Efficient Synthesis, Structural Characterization, Antibacterial Assessment, ADME-Tox Analysis, Molecular Docking and Molecular Dynamics Simulations of New Functionalized Isoxazoles
Abstract
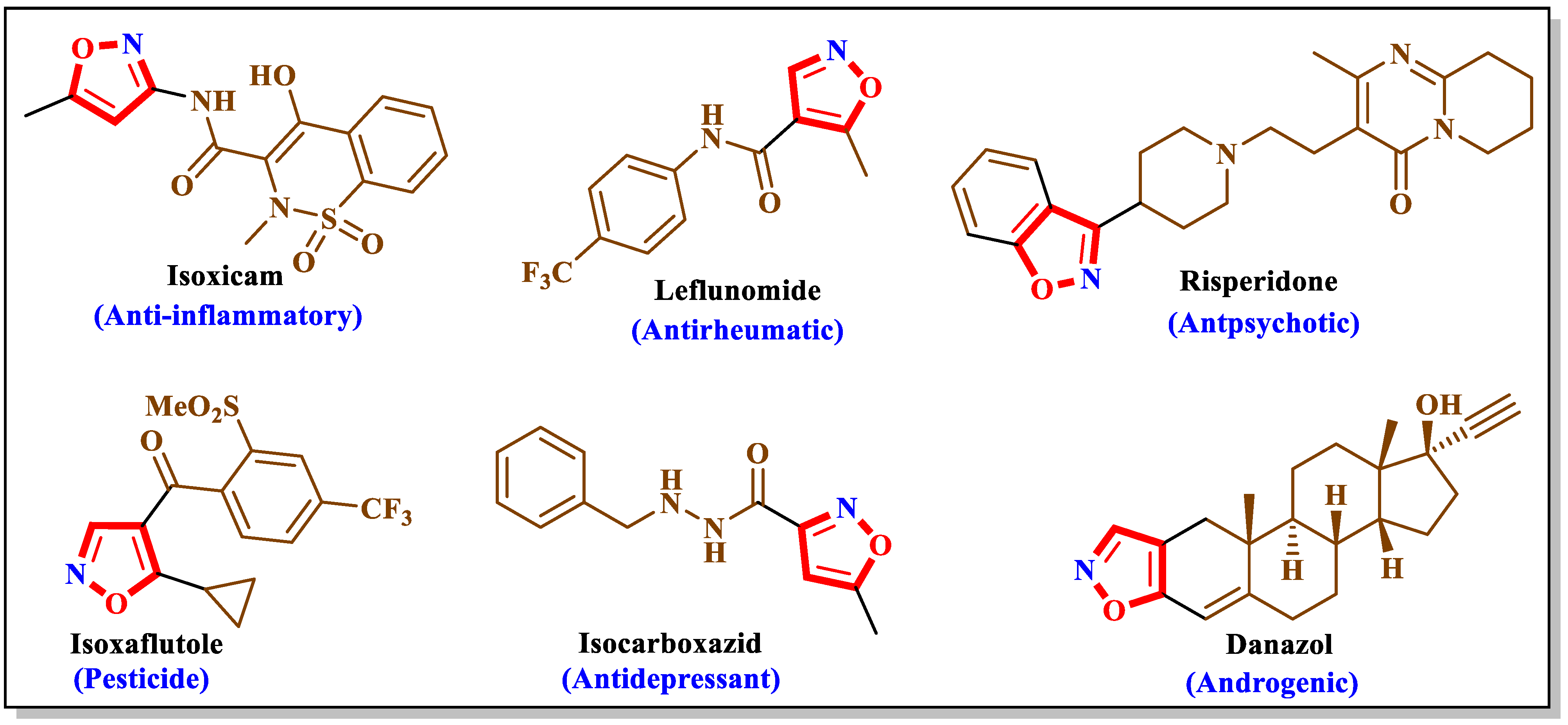
1. Introduction
2. Results and Discussion
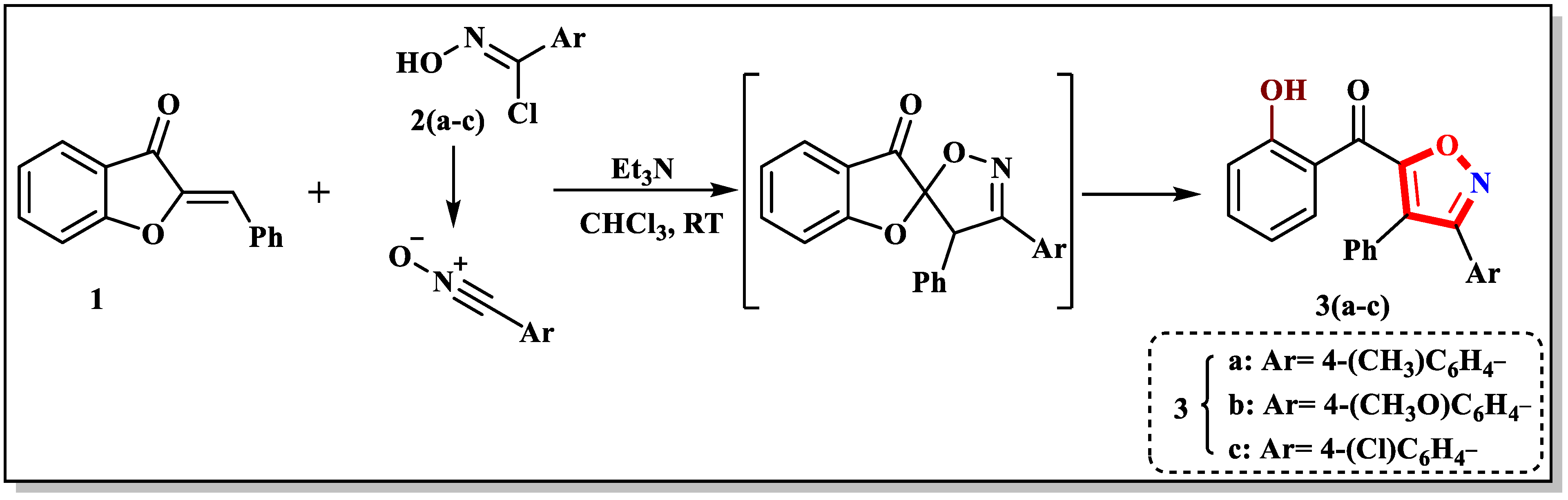
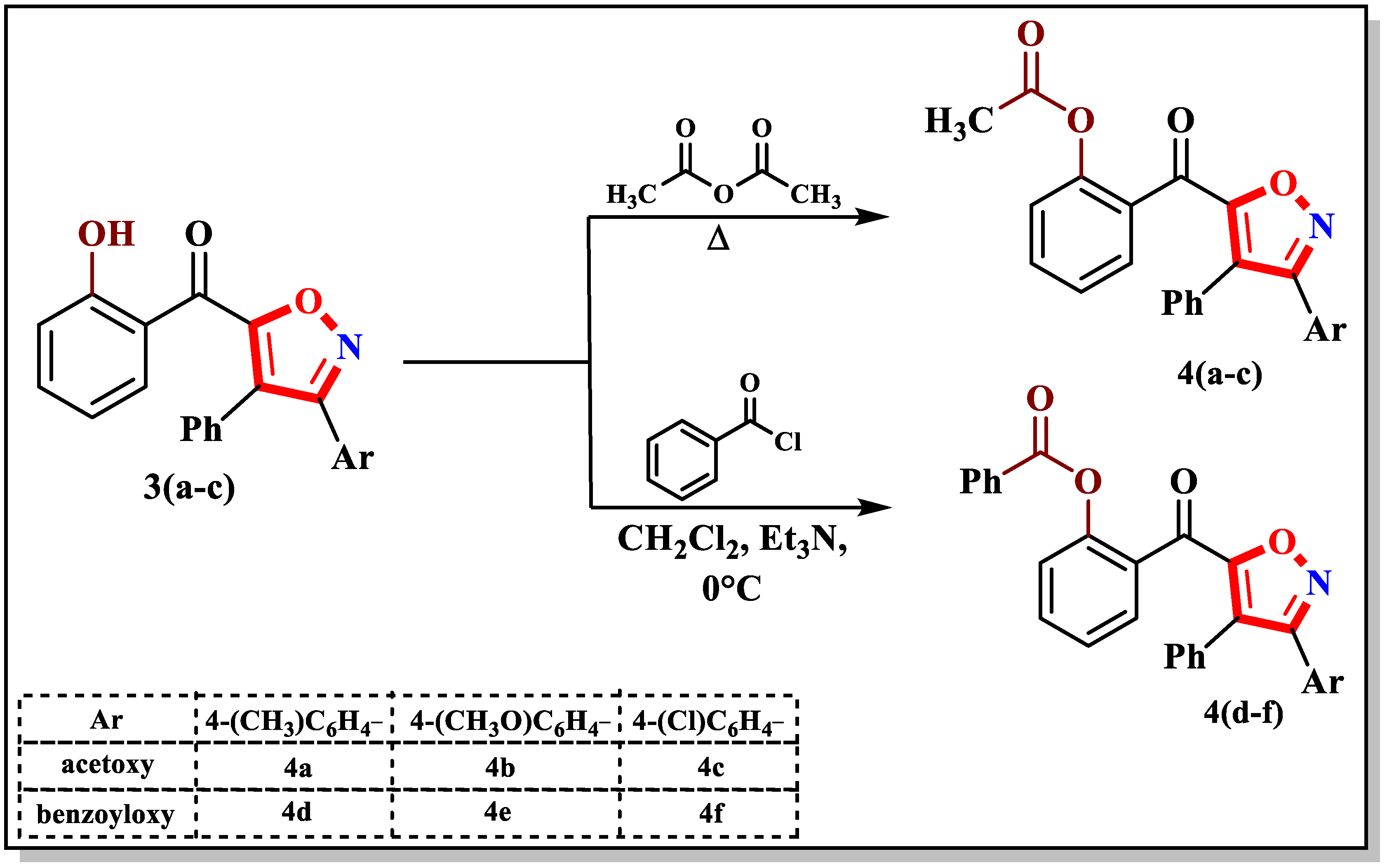
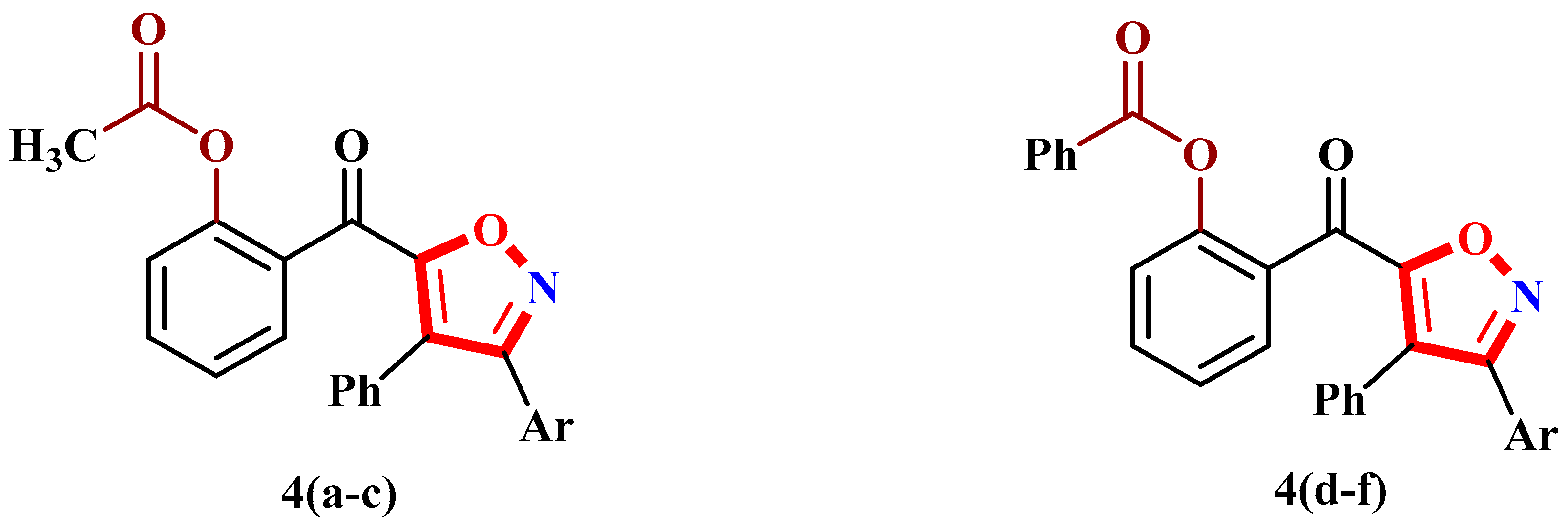
2.1. Syntheses of Functionalized Isoxazoles 4 and 5
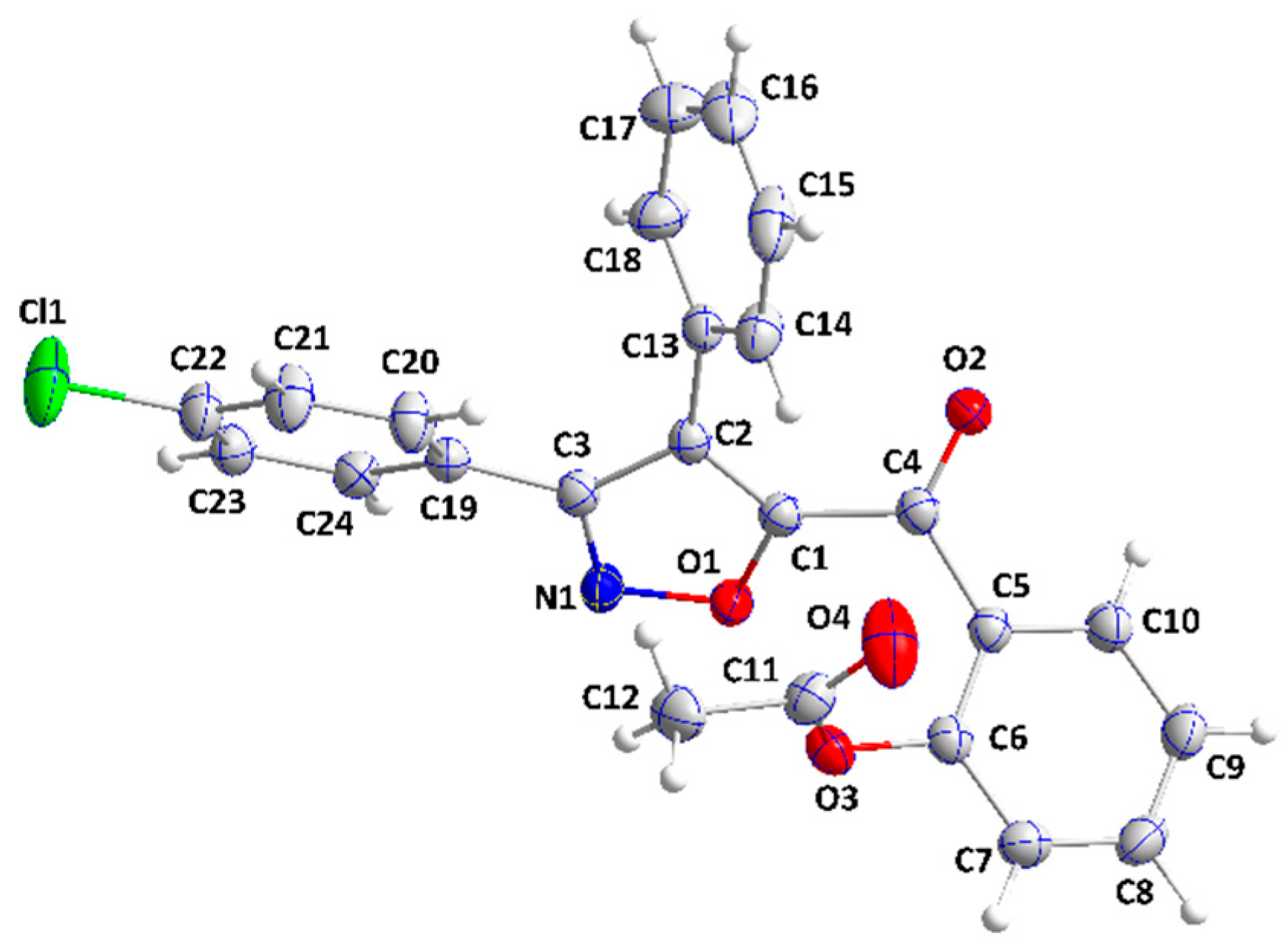
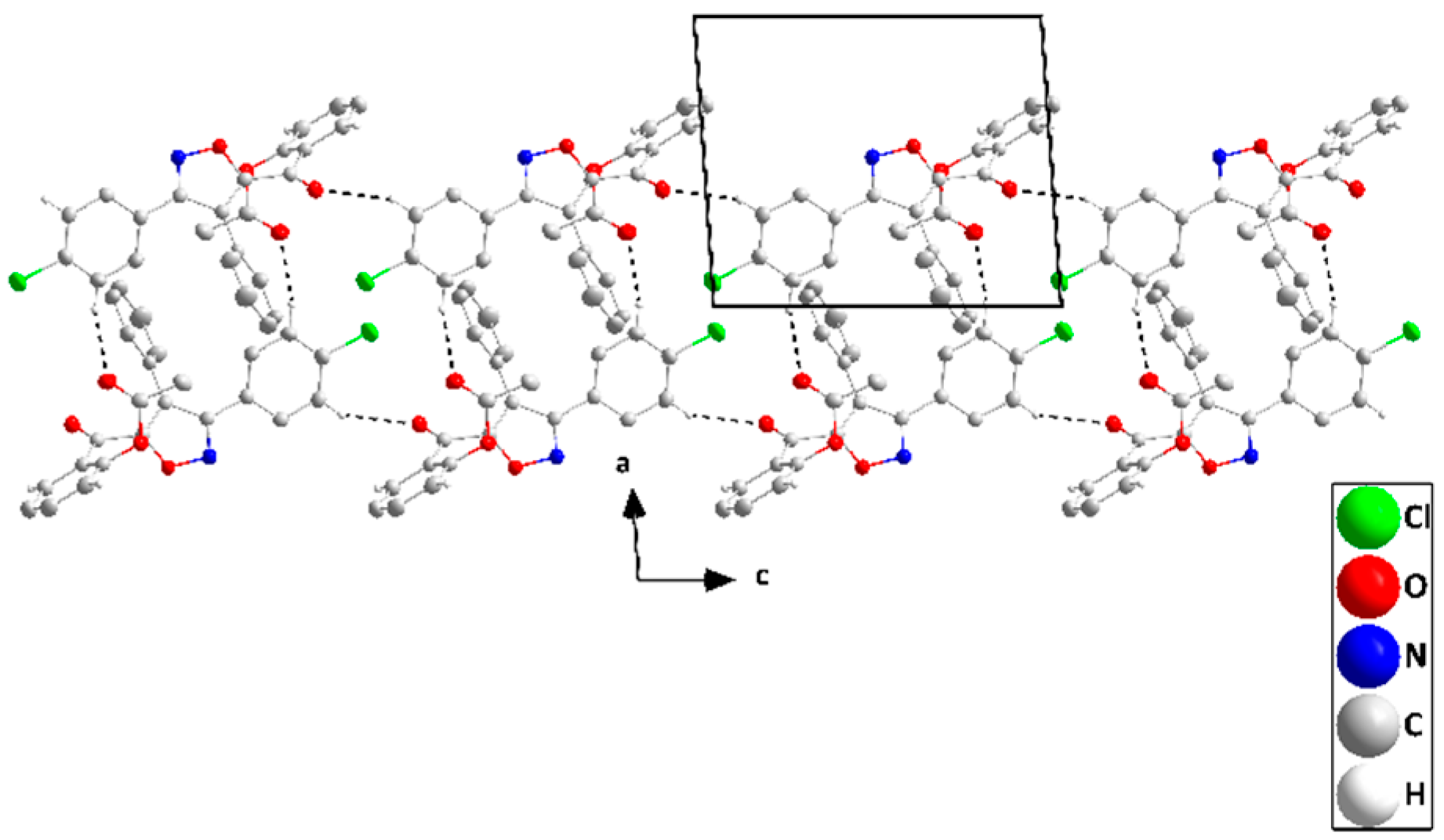
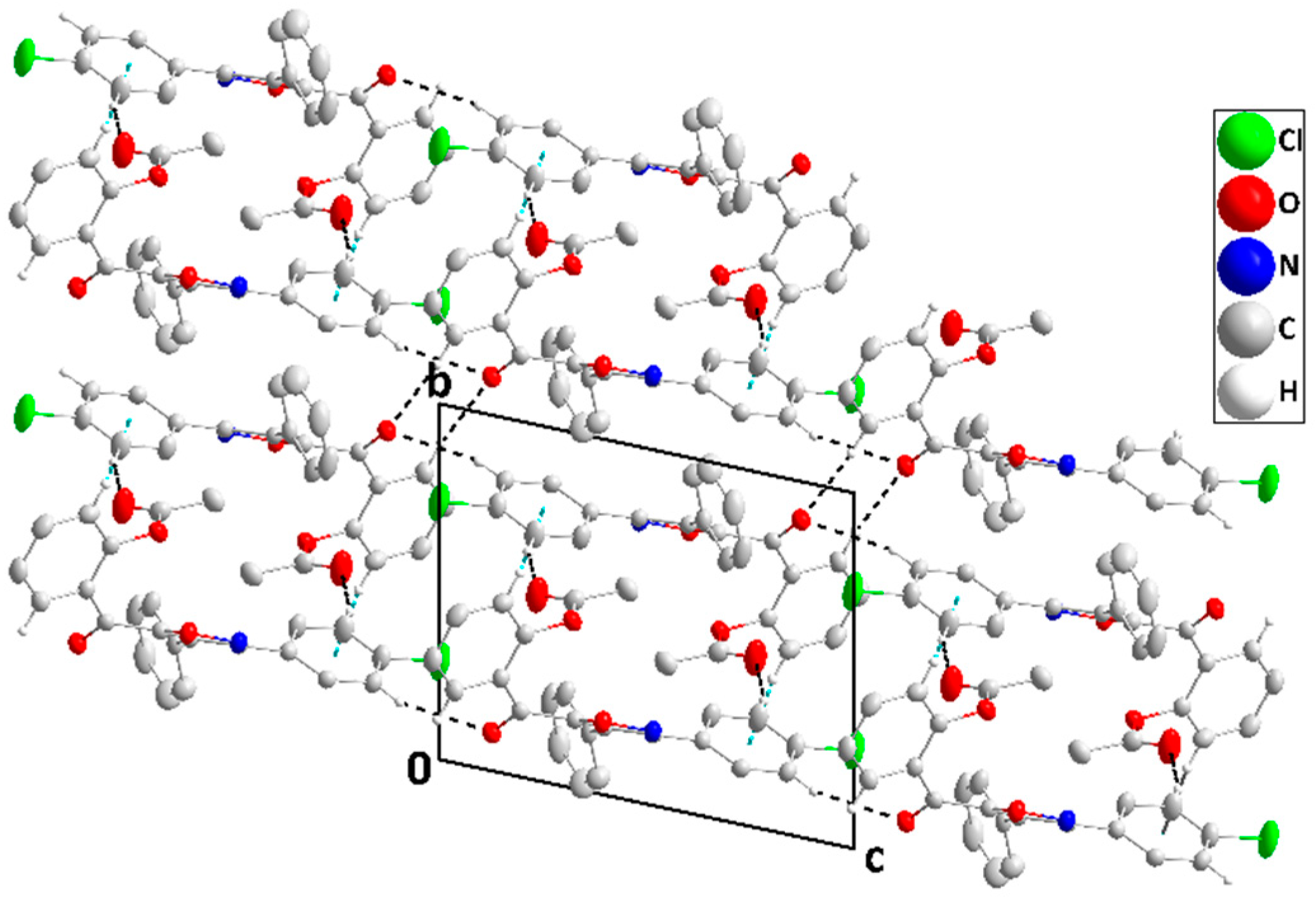
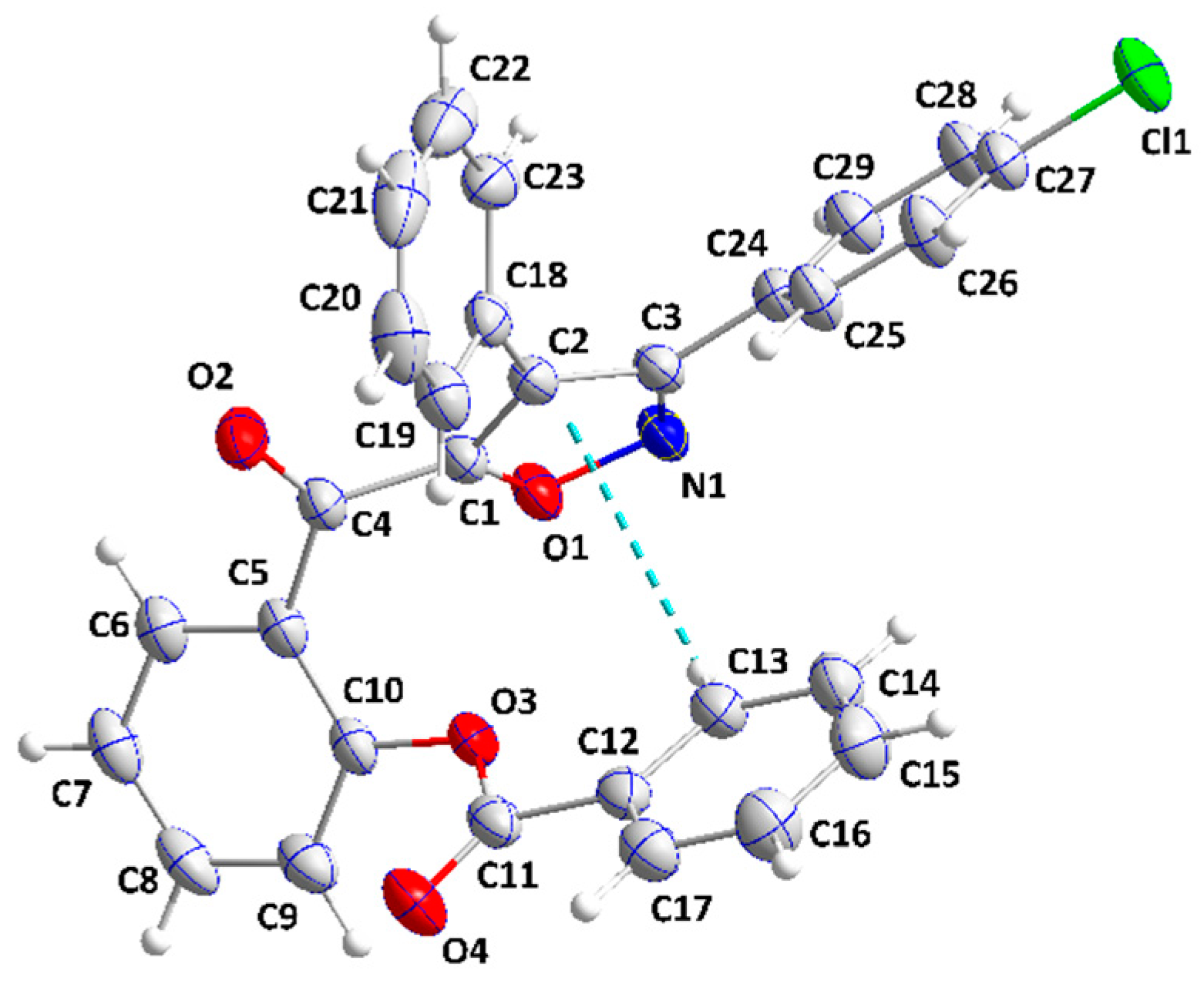
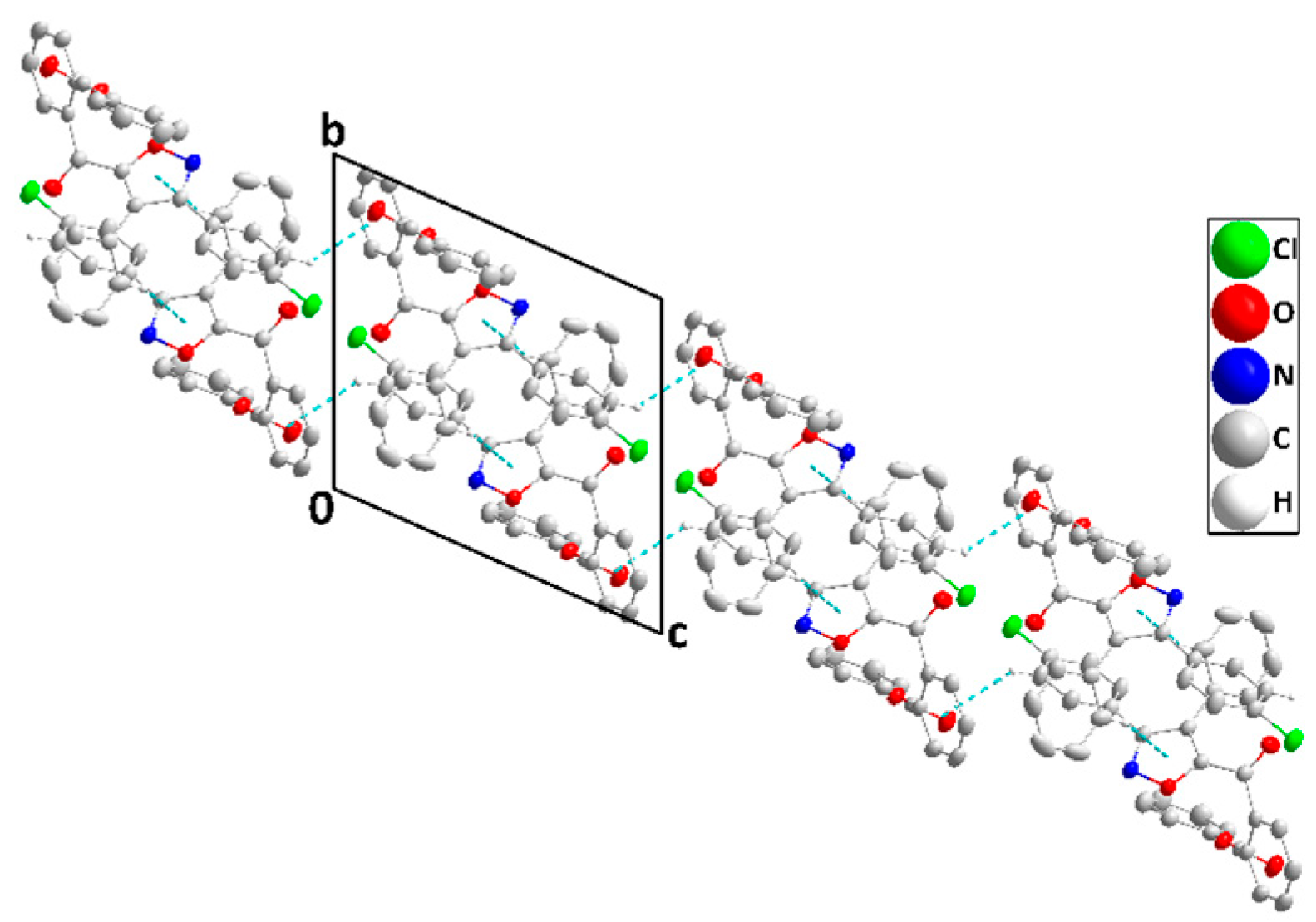
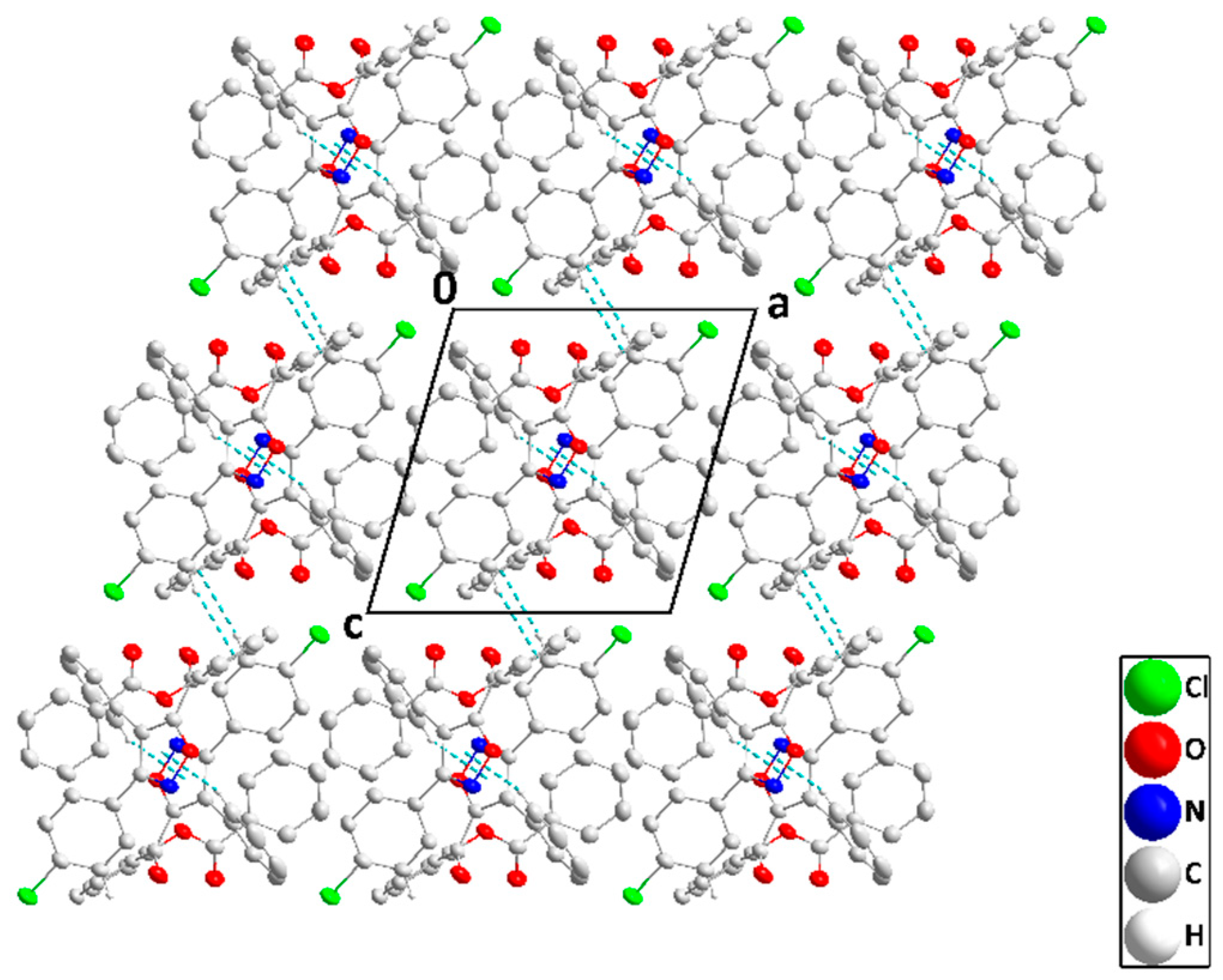
2.2. X-ray Diffraction Data and Structural Determination of 4c and 4f
2.3. Antibacterial Screening of Isoxazoles 4(a–f)
2.4. Molecular Docking Studies
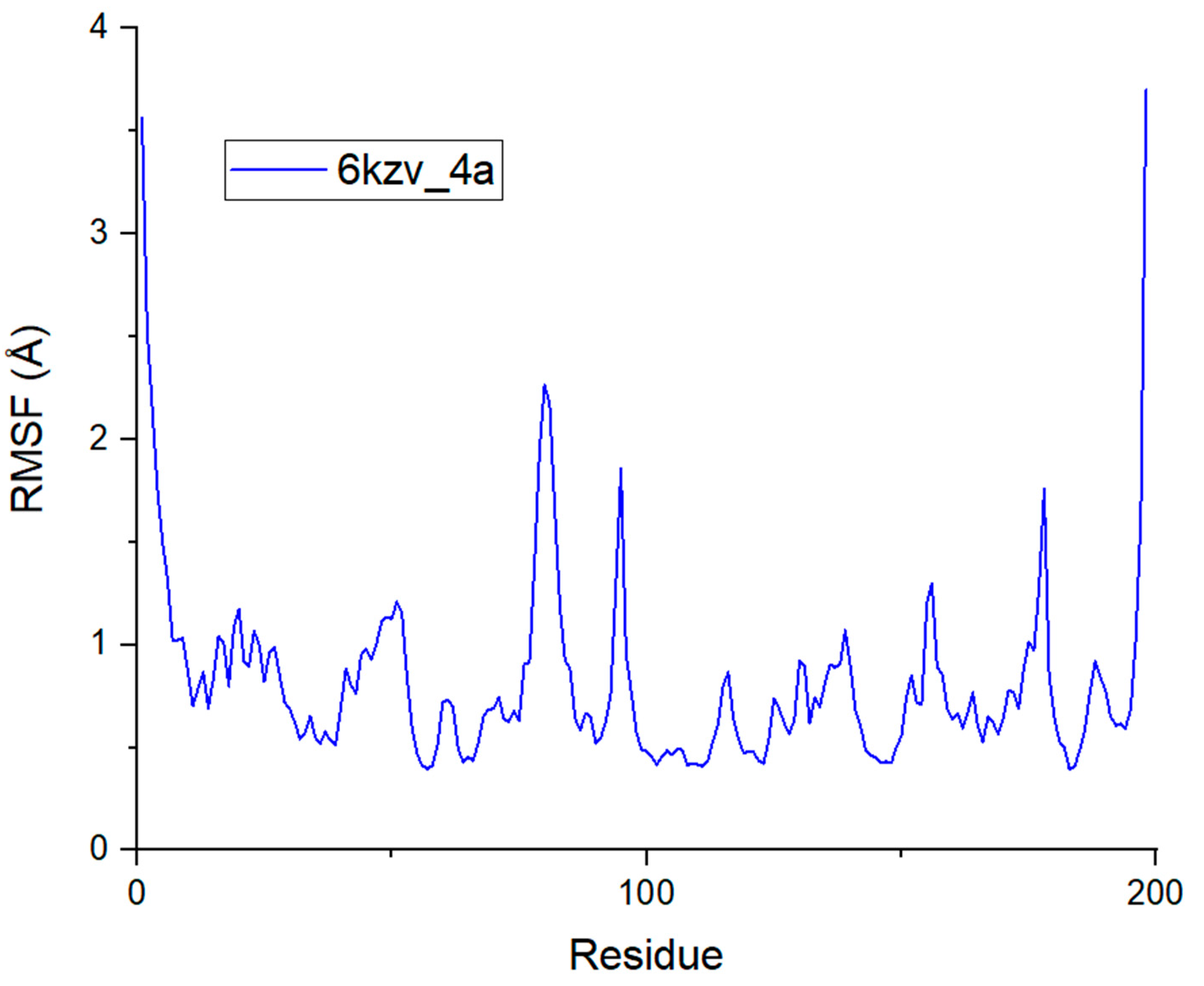
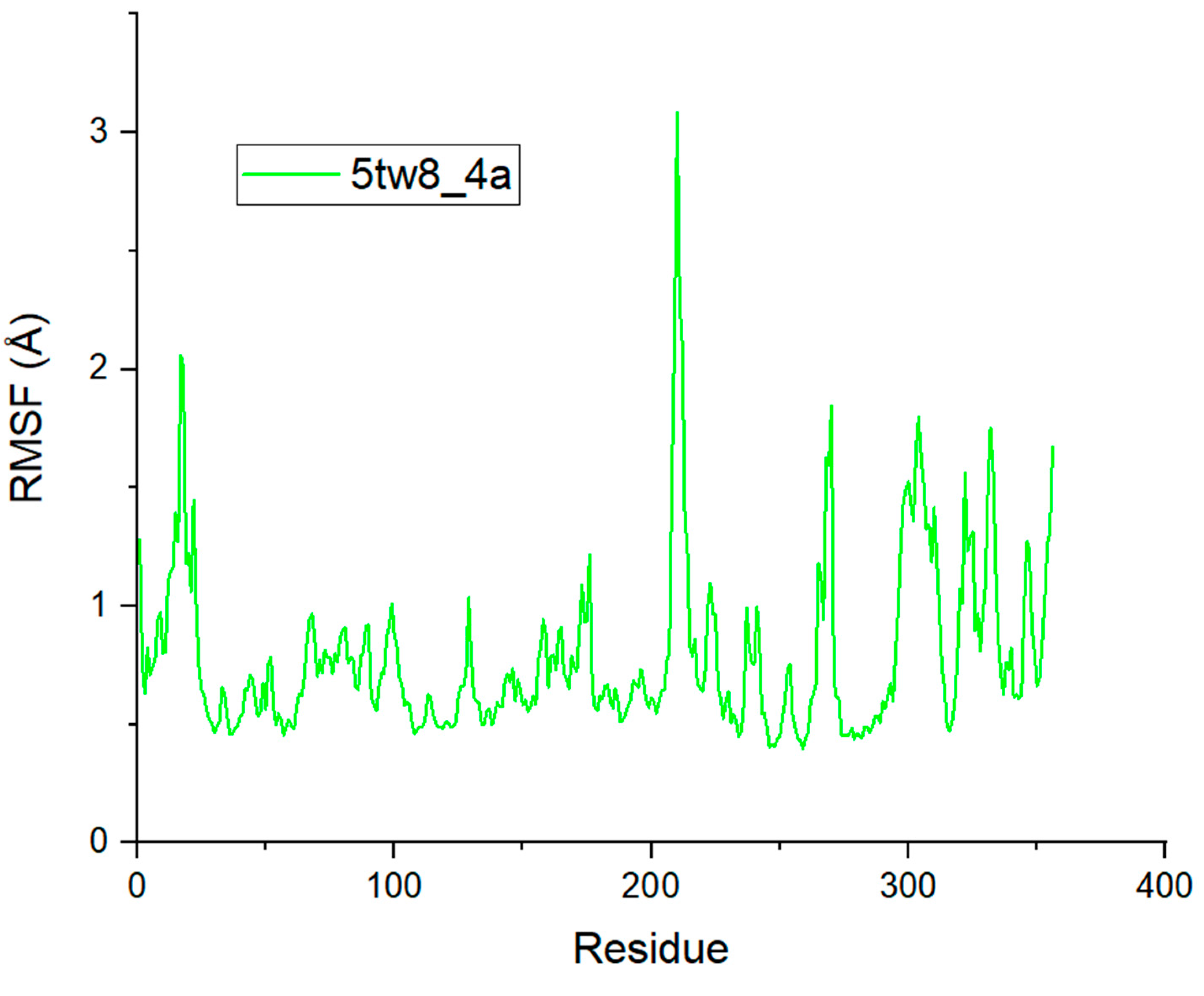
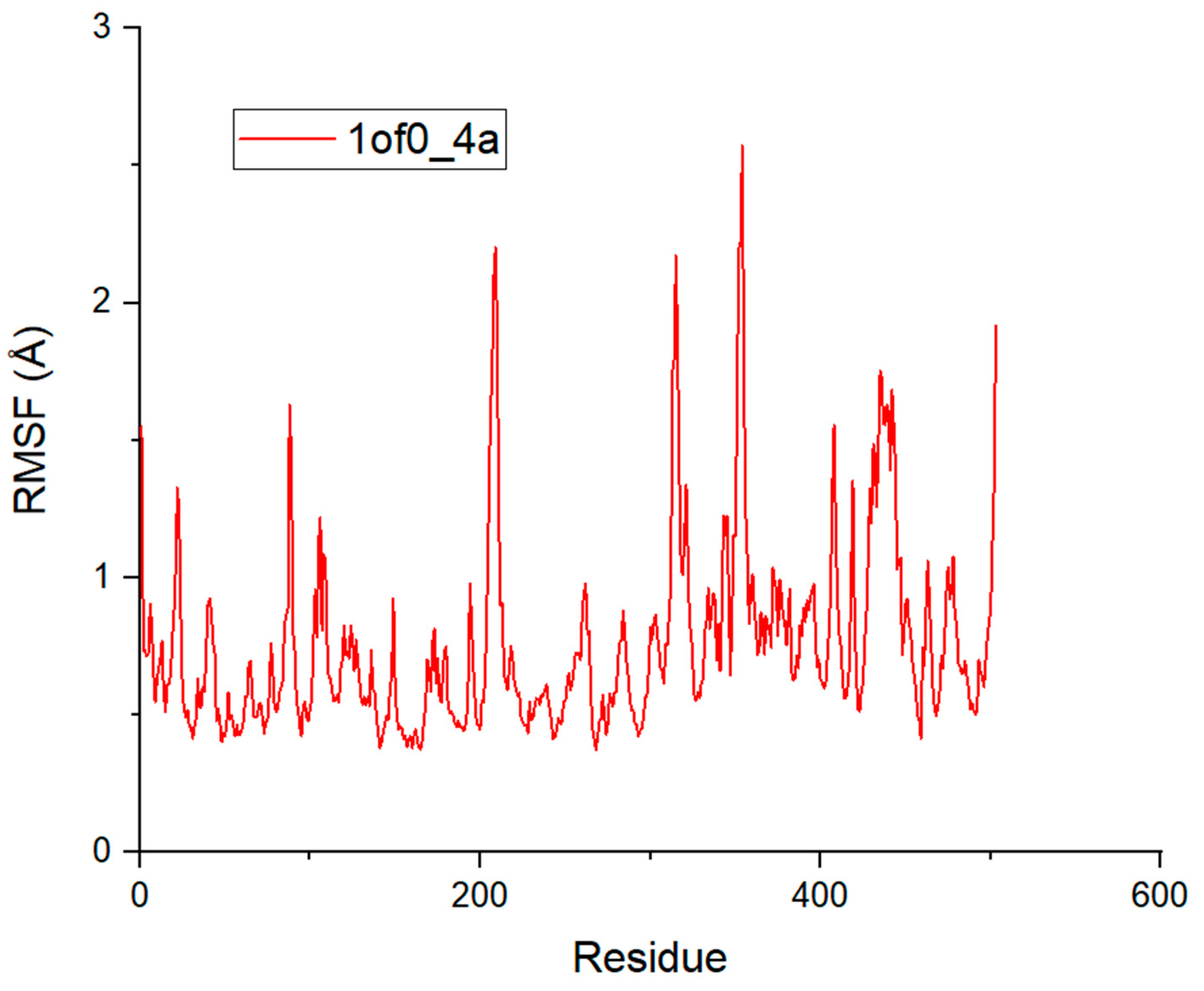
2.5. Molecular Dynamics Simulation Analysis
2.6. ADME-Tox Analyses
3. Summary
4. Materials and Methods
4.1. Chemistry
4.2. Antibacterial Screening Protocol
4.3. Molecular Docking
- ➢
- Escherichia coli (PDB ID: 6kzv) [51]:
- Target protein: Gyrase A (DNA gyrase subunit A).
- Biological role: Gyrase A is essential for bacterial survival as it catalyzes the negative supercoiling of DNA, which is crucial for DNA replication and transcription. Inhibiting this enzyme can prevent DNA replication, leading to cell death.
- ➢
- Staphylococcus aureus (PDB ID: 5tw8) [52]:
- Target protein: PBP2a (Penicillin-binding protein 2a).
- Biological role: PBP2a is involved in bacterial cell wall synthesis. This protein confers resistance to β-lactam antibiotics by preventing these antibiotics from binding to penicillin-binding proteins, allowing the bacteria to continue synthesizing its cell wall despite the presence of the antibiotic.
- ➢
- Bacillus subtilis (PDB ID: 1of0) [53]:
- Target protein: endospore coat protein.
- Biological role: This protein is involved in the formation of the endospore coat, a resistant structure that protects bacterial spores under extreme environmental conditions. Inhibiting this protein can disrupt spore formation, reducing bacterial survival in adverse conditions.
4.4. Molecular Dynamics (MD)
4.5. In Silico Pharmacokinetics ADMET
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Morens, D.M.; Fauci, A.S. Emerging Infectious Diseases: Threats to Human Health and Global Stability. PLoS Pathog. 2013, 9, e1003467. [Google Scholar] [CrossRef] [PubMed]
- Bloom, D.E.; Cadarette, D. Infectious Disease Threats in the Twenty-First Century: Strengthening the Global Response. Front. Immunol. 2019, 10, 549. [Google Scholar] [CrossRef] [PubMed]
- Davies, J. Origins and Evolution of Antibiotic Resistance. Microbiologia 1996, 12, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.-H.; Cohen, T.; Grad, Y.H.; Hanage, W.P.; O’Brien, T.F.; Lipsitch, M. Origin and Proliferation of Multiple-Drug Resistance in Bacterial Pathogens. Microbiol. Mol. Biol. Rev. 2015, 79, 101–116. [Google Scholar] [CrossRef] [PubMed]
- Vivas, R.; Barbosa, A.A.T.; Dolabela, S.S.; Jain, S. Multidrug-Resistant Bacteria and Alternative Methods to Control Them: An Overview. Microb. Drug Resist. 2019, 25, 890–908. [Google Scholar] [CrossRef] [PubMed]
- Dadgostar, P. Antimicrobial Resistance: Implications and Costs. Infect. Drug Resist. 2019, 12, 3903–3910. [Google Scholar] [CrossRef] [PubMed]
- Molalign, S.; Wencheko, E. Risk Factors of Mortality in Patients with Multi-Drug Resistant TB. Ethiop. J. Health Dev. 2015, 29, 82–88. [Google Scholar]
- Assefa, M. Multi-Drug Resistant Gram-Negative Bacterial Pneumonia: Etiology, Risk Factors, and Drug Resistance Patterns. Pneumonia 2022, 14, 4. [Google Scholar] [CrossRef] [PubMed]
- Verma, R.; Verma, S.K.; Rakesh, K.P.; Girish, Y.R.; Ashrafizadeh, M.; Sharath Kumar, K.S.; Rangappa, K.S. Pyrazole-Based Analogs as Potential Antibacterial Agents against Methicillin-Resistance Staphylococcus Aureus (MRSA) and Its SAR Elucidation. Eur. J. Med. Chem. 2021, 212, 113134. [Google Scholar] [CrossRef]
- Wu, G.; Zhao, T.; Kang, D.; Zhang, J.; Song, Y.; Namasivayam, V.; Kongsted, J.; Pannecouque, C.; De Clercq, E.; Poongavanam, V.; et al. Overview of Recent Strategic Advances in Medicinal Chemistry. J. Med. Chem. 2019, 62, 9375–9414. [Google Scholar] [CrossRef]
- Kidwai, M.; Venktaramanan, R.; Mohan, R.; Sapra, P. Cancer Chemotherapy and Heterocyclic Compounds. Curr. Med. Chem. 2012, 9, 1209–1228. [Google Scholar] [CrossRef] [PubMed]
- Sadek, K.U.; Mekheimer, R.A.; Abd-Elmonem, M.; Abo-Elsoud, F.A.; Hayallah, A.M.; Mostafa, S.M.; Abdellattif, M.H.; Abourehab, M.A.S.; Farghaly, T.A.; Elkamhawy, A. Recent Developments in the Synthesis of Hybrid Heterocycles, a Promising Approach to Develop Multi-Target Antibacterial Agents. J. Mol. Struct. 2023, 1286, 135616. [Google Scholar] [CrossRef]
- Javahershenas, R.; Nikzat, S. Recent Developments Using Malononitrile in Ultrasound-Assisted Multicomponent Synthesis of Heterocycles. Ultrason. Sonochem. 2023, 102, 106741. [Google Scholar] [CrossRef] [PubMed]
- Dadiboyena, S.; Nefzi, A. Synthesis of Functionalized Tetrasubstituted Pyrazolyl Heterocycles—A Review. Eur. J. Med. Chem. 2011, 46, 5258–5275. [Google Scholar] [CrossRef] [PubMed]
- Kaur, N.; Grewal, P.; Bhardwaj, P.; Verma, Y.; Ahlawat, N. Synthesis of Higher-Membered Heterocycles. Synth. Commun. 2023, 53, 443–475. [Google Scholar] [CrossRef]
- Lamberth, C. Oxazole and Isoxazole Chemistry in Crop Protection. J. Heterocycl. Chem. 2018, 55, 2035–2045. [Google Scholar] [CrossRef]
- Guo, K.L.; Zhao, L.X.; Wang, Z.W.; Gao, Y.C.; Li, J.J.; Gao, S.; Fu, Y.; Ye, F. Design, Synthesis, and Bioevaluation of Substituted Phenyl Isoxazole Analogues as Herbicide Safeners. J. Agric. Food Chem. 2020, 68, 10550–10559. [Google Scholar] [CrossRef]
- Bumagin, N.A.; Potkin, V.I. Functionalized Isoxazole and Isothiazole Ligands: Design, Synthesis, Palladium Complexes, Homogeneous and Heterogeneous Catalysis in Aqueous Media. Russ. Chem. Bull. 2016, 65, 321–332. [Google Scholar] [CrossRef]
- Díaz-Trelles, R.; Novelli, A.; Fernández-Sánchez, M.T. RNA Synthesis-Dependent Potentiation of α-Amino-3-Hydroxy-5-Methyl-4-Isoxazole Propionate Receptor-Mediated Toxicity by Antihistamine Terfenadine in Cultured Rat Cerebellar Neurons. Neurosci. Lett. 2003, 345, 136–140. [Google Scholar] [CrossRef]
- Trefzger, O.S.; Barbosa, N.V.; Scapolatempo, R.L.; das Neves, A.R.; Ortale, M.L.F.S.; Carvalho, D.B.; Honorato, A.M.; Fragoso, M.R.; Shuiguemoto, C.Y.K.; Perdomo, R.T.; et al. Design, Synthesis, Antileishmanial, and Antifungal Biological Evaluation of Novel 3,5-Disubstituted Isoxazole Compounds Based on 5-Nitrofuran Scaffolds. Arch. Pharm. 2019, 353, 1900241. [Google Scholar] [CrossRef]
- Ni, T.; Chi, X.; Xie, F.; Li, L.; Wu, H.; Hao, Y.; Wang, X.; Zhang, D.; Jiang, Y. Design, Synthesis, and Evaluation of Novel Tetrazoles Featuring Isoxazole Moiety as Highly Selective Antifungal Agents. Eur. J. Med. Chem. 2023, 246, 115007. [Google Scholar] [CrossRef]
- Pradeep Kumar, M.; Ayodhya, D.; Rambabu, A. Shivaraj Synthesis, Characterization, X-ray Crystal Structure, Antioxidant, Antimicrobial, and DNA Binding Interaction Studies of Novel Copper (II)-Isoxazole Binary Complexes. Results Chem. 2023, 5, 100846. [Google Scholar] [CrossRef]
- Khanage, S.; Mohite, P.; Pandhare, R.; Raju, A. 1,2,4-Triazol Içeren Izoksazol Türevlerinin Sentezi Ve Farmakolojik Etkisi. Marmara Pharm. J. 2012, 16, 134–140. [Google Scholar] [CrossRef]
- Sysak, A.; Obmińska-Mrukowicz, B. Isoxazole Ring as a Useful Scaffold in a Search for New Therapeutic Agents. Eur. J. Med. Chem. 2017, 137, 292–309. [Google Scholar] [CrossRef]
- Liu, T.; Huang, B.; Zhan, P.; De Clercq, E.; Liu, X. Discovery of Small Molecular Inhibitors Targeting HIV-1 Gp120-CD4 Interaction Drived from BMS-378806. Eur. J. Med. Chem. 2014, 86, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, N.; Mishra, P. The Synthetic and Therapeutic Expedition of Isoxazole and Its Analogs. Med. Chem. Res. 2018, 27, 1309–1344. [Google Scholar] [CrossRef] [PubMed]
- Mączyński, M.; Artym, J.; Kociȩba, M.; Kochanowska, I.; Ryng, S.; Zimecki, M. Anti-Inflammatory Properties of an Isoxazole Derivative—MZO-2. Pharmacol. Rep. 2016, 68, 894–902. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, R.; Vyas, A.; El-Bahy, S.M.; Hessien, M.M.; Mersal, G.A.M.; Ibrahim, M.M.; Dogra, R.; Kumar, B. Rationale Design, Synthesis, Pharmacological and In-Silico Investigation of Indole-Functionalized Isoxazoles as Anti-Inflammatory Agents. ChemistrySelect 2022, 7, e202200800. [Google Scholar] [CrossRef]
- Arya, G.C.; Kaur, K.; Jaitak, V. Isoxazole Derivatives as Anticancer Agent: A Review on Synthetic Strategies, Mechanism of Action and SAR Studies. Eur. J. Med. Chem. 2021, 221, 113511. [Google Scholar] [CrossRef]
- Hawash, M.; Jaradat, N.; Eid, A.M.; Abubaker, A.; Mufleh, O.; Al-Hroub, Q.; Sobuh, S. Synthesis of Novel Isoxazole–Carboxamide Derivatives as Promising Agents for Melanoma and Targeted Nano-Emulgel Conjugate for Improved Cellular Permeability. BMC Chem. 2022, 16, 47. [Google Scholar] [CrossRef]
- Ye, F.; Zhai, Y.; Kang, T.; Wu, S.L.; Li, J.J.; Gao, S.; Zhao, L.X.; Fu, Y. Rational Design, Synthesis and Structure-Activity Relationship of Novel Substituted Oxazole Isoxazole Carboxamides as Herbicide Safener. Pestic. Biochem. Physiol. 2019, 157, 60–68. [Google Scholar] [CrossRef]
- Lin, X.; Li, Y.; Zhong, W.; Hong, T.; Li, L.; Song, S.; He, D. Synthesis, Bioactivity, and QSAR Study of 3,4-Dichlorophenyl Isoxazole-Substituted Stilbene Derivatives against the Phytopathogenic Fungus Botrytis Cinerea. J. Agric. Food Chem. 2021, 69, 9520–9528. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhao, Y.; Li, P.; He, Y. Design, Synthesis, and Insecticidal Activity of Novel Isoxazole Derivatives Containing Bisamide Moiety. J. Heterocycl. Chem. 2019, 56, 3042–3047. [Google Scholar] [CrossRef]
- Lasri, M.; Bimoussa, A.; Ait-karra, A.; Laamari, Y.; Zakir, O.; Idouhli, R.; Maatallah, M.; Eddine, K.M.; Auhmani, A.; Itto, M.Y.A.; et al. Synthesis and Evaluation of Benzo[1,2,3]selenadiazole-Isoxazoles as Corrosion Inhibitors for Copper in NaCl: An Integrated Experimental and Theoretical Approach. Colloids Surf. A Physicochem. Eng. Asp. 2024, 695, 134227. [Google Scholar] [CrossRef]
- Hu, F.; Szostak, M. Recent Developments in the Synthesis and Reactivity of Isoxazoles: Metal Catalysis and Beyond. Adv. Synth. Catal. 2015, 357, 2583–2614. [Google Scholar] [CrossRef]
- Kohler, E.P.; Davis, A.R. Isoxazoline Oxides. X. Reduction. J. Am. Chem. Soc. 1930, 52, 4520–4528. [Google Scholar] [CrossRef]
- Akella, L.B.; DeCaprio, D. Cheminformatics Approaches to Analyze Diversity in Compound Screening Libraries. Curr. Opin. Chem. Biol. 2010, 14, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Begam, B.F.; Kumar, J.S. A Study on Cheminformatics and Its Applications on Modern Drug Discovery. Procedia Eng. 2012, 38, 1264–1275. [Google Scholar] [CrossRef]
- Altındağ, F.D.; Sağlık, B.N.; Acar Çevik, U.; Işıkdağ, İ.; Özkay, Y.; Karaca Gençer, H. Novel Imidazole Derivatives as Antifungal Agents: Synthesis, Biological Evaluation, ADME Prediction and Molecular Docking Studies. Phosphorus Sulfur Silicon Relat. Elem. 2019, 194, 887–894. [Google Scholar] [CrossRef]
- Patil, M.; Poyil, A.N.; Joshi, S.D.; Patil, S.A.; Patil, S.A.; Bugarin, A. Synthesis, Molecular Docking Studies, and Antimicrobial Evaluation of New Structurally Diverse Ureas. Bioorg. Chem. 2019, 87, 302–311. [Google Scholar] [CrossRef]
- Morris, C.J.; Corte, D. Della Using Molecular Docking and Molecular Dynamics to Investigate Protein-Ligand Interactions. Mod. Phys. Lett. B 2021, 35, 2130002. [Google Scholar] [CrossRef]
- Chalkha, M.; Chebbac, K.; Nour, H.; Asmae, N.; El Moussaoui, A.; Tüzün, B.; Bourhia, M.; Chtita, S.; Bakhouch, M.; Laaroussi, H.; et al. In Vitro and In Silico Evaluation of the Antimicrobial and Antioxidant Activities of Spiropyrazoline Oxindole Congeners. Arab. J. Chem. 2023, 17, 105465. [Google Scholar] [CrossRef]
- Kanzouai, Y.; Chalkha, M.; Hadni, H.; Laghmari, M.; Bouzammit, R.; Nakkabi, A.; Benali, T.; Tüzün, B.; Akhazzane, M.; El Yazidi, M.; et al. Design, Synthesis, in-Vitro and in-Silico Studies of Chromone-isoxazoline Conjugates as Anti-bacterial Agents. J. Mol. Struct. 2023, 129, 136205. [Google Scholar] [CrossRef]
- Rhazi, Y.; Chalkha, M.; Nakkabi, A.; Hammoudan, I.; Akhazzane, M.; Bakhouch, M.; Chtita, S.; El Yazidi, M. Novel Quinazolinone–Isoxazoline Hybrids: Synthesis, Spectroscopic Characterization, and DFT Mechanistic Study. Chemistry 2022, 4, 969–982. [Google Scholar] [CrossRef]
- Chalkha, M.; Moussaoui, A.E.; Hadda, T.B.; Berredjem, M.; Bouzina, A.; Almalki, F.A.; Saghrouchni, H.; Bakhouch, M.; Saadi, M.; El Ammari, L.; et al. Crystallographic Study, Biological Evaluation and DFT/POM/Docking Analyses of Pyrazole Linked Amide Conjugates: Identification of Antimicrobial and Antitumor Pharmacophore Sites. J. Mol. Struct. 2022, 1252, 131818. [Google Scholar] [CrossRef]
- Arzine, A.; Abchir, O.; Chalkha, M.; Chebbac, K.; Rhazi, Y.; Barghady, N.; Yamari, I.; Moussaoui, A.E.L.; Nakkabi, A.; Akhazzane, M.; et al. Design, Synthesis, In-Vitro, In-Silico and DFT Studies of Novel Functionalized Isoxazoles as Antibacterial and Antioxidant Agents. Comput. Biol. Chem. 2023, 108, 107993. [Google Scholar] [CrossRef] [PubMed]
- Elyazidi, M.; Bougrin, K.; Daou, B.; Doua, H.; Soufiaoui, M. Microwave-Assisted Synthesis of New Spiro-isoxazolino-indol-3-ones and 5-Aroylisoxazoles on KF-Al2O3 under Solvent-Free Conditions. J. Soc. Chim. Tunis. 2003, 6, 239–245. [Google Scholar]
- Chalkha, M.; Ameziane el Hassani, A.; Nakkabi, A.; Tüzün, B.; Bakhouch, M.; Benjelloun, A.T.; Sfaira, M.; Saadi, M.; Ammari, L.E.; Yazidi, M. El Crystal Structure, Hirshfeld Surface and DFT Computations, along with Molecular Docking Investigations of a New Pyrazole as a Tyrosine Kinase Inhibitor. J. Mol. Struct. 2023, 1273, 134255. [Google Scholar] [CrossRef]
- Madhavan, S.; Keshri, S.K.; Kapur, M. Transition Metal-Mediated Functionalization of Isoxazoles: A Review. Asian J. Org. Chem. 2021, 10, 3127–3165. [Google Scholar] [CrossRef]
- Hadni, H.; Elhallaoui, M. Discovery of Anti-Colon Cancer Agents Targeting Wild-Type and Mutant P53 Using Computer-Aided Drug Design. J. Biomol. Struct. Dyn. 2023, 41, 10171–10189. [Google Scholar] [CrossRef]
- Islam, A.U.; Hadni, H.; Ali, F.; Abuzreda, A.; Kawsar, S.M.A. Synthesis, Antimicrobial Activity, Molecular Docking, Molecular Dynamics Simulation, and ADMET Properties of the Mannopyranoside Derivatives as Antimicrobial Agents. J. Taibah Univ. Sci. 2024, 18, 2327101. [Google Scholar] [CrossRef]
- Yamada, M.; Hatsuta, K.; Niikawa, M.; Imaishi, H. Detoxification of Aflatoxin B1 Contaminated Maize Using Human CYP3A4. J. Microbiol. Biotechnol. 2020, 30, 1207–1213. [Google Scholar] [CrossRef]
- Ushiyama, F.; Amada, H.; Takeuchi, T.; Tanaka-Yamamoto, N.; Kanazawa, H.; Nakano, K.; Mima, M.; Masuko, A.; Takata, I.; Hitaka, K.; et al. Lead Identification of 8-(Methylamino)-2-oxo-1,2-dihydroquinoline Derivatives as DNA Gyrase Inhibitors: Hit-to-Lead Generation Involving Thermodynamic Evaluation. ACS Omega 2020, 5, 10145–10159. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.A.N.; Chatterjee, S.S.; Hamilton, S.M.; Eltis, L.D.; Chambers, H.F.; Strynadka, N.C.J. Structural and Kinetic Analyses of Penicillin-Binding Protein 4 (PBP4)-Mediated Antibiotic Resistance in Staphylococcus aureus. J. Biol. Chem. 2018, 293, 19854–19865. [Google Scholar] [CrossRef] [PubMed]
- Enguita, F.J.; Marçal, D.; Martins, L.O.; Grenha, R.; Henriques, A.O.; Lindley, P.F.; Carrondo, M.A. Substrate and Dioxygen Binding to the Endospore Coat Laccase from Bacillus Subtilis. J. Biol. Chem. 2004, 279, 23472–23476. [Google Scholar] [CrossRef]
- El Rhabori, S.; El Aissouq, A.; Daoui, O.; Elkhattabi, S.; Chtita, S.; Khalil, F. Design of New Molecules against Cervical Cancer Using DFT, Theoretical Spectroscopy, 2D/3D-QSAR, Molecular Docking, Pharmacophore and ADMET Investigations. Heliyon 2024, 10, e24551. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable Molecular Dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed]
- Hadni, H.; Fitri, A.; Touimi Benjelloun, A.; Benzakour, M.; Mcharfi, M.; Benbrahim, M. Identification of Terpenoids as Potential Inhibitors of SARS-CoV-2 (Main Protease) and Spike (RBD) via Computer-Aided Drug Design. J. Biomol. Struct. Dyn. 2023, 1–14. [Google Scholar] [CrossRef]
- Beney, C.; Mariotte, A.; Boumendjel, A. An Efficient Synthesis of 4,6-Dimethoxyaurones. Heterocycles 2001, 55, 967–972. [Google Scholar] [CrossRef]
- Masoomi, S.; Alipour, E.; Ali, M.; Reza, A.; Shafiee, A. Synthesis of Novel 2-Benzylidenebenzofuran-3(2H)-one Derivatives. Iran. J. Org. Chem. 2011, 3, 733–736. [Google Scholar]
- Bruker APEX4, SAINT & SHELXTL; Bruker AXS LLC: Madison, WI, USA, 2021.
- Krause, L.; Herbst-Irmer, R.; Sheldrick, G.M.; Stalke, D. Comparison of Silver and Molybdenum Microfocus X-ray Sources for Single-Crystal Structure Determination. J. Appl. Crystallogr. 2015, 48, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. SHELXT—Integrated Space-Group and Crystal-Structure Determination. Acta Crystallogr. Sect. A Found. Crystallogr. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. Crystal Structure Refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Brandenburg, K.; Putz, H. DIAMOND; Crystal Impact GbR: Bonn, Germany, 2012. [Google Scholar]
- Chalkha, M.; Nour, H.; Chebbac, K.; Nakkabi, A.; Bahsis, L.; Bakhouch, M.; Akhazzane, M.; Bourass, M.; Chtita, S.; Bin Jardan, Y.A.; et al. Synthesis, Characterization, DFT Mechanistic Study, Antimicrobial Activity, Molecular Modeling, and ADMET Properties of Novel Pyrazole-Isoxazoline Hybrids. ACS Omega 2022, 7, 46731–46744. [Google Scholar] [CrossRef]
- Chebbac, K.; Benziane Ouaritini, Z.; El Moussaoui, A.; Chalkha, M.; Lafraxo, S.; Bin Jardan, Y.A.; Nafidi, H.A.; Bourhia, M.; Guemmouh, R. Antimicrobial and Antioxidant Properties of Chemically Analyzed Essential Oil of Artemisia annua L. (Asteraceae) Native to Mediterranean Area. Life 2023, 13, 807. [Google Scholar] [CrossRef]


| N° | Ar | Formula (M. g/mol) | M.p (°C) | Yield a (%) | 1H-NMR (ppm) | 13C-NMR (ppm) | IR (cm−1) | MS (m/z) [M+H]+ |
|---|---|---|---|---|---|---|---|---|
| CH3 | C=O(ketone) | C=O(ketone) | ||||||
| OCH3 | C=O(ester) | C=O(ester) | ||||||
| CH3(ester) | C=N | C=N | ||||||
| 4a | 4-(CH3)C6H4 | C25H19NO4 (397.43) | 142–144 | 95 | 3.38 | 181.78 | 1667 | 398.41 |
| --- | 168.91 | 1753 | ||||||
| 2.20 | 162.30 | 1603 | ||||||
| 4b | 4-(CH3O)C6H4 | C25H19NO5 (413.42) | 140–142 | 90 | --- | 181.79 | 1666 | 414.36 |
| 3.83 | 168.92 | 1756 | ||||||
| 2.20 | 160.92 | 1604 | ||||||
| 4c | 4-(Cl)C6H4 | C24H16ClNO4 (417.84) | 128–130 | 85 | --- | 181.55 | 1677 | 417.34 |
| --- | 168.95 | 1764 | ||||||
| 2.21 | 161.39 | 1609 | ||||||
| 4d | 4-(CH3)C6H4 | C30H21NO4 (459.50) | 118–120 | 92 | 2.23 | 182.07 | 1667 | 461.42 |
| ---- | 164.36 | 1754 | ||||||
| ---- | 161.89 | 1600 | ||||||
| 4e | 4-(CH3O)C6H4 | C30H21NO5 (475.50) | 124–126 | 89 | ---- | 182.08 | 1670 | 476.43 |
| 3.9 | 164.35 | 1741 | ||||||
| ---- | 161.99 | 1597 | ||||||
| 4f | 4-(Cl)C6H4 | C29H18ClNO4 (479.91) | 126–130 | 91 | ----- | 181.88 | 1677 | 481.36 |
| ----- | 164.30 | 1760 | ||||||
| ----- | 161.39 | 1609 |
| D—H···A | D—H | H···A | D···A | D—H···A |
| C7—H7···Cg4 i | 0.95 | 2.77 | 3.5994 (17) | 146 |
| C10—H10···O2 ii | 0.95 | 2.43 | 3.3645 (18) | 170 |
| C21—H21···O4 iii | 0.95 | 2.36 | 3.289 (2) | 164 |
| C23—H23···O2 iv | 0.95 | 2.58 | 3.4615 (17) | 154 |
| D—H···A | D—H | H···A | D···A | D—H···A |
| C13—H13···Cg1 | 0.95 | 2.87 | 3.534 (2) | 128 |
| C23—H23···Cg1 i | 0.95 | 2.87 | 3.744 (2) | 154 |
| C28—H28···Cg2 ii | 0.95 | 2.76 | 3.532 (2) | 140 |
| Zone of Inhibition (ZI) in mm a | |||||
|---|---|---|---|---|---|
| Compounds | Tested Bacteria | ||||
| N° | Ar | Group | Escherichia coli | Bacillus subtilis | Staphylococcus aureus |
| 4a | 4-(CH3)C6H4 | acetoxy | 17.5 ± 1.20 | 14.5 ± 0.80 | 9.5 ± 1.40 |
| 4b | 4-(CH3O)C6H4 | acetoxy | 12.5 ± 1.40 | - | - |
| 4c | 4-(Cl)C6H4 | acetoxy | 13.5 ± 1.50 | 11 ± 1.20 | 10.5 ± 1.50 |
| 4d | 4-(CH3)C6H4 | benzoyloxy | 11 ± 0.50 | 11.5 ± 1.15 | 12 ± 1.05 |
| 4e | 4-(CH3O)C6H4 | benzoyloxy | 14.5 ± 0.90 | 09 ± 0.60 | 13.75 ± 1.25 |
| 4f | 4-(Cl)C6H4 | benzoyloxy | 13.25 ± 0.40 | 11.25 ± 0.75 | 15.5 ± 1.55 |
| Ampicillin | NT | 16 ± 1.30 | 23 ± 2.60 | ||
| Streptomycin | 24 ± 1.60 | NT | NT | ||
| Compounds | Escherichia coli (6kzv) (kcal/mol) | Staphylococcus aureus (5tw8) (kcal/mol) | Bacillus subtilis (1of0) (kcal/mol) |
|---|---|---|---|
| 4a | −10.82 | −9.01 | −9.21 |
| 4b | −10.18 | −8.16 | −8.58 |
| 4c | −9.38 | −10.02 | −8.75 |
| 4d | −10.24 | −10.45 | −10.47 |
| 4e | −10.07 | −10.43 | −10.32 |
| 4f | −10.85 | −10.34 | −11.17 |
| Ampicillin | −9.58 | −8.82 | |
| Streptomycin | −12.84 |
| Compounds | Absorption | Distribution | Metabolism | Excretion | Toxicity | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Intestinal Absorption (Human) | BBB Permeability | CNS Permeability | Substrate | Inhibitor | Total Clearance | AMES Toxicity | ||||||
| CYP | ||||||||||||
| 2D6 | 3A4 | 1A2 | 2C19 | 2C9 | 2D6 | 3A4 | ||||||
| Numeric (% Absorbed) | Numeric (LogBB) | Numeric (LogPS) | Categorical (Yes/No) | Numeric (Log mL/min/kg) | Categorical (Yes/No) | |||||||
| 4a | 99.42 | −0.626 | −1.646 | No | Yes | Yes | Yes | Yes | No | Yes | 0.283 | No |
| 4b | 100 | −0.873 | −1.921 | No | Yes | Yes | Yes | Yes | No | Yes | 0.314 | No |
| 4c | 97.961 | −0.826 | −1.606 | No | Yes | Yes | Yes | Yes | Yes | Yes | 0.15 | No |
| 4d | 99.926 | −0.637 | −1.364 | No | Yes | No | Yes | Yes | No | No | 0.502 | No |
| 4e | 100 | −0.884 | −1.639 | No | Yes | No | Yes | Yes | No | Yes | 0.486 | No |
| 4f | 98.467 | −0.837 | −1.324 | No | Yes | No | Yes | Yes | No | No | 0.11 | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arzine, A.; Hadni, H.; Boujdi, K.; Chebbac, K.; Barghady, N.; Rhazi, Y.; Chalkha, M.; Nakkabi, A.; Chkirate, K.; Mague, J.T.; et al. Efficient Synthesis, Structural Characterization, Antibacterial Assessment, ADME-Tox Analysis, Molecular Docking and Molecular Dynamics Simulations of New Functionalized Isoxazoles. Molecules 2024, 29, 3366. https://doi.org/10.3390/molecules29143366
Arzine A, Hadni H, Boujdi K, Chebbac K, Barghady N, Rhazi Y, Chalkha M, Nakkabi A, Chkirate K, Mague JT, et al. Efficient Synthesis, Structural Characterization, Antibacterial Assessment, ADME-Tox Analysis, Molecular Docking and Molecular Dynamics Simulations of New Functionalized Isoxazoles. Molecules. 2024; 29(14):3366. https://doi.org/10.3390/molecules29143366
Chicago/Turabian StyleArzine, Aziz, Hanine Hadni, Khalid Boujdi, Khalid Chebbac, Najoua Barghady, Yassine Rhazi, Mohammed Chalkha, Asmae Nakkabi, Karim Chkirate, Joel T. Mague, and et al. 2024. "Efficient Synthesis, Structural Characterization, Antibacterial Assessment, ADME-Tox Analysis, Molecular Docking and Molecular Dynamics Simulations of New Functionalized Isoxazoles" Molecules 29, no. 14: 3366. https://doi.org/10.3390/molecules29143366
APA StyleArzine, A., Hadni, H., Boujdi, K., Chebbac, K., Barghady, N., Rhazi, Y., Chalkha, M., Nakkabi, A., Chkirate, K., Mague, J. T., Kawsar, S. M. A., Al Houari, G., M. Alanazi, M., & El Yazidi, M. (2024). Efficient Synthesis, Structural Characterization, Antibacterial Assessment, ADME-Tox Analysis, Molecular Docking and Molecular Dynamics Simulations of New Functionalized Isoxazoles. Molecules, 29(14), 3366. https://doi.org/10.3390/molecules29143366










