., Fadeev, A., Chudinov, M., Rogacheva, E., Kraeva, L., Gureev, M., Porozov, Y., Dogonadze, M., & Vinogradova, T. (2024). Molecular Periphery Design Allows Control of the New Nitrofurans Antimicrobial Selectivity. Molecules, 29(14), 3364. https://doi.org/10.3390/molecules29143364







