Safflower Flavonoid 3′5′Hydroxylase Promotes Methyl Jasmonate-Induced Anthocyanin Accumulation in Transgenic Plants
Abstract
1. Introduction
2. Results
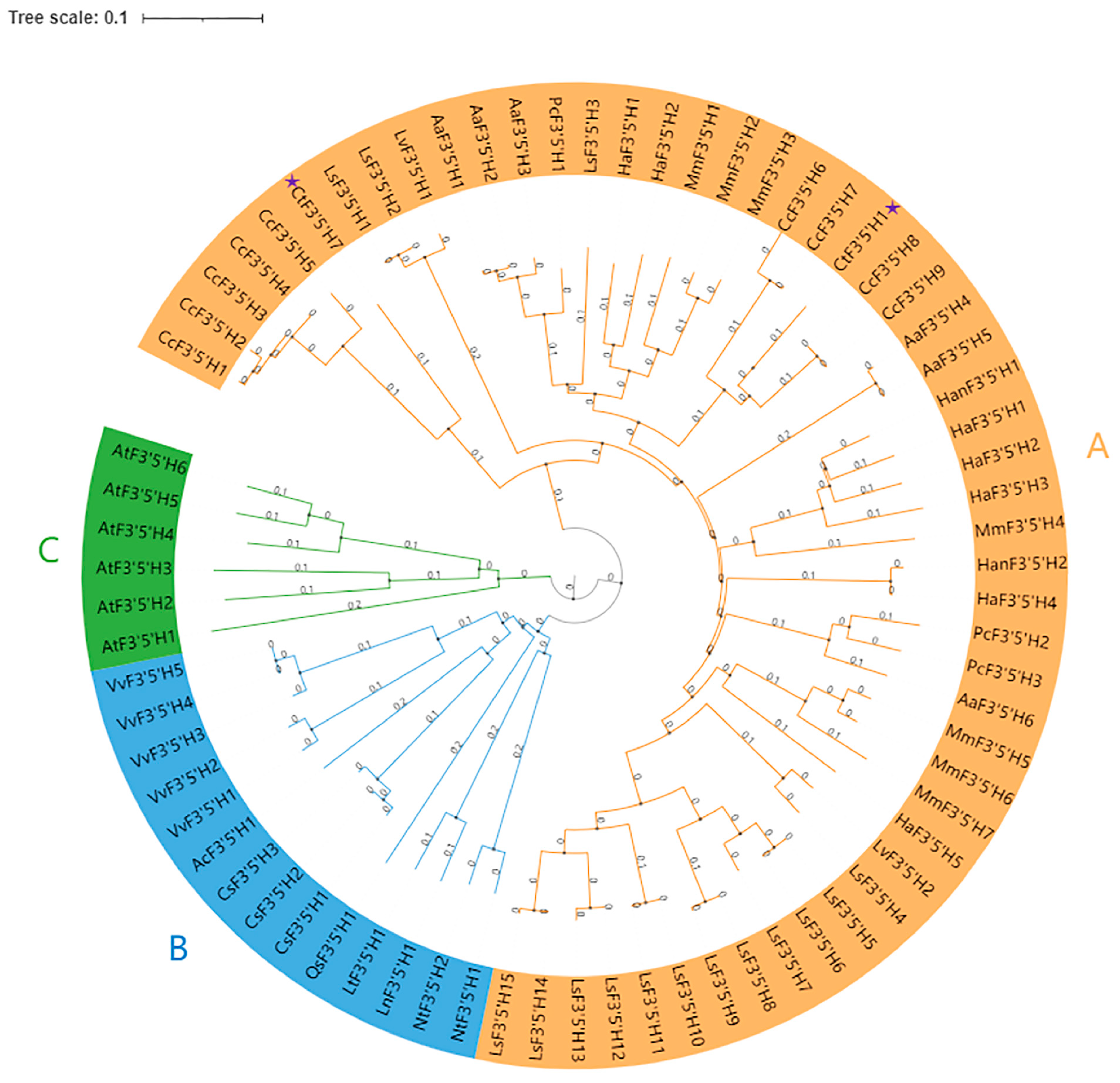
2.1. Identification, Physicochemical Properties, and Phylogenetic Analysis
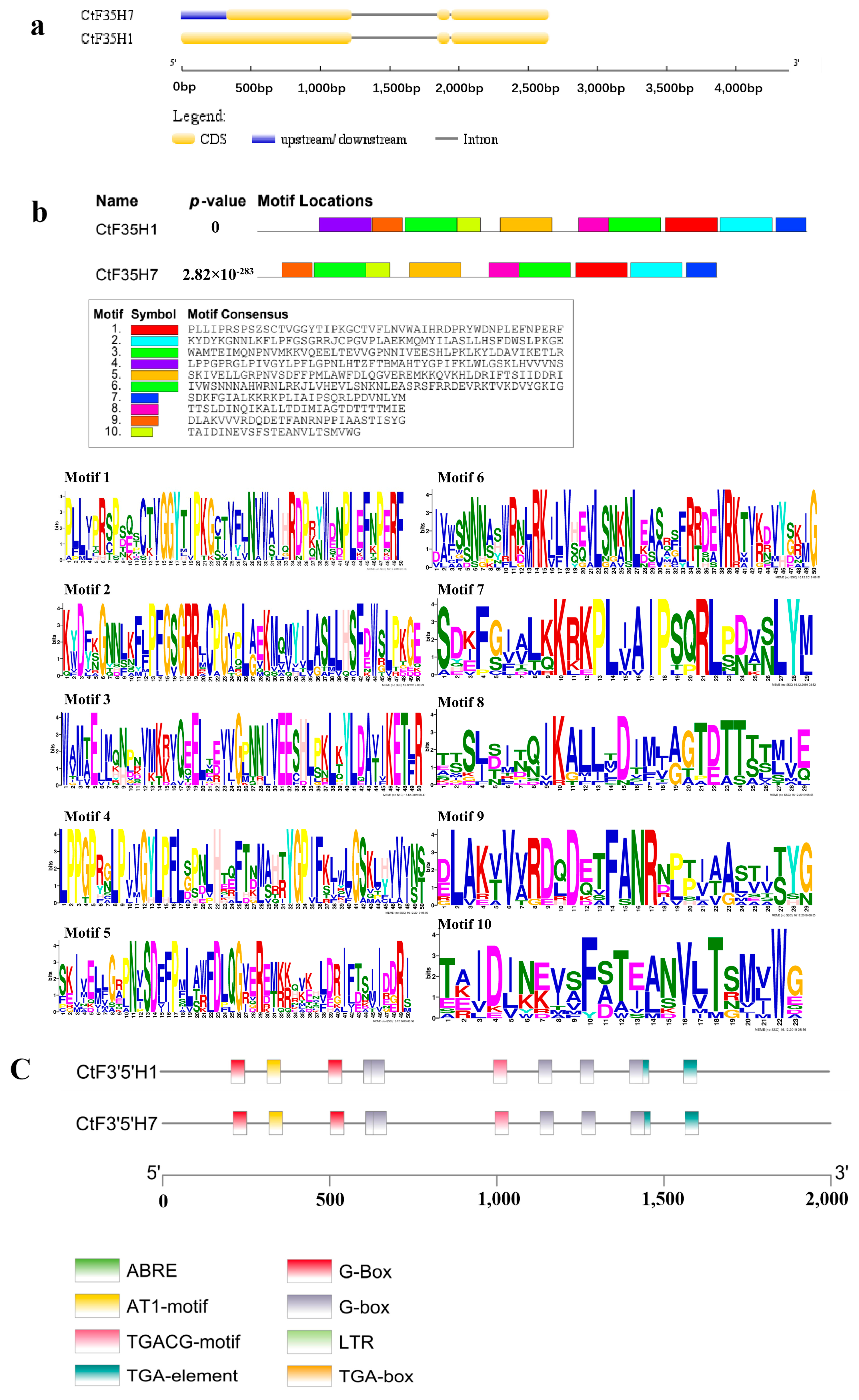
2.2. Gene Structure, Protein Motif, and Cis-Regulatory Units
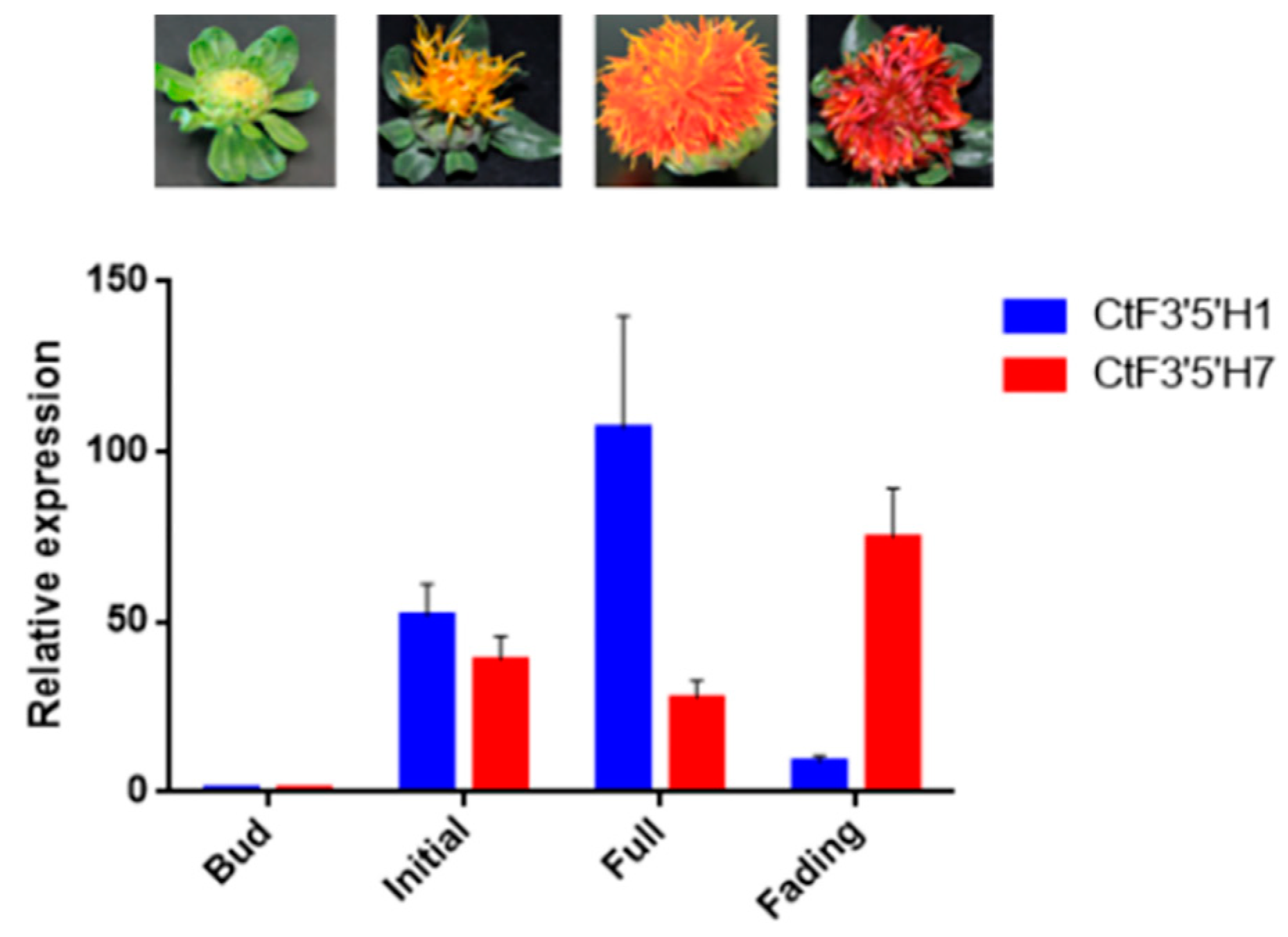
2.3. Tissue Specific Expression of Two CtF3′5′H Genes Revealed Differential Pattern of Transcriptional Regulation in Safflower
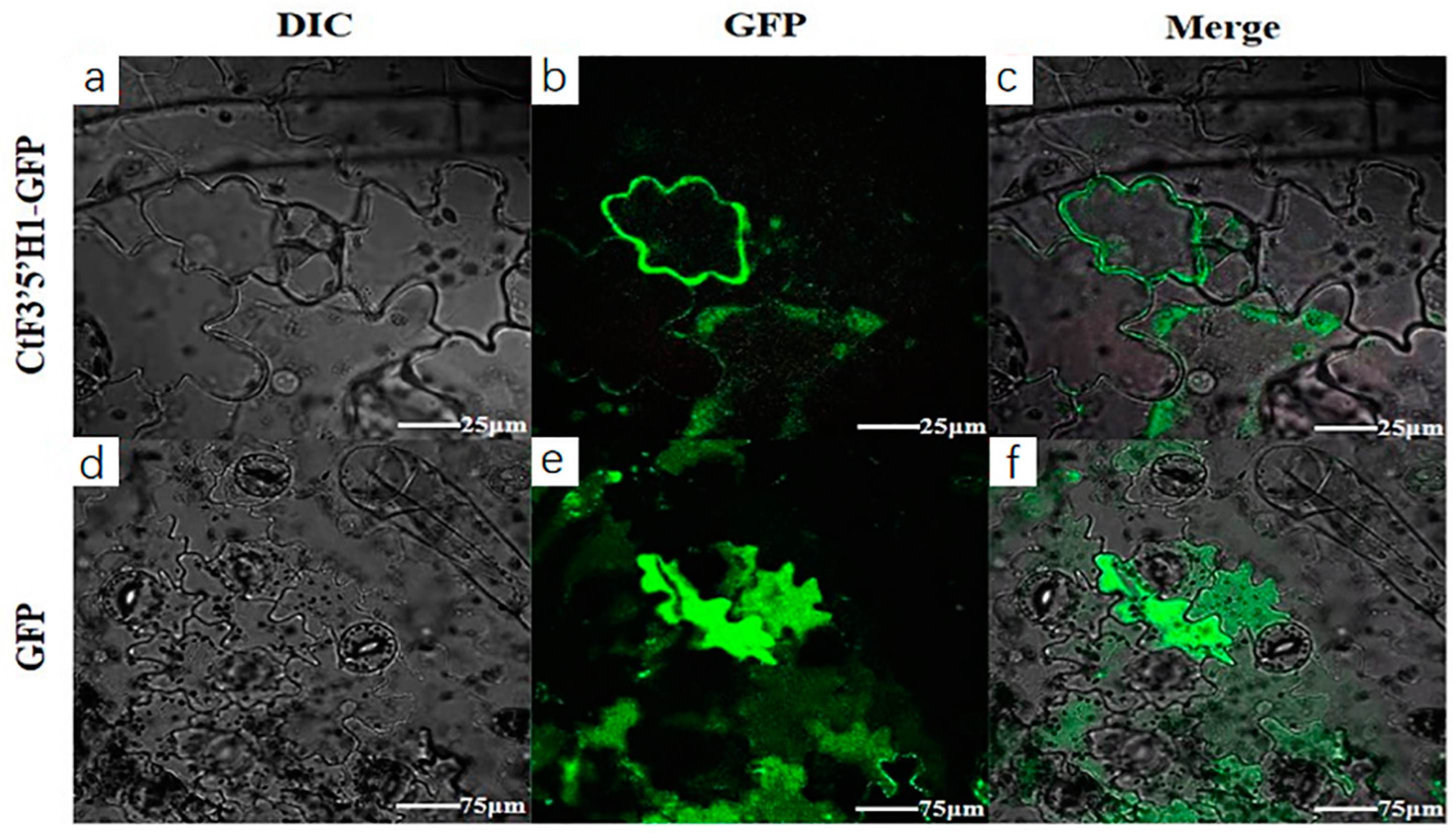
2.4. Molecular Cloning and Subcellular Localization
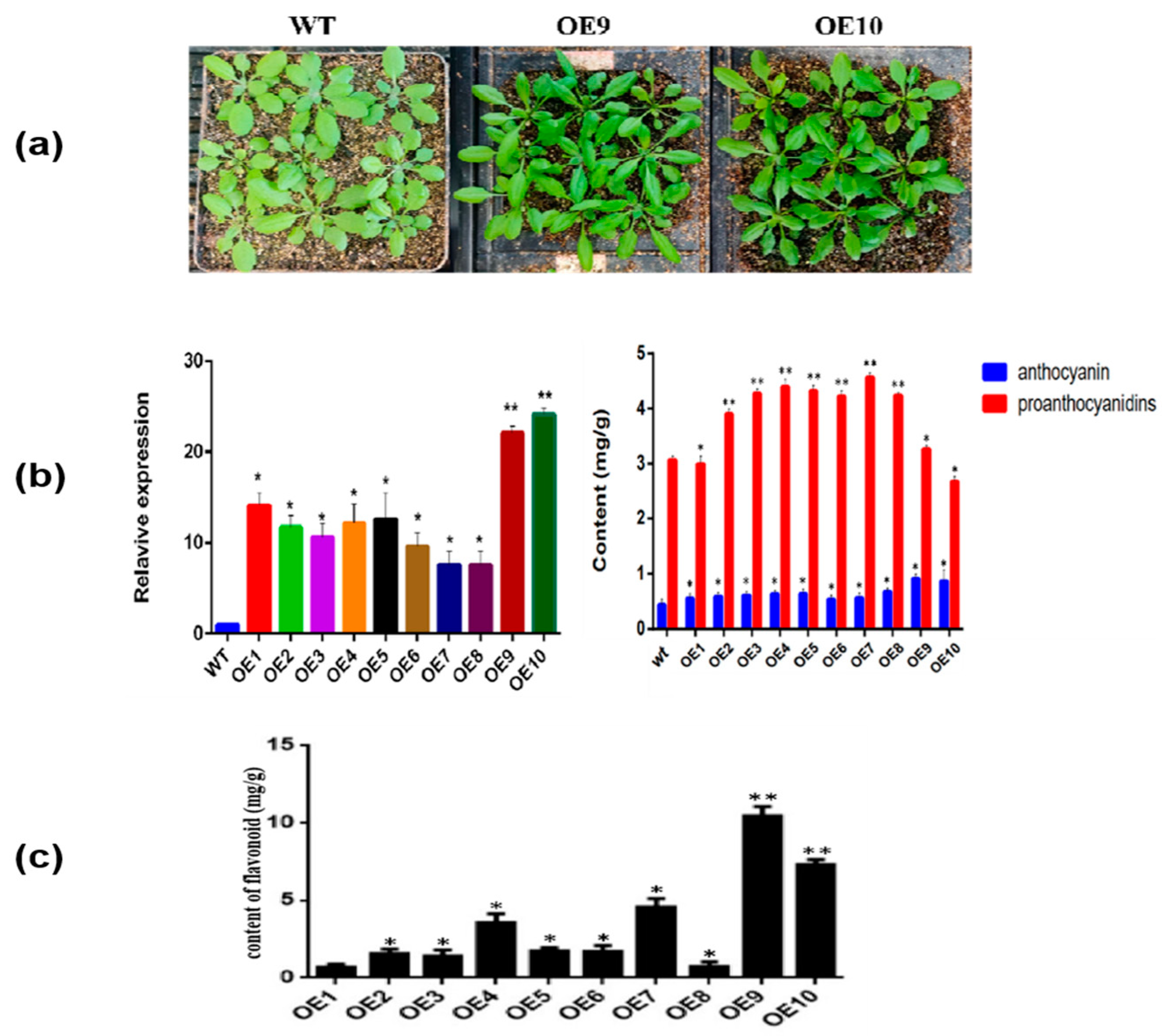
2.5. Generation of CtF3′5′H1 Overexpressed Arabidopsis Lines and Expression Analysis
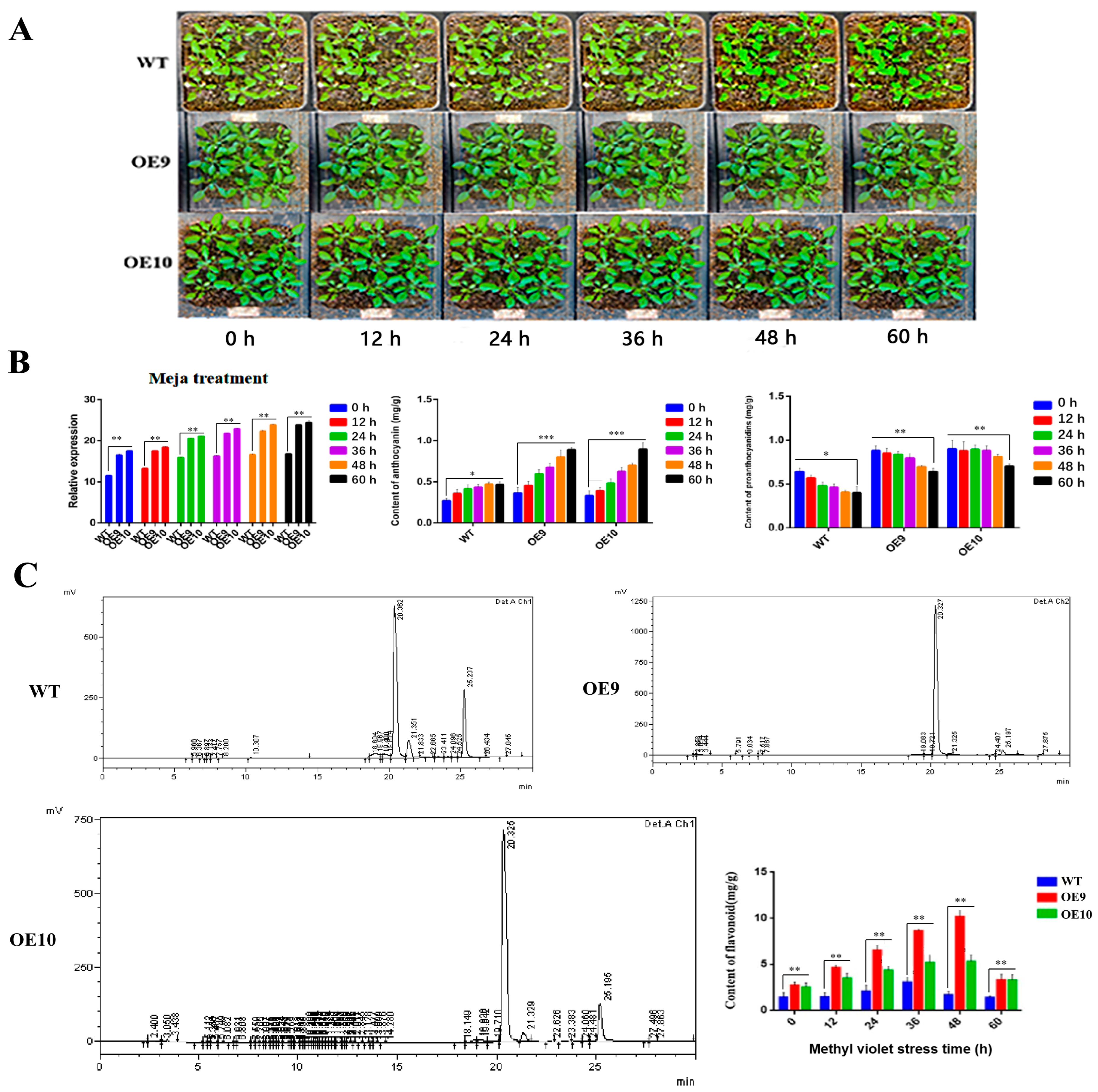
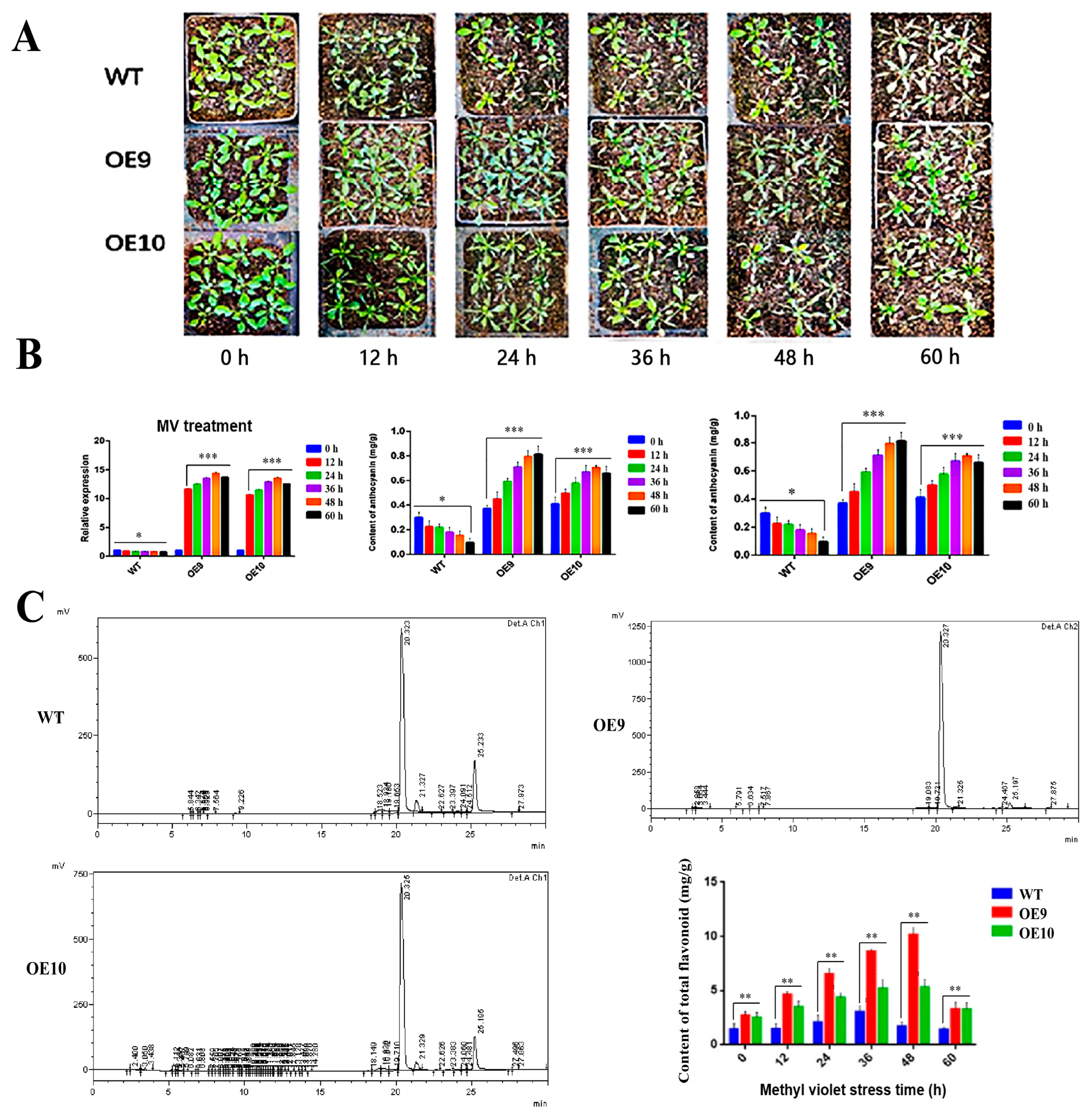
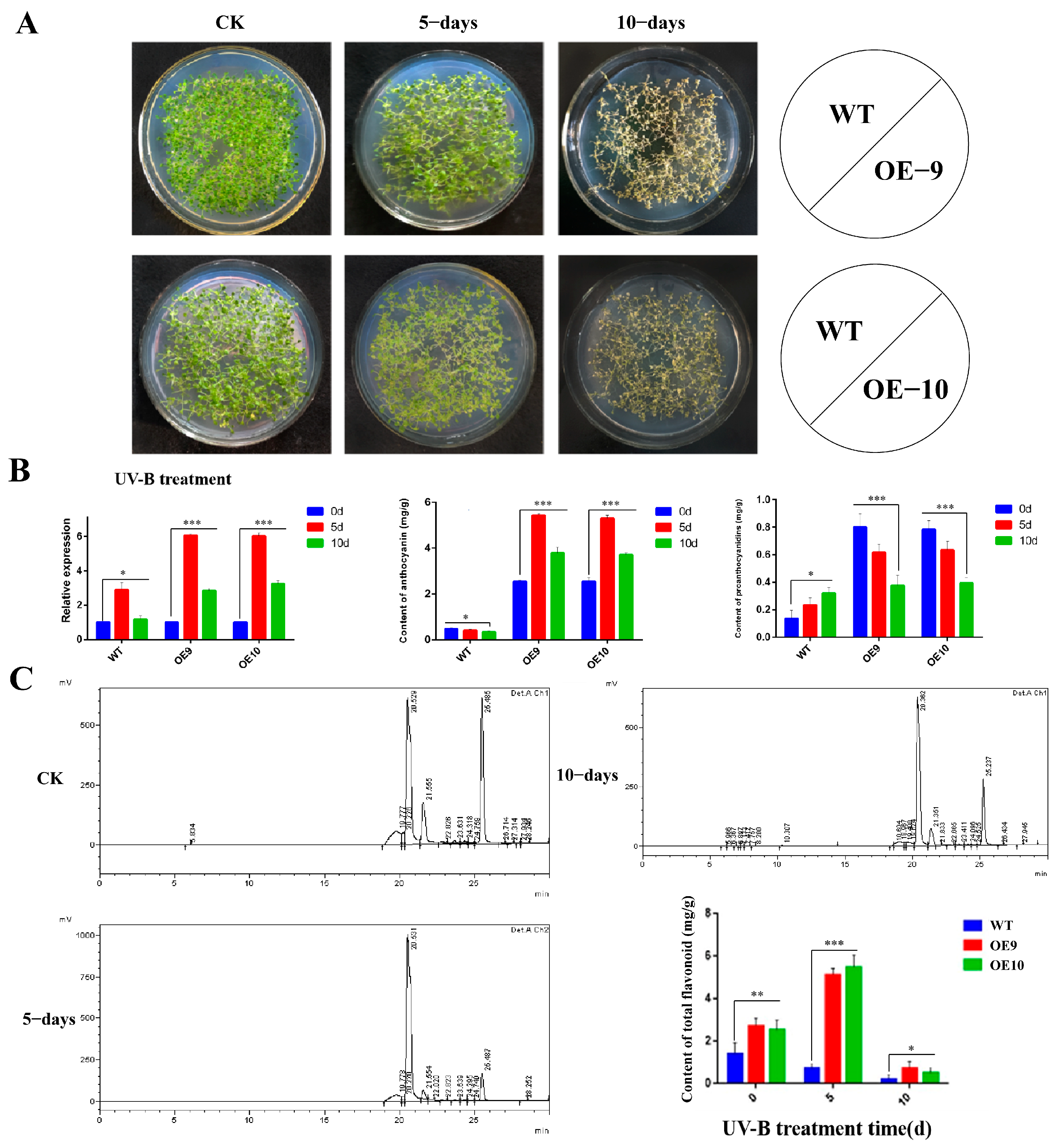
2.6. Relative Expression of CtF3′5′H1 and Determination of Metabolites in Transgenic Arabidopsis under Various Abiotic Stresses
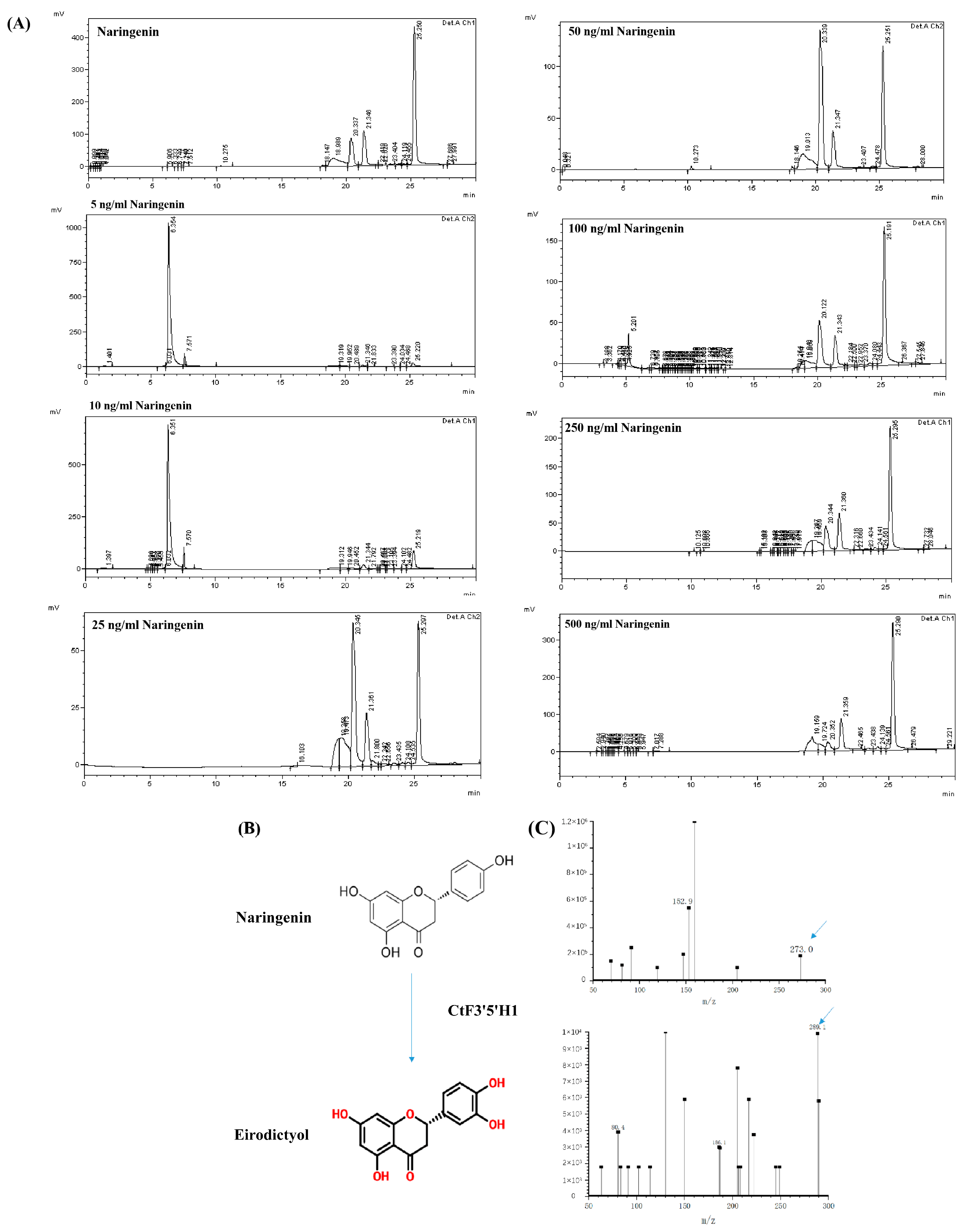
2.7. Recombinant CtF3′5′H1 Protein Possess In Vitro Catalytic Activity
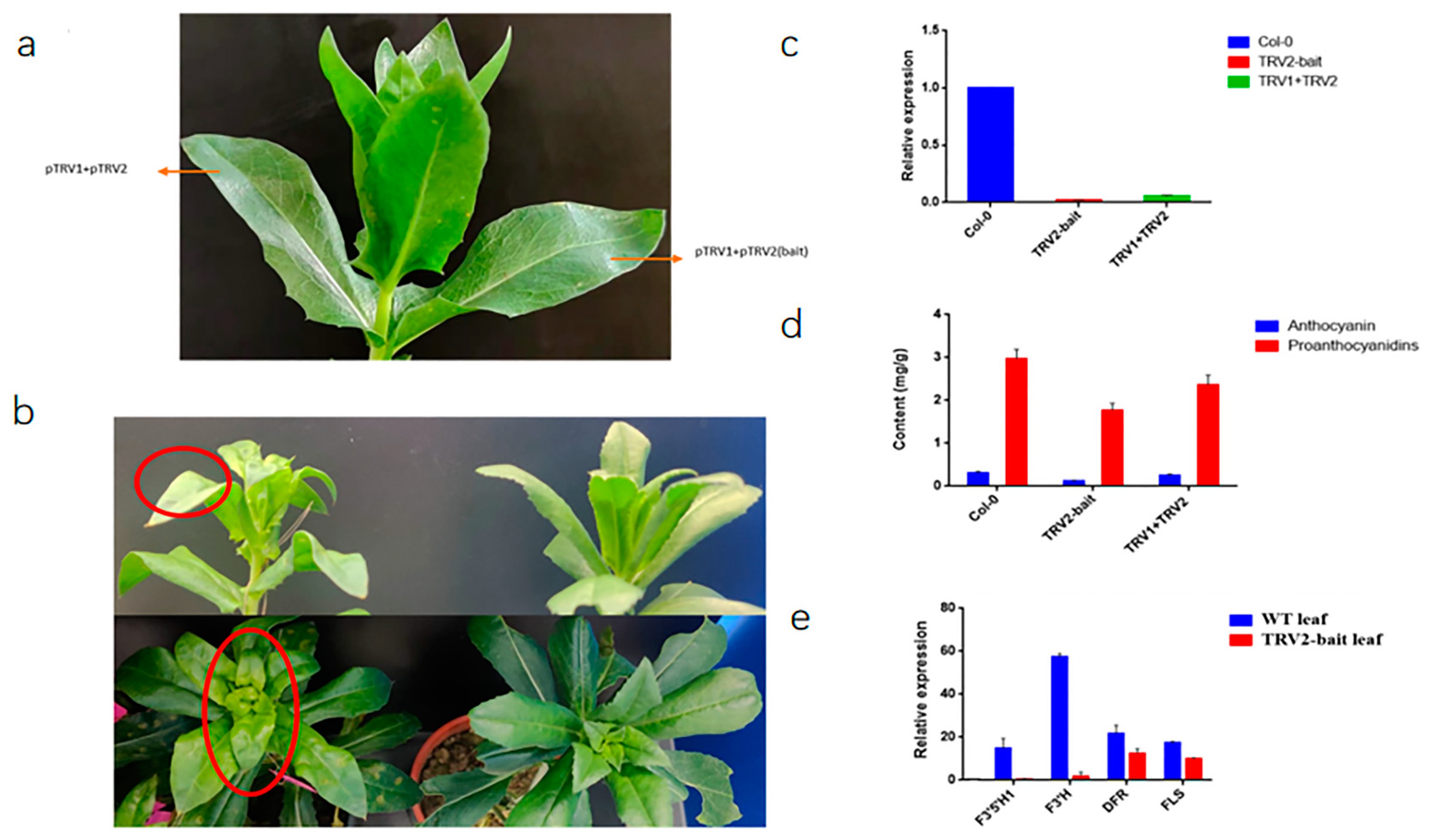
2.8. Virus-Induced Gene Silencing (VIGS) Assay of CtF3′5′H1 in Safflower
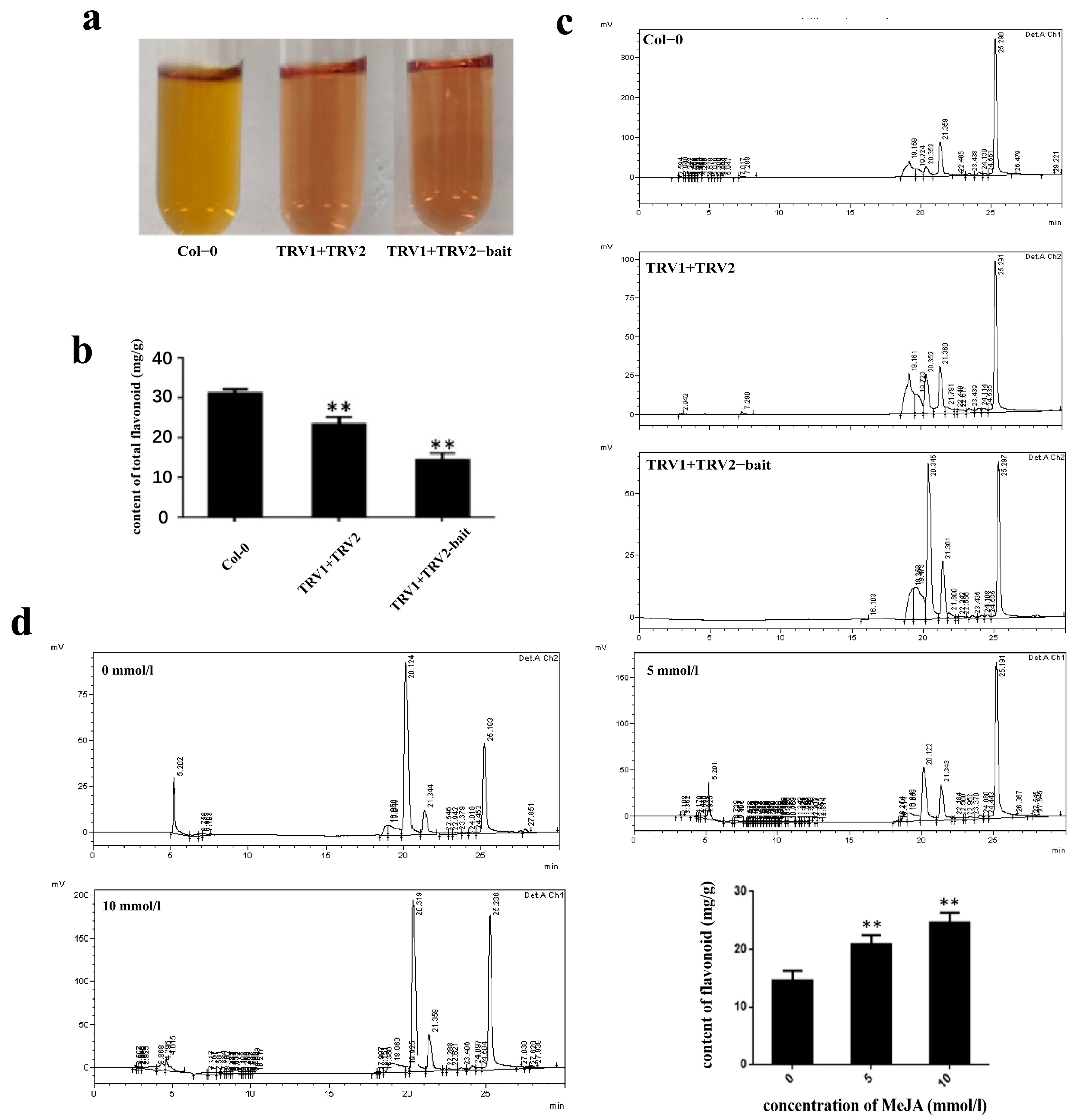
2.9. Quantification of Total Flavonoids Content during VIGS by HPLC under MeJA Stress
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Vectors and Strains
4.2. Genome-Wide Identification of CtF3′5′H Genes from Safflower
4.3. Phylogenetic Analysis
4.4. Gene Structure, Conserved Motifs, and Promoter Analysis
4.5. Expression Analysis of CtF3′5′H1 and CtF3′5′H7 in Safflower
4.6. Molecular Cloning of CtF3′5′H1 and GFP Translational Fusion Construction
4.7. Generation of Transgenic Arabidopsis via Floral Dip Transformation
4.8. Abiotic Stress Elicitation of CtF3′5′H1-Overexpressed Transgenic Arabidopsis
4.9. Quantitative Analysis of Anthocyanin, Proanthocyanidins and Total Flavonoid Content
4.10. Recombinant Protein Expression and Purification
4.11. In Vitro Enzyme Activity Assays of Recombinant CtF3′5′H1 Proteins
4.12. Suppression of Agrobacterium Tumefaciens Infection of Safflower
4.13. Safflower Leaves under Methyl Jasmonate Stress after Gene Silencing
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- An, X.-H.; Tian, Y.; Chen, K.-Q.; Liu, X.-J.; Liu, D.-D.; Xie, X.-B.; Cheng, C.-G.; Cong, P.-H.; Hao, Y.-J. MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol. 2015, 56, 650–662. [Google Scholar] [CrossRef]
- Balbi, V.; Devoto, A. Network in Arabidopsis thaliana: Crucial regulatory nodes and new physiological scenarios. New Phytol. 2008, 177, 301–318. [Google Scholar] [CrossRef] [PubMed]
- Bogs, J.; Ebadi, A.; McDavid, D.; Robinson, S.P. Identification of the flavonoid hydroxylases from grapevine and their regulation during fruit development. Plant Physiol. 2006, 140, 279–291. [Google Scholar] [CrossRef] [PubMed]
- Browse, J. Jasmonate passes muster: A receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 2009, 60, 183–205. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Qiao, L.; Wang, M.; He, B.; Lin, F.M.; Palmquist, J.; Huang, S.D.; Jin, H. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 2018, 360, 1126–1129. [Google Scholar] [CrossRef]
- Chen, J.; Wang, J.; Wang, R.; Xian, B.; Ren, C.; Liu, Q.; Wu, Q.; Pei, J. Integrated metabolomics and transcriptome analysis on flavonoid biosynthesis in safflower (Carthamus tinctorius L.) under MeJA treatment. BMC Plant Biol. 2020, 20, 353. [Google Scholar] [CrossRef]
- Bai, X.; Xie, T.; Guo, R.; Liu, J.; Yu, H.; Xie, L. Comparison of transcriptional expression patterns of phenols and carotenoids in ‘Kyoho’ grapes under a two-crop-a-year cultivation system. PLoS ONE 2019, 14, e0210322. [Google Scholar]
- Cristóbal, C.M.; Figueroa, N.E.; Poblete, L.A.; Oñate, F.A.; Schwab, W.; Figueroa, C.R. Methyl jasmonate treatment induces changes in fruit ripening by modifying the expression of several ripening genes in Fragaria chiloensis fruit. Plant Physiol. Biochem. 2013, 70, 433–444. [Google Scholar]
- Croft, K.D. The chemistry and biological effects of flavonoids and phenolic acids. Ann. N. Y. Acad. Sci. 1998, 854, 435–442. [Google Scholar] [CrossRef]
- Das, P.K.; Shin, D.H.; Choi, S.-B.; Yoo, S.-D.; Choi, G.; Park, Y.-I. Cytokinins enhance sugar-induced anthocyanin biosynthesis in Arabidopsis. Mol. Cells 2012, 34, 93–101. [Google Scholar] [CrossRef]
- Delgado, L.D.; Zúñiga, P.E.; Figueroa, N.E.; Pastene, E.; Escobar-Sepúlveda, H.F.; Figueroa, P.M.; Garrido-Bigotes, A.; Figueroa, C.R. Application of a JA-Ile Biosynthesis Inhibitor to Methyl Jasmonate-Treated Strawberry Fruit Induces Upregulation of Specific MBW Complex-Related Genes and Accumulation of Proanthocyanidins. Molecules 2018, 23, 1433. [Google Scholar] [CrossRef] [PubMed]
- De Vetten, N.; ter Horst, J.; van Schaik, H.P.; de Boer, A.; Mol, J.; Koes, R. A cytochrome b5 is required for full activity of flavonoid 3′, 5′-hydroxylase, a cytochrome P450 involved in the formation of blue flower colors. Proc. Natl. Acad. Sci. USA 1999, 96, 778–783. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Flores, G.; Blanch, G.P.; del Castillo, M.L.R. Technology, Postharvest treatment with (−) and (+)-methyl jasmonate stimulates anthocyanin accumulation in grapes. LWT-Food Sci. Technol. 2015, 62, 807–812. [Google Scholar] [CrossRef]
- Gourlay, G.; Constabel, C.P. Condensed tannins are inducible antioxidants and protect hybrid poplar against oxidative stress. Tree Physiol. 2019, 39, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Gao, L.; Ma, X.; Guo, F.; Ruan, H.; Bao, Y.; Xia, T.; Wang, Y. Functional analysis of flavonoid 3′-hydroxylase and flavonoid 3′, 5′-hydroxylases from tea plant (Camellia sinensis), involved in the B-ring hydroxylation of flavonoids. Gene 2019, 717, 144046. [Google Scholar] [CrossRef]
- Hammerbacher, A.; Raguschke, B.; Wright, L.P.; Gershenzon, J. Gallocatechin biosynthesis via a flavonoid 3′, 5′-hydroxylase is a defense response in Norway spruce against infection by the bark beetle-associated sap-staining fungus Endoconidiophora polonica. Phytochemistry 2018, 148, 78–86. [Google Scholar] [CrossRef]
- He, J.B.; Zhao, P.; Hu, Z.M.; Liu, S.; Kuang, Y.; Zhang, M.; Li, B.; Yun, C.H.; Qiao, X.; Ye, M. Molecular and Structural Characterization of a Promiscuous C-Glycosyltransferase from Trollius chinensis. Angew. Chem. 2019, 131, 11637–11644. [Google Scholar]
- Holton, T.A.; Brugliera, F.; Lester, D.R.; Tanaka, Y.; Hyland, C.D.; Menting, J.G.; Lu, C.-Y.; Farcy, E.; Stevenson, T.W.; Cornish, E.C. Cloning and expression of cytochrome P450 genes controlling flower colour. Nature 1993, 366, 276–279. [Google Scholar] [CrossRef]
- Huang, X.; Li, J.; Shang, H.; Meng, X. Effect of methyl jasmonate on the anthocyanin content and antioxidant activity of blueberries during cold storage. J. Sci. Food Agric. 2015, 95, 337–343. [Google Scholar] [CrossRef]
- Jin, J.-Q.; Ma, J.-Q.; Yao, M.-Z.; Ma, C.-L.; Chen, L. Functional natural allelic variants of flavonoid 3′, 5′-hydroxylase gene governing catechin traits in tea plant and its relatives. Planta 2017, 245, 523–538. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.S.; Griffiths, H.M.; De Jong, D.M.; Cheng, S.; Bodis, M.; De Jong, W.S. The potato P locus codes for flavonoid 3′, 5′-hydroxylase. Theor. Appl. Genetics 2005, 110, 269–275. [Google Scholar] [CrossRef]
- Kaltenbach, M.; Schröder, G.; Schmelzer, E.; Lutz, V.; Schröder, J. Flavonoid hydroxylase from Catharanthus roseus: cDNA, heterologous expression, enzyme properties and cell-type specific expression in plants. Plant J. 1999, 19, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Zhao, H.; Liu, X.; Liu, D.; Chen, J.; Li, Z.; Chu, S.; Kou, X.; Liao, S.; Deng, Y. Protective effect of hydroxysafflor yellow A on nephropathy by attenuating oxidative stress and inhibiting apoptosis in induced type 2 diabetes in rat. Oxidative Med. Cell. Longevity 2020, 2020, 7805393. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Lin, Y.; Zhang, Z.; Shen, J.; Yang, C.; Jiang, M.; Hou, Y. Systematic analysis of bacteriostatic mechanism of flavonoids using transcriptome and its therapeutic effect on vaginitis. Aging 2020, 12, 6292–6305. [Google Scholar] [CrossRef]
- Moreau, C.; Ambrose, M.J.; Turner, L.; Hill, L.; Ellis, T.N.; Hofer, J.M. The b gene of pea encodes a defective flavonoid 3′5-hydroxylase, and confers pink flower color. Plant Physiol. 2012, 159, 759–768. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Zhao, Y.; Tao, R.; Yin, L.; Gao, L.; Strid, Å.; Qian, M.; Li, J.; Li, Y.; Shen, J. Ethylene mediates the branching of the jasmonate-induced flavonoid biosynthesis pathway by suppressing anthocyanin biosynthesis in red Chinese pear fruits. Plant Biotechnol. J. 2020, 18, 1223–1240. [Google Scholar] [CrossRef]
- Olsen, K.M.; Hehn, A.; Jugdé, H.; Slimestad, R.; Larbat, R.; Bourgaud, F.; Lillo, C. Identification and characterisation of CYP75A31, a new flavonoid 3′5′-hydroxylase, isolated from Solanum lycopersicum. BMC Plant Biol. 2010, 10, 21. [Google Scholar] [CrossRef]
- Opriș, O.; Lung, I.; Soran, M.-L.; Ciorîță, A.; Copolovici, L. Investigating the effects of non-steroidal anti-inflammatory drugs (NSAIDs) on the composition and ultrastructure of green leafy vegetables with important nutritional values. Plant Physiol. Biochem. 2020, 151, 342–351. [Google Scholar] [CrossRef]
- Peng, Z.; Wang, Y.; Zuo, W.T.; Gao, Y.R.; Li, R.Z.; Yu, C.X.; Liu, Z.Y.; Zheng, Y.; Shen, Y.Y.; Duan, L.S. Integration of Metabolome and Transcriptome Studies Reveals Flavonoids, Abscisic Acid, and Nitric Oxide Comodulating the Freezing Tolerance in Liriope spicata. Front. Plant Sci. 2022, 12, 764625. [Google Scholar] [CrossRef]
- Qi, T.; Song, S.; Ren, Q.; Wu, D.; Huang, H.; Chen, Y.; Fan, M.; Peng, W.; Ren, C.; Xie, D. The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 2011, 23, 1795–1814. [Google Scholar] [CrossRef] [PubMed]
- Qian, M.; Yu, B.; Li, X.; Sun, Y.; Zhang, D.; Teng, Y. Isolation and expression analysis of anthocyanin biosynthesis genes from the red Chinese sand pear, Pyrus pyrifolia Nakai cv. Mantianhong, in response to methyl jasmonate treatment and UV-B/VIS conditions. Plant Mol. Biol. Report. 2014, 32, 428–437. [Google Scholar] [CrossRef]
- Reymond, P.; Farmer, E.E. Jasmonate and salicylate as global signals for defense gene expression. Curr. Opin. Plant Biol. 1998, 1, 404–411. [Google Scholar] [CrossRef] [PubMed]
- Rinaldo, A.R.; Cavallini, E.; Jia, Y.; Moss, S.M.; McDavid, D.A.; Hooper, L.C.; Robinson, S.P.; Tornielli, G.B.; Zenoni, S.; Ford, C.M. A grapevine anthocyanin acyltransferase, transcriptionally regulated by VvMYBA, can produce most acylated anthocyanins present in grape skins. Plant Physiol. 2015, 169, 1897–1916. [Google Scholar] [CrossRef] [PubMed]
- Rommens, C.M.; Richael, C.M.; Yan, H.; Navarre, D.A.; Ye, J.; Krucker, M.; Swords, K. Engineered native pathways for high kaempferol and caffeoylquinate production in potato. Plant Biotechnol. J. 2008, 6, 870–886. [Google Scholar] [CrossRef]
- Sanders, P.M.; Lee, P.Y.; Biesgen, C.; Boone, J.D.; Beals, T.P.; Weiler, E.W.; Goldberg, R.B. The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 2000, 12, 1041–1061. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, F.M.; Coelho, M.N.; do Nascimento José-Chagas, F.; do Carmo Malvar, D.; Kanashiro, A.; Cunha, F.Q.; Vianna-Filho, M.D.M.; da Cunha Pinto, A.; Vanderlinde, F.A.; Costa, S.S. Oral treatments with a flavonoid-enriched fraction from Cecropia hololeuca and with rutin reduce articular pain and inflammation in murine zymosan-induced arthritis. J. Ethnopharmacol. 2020, 260, 112841. [Google Scholar] [CrossRef]
- Tohge, T.; de Souza, L.P.; Fernie, A.R. Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J. Exp. Bot. 2017, 68, 4013–4028. [Google Scholar] [CrossRef]
- Wang, T.; Gao, X.; Chen, S.; Li, D.; Chen, S.; Xie, M.; Xu, Z.; Yang, G. Genome-wide identification and expression analysis of ethylene responsive factor family transcription factors in Juglans regia. PeerJ. 2021, 9, e12429. [Google Scholar] [CrossRef]
- Wang, Y.; Chai, C.; Khatabi, B.; Scheible, W.R.; Udvardi, M.K.; Saha, M.C.; Kang, Y.; Nelson, R.S. An Efficient Brome mosaic virus-Based Gene Silencing Protocol for Hexaploid Wheat (Triticum aestivum L.). Front. Plant Sci. 2021, 12, 685187. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, W.; Jiang, H.; Mao, Z.; Wang, N.; Jiang, S.; Xu, H.; Yang, G.; Zhang, Z.; Chen, X. The R2R3-MYB transcription factor MdMYB24-like is involved in methyl jasmonate-induced anthocyanin biosynthesis in apple. Plant Physiol. Biochem. 2019, 139, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C. Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 2007, 100, 681–697. [Google Scholar] [CrossRef] [PubMed]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Grzech, D.; Koudounas, K.; Stander, E.A.; Caputi, L.; Mimura, T.; Courdavault, V.; O’Connor, S.E. Improved virus-induced gene silencing allows discovery of a serpentine synthase gene in Catharanthus roseus. Plant Physiol. 2021, 187, 846–857. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wang, R.; Cheng, X.; Qu, H.; Qi, J.; Li, D.; Xing, Y.; Bai, Y.; Zheng, X. The vascular dilatation induced by hydroxysafflor yellow A (HSYA) on rat mesenteric artery through TRPV4-dependent calcium influx in endothelial cells. J. Ethnopharmacol. 2020, 256, 112790. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Li, F.-D.; Li, K.; Wang, Z.-L.; Wang, Y.-X.; He, J.-B.; Su, H.-F.; Zhang, Z.-Y.; Chi, C.-B.; Shi, X.-M. Functional characterization and structural basis of an efficient di-C-glycosyltransferase from Glycyrrhiza glabra. J. Am. Chem. Soc. 2020, 142, 3506–3512. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Ahmad, N.; Zhang, Q.; Wakeel Umar, A.; Wang, N.; Zhao, X.; Zhou, K.; Yao, N.; Liu, X. Safflower Flavonoid 3′5′Hydroxylase Promotes Methyl Jasmonate-Induced Anthocyanin Accumulation in Transgenic Plants. Molecules 2023, 28, 3205. https://doi.org/10.3390/molecules28073205
Zhang X, Ahmad N, Zhang Q, Wakeel Umar A, Wang N, Zhao X, Zhou K, Yao N, Liu X. Safflower Flavonoid 3′5′Hydroxylase Promotes Methyl Jasmonate-Induced Anthocyanin Accumulation in Transgenic Plants. Molecules. 2023; 28(7):3205. https://doi.org/10.3390/molecules28073205
Chicago/Turabian StyleZhang, Xinyue, Naveed Ahmad, Qingyu Zhang, Abdul Wakeel Umar, Nan Wang, Xu Zhao, Kang Zhou, Na Yao, and Xiuming Liu. 2023. "Safflower Flavonoid 3′5′Hydroxylase Promotes Methyl Jasmonate-Induced Anthocyanin Accumulation in Transgenic Plants" Molecules 28, no. 7: 3205. https://doi.org/10.3390/molecules28073205
APA StyleZhang, X., Ahmad, N., Zhang, Q., Wakeel Umar, A., Wang, N., Zhao, X., Zhou, K., Yao, N., & Liu, X. (2023). Safflower Flavonoid 3′5′Hydroxylase Promotes Methyl Jasmonate-Induced Anthocyanin Accumulation in Transgenic Plants. Molecules, 28(7), 3205. https://doi.org/10.3390/molecules28073205







