Abstract
Kinases are among the most important families of biomolecules and play an essential role in the regulation of cell proliferation, apoptosis, metabolism, and other critical physiological processes. The dysregulation and gene mutation of kinases are linked to the occurrence and development of various human diseases, especially cancer. As a result, a growing number of small-molecule drugs based on kinase targets are being successfully developed and approved for the treatment of many diseases. The indole/azaindole/oxindole moieties are important key pharmacophores of many bioactive compounds and are generally used as excellent scaffolds for drug discovery in medicinal chemistry. To date, 30 ATP-competitive kinase inhibitors bearing the indole/azaindole/oxindole scaffold have been approved for the treatment of diseases. Herein, we summarize their research and development (R&D) process and describe their binding models to the ATP-binding sites of the target kinases. Moreover, we discuss the significant role of the indole/azaindole/oxindole skeletons in the interaction of their parent drug and target kinases, providing new medicinal chemistry inspiration and ideas for the subsequent development and optimization of kinase inhibitors.
1. Introduction
Kinases are a class of biochemical molecules that can transfer phosphate groups from high-energy donor molecules, such as ATP, to specific target molecules (substrates), a process called phosphorylation that ultimately leads to the altered biological function of the target protein [1]. Abnormal expression of many kinases leads to inflammatory diseases [2], metabolic diseases [3], cancer [4], etc. Therefore, kinases are extremely significant targets for drug development.
The structure of kinase was first described by Knighton et al. in 1991 [4]. The conserved structure of the catalytic structural domain of kinase consists mainly of N-terminal and C-terminal lobes connected by a short loop in the hinge region [5,6]. The N-lobe flap consists of five antiparallel β-strands and one Cα-helix, whereas the C-terminal flap consists of eight α-helices and four β-strands. The region between the N-terminal and C-terminal lobes and the hinge region forms a deep hydrophobic cleft containing the ATP-binding site. The adenine group of ATP forms several critical hydrogen bond interactions with the backbone atoms of the hinge residues, thereby facilitating the stable binding of ATP within the catalytic pocket. In addition, the phosphate binding loop forms the ceiling of the ATP binding site and clamps on the phosphate group to allow for targeted catalysis. The protein substrate binding site is located within the C-lobe, where the activation loop is also located in the C-terminal lobe. Many kinases are phosphorylated within this loop and then undergo conformational changes to activate the kinase and allow access to the substrate binding site. In addition to the catalytic structural domain, kinases may also contain other regulatory structural domains, and ATP is also coordinated by a glycine-rich ring (G-loop), a highly flexible region that is present in the β-sheet structures. These regulatory structural domains play different roles in the kinome, including regulating catalytic activity, recruiting substrates, controlling localization, and acting as scaffolding sites for other proteins [7,8,9]. The FDA approved Imatinib in 2001 as the first marketed targeted tyrosine kinase (ABL) inhibitor for the treatment of chronic myeloid leukemia (CML), which was a breakthrough in molecularly targeted cancer therapy [10]. Since then, research on kinase-related signaling pathways and the development of related targeted drugs have been proceeding promptly; until 2021, 87 small-molecule kinase inhibitors have been approved for the treatment of a variety of diseases, including tumors [11,12].
In order to better understand the mechanism of action of small-molecule kinases inhibitors, researchers have classified these inhibitors into four types based on their binding models. Type I inhibitors, which typically consist of a heterocyclic system occupying an adenine binding site and side chains occupying adjacent hydrophobic regions, target the ATP pocket of the kinase in its active state. Type II inhibitors bind to the ATP-binding site and an additional back hydrophobic pocket in the inactive state. Type III inhibitors are allosteric kinase inhibitors that bind to allosteric sites in the vicinity of the ATP site that do not compete with ATP. Type IV inhibitors are also allosteric kinase inhibitors that bind to allosteric sites in the structural domain of the kinase away from the ATP pocket [12,13,14]. Type III and IV inhibitors are classified primarily based on the distance of small-molecule allosteric kinase inhibitors from the ATP-binding pocket to the allosteric site [15].
This perspective focuses on ATP-competitive kinase inhibitors (type I and type II), and it is found that indole/azaindole/oxindole parent cores appear in numerous ATP-competitive kinase inhibitors [16,17,18]. This may be due to the fact that indole/azaindole/oxindole-containing fragments can better fit Lipinski’s rule than other bicyclic fused heterocycles. The indole/azaindole/oxindole scaffold competitively occupies the ATP-binding pocket of the kinase and interacts with key residues in the binding site with non-covalent bonding interactions such as hinge hydrogen bonds, pi–pi stacking, pi–cation, etc., thereby inhibiting the kinase activity and thus regulating kinase-related signaling pathways. This paper provides a detailed review of the discovery of thirty approved, three in clinical trials, and one in preclinical studied ATP-competitive small-molecule kinase inhibitors containing indoles/azaindole/oxindole scaffolds and their binding patterns to the ATP-binding pockets of their target kinases. The profiling of these binding models based on X-ray crystal structures provides structural insights for the design of more desirable and selective ATP-competitive small-molecule inhibitors. All of the 3D diagrams of the binding model analyses were rendered by PyMOL (The PyMOL Molecular Graphics System, Version 2.3 Schrödinger, LLC, New York, NY, USA). Among them, the violet dashed lines denote hydrogen bonds, the orange dashed lines represent salt bridges, the red dashed lines represent pi–cation interactions, the green dashed lines indicate pi–pi stacking, key residues that have non-covalent interactions with ligands are represented in yellow sticks, and proteins are shown as light red cartoons. In addition, all 2D interaction illustrations were generated using PoseView [19,20], where the hinge hydrogen bonds are highlighted in violet, and the hinge residues and the indole/azaindole/oxindole skeletons are highlighted in yellow.
2. Indole/Azaindole/Oxindole-Based Approved ATP-Competitive Kinase Drugs
2.1. The Breakpoint Cluster Region Abelson (Bcr-Abl) Inhibitors
The breakpoint cluster region Abelson (Bcr-Abl) tyrosine kinase (TK) is a constitutively activated cytoplasmic TK that is an underlying cause of CML [21]. In 2001, the Bcr-Abl inhibitor Imatinib was used as first-line therapy for the treatment of CML [18]. However, clinical findings suggest that patients may develop drug resistance, especially in the advanced stages of the disease [22,23,24]. Mutations in the structural domain of the kinase are the main reason for Bcr-Abl inhibitor resistance, and one of the most common mutations in Bcr-Abl is the substitution of threonine 315 (located in the center of the Imatinib binding site) by isoleucine [25,26,27]. The drug resistance problem has prompted researchers to search for lines of Bcr-Abl inhibitors.
In this section, we concentrate on ATP-competitive inhibitors that target Bcr-Abl and contain azaindole scaffold (Figure 1). 1e (Ponatinib) was approved for second-line treatment of chronic granulocytic leukemia and Ph + ALL in the US in December 2012 and in the EU in July 2013 [28]. 1e has been shown to be a potent inhibitor of Bcr-Abl. It has demonstrated clinical activity in both Bcr-Abl wild-type and mutant CML, including anti-T315I mutation activity [21]. Here, we briefly describe the development history of 1e, where researchers targeted the inactive DFG domain of ABL proteins based on the 9-[anyenyl]purine core (Figure 1). Compound 1a had a weak inhibitory effect on ABL T315I with an IC50 of 14,142 nM. To further reduce the steric clash with the main backbone of the isoleucine side chain, the double bond linker of 1a was replaced with a triple bond to obtain compound 1b (T315I kinase IC50 = 524 nM). Compared to 1a, 1b showed a significant increase in kinase activity and anti-proliferative activity (tested in Ba/F3 cells). However, 1b showed poor pharmacokinetics (PK) performance in rats. To further improve the bioavailability of compounds, the researchers further modified the structure of 1b to obtain compound 1c (T315I kinase IC50 = 102 nM). The experimental results showed that the PK properties of 1c were considerably improved with an oral bioavailability (F) of 42.4% at dose of 15 mg/kg in rats, but the anti-proliferative activity was low against ABL T315I Ba/F3 cells (IC50 = 471 nM). To further enhance the anti-proliferative potency, compound 1d (T315I kinase IC50 = 56 nM) was obtained by introducing N-methylpiperazine at C-4 on the 3-trifluoromethylbenzene ring. 1d showed a 2-fold increase in anti-T315I mutant kinase activity and an approximately 20-fold increase in potency (IC50 = 26 nM) in inhibiting the growth of Ba/F3 cells of Bcr-Abl T315I, and 1d also exhibited desirable PK properties (F = 29.0%). Encouraged by the overall performance of 1d, structure–activity relationships (SARs) were further explored around 1d by introducing an additional N atom to the pyridine ring to reduce the lipophilicity of 1d (cLogP = 6.69), targeting the final compound 1e (cLogP ≈ 4.69, T315I kinase IC50 = 40 nM; T315I Ba/F3 IC50 = 1.2 nM). Compared to 1d, 1e not only showed better enzymatic activity levels and anti-cell proliferation activity but also showed more desirable results in PK properties [29]. The X-ray structure of the Ponatinib-Bcr-Abl complex shows that 1e can occupy the ATP-binding pocket properly, where N-3 in imidazo [1,2-b]pyridazine can form hydrogen bond interactions with residue MET318 at a distance of 1.9 Å. The -NH and -C=O of the amide group can form hydrogen bonds with the carbonyl oxygen atom of GLU286 and the -NH of ASP381 at distances of 2.6 and 1.8 Å, respectively. The N atom of the N-methylpiperazine portion can form hydrogen bonds with residues HIS316 and ILE360 at distances of 3.4 and 2.9 Å, respectively, and also form salt bridges with ASP381.
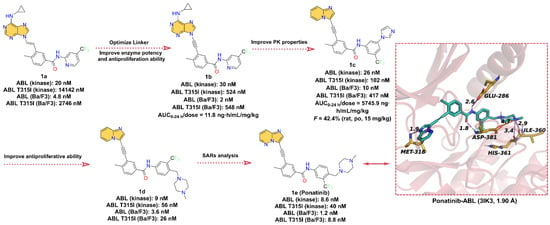
Figure 1.
Key medicinal chemistry optimization leading to Ponatinib and its binding model with Bcr-Abl.
Taken together, 1e is a potent Bcr-Abl inhibitor that has been approved for the treatment of CML patients who are resistant or intolerant to Imatinib, Dasatinib, or Nilotinib, and studies have confirmed that 1e can be effective in a variety of other cancers [30]. In the future, the clinical safety and patient tolerability profile of 1e need to be further explored.
2.2. Bruton’s Tyrosine Kinase (BTK) Inhibitors
Bruton’s tyrosine kinase (BTK), a member of non-receptor tyrosine kinase belonging to the Tec family kinases [31], is a vital component in the B-cell receptor signaling pathway. BTK is widely expressed in different types of malignant hematological diseases and is involved in the proliferation, differentiation, and apoptosis of B cells [32]. BTK has emerged as a promising target for the treatment of multiple diseases, especially B-cell-related malignancies, and the first-, second-, and third-generation BTK inhibitors have been developed over the past decades [33,34]. Small-molecule BTK inhibitors include both covalent and non-covalent inhibitors, of which the first- and second generations are predominantly covalent, while the third generation is mostly non-covalent.
Here, we focus on small-molecule BTK inhibitors with azaindole or oxindole core in clinical applications (Figure 2). 2b (Ibrutinib, PCI-32765), the first-in-class BTK inhibitor, is an orally available, highly potent, and irreversible ATP-competitive kinase inhibitor for the treatment of mantle cell lymphoma (MCL) and chronic lymphocytic leukemia (CLL) [35,36]. The development of 2b was first reported by Pan. et al. [37], who identified compound 2a had moderate inhibitory activity against BTK with an IC50 of 8.2 nM but was poorly selective for BTK. To further give compounds with potent kinase activity and better selectivity for BTK, a diverse array of Michael acceptor groups was introduced into 2a. Encouragingly, 2b exhibits the strongest inhibition of BTK kinase with an IC50 of 0.5 nM and a significant improvement in selectivity for BTK. 2b, which contains an acrylamide motif, can form a covalent bond with the residue CYS481 to enhance BTK selectively. In addition to the formation of a covalent bond, 2b can also form non-covalent bond interactions with key residues around the ATP-binding pocket of BTK. As shown in Figure 2, the cocrystal structure of 2b with BTK [38] reveals that the 4-amino-1H-pyrazolo [3,4-d]pyrimidin scaffold can form tridentate hydrogen bonds with the hinge residues THR474, GLU475 and MET477, with distances of 2.8, 2.0, and 2.1 Å, respectively. In addition, the phenyl of 2b occupies the hydrophobic pocket in the N-lobe of BTK and displays edge-to-face pi–pi interactions with residue PHE540. Overall, the azaindole core of 2b is able to form hinge hydrogen bonds with key residues in the hinge region, stabilizing it at the ATP-binding site.
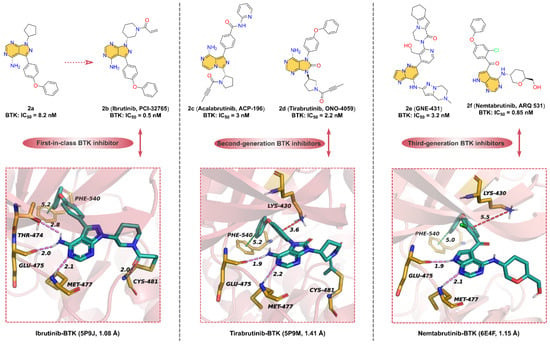
Figure 2.
Representative azaindole/oxindole core-containing BTK inhibitor in clinical applications (2e, GNE-431, has not yet entered into clinical studies).
Subsequent studies have found that oral 2b has serious adverse effects, limiting its application in cancer therapeutics [39,40,41]. To overcome the drug resistance and off-target side effects of 2b, selective second-generation BTK inhibitors have been developed. 2c (Acalabrutinib, ACP-196), a second generation BTK inhibitor, has better selectivity and safety than the first-in-class BTK inhibitor 2b and improved off-target effects [42]. 2c is structurally related to 2b, used in the treatment of a variety of hematologic malignancies and solid tumors, as well as potentially in the treatment of rheumatoid arthritis (RA) [43]. The key structural components of 2c include the 8-amino-imidazo [1,5-a]pyrazin scaffold involved in the formation of hinge hydrogen bonds, the 2-pyridylbenzoamide moiety forming hydrophobic interactions at the binding site, and 2-butylamide part covalently binding to Cys481. Unlike the acrylamide functional group in the previously reported inhibitors, the covalent motif of 2c is 2-butylamide. It is speculated that due to this unique active 2-butylamide moiety, 2c exhibits lower reactivity than acrylamides 2b, which may help to minimize the inhibition of off-target cysteine kinases [42]. Compared with 2b, 2c only inhibited BTK, BMX, and ERBB4 at clinically relevant concentrations, which explains the higher specificity of the BTK inhibitor [44].
2d (Tirabrutinib, ONO-4059), an analog of compound 2b scaffold, is a potent, second-generation inhibitor of BTK (IC50 = 2.2 nM) [45]. 2d can inhibit BTK activity by blocking the autophosphorylation of residue TYR223 [33]. Although 2d and 2b display similar binding models, 2d is significantly more selective and effective. As depicted in Figure 2, the parent core of the molecular structure of 2d is a 6-5-membered, fused, heterocyclic ring (oxindole moiety) that can form hydrogen bonds with hinge residues Glu475 and Met477 with distances of 1.9 and 2.2 Å, respectively. Furthermore, another model for the interaction between 2d and BTK is the covalent interaction between 2-butylamide and the residue CYS481. The phenoxyphenyl extends into the hydrophobic pocket and forms pi–pi interactions with residue PHE540, such as 2b.
2e (GNE-431) is a potent, selective, and noncovalent BTK inhibitor with IC50 of 3.2 nM and 2.5 nM for wild-type BTK and C481S mutants, respectively [46,47]. 2e, unlike covalent inhibitors such as 2b, does not form a covalent bond with CYS481 and potently inhibits 2b-resistant BTK C481S mutant in vitro and in vivo. The computational binding model of BTK to 2e suggests that the imidazo [1,2-b]pyridazin-8-amine moiety can form hinge hydrogen bonds with the hinge residues and that the tetrahydropyrazolo [1,5-a]pyrazine group has a distinctly different binding orientation than other BTK inhibitors [46]. Due to the unique binding pose of 2e, this allows it to inhibit wild-type BTK and the mutants, such as C481S, C481R, T474I, and T474M (IC50 = 7.5–10 nM). These mutant residues have no spatial overlap with 2e at the binding site.
In response to emerging drug resistance of 2b, a third-generation orally bioavailable reversible BTK inhibitor, 2f (Nemtabrutinib, ARQ 531), has been identified [48]. 2f shows potent inhibition of both wild-type and C481S-mutant BTK with IC50 of 0.85 and 0.39 nM, respectively. Additionally, 2f has significant anti-proliferative activity in vitro against many hematological malignant cell lines, including Ibrutinib-resistant cell lines [49,50]. As shown in Figure 2, the crystal structure of BTK with 2f [49] at 1.15 Å resolution is similar to that of BTK with 2b. The pyrrolo [2,3-d]pyrimidine core can form hinge hydrogen bonds with key residues GLU475 and MET477 in the hinge region. Similar to 2b, phenoxyphenyl group of 2f occupies the hydrophobic pocket of the ATP-binding site, forming pi–pi stacking interactions and pi–cation interactions, respectively. It is worth mentioning that 2f does not form a covalent bond with CYS481; the polar tetrahydropyran methanol motif is exposed in the solvent region, indicating that the C481S-mutat BTK does not affect the binding of 2f.
2.3. Cyclin-Dependent Kinases 4/6 (CDK4/6) Inhibitors
Cyclin-dependent kinases 4/6 (CDK4/6) are members of the serine/threonine kinases family that play pivotal roles in the biological processes that regulate the cell cycle. Aberrant activation or dysregulation of CDK 4/6 is closely associated with cancer development and progression [51]. First-generation pan-CDK inhibitors and second-generation multi-CDK inhibitors are poorly selective and have highly toxic side effects [52]. As a result, a new generation of selective CDK4/6 inhibitors (including Abemaciclib, Palbociclib, and Ribociclib, as shown in Figure 3) have been developed, aiming to improve the selectivity and reduce the adverse side effects of the drugs [53].
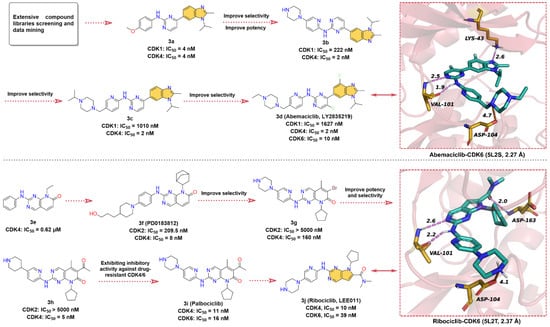
Figure 3.
Key medicinal chemistry optimization of Abemaciclib and Ribociclib along with the binding model with CDK6.
The hit compound 2-anilino-2,4-pyrimidinyl-benzimidazole scaffold (3a), which showed strong inhibitory activity against both CDK1 and CDK4 with IC50 value of 4 nM, was obtained through screening and data mining of extensive chemical libraries [54]. Subsequently, 3a was subjected to SARs studies to improve its inhibitory potency and selectivity against CDK4. This process mainly involves the replacement of the phenyl ring in aniline with pyridine and the introduction of the piperazine ring to obtain 3b, which improves the selectivity for CDK4 and its IC50 for CDK1 inhibition increases from 4 nM to 222 nM. Modification of the piperazine fragment of 3b gives 3c (CDK4 IC50 = 2 nM, CDK1 IC50 = 1010 nM), whose activity against CDK4 is maintained while selectivity is further improved. To enhance the specificity of 3c for CDK4/6, the introduction of fluorine atoms on pyrimidines and benzimidazoles resulted in the approved drug 3d (Abemaciclib, LY2835219). 3d has a greatly reduced inhibitory activity against CDK1 (IC50 = 1627 nM) and shows potent inhibitory efficacy on CDK4/6 with IC50 values of 2 and 10 nM, respectively [55]. Crystal structure analysis of 3d with CDK6 (Figure 3, upper right) yields that the 2-amino-pyrimidine fragment forms bidentate hydrogen bonds with the hinge residue VAL101 (1.9 & 2.5 Å), and the benzimidazole moiety binds to the hydrophobic region of the ATP-binding site and forms a hydrogen bond interaction with LYS43 at a distance of 2.6 Å [56]. In addition, the piperazine moiety of 3d is exposed to the solvent region, forming a salt bridge interaction with ASP104. These diverse non-covalent bond interactions enable 3d to bind competitively to the ATP-binding site of CDK6.
Pyrido [2,3-d]pyrimidin-7-one analogs are reported to have good inhibitory activity against CDKs, among which, 2-phenylamino-pyrido [2,3-d]pyrimidin-7-one (3e) shows moderate inhibitory activity against CDK4 with an IC50 of 0.62 μM [57]. Chemical modification of 3e resulted in the identification of 3f (PD0183812) [58], which shows potent inhibition and high selectivity for both CDK4 (IC50 = 8 nM) and CDK6 (IC50 = 13 nM), but poor selectivity over other CDK isoforms (CDK2, IC50 = 209.5 nM). SARs studies have shown that the selectivity of the 2-aminopyridine fragment on the quinazoline core is higher than that of the aniline moiety. Thus, the modification of the aniline component of 3f leads to 3g with higher selectivity against CDK4/6. Intriguingly, replacing the -Br on the quinazoline scaffold with an acetyl group gives 3h with a 32-fold increase in its inhibitory activity against CDK4. Replacement of the piperidine moiety of 3h with piperazine affords 3i (Palbociclib) [59], a highly specific CDK4/6 inhibitor with IC50 values of 11 and 16 nM, respectively. In 2010, Novartis, in collaboration with Astex, reported 3j (Ribociclib, LEE011), an orally active, highly specific CDK4/6 inhibitor with IC50 values of 10 and 39 nM, respectively [60,61]. To the best of our knowledge, the development course of 3j has not been disclosed [55,62]. Given the structural similarity and the chronological order of launch, we assume that 3j was designed with 3i as the lead compound. In short, the 6,6-membered fused quinazoline ring of 3i is reduced to a 6,5-membered fused pyrrolo [2,3-d]pyrimidine scaffold. The X-ray cocrystal structure of human CDK6 and 3j (Figure 3, lower right) illustrates that the pyrrolo [2,3-d]pyrimidine group can form a hinge hydrogen bond with VAL101, and the -NH of the pyrimidine amino group also forms a hydrogen bond with hinge residue VAL101. Same as with 3i, the two side chains of 3j can form salt bridge interaction and hydrogen bond interaction with ATP-binding site residues ASP104 and ASP163, respectively.
2.4. Colony-Stimulating Factor 1 Receptor (CSF1R) Inhibitors
Receptor tyrosine kinases (RTKs) play an imperative role in maintaining homeostasis in vivo. RTKs provide important pathways for cellular communication, maintain signal transduction for normal cellular processes, and provide surface receptors for many hormones and growth factors [63]. The human genome contains 58 RTKs, which are divided into different groups according to the homology of their active sites and the similarity of ligands [64]. Mutations in receptor structure and ligand overexpression facilitate the emergence and development of several types of cancers. Colony-stimulating factor 1 receptor (CSF1R), also known as c-FMS, CD115, or M-CSFR, is one of the most important receptors in type III RTKs and is increasingly becoming a promising target for cancer therapy [65]. Activation of CSF1R promotes survival, proliferation, and differentiation of macrophage and monocyte lineages. Monocytes in the tumor microenvironment can differentiate into tumor-associated macrophages (TAMs) that allow cancer cell growth, metastasis, angiogenesis, and especially local immunosuppression. Tumor-released colony-stimulating factor 1 (CSF1) binds to CSF1R and promotes macrophage proliferation, survival, and differentiation. Overexpression of CSF1 and CSF1R in a variety of tumors, including breast cancer, is associated with poor patient prognosis, thus suggesting that overexpression of CSF1R is associated with diseases such as cancer [66,67,68,69]. Therefore, the development of kinase inhibitors targeting CSF1R could be an effective cancer treatment strategy [70].
4e (Pexidartinib, Figure 4) is an oral antitumor small-molecule drug that potently inhibits CSF1R with an IC50 of 13 nM for the treatment of tenosynovial giant-cell tumors [71,72]. 4a (7-azaindole) is identified as a unique kinase inhibitor scaffold by screening over 20,000 low-molecular-weight compounds against multiple kinases [73,74]. 4b (PLX070) is derived by introducing a 3-methoxybenzyl group at the 3-position of 4a. The crystal structure shows that 4b is able to bind at the ATP-binding site of FGFR1 (fibroblast growth factor receptor 1, IC50 = 1.9 μM) and that the 7-azaindole moiety can form two hydrogen bonds with the hinge residues, indicating that 4b is an effective ATP-competitive kinase inhibitor designed building block [74]. The researchers maintained the 3-methylene linker of 4b and employed pyridine in place of the benzene ring to provide the endocyclic nitrogen as a hydrogen bond acceptor to replace the methoxy of the exocyclic ring and introduced trifluoromethyl-benzylamine to obtain 4c (PLX647) (IC50 = 28 nM for CSF1R) [75]. To further optimize the structure, 4d (PLX647-OMe) is obtained by introducing a methoxy group at the 5-position of the 7-azaindole of 4c. The inhibitory efficacy of 4d against SCF1R was slightly reduced (IC50 = 62 nM), but the aqueous solubility is significantly enhanced, from 14 μM for 4c to 77 μM [75]. To further optimize the structure of 4d to enhance the inhibitory potency of SCF1R, 4e (Pexidartinib) is obtained by replacing the -OCH3 group with -Cl, and the inhibition of CSF1R kinase is improved with an IC50 of 13 nM [70]. As shown in Figure 4, 4e binds into the ATP-binding pocket of CSF1R and makes direct contact with surrounding key residues [72]. Specifically, the -NH and -N on the 7-azaindole scaffold form two hydrogen bonds with hinge residues GLU664 and CYS666 at distances of 1.8 and 2.1 Å, respectively. In addition to the pi–cation interaction between the middle pyridine group and residue LYS616, the polar nitrogen of the pyridine is also involved in the formation of a hydrogen bond with residue ASP796 at a distance of 2.1 Å. Additionally, the 3-fluoromethylpyridine block in the tail can interact with the indole group of the residues TRP550 to form face-to-face pi–pi interactions.
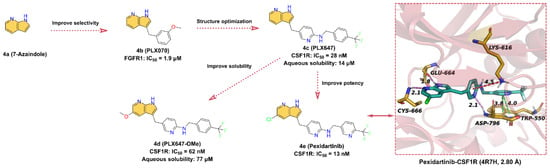
Figure 4.
Key medicinal chemistry optimization of Pexidartinib along with the binding model with CSF1R.
2.5. Human Epidermal Growth Factor Receptor (HER) Inhibitors
Functional activation of the human epidermal growth factor receptor 2 (HER2) gene promotes the development of cancer. The main mechanism of HER2 activation in human breast and gastric cancers is HER2 gene amplification, leading to overexpression of its protein on the cell membrane, which is associated with disease recurrence and short overall patient survival [76,77]. Therefore, the development of specific HER2 antagonists is of great therapeutic importance in oncology [76]. Due to the high expression of HER2 in tumors such as breast cancer, several novel therapeutic strategies, including the administration of small-molecule inhibitors and monoclonal antibodies, have significantly improved patient survival. Currently, the main small-molecule drugs that effectively target HER2 are ATP-competitive inhibitors, including Tucatinib, Lapatinib, Neratinib (HKI-272), and Pyrotinib, which compete with ATP to block phosphorylation and activate downstream signaling cascades [78]. Notably, Tucatinib (5f) is a selective HER2 inhibitor containing azaindole scaffold, which is also the focus of the next discussion (Figure 5).
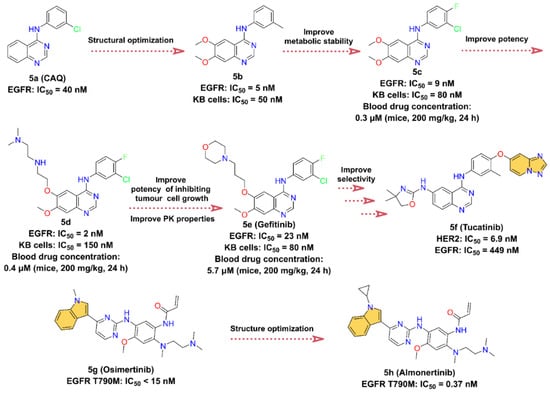
Figure 5.
Key medicinal chemistry optimization of Tucatinib, Osimertinib, and Almonertinib.
In the course of investigating the catalytic mechanism of epidermal growth factor receptor (EGFR) tyrosine kinase using a structure-based searching approach, a potent ATP-competitive inhibitor of EGFR, 4-(3-chloroanilino)quinazoline 5a (CAQ), with a Ki of 16 nM and an IC50 of 40 nM, was identified [79,80]. 5a is the first representative of a new structural class of anilinoquinazoline tyrosine kinase inhibitors [79]. We speculate that the optimization of the structure of 5a yields 5b with stronger enzyme inhibitory activity (IC50 = 5 nM) and cell proliferation inhibitory activity (KB cells, IC50 = 50 nM). However, the phenylmethyl of 5b is easily oxidized by metabolism in vivo. To improve the metabolic stability of 5b, an F atom is introduced at the 4-position of the phenyl group, and the methyl is replaced with a Cl atom to close the metabolic site to obtain 5c. 5c inhibits EGFR with an IC50 value of 9 nM, with a slight loss of inhibitory potency but improved PK properties, such as its half-life (t1/2) is increased by approximately three times [81]. Optimization of the 6-position of the quinazoline ring at 5c gives 5d, which shows a stronger inhibitory effect for EGFR with an IC50 of 2 nM, but the proliferation inhibitory activity against KB cells is reduced (IC50 = 150 nM). In addition, there is a slight improvement in the PK properties at 5d, with blood drug concentrations in mice at 24 h elevated from 0.3 μM at 5c to 0.4 μM at a dose of 200 mg/kg. To further enhance the cell proliferation inhibitory activity as well as the bioavailability of 5d, the morpholine group is introduced to obtain 5e (Gefitinib). The blood concentration of 5e is further increased at 5.7 μM after 24 h of administration, while the IC50 of 5e inhibition on KB cells is 80 nM, which is also enhanced. 5e has good oral bioavailability and is a first-line treatment for patients with metastatic non-small-cell lung cancer (NSCLC) [81,82,83]. 5f (Tucatinib) is initially identified through a small-molecule discovery effort on HER2 and EGFR inhibitors. It was discovered by Array BioPharma Inc. and received FDA approval in 2020 to treat advanced unresectable or metastatic HER2-positive breast cancer [84]. 5f is a highly selective HER2 inhibitor containing a quinazoline core such as 5e, with IC50 values of 6.9 nM for HER2 and 449 nM for EGFR. Therefore, it is speculated that 5f is optimized from 5e [85,86].
EGFR is also known as HER1, and its overexpression plays an important role in a variety of cancers, such as squamous-cell carcinoma of the lung, glioblastoma, and epithelial tumors of the head and neck [87]. 5g (Osimertinib) is a third-generation EGFR inhibitor developed by AstraZeneca to overcome EGFR T790M mutation-related resistance [88]. It is highly selective for EGFR-activating mutations and EGFR T790M mutation [89]. In November 2015, it was approved by the FDA to treat metastatic EGFR T790M mutation-positive NSCLC, with IC50 less than 15 nM for EGFR T790M [88,90,91]. Innovative optimization on the structure of 5g, 5h (Almonertinib) is obtained by substituting the -CH3 on the N atom of the indole group and introducing cyclopropyl [92,93]. Surprisingly, the problem of high toxicity and poor selectivity of 5g’s metabolites is solved [92,94]. In March 2020, 5h was approved by the National Medical Products Administration (NMPA) in China for the treatment of advanced EGFR T790M + NSCLC [93]. It is the third-generation EGFR inhibitor developed by Jiangsu Hansoh Pharmaceutical Co. [93], with IC50 values of 0.37 ± 0.04 nM, 0.21 ± 0.10 nM, 0.29 ± 0.10 nM, and 3.39 ± 0.53 nM for T790M, Del19/T790M, L858R/T790M, and EGFR WT, respectively [92].
2.6. Janus Kinases (JAK) Inhibitors
Janus kinases (JAK) belong to the family of non-receptor kinase tyrosine kinases, which consists of four members together, including JAK1, JAK2, JAK3, and TYK2 [95]. JAK1, JAK2, and TYK2 are commonly expressed, while JAK3 is mainly in hematopoietic cells [96]. In the 1990s, JAKs were found to have a role in cytokine signaling pathways [97]. JAKs can transmit signals from cell membrane receptors to the signal transducer and activator of the transcription (STAT) family, the JAK/STAT signaling pathway, and are closely associated with cancer [98] and inflammatory diseases [99]. In 2005, researchers identified in patients with myeloproliferative neoplasms V617F mutations in JAK2, including myelofibrosis, true erythroblastosis, and primary thrombocytopenia [100]. Studies have shown that excessive activation of the JAK/STAT pathway has been detected in several solid cancers (lung, breast, head, and neck) and hematologic malignancies (multiple myeloma, lymphoma, and non-lymphoma), as well as in acute leukemia [101]. Increasing evidence suggests that inhibition of JAK expression can treat inflammation or cancer that is dependent on JAK/STAT pathway activation, and such, this has attracted an increasing number of scholars and pharmaceutical companies to invest in the development of small-molecule inhibitors targeting JAK [102].
Herein, we describe small-molecule JAK inhibitors containing azaindole core in clinical applications (Figure 6). To date, there are six JAK inhibitors with azaindoles structural motifs have been applied in the clinic, namely Ruxolitinib, Baricitinib, Tofacitinib, Delgocitinib, Peficitinib, and Filgotinib. Among them, 6a (Ruxolitinib), the first FDA-approved JAK1/2 inhibitor, kills tumor cells by enhancing apoptosis and inducing autophagy [103]. However, some side effects, such as anemia, thrombocytopenia, and neutropenia, may occur as 6a also inhibits JAK2 [104]. As shown in Figure 6, 6a has a pyrrolo [2,3-d]pyrimidin core, in which the -NH of the pyrrole can form a hinge hydrogen bond with residue GLU930 at a distance of 1.9 Å, while the N atom on the pyrimidine group can form a hydrogen bond with hinge residue LEU932 at a distance of 2.2 Å. Overall, the azaindole fragment of 6a is able to form hinge hydrogen bonds with key residues in the ATP-binding site of JAK2, thereby competing with ATP for the binding pocket.
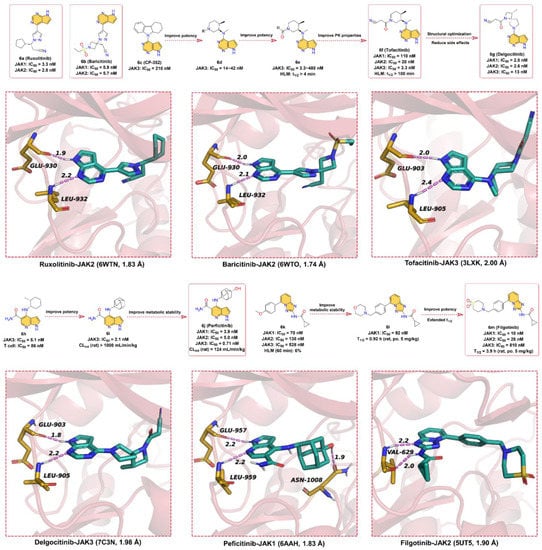
Figure 6.
Key medicinal chemistry optimization of JAKs inhibitors along with the binding models with JAKs.
6b (Baricitinib) is a JAK1 and JAK2 inhibitor developed by Eli Lilly and Company for the treatment of RA [105], atopic dermatitis, and systemic lupus erythematosus. 6b has an IC50 of 5.9 nM and 5.7nM for in vitro kinase activity testing on JAK1 and JAK2, respectively, and does not inhibit c-Met or CHK2. Attempts have been made to approve 6b as a therapeutic agent for RA in Japan and the United States, but the US FDA is currently unable to approve the drug because further clinical data are needed to determine the appropriate dose and to better characterize the safety of the compound [106]. As shown in Figure 6, the chemical parent core of 6b is also the pyrrolo [2,3-d]pyrimidin scaffold, and the binding pattern between 6b and JAK2 is similar to that of 6a; that is, the parent scaffold of 6b forms bidentate hydrogen bond interactions with key residues GLU930 and LEU932 in the hinge region of JAK2.
In 1996, Pfizer screened its compound library for small-molecule inhibitors targeting JAK3 and identified the pyrrolo [2,3-d]pyrimidin-type derivative 6c (CP-352), which has a JAK3 kinase inhibition of 210 nM and a short half-life in human liver microsomes [107]. To further enhance the potency and extend the half-life in human liver microsomes, a variety of substituents, including cyclic, lipophilic, and amino groups, are used to explore compounds with better JAK3 inhibitory activity. Surprisingly, N-methylcycloalkyl analogs (6d) showed better enzymatic activity against JAK1 and some improvement in T-cell activity, suggesting that simultaneous inhibition of JAK1 and JAK3 may be the key to improving cellular potency. The amide group with better hydrophilicity was introduced to 6d as a linker to obtain 6e (human liver microsome, HLM, t1/2 > 4min). To further improve the PK properties, a small polar cyano group was introduced next to the amide group to finally give 6f (Tofacitinib, HLM, t1/2 > 100 min), which can maintain a favorable balance in terms of JAK kinases selectivity and druggability [107]. The IC50 values of 6f for JAK3, JAK2, and JAK1 kinases inhibitory effect are 3.3, 20, and 110 nM, respectively, and 6f citrate has anti-infective activity [108]. 6f, a pan-JAK inhibitor developed by Pfizer, was approved by the US FDA in 2012 for the clinical treatment of moderate-to-severe RA that cannot be treated with methotrexate [16]. As depicted in Figure 6, the pyrrolo [2,3-d]pyrimidin scaffold of 6f occupies the ATP-binding pocket of JAK3 and forms two hydrogen bonds with hinge residues GLU903 and LEU905 at distances of 2.0 and 2.4 Å, respectively.
In the clinical phase II trial, 6f showed adverse effects such as headache and nausea [109]. The researchers developed 6g (Delgocitinib) based on the structure of 6f to reduce these side effects. 6g is a potent, orally available, pan-JAK inhibitor with IC50 values of 2.8, 2.6, 13, and 58 nM for JAK1, JAK2, JAK3, and Tyk2, respectively, which was approved for the treatment of atopic dermatitis in Japan [110]. As depicted by the X-ray cocrystal structure of JAK with 6g, the -NH of pyrrole in the pyrrolo [2,3-d]pyrimidin core of 6g can form a hinge hydrogen bond with residue GLU903 at a distance of 1.8 Å, and the N atom of pyrimidine serves as hydrogen bond acceptor to form hydrogen bond interactions with hinge residue LEU905 at a distance 2.2 Å.
In 2007, the patent reported that 6j (Peficitinib) is a particularly potent JAK inhibitor against JAK3 with an IC50 of 0.71 nM, while the inhibitory activity against JAK1, 2, and TYK2 kinases is 3.9, 5.0, and 4.8 nM, respectively [111]. 6j has reached late-stage clinical trials for the treatment of RA [112]. 6j was optimized from the lead compound 6h, which exhibited potent JAK3 inhibitory activity (IC50 = 5.1 nM) and moderate T-cell proliferation inhibitory activity (IC50 = 86 nM). However, 6h showed poor metabolic stability in liver microsomes and in vivo PK profiles. To further improve the inhibitory potency of JAK3, 6i was obtained by replacing cyclohexane with adamantane moiety. 6i showed stronger inhibition of JAK3 with an IC50 of 2.1 nM, but its PK properties were poor, and its plasma clearance (CL) was greater than 1000 mL/min/kg in rats. Finally, the polar group -OH was introduced in the C4-adamantyl of 6i to obtain 6j to reduce lipophilicity and thus improve metabolic stability (CL = 124 mL/min/kg). According to the binding model between 6j and JAK1, the -NH and N-atom on the pyrrolo [2,3-d]pyridine substructure can form bidentate hinge hydrogen bonds with residues GLU957 and LEU959, both at a distance of 2.2 Å. Unlike other JAK inhibitors, the -OH on adamantane can form a strong hydrogen bond interaction with residue ASN1008 with a distance of 1.9 Å. It is speculated that this may account for the particularly good activity of 6j against JAK3.
In 2013, researchers obtained hit compound 6k against the JAK family by high-throughput screening, but with low to medium inhibitory activity (JAK1 IC50 = 70 ± 14 nM, JAK2 IC50 = 138 ± 22 nM, JAK3 IC50 = 528 ± 82 nM, TYK2 IC50 = 519 ± 55 nM) [113]. Moreover, 6k shows low metabolic stability; only 6% of the prototype drug remains in human liver microsomes at 60 min. In the modification of 6k, the researchers found that the potency of enzyme activity was increased when the substituent on the phenyl ring was large enough to contact the glycine-rich ring. When the methoxy on the benzene ring is replaced with morpholine (6l), it is found that the t1/2 in human microsomes could be prolonged, but the inhibitory effect on JAK1 remains low (IC50 = 92 ± 12 nM). Encouragingly, the replacement of the morpholine with cyclic sulfone fragment resulted in 6m (Filgotinib), which has greatly improved potency against JAK1 (IC50 = 10 ± 0.8 nM) and also possessed more desirable PK properties (t1/2 = 3.9 h) [114]. Based on the X-ray cocrystal structure, 6m binds into the ATP-binding site, the N-atom in triazolo [1,5-a]pyridine scaffold, and the -NH in the neighboring amide form bidentate hinge hydrogen bond interactions with the residue VAL629 at distances of 2.2 and 2.0 Å, respectively. The benzene ring forms hydrophobic interactions, while the partial fragment of thiomorpholine dioxide extends to the solvent-exposed region. The favorable PK profile, especially the oral availability in different animal species, as well as the activity in preclinical models, led to the development of this inhibitor in clinical application [115,116].
Briefly, all six of these JAK inhibitors are ATP-competitive inhibitors that bind to the ATP-binding pockets. Current clinical JAK small-molecule inhibitors have progressed to higher levels of potency against JAK, and these pan-JAK inhibitors have been found to cause side effects in patients with different conditions in the clinic. Further improvements in the selectivity of JAK kinases are essential to reduce side effects, while attention should also be paid to the PK of the drug in vivo, which will be the focus of the development of the next generation of JAK kinase inhibitors.
2.7. BRAF Inhibitors
BRAF is a member of the serine/threonine kinase RAF family (ARAF, BRAF, and CRAF) and is an important part of the RAS/RAF/MEK/ERK mitogen-activated protein kinase (MAPK) signaling pathway [117,118]. Among these, the MAPK cascade is a key signaling pathway involved in the regulation of normal cell proliferation, survival, and differentiation [119], and oncogenic mutations of this pathway are commonly observed in numerous cancers [120]. Since RAS mutations are found in approximately 30% of cancers, RAS is considered a central therapeutic target of this pathway, while the physiological signaling of RAS is mainly caused by BRAF [121]. Studies have demonstrated that regulation of the MAPK signaling pathway by targeting BRAF kinase is already the standard of care for patients with metastatic melanoma containing BRAF mutations [122,123]. Since the identification of BRAF-V600E as a drug target, many researchers have been attracted to developing specific kinase inhibitors against RAF.
Currently, the inhibitors targeting RAF have been developed into the third generation [124]. In this section, our perspective is focused on 7c (Vemurafenib, PLX4032), an ATP-competitive RAF inhibitor containing azaindole scaffold, which was approved by the US FDA in 2011 for the treatment of patients with BRAF-V600E metastatic melanoma (Figure 7). 7c has high activity against BRAF-V600E with an IC50 of 31 nM and effectively inhibits ERK phosphorylation in tumor cell lines bearing BRAF-V600E [125]. The discovery of 7c was based on a 7-azaindole(4a) scaffold obtained from kinases screening experiments of a library of more than 20,000 compounds [74]. Subsequently, a library of mono- and disubstituted derivatives was constructed using 4a as the parent scaffold, and screening of this compound library yielded that 7b (PLX4720) exhibited excellent potency against BRAF-V600E with an IC50 of 13 nM and high selectivity for other kinases. The IC50 for the in vitro proliferation inhibitory activity of 7b against melanoma A375 (BRAF-V600E) cells was 500 nM. We presume that in order to further obtain drugs with better anticancer activity, the researchers used a rational structure-based drug design approach to introduce p-chlorophenyl moiety at the 5-position of the azaindole scaffold to obtain 7c, which showed significantly enhanced anticancer effects on A375 cells (IC50 = 310 nM) [126]. As shown in Figure 7, 7c is able to well occupy the ATP-binding pocket of BRAF, where the hydrogen bonds of azaindoles to the hinge residues anchor the structure [127]. Specifically, N1 serves as a hydrogen bond donor to form a hydrogen bond with the backbone carbonyl of GLN530, while N7 provides a hydrogen bond acceptor to produce a hydrogen bond with the backbone amide of CYS532 at distances of 2.0 and 2.1 Å, respectively. 3-chlorobenzene extends toward the activation segment; the N-(2,4-difluorophenyl)propanesulfonamide fragment, on the other hand, is binding in the hydrophobic region, where the two O atoms of the sulfanedione group can form three hydrogen bonds with residues LYS483, PHE595, and GLY596 at distances of 2.6, 1.9, and 2.3 Å, respectively.
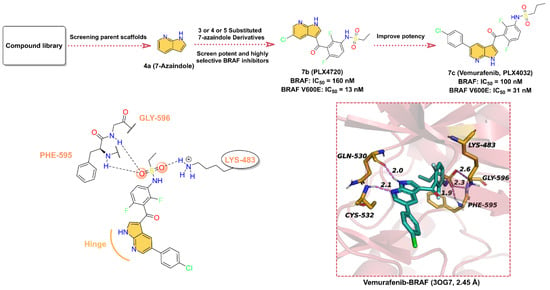
Figure 7.
Key medicinal chemistry optimization of BRAF inhibitors along with the binding models with BRAF.
In conclusion, 7c is a very effective drug for the treatment of unresectable metastatic melanoma, with clinical manifestations of tumor regression in 85% of patients, but half of the patients also experienced resistance problems [128]. It has been reported that increased RAF dimerization may be responsible for the clinical resistance to RAF inhibitors [129,130,131]. Since compensatory survival signals occur upon BRAF inhibition, it has been shown that resistance in these cell lines can be overcome in vitro when MEK inhibitors are used in combination [132,133]. Therefore, the future use of 7c in combination with other kinase inhibitors to address its clinical resistance is possible. In the future, further structural optimization of existing ATP-competitive inhibitors to obtain safe, efficient, and potent RAF inhibitors can still be continued. Meanwhile, the development of allosteric inhibitors of RAF is also a promising area of interest.
2.8. Phosphatidylinositol 3 Kinases (PI3Ks) Inhibitors
Human cells express three classes of phosphatidylinositol 3 kinases (PI3Ks), class I, II, and III, of which class I is the most widely studied and mainly includes PI3Kα, PI3Kβ, PI3Kγ, and PI3Kδ [134]. PI3Kα and PI3Kβ are commonly expressed, PI3Kδ and PI3Kγ are mainly expressed by leukocytes, and the activation of PI3Kγ is driven by activation of the G protein-coupled receptor (GPCR) with a more extensive expression pattern than PI3Kδ [135,136]. PI3Kδ has an impact on both the proliferation and function of B cells, providing ideas for the treatment of B cell-mediated malignancies [137]. PI3Ks are overexpressed in many types of tumor tissues, such as breast cancer, NSCLC, and colorectal cancer, and the development of PI3Ks inhibitors could be an effective strategy for tumor therapy [138]. PI3Ks inhibitors mainly cover pan-PI3K inhibitors and isoform-selective PI3K inhibitors [139]. The majority of pan-PI3K inhibitors are Pictilisib (GDC-0941), Buparlisib (BKM120), and Pilaralisib (XL147), while the major isoform-selective PI3K inhibitors include Umbralisib, Duvelisib, and Idelalisib [139,140,141,142]. In the following text, we focus on three isoform-selective PI3Ks inhibitors with azaindole cores (Figure 8).
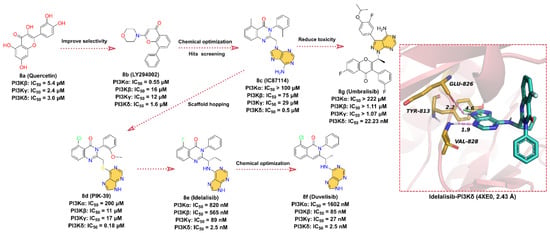
Figure 8.
Key medicinal chemistry optimization of PI3K inhibitors along with the binding models with PI3K.
In the early 1990s, Eli Lilly and Company obtained the PI3K inhibitor 8a (quercetin) with IC50 values of 5.4, 2.4, and 3.0 μM for PI3Kβ, γ, and δ isoform, respectively, by screening a chemical library [143]. In order to improve the selectivity of 8a, more druggable substituents are used to substitute catechol moiety, and the ATP-competitive PI3Ks inhibitor 8b (LY294002) is finally obtained with IC50 values of 0.55, 16, 12, and 1.6 μM for PI3Kα, β, γ, and δ isoform, respectively. The aromatic ring linked to the morpholine scaffold in 8b is embedded as a core pharmacophore in many PI3K inhibitors and is one of the most widely used tool compounds in biological research. In 2003, scientists from ICOS discovered 8c (IC87114), an inhibitor that selectively inhibits PI3Kδ with an IC50 of 0.5 μM, by screening the SARs-optimized chemical library. Later, structural optimization of 8c by scaffold hopping approach was performed to obtain 8d (PIK-39). A comprehensive analysis of the crystal structure of 8d complexed with PI3Kγ revealed that the binding conformation of this compound is distinct from the flat orientation of other PI3K inhibitors [144]. The isoquinolone moiety of 8d projects upward to the roof of the ATP-binding pocket, and the kinase undergoes a conformational rearrangement in order to adapt to the inhibitor, where Met 804 shifts from an “up” position to a “down” position and forms an inducible drug-binding pocket. It is the unique binding model of 8d to PI3K kinase that makes 8d more selective for PI3Kδ kinase compared to other isoforms (IC50 = 0.18 μM) [136,143,144]. No detailed report of the development history of 8e (Idelalisib) is available to date, but it binds in a similar conformation to 8d at the ATP-binding site of the catalytic subunit of PI3Kδ [136], derived from 8c [145,146]. Therefore, we presume that 8e is designed based on the structure of 8d. 8e is an orally available, highly selective, first-in-class PI3Kδ inhibitor with an IC50 of 2.5 nM for the inhibition of p110δ, the catalytic subunit of PI3Kδ, and exhibits 40- to 300-fold selectivity for p110δ than for p110α/β/γ [147,148]. 8e was approved by the FDA in July 2014 for the treatment of relapsed chronic lymphocytic leukemia (CLL), relapsed follicular B-cell non-Hodgkin lymphoma (NHL), and relapsed small lymphocytic leukemia (SLL) [140]. The cocrystal structure of 8e with p110δ suggests that 8e has a similar binding orientation to 8d. The purine scaffold forms hydrogen bond interactions with key residues GLU826 and VAL828 in the hinge region of the ATP-binding site at distances of 2.2 and 1.9 Å, respectively. Moreover, the imidazole group of the purine scaffold can also form face-to-edge pi–pi interactions with residue TYR813.
8f (Duvelisib) is an important dual PI3Kδ/γ kinase inhibitor developed by Verastem that inhibits the PI3K catalytic subunits p110α, p110β, p110δ, and p110γ with IC50 values of 1602, 85, 2.5, and 27 nM, respectively [142,149]. In September 2018, the FDA approved 8f for the treatment of patients with CLL/SLL or relapsed/refractory follicular lymphoma (FL) [142]. 8f is structurally similar to 8e and is also an ATP-competitive inhibitor discovered through a structure-based optimization approach [84,150,151,152]. The -F on 8e’s isoquinolone is substituted with -Cl and the ethyl group is reduced to methyl to obtain the dual potent inhibitor of PI3Kδ/γ 8f. Although no crystal structure of 8f with PI3K has been reported, we suppose that 8f shares the same binding pattern as 8e and that the purine scaffold can form hydrogen bond interactions with key residues in the hinge region of the ATP-binding site. Additionally, the binding orientation of 8f is similar to that of 8d.
8g (Umbralisib) is a novel, orally available, selective, next-generation PI3Kδ inhibitor developed by TG Therapeutics with an IC50 of 22.23 nM [141,153]. 8g was approved by the FDA in February 2021 for the treatment of several B-cell malignancies, including FL and relapsed/refractory marginal zone lymphoma (MZL) [141]. 8g is structurally distinct from other PI3Kδ inhibitors. Based on the timeline of the drug discovery and the characteristics of the structure, we can infer that 8g may also be optimized from 8c. Since the N atom in the isoquinolone scaffold causes hepatotoxicity, the N is replaced by C or O atoms, which alters the toxicity distribution of 8g [154]. Although the binding model of 8g to PI3Kδ has not been released, the purine moiety is predicted to bind in the hinge region of the ATP-binding site and makes contact with key hinge residues based on its structural features. The 4-benzopyrone takes a similar orientation to 8e, extending to the roof of the ATP-binding pocket, while the newly introduced 3-fluoro-4-isopropoxyphenyl binds to the internal hydrophobic pocket.
2.9. Tropomyosin-Related Kinases (TRK) Inhibitors
Neurotrophins binding to tropomyosin-related kinases (TRK) induce receptor dimerization, phosphorylation, and activation of downstream signaling cascades through PI3K, RAS/MAPK/ERK, and plc-γ pathways, which are associated with cell proliferation, differentiation, apoptosis, and survival of neurons and other cell types [155]. The TRK family includes TRKA, TRKB, and TRKC proteins, encoded by the neurotrophic receptor tyrosine kinase 1 (NTRK1), NTRK2, and NTRK3 genes, respectively, and these NTRK gene fusions are oncogenic drivers of various adult and pediatric tumor types [156,157]. The first-generation TRK inhibitors Larotrectinib and Entrectinib were approved by the FDA for the treatment of TRK fusion-positive cancers in November 2018 and August 2019, respectively [158,159,160]. In comparison, second-generation TRK inhibitors such as Selitrectinib and Repotrectinib are being investigated in clinical trials with the aim of addressing resistance to TRK mutations [161].
A TRKB inhibitor with novel benzonitrile-substituted imidazopyrazine 9a is screened by a biochemical inhibition assay (Scintillation Proximity Assay) with an IC50 of 83 nM (Figure 9). The structure of 9a is used as a starting point for further optimization of the TRK inhibitor [162]. In order to obtain inhibitors with better potency and PK properties, the electron-withdrawing groups such as -F is introduced at the 3-position of the phenylbenzylamine to obtain 9b, which inhibits TRKA, TRKB, and TRKC with IC50 of 50, 21 nM, and 6 nM, respectively. Surprisingly, the rigidification of the phenylbenzylamine fraction by reducing the conformational entropy in a cyclic manner favors enhanced potency, with five-membered heterocyclic and R-enantiomeric showing the best performance. The 9c with better potency is then obtained by replacing the 3-cyano-phenyl group with 2-pyridine moiety, and the IC50 for the inhibition of TRKA, TRKB, and TRKC are 3, 1, and 1 nM, respectively. According to the X-ray crystal structure, 9c acts as an ATP-competitive inhibitor and binds to the ATP-binding site of TRKA. The -N on the imidazole of the imidazolopyrazine scaffold forms a hydrogen bond interaction with the hinge residue MET592 at a distance of 2.1 Å. The 3-fluorophenyl ring is bound in the hydrophobic region of the active site, while the pyridine group is in the solvent-exposed region. Further modification of the pyridine moiety in the solvent region leads to 9d, which inhibits TRKA, TRKB, and TRKC with IC50 of 4, 4, and 2 nM, respectively. 9d is further used for in vivo pharmacodynamic studies due to its pan-TRK potency, relatively low brain exposure, and overall acceptable PK properties (F = 27%) in vivo [162]. 9e (Larotrectinib) is a highly selective TRK inhibitor developed by Loxo Oncology in collaboration with Bayer AG for the treatment of adult and pediatric patients with NTRK gene fusion-positive cancers, with IC50 of 1–20 nM for inhibition of TRKA, B, and C [159,163]. Based on the structural features, we speculate that 9e was further optimized using 9d as the lead compound. The problem of TRK resistance mutations has increasingly occurred in subsequent clinical practice. To overcome the challenge of acquired resistance, researchers have developed next-generation TRK inhibitors, of which 9f (Selitrectinib, LOXO-195) and 9g (Repotrectinib, TPX-0005) are two representatives [164]. Rational macrocyclization is an effective way to improve the activity of the compound on the target. Not only does it result in a relative reduction in molecular weight, but the increased rigidity of the structure also improves the potential for membrane permeability. 9f and 9g are optimized by macrocyclization based on the structure of 9e. Using the cocrystal structure of 9g and RTKA as an example, the -N on the pyrazole ring of the pyrimidinopyrazole scaffold still forms a hinge hydrogen bond with MET592 at a distance of 2.0 Å. When residue G595 of TRKA is mutated to G595R, the hinge hydrogen bond remains as expected, but the macrocyclized 9g does not clash in space with the mutated G595R, indicating that the cyclized molecule is able to overcome the acquired drug resistance mutation of RTK.
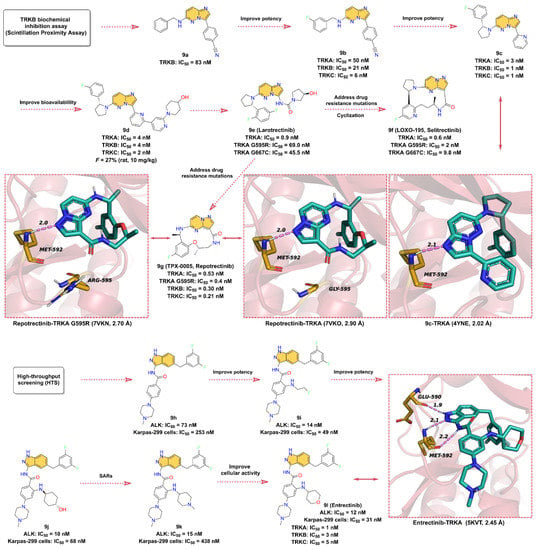
Figure 9.
Key medicinal chemistry optimization of TRK inhibitors together with the binding models with TRK.
9l (Entrectinib, Figure 9) is a potent, orally available, brain penetrant RTK inhibitor that targets TRKA, TRKB, TRKC, ROS1 (c-ros Oncogene 1 Kinase), and ALK (Anaplastic lymphoma kinase) with IC50 values of 1, 3, 5, 7, and 12 nM, respectively [165,166]. Interestingly, the development process of 9l initially started with the target of ALK kinase [167,168]. The researchers use a high-throughput screening experiment to obtain compound 9h, a 3-amino-5-substituted indazole derivative, which shows good inhibitory activity against ALK with an IC50 of 73 nM and also exhibits promising anti-proliferative activity against the ALK-dependent ALCL Karpas-299 cell lines with an IC50 of 253 nM. Optimization of the benzene ring attached to methylpiperazine using different substituents to obtain compound 9i, which shows a significant increase in the inhibitory potency against ALK with an IC50 of 14 nM, as well as a remarkably elevated proliferation inhibitory activity against the Karpas-299 cells with IC50 of 49 nM. Further substitution of 9i with 4-amino-4-cyclohexanol to afford 9j, which further enhances its inhibitory potency on ALK (IC50 = 10 nM). In addition, 9k and 9l are obtained using 4-amino-N-methylpiperidinyl and 4-aminotetrahydropyranyl substitutions, respectively, both with good biochemical potency, and their IC50 values for ALK inhibition are 15 nM and 12 nM, respectively. However, 9k exhibits (Karpas-299 cells, IC50 = 438 nM) worse cellular activity than 9l (Karpas-299 cells, IC50 = 31 nM), which might be due to the low cell permeability. As mentioned earlier, 9l is also a potent inhibitor of TRKs. The crystallographic structure reveals that 9l binds in the ATP-binding pocket of TRKA and contacts with key residues around the binding site. In particular, the three N atoms on 3-aminoindazole scaffold can serve as hydrogen bond donors and acceptors to form three hydrogen bond interactions with the hinge residues GLU590 and MET592 at distances of 1.9, 2.1, and 2.2 Å, respectively.
2.10. Vascular Endothelial Growth Factor Receptors (VEGFRs) Inhibitors
Tumor angiogenesis is a prerequisite for tumor growth and metastasis [169], which is regulated by a number of pro- and anti-angiogenic factors produced by the host or tumor cells, including vascular endothelial growth factor (VEGF) and other cytokines [170,171]. Vascular endothelial growth factor receptors (VEGFRs) are members of the RTKs family and are classified into three isoforms, VEGFR1, VEGFR2, and VEGFR3 [172]. VEGFR1 and VEGFR2 play important roles in angiogenesis, including tumor angiogenesis, while VEGFR3 is associated with lymphangiogenesis [173]. Compared to normal endothelial cells, VEGFR2 activation by VEGF triggers a phosphorylation process that leads to greatly enhanced proliferation and migration of endothelial cells [173,174]. EGFR2 is often overexpressed in tumor endothelial cells, so treatment of tumors can be achieved by inhibiting angiogenesis [175]. Selective inhibition of VEGFR kinases can inhibit angiogenesis and is currently a very successful clinical strategy in cancer therapy [176,177]. VEGFR kinase inhibitors are mainly classified into two types according to the binding model. Type I kinase inhibitors compete directly with ATP for binding at the ATP-binding site in the active form of the kinase, while type II kinase inhibitors bind to the adenine-binding and an additional hydrophobic back pocket at the ATP-binding site in the inactive form. More than 10 small-molecule VEGFR2 kinase inhibitors have been approved for various cancer therapies [176]. Here, we only discuss VEGFR2 inhibitors that contain the core structure of azaindole or oxindole, including the type I inhibitors Anlotinib, Sunitinib, Nintedanib, and Pazopanib, and the type II inhibitor Axitinib (Figure 10).
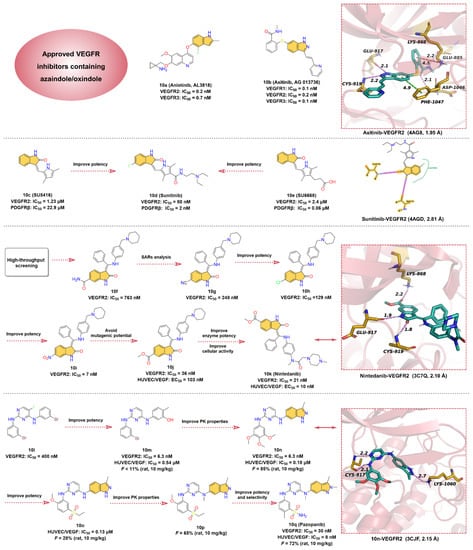
Figure 10.
Key medicinal chemistry optimization of VEGFR inhibitors along with the binding models with VEGFR2.
Quinoline and indole fragments have a variety of biological activities and play a crucial role in the development of anticancer drugs. The VEGFR2 kinase inhibitor 10a (Anlotinib, AL3818) is one such example [178], which exhibits potent inhibition of VEGFR2 and VEGFR3 with IC50 values of 0.2 and 0.7 nM, respectively. 10a, co-developed by Jiangsu Chia-Tai Tianqing Pharmaceutical and Advenchen Laboratories, was approved in 2018 by NMPA for the treatment of patients with locally advanced or metastatic NSCLC [179,180]. Preclinical studies have shown that 10a inhibits VEGF/PDGF-BB/FGF-2-induced cell migration, angiogenesis, and capillary-like tube formation in endothelial cells and has a broad inhibitory effect on tumor angiogenesis and growth [181,182]. To date, no cocrystal structure of 10a with VEGFR2 has been reported, but a molecular docking study has shown that 10a can bind to the ATP-binding pocket of VEGFR2 [183]. The hinge residues GLU917 and CYS919 can form hydrogen bond interactions with the quinoline fragment of 10a, while the indole moiety is located deep in the hydrophobic region of the ATP-binding pocket. Thus, we conclude that 10a is a type I VEGFR inhibitor.
Renal cell carcinoma (RCC) is the most common form of kidney cancer. More than 200,000 patients are diagnosed with this disease worldwide each year, and approximately 100,000 deaths occur each year [184,185]. Starting with the pyrazole-styryl derivative, researchers obtained the small-molecule indazole derivative 10b (Axitinib, AG-013736), a type II inhibitor of VEGFR kinases, using a truncated lead compound strategy and the introduction of a conformational constraint strategy for rational structure-based drug design [186,187]. 10b is a potent and selective inhibitor of VEGFR1, 2, and 3 with IC50 values of 0.1, 0.2, and 0.1 nM, respectively [188]. 10b is a highly potent and selective drug compared to other approved RCC active agents and is currently approved for the treatment of RCC [189,190]. The cocrystal structure reveals that the indazole moiety of 10b forms two hydrogen bonds with the hinge residues GLU917 and CYS919 at distances of 2.1 and 2.2 Å, respectively. Moreover, the indazole core also establishes a pi–pi stacking interaction with the residue PHE1047. While the N-methylbenzamide motif extends deep into the back hydrophobic pocket of the ATP-binding site, forming hydrogen bonds and pi–cation interactions with residues ASP1046, GLU885, and LYS868, respectively. Additionally, the pyridine-vinyl part of 10b is in the hinge region and forms van der Waals interactions with surrounding residues.
10d (Sunitinib), a multi-target RTK inhibitor, was FDA-approved for the treatment of RCC, gastrointestinal stromal tumors, and progressive neuroendocrine tumors of pancreatic origin in 2006 [191,192]. Researchers have conducted extensive medicinal chemistry investigations on existing selective VEGFR2 inhibitors and selective platelet-derived growth factor receptor β (PDGFRβ) inhibitors in an effort to obtain potent antitumor drugs that can inhibit both VEGFR2 and PDGFRβ [193]. 10c (SU5416) is a selective VEGFR2 inhibitor with an IC50 of 1.23 μM for VEGFR2 and 22.9 μM for PDGFRβ. 10e (SU6668) is a selective PDGFRβ inhibitor with an IC50 of 0.06 μM for PDGFRβ and 2.4 μM for VEGFR2. Based on the structures of 10c and 10e, the researchers modified the C-4 position on the pyrrole ring to obtain 10d. The SAR implied that 10d exhibited potent inhibitory activity against both VEGFR2 and PDGFRβ with IC50 values of 80 and 2 nM, respectively. The crystal structure of VEGRF2 in complex with 10d reveals that the indolin-2-one core forms two hinge hydrogen bonds with residues GLU917 and CYS919, which further anchor 10d at the ATP-binding pocket.
10k (Nintedanib), a potent triple angiokinase (VEGFR/PDGFR/FGFR) inhibitor, is approved by FDA to treat idiopathic pulmonary fibrosis by blocking fibroblast proliferation and reducing extracellular matrix deposition [194,195]. In particular, 10k has the most potent kinase inhibitory effect on VEGFR2 with an IC50 of 21 nM [196]. The researchers obtained the hit compound 10f against VEGFR2 with an IC50 of 763 nM through high-throughput screening methods for selective testing of derivatives of related kinases [197]. It was found that the vertical conformation of the central benzene ring and the benzopyrrole scaffold can increase the solubility of the compound, so these key fragments are retained in the subsequent structure optimization process [194]. SARs studies on 10f produced a more active compound 10g, with an IC50 of 248 nM against VEGFR2 [195]. After replacing the -CN group on benzopyrrole with -Cl, the potency of compound 10h was further enhanced with an IC50 of 129 nM. Interestingly, the -NO2 moiety has the strongest inhibitory effect on VEGFR2 with an IC50 of 7 nM (10i) but is eventually terminated due to a mutagenic potential. Despite the risk of degradation by esterase metabolism by replacing the -NO2 group with -COOCH3, 10j yields an effective inhibition for VEGFR2 with an IC50 of 36 nM and moderate cytotoxicity (HUVEC/VEGF: EC50 = 103 nM). In addition, the piperidine fragment is optimized to fine-tune properties such as cellular activity and solubility, resulting in 10k (HUVEC/VEGF: EC50 = 10 nM), a potent and selective VEGFR2 inhibitor [197]. Based on the X-ray cocrystal structure, it can be concluded that 10k binds into the ATP-binding site of VEGFR2. The -NH and -CO groups on the indolinone scaffold can form two strong hydrogen bonds with the backbone carbonyl oxygen of GLU917 and the backbone nitrogen of CYS919 in the hinge region at distances of 1.9 and 1.8 Å, respectively. The phenyl group binds in the hydrophobic pocket of the ATP-binding site, forming hydrophobic interactions with the surrounding residues, while N-methylpiperazine group points into the solvent region of the active site, forming ionic and polar interactions.
10q (Pazopanib) is an oral multi-kinases inhibitor that primarily inhibits the VEGFRs with IC50 of 10, 30, and 47 nM for VEGFR1, 2, and 3, respectively, and is currently approved for the treatment of advanced soft-tissue sarcoma (STS) and RCC [198,199]. Compound 10l with an inhibitory effect on VEGFR2 (IC50 = 400 nM) is obtained as a hit compound by screening the compound library [199]. Substitution of -Br in 10l using -OH group gives 10m, which inhibits VEGFR2 with an IC50 of 6.3 nM. It is presumed that the reason for this hundred-fold increase in activity may be attributed to the newly introduced -OH can form hydrogen bond interactions with key residues in the binding pocket [199]. Replacement of the benzene ring of 10m with a 3-methylindazole heterocycle affords 10n, which has significantly improved PK properties. The clearance of 10n in rats was 16 mL/min/kg at an administered dose of 10 mg/kg, with an oral bioavailability of 85%. The cocrystal structure of VEGFR2 in complex with 10n demonstrates that the 3-methylindazole motif can form a hydrogen bond with residue LYS1060 at a distance of 2.7 Å, and the 2-aminopyrimidine segment can form bidentate hydrogen bonds with the hinge residue CYS917 at distances of 2.1 and 2.2 Å, respectively (Figure 10, lower right). To further enhance the potency, the introduction of 5-sulfone in aniline to obtain 10o leads to a moderate increase in the potency of the enzyme and cellular assay with IC50 of 130 nM in human umbilical vein endothelial cells (HUVEC). Methylation of the C-4 amino of the pyrimidine group to obtain 10p can significantly improve the PK properties. To reduce the risk of nitrogen on the indazole heterocycle binding to heme iron of cytochrome P450 enzymes, its methylation to increase the steric resistance of the heterocycle and also optimization of the substituents on the aniline yields 10q with desirable selectivity and cellular potency with an IC50 of 30 nM against VEGFR2 and 8 nM against HUVEC/VEGFR.
2.11. Others
Additionally, there are several other ATP-competitive small-molecule inhibitors containing azaindoles that have been approved in clinical, including Selpercatinib, Avapritinib, and Capmatinib (Figure 11).
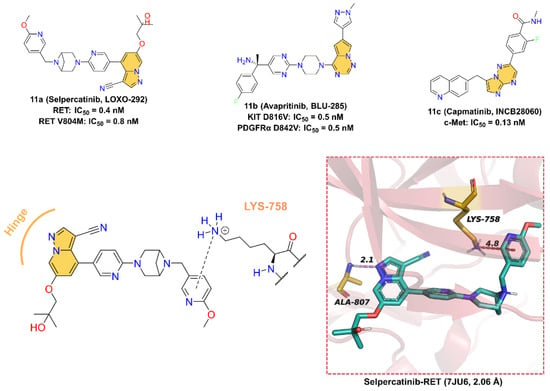
Figure 11.
Two-dimensional structures of Selpercatinib, Avapritinib, and Capmatinib together with the binding pattern of Selpercatinib and RET.
11a (Selpercatinib, LOXO-292), developed by Loxo Oncology for the treatment of various solid tumors, including NSCLC and thyroid cancer, is a highly selective receptor tyrosine kinase rearranged during transfection (RET) inhibitor with IC50 values of 0.4 nM and 0.8nM for RET wild-type and mutant-type (V804M), respectively [200,201]. The cocrystal structure of RET in a complex with 11a reveals that 11a binds at the ATP-binding site. The pyrazolo [1,5-a]pyridine scaffold forms a hinge hydrogen bond with the backbone -NH of residue ALA807 at a distance of 2.1 Å, and the 2-methoxypyridine group inserts into the deep hydrophobic pocket forming a pi–cation interaction with residue LYS758.
11b (Avapritinib, BLU-285), developed by Blueprint Medicines for the treatment of gastrointestinal stromal tumors and systemic mastocytosis, is a potent, selective, orally active inhibitor of KIT and PDGFRα activation loop mutant kinases with IC50 values of 0.5 and 0.5 nM for KIT D816V and PDGFRα D842V mutant, respectively [202,203]. A molecular docking study was used to investigate the potential binding interactions between 11b and KIT D816V. According to the results, 11b binds at the ATP-binding site, and the pyrrolo [2,1-f][1,2,4]triazine scaffold makes one hinge hydrogen bond with the backbone of residue CYS673 at a distance of 2.4 Å [204].
11c (Capmatinib, INCB28060) is a potent, highly selective, ATP-competitive, and reversible c-Met kinase inhibitor with an average IC50 value of 0.13 nM [205]. On May 6, 2020, 11c received its first worldwide U.S. approval for the treatment of adults with metastatic NSCLS [206,207,208,209]. 11c was first reported by patent (US8420645B2) as a c-MET kinase inhibitor with an imidazo [1,2-b][1,2,4]triazin scaffold. However, the crystal structure of 11c with c-Met has not been disclosed so far, and therefore, the binding model to the protein cannot be accurately determined. Nevertheless, we can speculate that the imidazolotriazine core is an indispensable pharmacophore for binding to the ATP-binding site of c-Met.
3. Conclusions
Over the past 20 years, protein kinases have been attracting great interest, and significant achievements have already been made in developing ATP-competitive kinase inhibitors. A variety of kinase inhibitors investigated in preclinical and clinical studies have indole/azaindole/oxindole scaffolds. In particular, thirty indole/azaindole/oxindole-containing ATP-competitive kinase inhibitors have been approved as therapeutic agents for various diseases. This paper describes the discovery and optimization process of these thirty approved, another three in clinical trials, an additional one in preclinical studied indole/azaindole/oxindole-based ATP-competitive kinase inhibitors, including Bcr-Abl, BTK, CDK4/6, CSF1R, HER, JAK, BRAF, PI3K, TRK, VEGFR, and other kinase inhibitors (Table 1). Meanwhile, the drug mutant routes of these selected drugs were summarized to verify the potential contribution of indole-like structural fragments to their overall biological activity and therapeutic effect. We also explore the interactive bonding between the inhibitors and amino acid residues at the APT-binding sites of their target kinase based on the corresponding protein crystal complex. It is found that indole/azaindole/oxindole scaffolds can form hinge hydrogen bonds and other non-covalent bond interactions with residues in the hinge region of the kinase’s APT-binding site to better bind to the ATP-binding pocket. Convincingly, indole/azaindole/oxindole structures play an indispensable role in occupying the ATP pocket. This implies that these heterocyclic systems are a privileged scaffold in the quest for new ATP-competitive kinase inhibitors.

Table 1.
Indole/azaindole/oxindole-containing ATP-competitive kinase inhibitors in clinical applications.
At present, kinase inhibitors remain the first choice for many diseases. However, mutations in the residues of the kinase hinge region generally lead to drug resistance that reduces the potency and selectivity of the inhibitor. Additionally, the period from R&D to market for small-molecule inhibitors is long, and the investment is enormous. Therefore, the authors hope this review will help researchers quickly develop more selective and potent indole/azaindole/oxindole-based kinase inhibitors combined with artificial intelligence technology.
Author Contributions
Conceptualization, M.F. and F.H.; methodology, H.Z., F.H., and G.G.; software, F.H. and H.Z.; validation, F.H.; formal analysis, S.L. and Q.W.; investigation, F.H., H.Z., G.G., S.L., and Q.W.; data curation, F.H., H.Z., and G.G.; writing—original draft preparation, F.H., H.Z., and G.G.; writing—review and editing, M.F. and F.H.; visualization, H.H. and Z.W.; supervision, M.F. and X.W.; project administration, X.W. and M.F.; financial support, X.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Natural Science Foundation of Fujian Province of China, grant number 2022Y0001; and the Special fund for marine development of Fujian province, grant number FJHJF-L-2021-5.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Peterson, R.T.; Schreiber, S.L. Kinase phosphorylation: Keeping it all in the family. Curr. Biol. 1999, 9, R521–R524. [Google Scholar] [CrossRef] [PubMed]
- Dinarello, C.A. Interleukin-1 in the pathogenesis and treatment of inflammatory diseases. Blood 2011, 117, 3720–3732. [Google Scholar] [CrossRef] [PubMed]
- Day, E.A.; Ford, R.J.; Steinberg, G.R. AMPK as a Therapeutic Target for Treating Metabolic Diseases. Trends Endocrinol. Metab. 2017, 28, 545–560. [Google Scholar] [CrossRef] [PubMed]
- Kannaiyan, R.; Mahadevan, D. A comprehensive review of protein kinase inhibitors for cancer therapy. Expert Rev. Anticancer. Ther. 2018, 18, 1249–1270. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, S.R.; Till, J.H. Protein tyrosine kinase structure and function. Annu. Rev. Biochem. 2000, 69, 373–398. [Google Scholar] [CrossRef]
- Hanks, S.K.; Hunter, T. Protein kinases 6. The eukaryotic protein kinase superfamily: Kinase (catalytic) domain structure and classification. FASEB J. 1995, 9, 576–596. [Google Scholar] [CrossRef]
- Deshmukh, K.; Anamika, K.; Srinivasan, N. Evolution of domain combinations in protein kinases and its implications for functional diversity. Prog. Biophys. Mol. Biol. 2010, 102, 1–15. [Google Scholar] [CrossRef]
- Endicott, J.A.; Noble, M.E.; Johnson, L.N. The structural basis for control of eukaryotic protein kinases. Annu. Rev. Biochem. 2012, 81, 587–613. [Google Scholar] [CrossRef]
- Dohlman, H.G. A scaffold makes the switch. Sci. Signal. 2008, 1, pe46. [Google Scholar] [CrossRef]
- Cohen, P.; Cross, D.; Jänne, P.A. Kinase drug discovery 20 years after imatinib: Progress and future directions. Nat. Rev. Drug Discov. 2021, 20, 551–569. [Google Scholar] [CrossRef]
- Roskoski, R., Jr. Properties of FDA-approved small molecule protein kinase inhibitors: A 2022 update. Pharmacol. Res. 2022, 175, 106037. [Google Scholar] [CrossRef]
- Attwood, M.M.; Fabbro, D.; Sokolov, A.V.; Knapp, S.; Schioth, H.B. Trends in kinase drug discovery: Targets, indications and inhibitor design. Nat. Rev. Drug Discov. 2021, 20, 839–861. [Google Scholar] [CrossRef]
- Munoz, L. Non-kinase targets of protein kinase inhibitors. Nat. Rev. Drug Discov. 2017, 16, 424–440. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, P.L.; Gray, N.S. Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer 2009, 9, 28–39. [Google Scholar] [CrossRef]
- Lu, X.; Smaill, J.B.; Ding, K. New Promise and Opportunities for Allosteric Kinase Inhibitors. Angew. Chem. Int. Ed. Engl. 2020, 59, 13764–13776. [Google Scholar] [CrossRef]
- Döker, S.; Dewenter, M.; El-Armouche, A. Tofacitinib. Dtsch. Med. Wochenschr. 2014, 139, 1003–1008. [Google Scholar]
- Yang, H.; Higgins, B.; Kolinsky, K.; Packman, K.; Go, Z.; Iyer, R.; Kolis, S.; Zhao, S.; Lee, R.; Grippo, J.F.; et al. RG7204 (PLX4032), a selective BRAFV600E inhibitor, displays potent antitumor activity in preclinical melanoma models. Cancer Res. 2010, 70, 5518–5527. [Google Scholar] [CrossRef]
- Massaro, F.; Molica, M.; Breccia, M. Ponatinib: A Review of Efficacy and Safety. Curr. Cancer Drug Targets 2018, 18, 847–856. [Google Scholar] [CrossRef]
- Stierand, K.; Maass, P.C.; Rarey, M. Molecular complexes at a glance: Automated generation of two-dimensional complex diagrams. Bioinformatics 2006, 22, 1710–1716. [Google Scholar] [CrossRef]
- Fricker, P.C.; Gastreich, M.; Rarey, M. Automated drawing of structural molecular formulas under constraints. J. Chem. Inf. Comp. Sci. 2004, 44, 1065–1078. [Google Scholar] [CrossRef]
- Carofiglio, F.; Lopalco, A.; Lopedota, A.; Cutrignelli, A.; Nicolotti, O.; Denora, N.; Stefanachi, A.; Leonetti, F. Bcr-Abl Tyrosine Kinase Inhibitors in the Treatment of Pediatric CML. Int. J. Mol. Sci. 2020, 21, 4469. [Google Scholar] [CrossRef] [PubMed]
- von Bubnoff, N.; Veach, D.R.; van der Kuip, H.; Aulitzky, W.E.; Sänger, J.; Seipel, P.; Bornmann, W.G.; Peschel, C.; Clarkson, B.; Duyster, J. A cell-based screen for resistance of Bcr-Abl-positive leukemia identifies the mutation pattern for PD166326, an alternative Abl kinase inhibitor. Blood 2005, 105, 1652–1659. [Google Scholar] [CrossRef] [PubMed]
- Lahaye, T.; Riehm, B.; Berger, U.; Paschka, P.; Müller, M.C.; Kreil, S.; Merx, K.; Schwindel, U.; Schoch, C.; Hehlmann, R.; et al. Response and resistance in 300 patients with BCR-ABL-positive leukemias treated with imatinib in a single center: A 4.5-year follow-up. Cancer 2005, 103, 1659–1669. [Google Scholar] [CrossRef] [PubMed]
- Nicolini, F.E.; Chabane, K.; Tigaud, I.; Michallet, M.; Magaud, J.P.; Hayette, S. BCR-ABL mutant kinetics in CML patients treated with dasatinib. Leuk. Res. 2007, 31, 865–868. [Google Scholar] [CrossRef] [PubMed]
- Melo, J.V.; Chuah, C. Resistance to imatinib mesylate in chronic myeloid leukaemia. Cancer Lett. 2007, 249, 121–132. [Google Scholar] [CrossRef]
- Mughal, T.I.; Goldman, J.M. Emerging strategies for the treatment of mutant Bcr-Abl T315I myeloid leukemia. Clin. Lymphoma Myeloma 2007, 7, S81–S84. [Google Scholar] [CrossRef]
- Deininger, M.; Buchdunger, E.; Druker, B.J. The development of imatinib as a therapeutic agent for chronic myeloid leukemia. Blood 2005, 105, 2640–2653. [Google Scholar] [CrossRef]
- Tan, F.H.; Putoczki, T.L.; Stylli, S.S.; Luwor, R.B. Ponatinib: A novel multi-tyrosine kinase inhibitor against human malignancies. Onco. Targets Ther. 2019, 12, 635–645. [Google Scholar] [CrossRef]
- Huang, W.S.; Metcalf, C.A.; Sundaramoorthi, R.; Wang, Y.; Zou, D.; Thomas, R.M.; Zhu, X.; Cai, L.; Wen, D.; Liu, S. Discovery of 3-[2-(Imidazo [1,2-b]pyridazin-3-yl)ethynyl]-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzamide (AP24534), a Potent, Orally Active Pan-Inhibitor of Breakpoint Cluster Region-Abelson (BCR-ABL) Kinase Including the. J. Med. Chem. 2010, 53, 4701. [Google Scholar]
- Kantarjian, H.M.; Jabbour, E.; Deininger, M.; Abruzzese, E.; Apperley, J.; Cortes, J.; Chuah, C.; DeAngelo, D.J.; DiPersio, J.; Hochhaus, A.; et al. Ponatinib after failure of second-generation tyrosine kinase inhibitor in resistant chronic-phase chronic myeloid leukemia. Am. J. Hematol. 2022, 97, 1419–1426. [Google Scholar] [CrossRef]
- Lewis, C.M.; Broussard, C.; Czar, M.J.; Schwartzberg, P.L. Tec kinases: Modulators of lymphocyte signaling and development. Curr. Opin. Immunol. 2001, 13, 317–325. [Google Scholar] [CrossRef]
- Good, L.; Benner, B.; Carson, W.E. Bruton’s tyrosine kinase: An emerging targeted therapy in myeloid cells within the tumor microenvironment. Cancer Immunol. Immun. 2021, 70, 2439–2451. [Google Scholar] [CrossRef]
- Liang, C.Y.; Tian, D.N.; Ren, X.D.; Ding, S.J.; Jia, M.Y.; Xin, M.H.; Thareja, S. The development of Bruton’s tyrosine kinase (BTK) inhibitors from 2012 to 2017: A mini-review. Eur. J. Med. Chem. 2018, 151, 315–326. [Google Scholar] [CrossRef]
- Ran, F.S.; Liu, Y.; Wang, C.; Xu, Z.Y.; Zhang, Y.A.; Liu, Y.; Zhao, G.S.; Ling, Y. Review of the development of BTK inhibitors in overcoming the clinical limitations of ibrutinib. Eur. J. Med. Chem. 2022, 229, 114009. [Google Scholar] [CrossRef]
- Honigberg, L.A.; Smith, A.M.; Sirisawad, M.; Verner, E.; Loury, D.; Chang, B.; Li, S.; Pan, Z.Y.; Thamm, D.H.; Miller, R.A.; et al. The Bruton tyrosine kinase inhibitor PCI-32765 blocks B-cell activation and is efficacious in models of autoimmune disease and B-cell malignancy. Proc. Natl. Acad. Sci. USA 2010, 107, 13075–13080. [Google Scholar] [CrossRef]
- Feng, Y.F.; Duan, W.M.; Cu, X.C.; Liang, C.Y.; Xin, M.H. Bruton’s tyrosine kinase (BTK) inhibitors in treating cancer: A patent review (2010–2018). Expert Opin. Ther. Pat. 2019, 29, 217–241. [Google Scholar] [CrossRef]
- Pan, Z.Y.; Scheerens, H.; Li, S.J.; Schultz, B.E.; Sprengeler, P.A.; Burrill, L.C.; Mendonca, R.V.; Sweeney, M.D.; Scott, K.C.K.; Grothaus, P.G.; et al. Discovery of selective irreversible inhibitors for Bruton’s tyrosine kinase. Chemmedchem 2007, 2, 58–61. [Google Scholar] [CrossRef]
- Bender, A.T.; Gardberg, A.; Pereira, A.; Johnson, T.; Wu, Y.; Grenningloh, R.; Head, J.; Morandi, F.; Haselmayer, P.; Liu-Bujalski, L. Ability of Bruton’s Tyrosine Kinase Inhibitors to Sequester Y551 and Prevent Phosphorylation Determines Potency for Inhibition of Fc Receptor but not B-Cell Receptor Signaling. Mol. Pharmacol. 2017, 91, 208–219. [Google Scholar] [CrossRef]
- Jain, P.; Keating, M.; Wierda, W.; Estrov, Z.; Ferrajoli, A.; Jain, N.; George, B.; James, D.; Kantarjian, H.; Burger, J.; et al. Outcomes of patients with chronic lymphocytic leukemia after discontinuing ibrutinib. Blood 2015, 125, 2062–2067. [Google Scholar] [CrossRef]
- Jain, P.; Thompson, P.A.; Keating, M.; Estrov, Z.; Ferrajoli, A.; Jain, N.; Kantarjian, H.; Burger, J.A.; O’Brien, S.; Wierda, W.G. Long-term outcomes for patients with chronic lymphocytic leukemia who discontinue ibrutinib. Cancer 2017, 123, 2268–2273. [Google Scholar] [CrossRef]
- Mato, A.R.; Nabhan, C.; Thompson, M.C.; Lamanna, N.; Brander, D.M.; Hill, B.; Howlett, C.; Skarbnik, A.; Cheson, B.D.; Zent, C.; et al. Toxicities and outcomes of 616 ibrutinib-treated patients in the United States: A real-world analysis. Haematologica 2018, 103, 874–879. [Google Scholar] [CrossRef] [PubMed]
- Barf, T.; Covey, T.; Izumi, R.; van de Kar, B.; Gulrajani, M.; van Lith, B.; van Hoek, M.; de Zwart, E.; Mittag, D.; Demont, D.; et al. Acalabrutinib (ACP-196): A Covalent Bruton Tyrosine Kinase Inhibitor with a Differentiated Selectivity and In Vivo Potency Profile. J. Pharmacol. Exp. Ther. 2017, 363, 240–252. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhang, M.; Liu, D. Acalabrutinib (ACP-196): A selective second-generation BTK inhibitor. J. Hematol. Oncol. 2016, 9, 21. [Google Scholar] [CrossRef] [PubMed]
- da Cunha-Bang, C.; Niemann, C.U. Targeting Bruton’s Tyrosine Kinase Across B-Cell Malignancies. Drugs 2018, 78, 1653–1663. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, M.; Liu, D. Bruton tyrosine kinase inhibitor ONO/GS-4059: From bench to bedside. Oncotarget 2017, 8, 7201–7207. [Google Scholar] [CrossRef]
- Johnson, A.R.; Kohli, P.B.; Katewa, A.; Gogol, E.; Belmont, L.D.; Choy, R.; Penuel, E.; Burton, L.; Eigenbrot, C.; Yu, C.; et al. Battling Btk Mutants With Noncovalent Inhibitors That Overcome Cys481 and Thr474 Mutations. ACS Chem. Biol. 2016, 11, 2897–2907. [Google Scholar] [CrossRef]
- Guo, X.F.; Yang, D.Y.; Fan, Z.J.; Zhang, N.L.; Zhao, B.; Huang, C.; Wang, F.J.; Ma, R.J.; Meng, M.; Deng, Y.C. Discovery and structure-activity relationship of novel diphenylthiazole derivatives as BTK inhibitor with potent activity against B cell lymphoma cell lines. Eur. J. Med. Chem. 2019, 178, 767–781. [Google Scholar] [CrossRef]
- Musumeci, F.; Sanna, M.; Greco, C.; Giacchello, I.; Fallacara, A.L.; Amato, R.; Schenone, S. Pyrrolo[2,3-d]pyrimidines active as Btk inhibitors. Expert Opin. Ther. Pat. 2017, 27, 1305–1318. [Google Scholar] [CrossRef]
- Reiff, S.D.; Mantel, R.; Smith, L.L.; Greene, J.T.; Muhowski, E.M.; Fabian, C.A.; Goettl, V.M.; Tran, M.; Harrington, B.K.; Rogers, K.A.; et al. The BTK Inhibitor ARQ 531 Targets Ibrutinib-Resistant CLL and Richter Transformation. Cancer Discov. 2018, 8, 1300–1315. [Google Scholar] [CrossRef]
- Eathiraj, S.; Yu, Y.; Savage, R.; Woyach, J.A.; Reiff, S.D.; Johnson, A.J.; Schwartz, B. ARQ 531, a potent reversible BTK inhibitor, exhibits potent antitumor activity in ibrutinib-resistant diffuse large B-cell lymphoma. Cancer Res. 2018, 78, 1963. [Google Scholar] [CrossRef]
- Chong, Q.-Y.; Kok, Z.-H.; Bui, N.-L.-C.; Xiang, X.; Wong, A.L.-A.; Yong, W.-P.; Sethi, G.; Lobie, P.E.; Wang, L.; Goh, B.-C. A unique CDK4/6 inhibitor: Current and future therapeutic strategies of abemaciclib. Pharmacol. Res. 2020, 156, 104686. [Google Scholar] [CrossRef]
- Asghar, U.; Witkiewicz, A.K.; Turner, N.C.; Knudsen, E.S. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat. Rev. Drug Discov. 2015, 14, 130–146. [Google Scholar] [CrossRef]
- O’Leary, B.; Finn, R.S.; Turner, N.C. Treating cancer with selective CDK4/6 inhibitors. Nat. Rev. Clin. Oncol. 2016, 13, 417–430. [Google Scholar] [CrossRef]
- Gelbert, L.M.; Cai, S.; Lin, X.; Sanchez-Martinez, C.; del Prado, M.; Lallena, M.J.; Torres, R.; Ajamie, R.T.; Wishart, G.N.; Flack, R.S.; et al. Preclinical characterization of the CDK4/6 inhibitor LY2835219: In-vivo cell cycle-dependent/independent anti-tumor activities alone/in combination with gemcitabine. Investig. New Drugs 2014, 32, 825–837. [Google Scholar] [CrossRef]
- Sánchez-Martínez, C.; Gelbert, L.M.; Lallena, M.J.; de Dios, A. Cyclin dependent kinase (CDK) inhibitors as anticancer drugs. Bioorganic Med. Chem. Lett. 2015, 25, 3420–3435. [Google Scholar] [CrossRef]
- Chen, P.; Lee, N.V.; Hu, W.Y.; Xu, M.R.; Ferre, R.A.; Lam, H.; Bergqvist, S.; Solowiej, J.; Diehl, W.; He, Y.A.; et al. Spectrum and Degree of CDK Drug Interactions Predicts Clinical Performance. Mol. Cancer Ther. 2016, 15, 2273–2281. [Google Scholar] [CrossRef]
- Barvian, M.; Boschelli, D.H.; Cossrow, J.; Dobrusin, E.; Fattaey, A.; Fritsch, A.; Fry, D.; Harvey, P.; Keller, P.; Garrett, M.; et al. Pyrido[2,3-d]pyrimidin-7-one Inhibitors of Cyclin-Dependent Kinases. J. Med. Chem. 2000, 43, 4606–4616. [Google Scholar] [CrossRef]
- Fry, D.W.; Bedford, D.C.; Harvey, P.H.; Fritsch, A.; Keller, P.R.; Wu, Z.P.; Dobrusin, E.; Leopold, W.R.; Fattaey, A.; Garrett, M.D. Cell cycle and biochemical effects of PD 0183812-a potent inhibitor of the cyclin D-dependent kinases CDK4 and CDK6. J. Biol. Chem. 2001, 276, 16617–16623. [Google Scholar] [CrossRef]
- Fry, D.W.; Harvey, P.J.; Keller, P.R.; Elliott, W.L.; Meade, M.A.; Trachet, E.; Albassam, M.; Zheng, X.X.; Leopold, W.R.; Pryer, N.K.; et al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol. Cancer Ther. 2004, 3, 1427–1437. [Google Scholar] [CrossRef]
- Mullard, A. FDA approves Novartis’s CDK4/6 inhibitor. Nat. Rev. Drug Discov. 2017, 16, 229. [Google Scholar] [CrossRef]
- Marra, A.; Curigliano, G. Are all cyclin-dependent kinases 4/6 inhibitors created equal? NPJ Breast Cancer 2019, 5, 27. [Google Scholar] [CrossRef]
- Shi, Z.; Tian, L.; Qiang, T.; Li, J.; Xing, Y.; Ren, X.; Liu, C.; Liang, C. From Structure Modification to Drug Launch: A Systematic Review of the Ongoing Development of Cyclin-Dependent Kinase Inhibitors for Multiple Cancer Therapy. J. Med. Chem. 2022, 65, 6390–6418. [Google Scholar] [CrossRef]
- Hubbard, S.R.; Miller, W.T. Receptor tyrosine kinases: Mechanisms of activation and signaling. Curr. Opin. Cell Biol. 2007, 19, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Choura, M.; Rebai, A. Receptor tyrosine kinases: From biology to pathology. J. Recept. Signal Transduct. Res. 2011, 31, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Im, D.; Jung, K.; Yang, S.; Aman, W.; Hah, J.M. Discovery of 4-arylamido 3-methyl isoxazole derivatives as novel FMS kinase inhibitors. Eur. J. Med. Chem. 2015, 102, 600–610. [Google Scholar] [CrossRef] [PubMed]
- Stanley, E.R.; Chitu, V. CSF-1 Receptor Signaling in Myeloid Cells. CSH Perspect. Biol. 2014, 6, a021857. [Google Scholar] [CrossRef]
- Sica, A.; Larghi, P.; Mancino, A.; Rubino, L.; Porta, C.; Totaro, M.G.; Rimoldi, M.; Biswas, S.K.; Allavena, P.; Mantovani, A. Macrophage polarization in tumour progression. Semin. Cancer Biol. 2008, 18, 349–355. [Google Scholar] [CrossRef]
- Achkova, D.; Maher, J. Role of the colony-stimulating factor (CSF)/CSF-1 receptor axis in cancer. Biochem. Soc. T 2016, 44, 333–341. [Google Scholar] [CrossRef]
- Chockalingam, S.; Ghosh, S.S. Macrophage colony-stimulating factor and cancer: A review. Tumor. Biol. 2014, 35, 10635–10644. [Google Scholar] [CrossRef]
- El-Gamal, M.I.; Al-Ameen, S.K.; Al-Koumi, D.M.; Hamad, M.G.; Jalal, N.A.; Oh, C.H. Recent Advances of Colony-Stimulating Factor-1 Receptor (CSF-1R) Kinase and Its Inhibitors. J. Med. Chem. 2018, 61, 5450–5466. [Google Scholar] [CrossRef]
- Butowski, N.; Colman, H.; De Groot, J.F.; Omuro, A.M.; Nayak, L.; Wen, P.Y.; Cloughesy, T.F.; Marimuthu, A.; Haidar, S.; Perry, A.; et al. Orally administered colony stimulating factor 1 receptor inhibitor PLX3397 in recurrent glioblastoma: An Ivy Foundation Early Phase Clinical Trials Consortium phase II study. Neuro-Oncol. 2016, 18, 557–564. [Google Scholar] [CrossRef]
- Tap, W.D.; Wainberg, Z.A.; Anthony, S.P.; Ibrahim, P.N.; Zhang, C.; Healey, J.H.; Chmielowski, B.; Staddon, A.P.; Cohn, A.L.; Shapiro, G.I.; et al. Structure-Guided Blockade of CSF1R Kinase in Tenosynovial Giant-Cell Tumor. N. Engl. J. Med. 2015, 373, 428–437. [Google Scholar] [CrossRef]
- Kumar, A.; Mandiyan, V.; Suzuki, Y.; Zhang, C.; Rice, J.; Tsai, J.; Artis, D.R.; Ibrahim, P.; Bremer, R. Crystal structures of proto-oncogene kinase Pim1: A target of aberrant somatic hypermutations in diffuse large cell lymphoma. J. Mol. Biol. 2005, 348, 183–193. [Google Scholar] [CrossRef]
- Tsai, J.; Lee, J.T.; Wang, W.; Zhang, J.; Cho, H.; Mamo, S.; Bremer, R.; Gillette, S.; Kong, J.; Haass, N.K.; et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc. Natl. Acad. Sci. USA 2008, 105, 3041–3046. [Google Scholar] [CrossRef]
- Zhang, C.; Ibrahim, P.N.; Zhang, J.Z.; Burton, E.A.; Habets, G.; Zhang, Y.; Powell, B.; West, B.L.; Matusow, B.; Tsang, G.; et al. Design and pharmacology of a highly specific dual FMS and KIT kinase inhibitor. Proc. Natl. Acad. Sci. USA 2013, 110, 5689–5694. [Google Scholar] [CrossRef]
- Slamon, D.J.; Clark, G.M.; Wong, S.G.; Levin, W.J.; Ullrich, A.; Mcguire, W.L. Human-Breast Cancer-Correlation of Relapse and Survival with Amplification of the Her-2 Neu Oncogene. Science 1987, 235, 177–182. [Google Scholar] [CrossRef]
- Vranic, S.; Beslija, S.; Gatalica, Z. Targeting HER2 expression in cancer: New drugs and new indications. Bosn. J. Basic Med. 2021, 21, 1–4. [Google Scholar] [CrossRef]
- Swain, S.M.; Shastry, M.; Hamilton, E. Targeting HER2-positive breast cancer: Advances and future directions. Nat. Rev. Drug Discov. 2022, 1–26. [Google Scholar] [CrossRef]
- Ward, W.H.J.; Cook, P.N.; Slater, A.M.; Davies, D.H.; Holdgate, G.A.; Green, L.R. Epidermal Growth-Factor Receptor Tyrosine Kinase-Investigation of Catalytic Mechanism, Structure-Based Searching and Discovery of a Potent Inhibitor. Biochem. Pharmacol. 1994, 48, 659–666. [Google Scholar] [CrossRef]
- Wakeling, A.E.; Barker, A.J.; Davies, D.H.; Brown, D.S.; Green, L.R.; Cartlidge, S.A.; Woodburn, J.R. Specific inhibition of epidermal growth factor receptor tyrosine kinase by 4-anilinoquinazolines. Breast Cancer Res. Tr. 1996, 38, 67–73. [Google Scholar] [CrossRef]
- Barker, A.J.; Gibson, K.H.; Grundy, W.; Godfrey, A.A.; Barlow, J.J.; Healy, M.P.; Woodburn, J.R.; Ashton, S.E.; Curry, B.J.; Scarlett, L.; et al. Studies leading to the identification of ZD1839 (IRESSA): An orally active, selective epidermal growth factor receptor tyrosine kinase inhibitor targeted to the treatment of cancer. Bioorg. Med. Chem. Lett. 2001, 11, 1911–1914. [Google Scholar] [CrossRef]
- Muhsin, M.; Graham, J.; Kirkpatrick, P. Gefitinib. Nat. Rev. Drug Discov. 2003, 2, 515–516. [Google Scholar] [CrossRef]
- Tamura, K.; Fukuoka, M. Gefitinib in non-small cell lung cancer. Expert Opin. Pharm. 2005, 6, 985–993. [Google Scholar] [CrossRef]
- Ayala-Aguilera, C.C.; Valero, T.; Lorente-Macias, A.; Baillache, D.J.; Croke, S.; Unciti-Broceta, A. Small Molecule Kinase Inhibitor Drugs (1995-2021): Medical Indication, Pharmacology, and Synthesis. J. Med. Chem. 2022, 65, 1047–1131. [Google Scholar] [CrossRef]
- Kulukian, A.; Lee, P.; Taylor, J.; Rosler, R.; de Vries, P.; Watson, D.; Forero-Torres, A.; Peterson, S. Preclinical Activity of HER2-Selective Tyrosine Kinase Inhibitor Tucatinib as a Single Agent or in Combination with Trastuzumab or Docetaxel in Solid Tumor Models. Mol. Cancer Ther. 2020, 19, 976–987. [Google Scholar] [CrossRef]
- Li, Y.J.; Li, J.X.; Zhou, H.X.; Liu, C.Y.L.; Liu, Z.; Ying, B.W.; Xie, Y.M.; Hu, M.X.; Gong, Y.L. Discovery of potent and selective HER2 PROTAC degrader based Tucatinib with improved efficacy against HER2 positive cancers. Eur. J. Med. Chem. 2022, 244, 114775. [Google Scholar]
- Wang, M.N.; Hu, Y.Z.; Yu, T.; Ma, X.L.; Wei, X.W.; Wei, Y.Q. Pan-HER-targeted approach for cancer therapy: Mechanisms, recent advances and clinical prospect. Cancer Lett. 2018, 439, 113–130. [Google Scholar] [CrossRef]
- Greig, S.L. Osimertinib: First Global Approval. Drugs 2016, 76, 263–273. [Google Scholar] [CrossRef]
- Leonetti, A.; Sharma, S.; Minari, R.; Perego, P.; Giovannetti, E.; Tiseo, M. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Brit. J. Cancer 2019, 121, 725–737. [Google Scholar] [CrossRef]
- Song, Z.D.; Ge, Y.; Wang, C.Y.; Huang, S.S.; Shu, X.H.; Liu, K.X.; Zhou, Y.W.; Ma, X.D. Challenges and Perspectives on the Development of Small-Molecule EGFR Inhibitors against T790M-Mediated Resistance in Non-Small-Cell Lung Cancer. J. Med. Chem. 2016, 59, 6580–6594. [Google Scholar] [CrossRef]
- Lamb, Y.N. Correction to: Osimertinib: A Review in Previously Untreated, EGFR Mutation-Positive, Advanced NSCLC. Target. Oncol. 2021, 16, 869. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.C.H.; Camidge, D.R.; Yang, C.T.; Zhou, J.Y.; Guo, R.H.; Chiu, C.H.; Chang, G.C.; Shiah, H.S.; Chen, Y.; Wang, C.C.; et al. Safety, Efficacy, and Pharmacokinetics of Almonertinib (HS-10296) in Pretreated Patients With EGFR-Mutated Advanced NSCLC: A Multicenter, Open-label, Phase 1 Trial. J. Thorac. Oncol. 2020, 15, 1907–1918. [Google Scholar] [CrossRef] [PubMed]
- Nagasaka, M.; Zhu, V.W.; Lim, S.M.; Greco, M.; Wu, F.Y.; Ou, S.H.I. Beyond Osimertinib: The Development of Third-Generation EGFR Tyrosine Kinase Inhibitors For Advanced EGFR plus NSCLC. J. Thorac. Oncol. 2021, 16, 740–763. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Luo, Y.L.; Li, Z.R.; Chen, C.; Fang, L. Structural modifications on indole and pyrimidine rings of osimertinib lead to high selectivity towards L858R/T790M double mutant enzyme and potent antitumor activity. Bioorganic Med. Chem. 2021, 36, 116094. [Google Scholar] [CrossRef] [PubMed]
- McInnes, I.B.; Szekanecz, Z.; McGonagle, D.; Maksymowych, W.P.; Pfeil, A.; Lippe, R.; Song, I.H.; Lertratanakul, A.; Sornasse, T.; Biljan, A.; et al. A review of JAK-STAT signalling in the pathogenesis of spondyloarthritis and the role of JAK inhibition. Rheumatology 2022, 61, 1783–1794. [Google Scholar] [CrossRef]
- Shawky, A.M.; Almalki, F.A.; Abdalla, A.N.; Abdelazeem, A.H.; Gouda, A.M. A Comprehensive Overview of Globally Approved JAK Inhibitors. Pharmaceutics 2022, 14, 1001. [Google Scholar] [CrossRef]
- Darnell, J.E., Jr.; Kerr, I.M.; Stark, G.R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 1994, 264, 1415–1421. [Google Scholar] [CrossRef]
- Stark, G.R.; Darnell, J.E., Jr. The JAK-STAT pathway at twenty. Immunity 2012, 36, 503–514. [Google Scholar] [CrossRef]
- Babon, J.J.; Lucet, I.S.; Murphy, J.M.; Nicola, N.A.; Varghese, L.N. The molecular regulation of Janus kinase (JAK) activation. Biochem. J. 2014, 462, 1–13. [Google Scholar] [CrossRef]
- Jones, A.V.; Kreil, S.; Zoi, K.; Waghorn, K.; Curtis, C.; Zhang, L.; Score, J.; Seear, R.; Chase, A.J.; Grand, F.H.; et al. Widespread occurrence of the JAK2 V617F mutation in chronic myeloproliferative disorders. Blood 2005, 106, 2162–2168. [Google Scholar] [CrossRef]
- Quintás-Cardama, A.; Verstovsek, S. Molecular pathways: Jak/STAT pathway: Mutations, inhibitors, and resistance. Clin. Cancer Res. 2013, 19, 1933–1940. [Google Scholar] [CrossRef]
- Philips, R.L.; Wang, Y.; Cheon, H.; Kanno, Y.; Gadina, M.; Sartorelli, V.; Horvath, C.M.; Darnell, J.E., Jr.; Stark, G.R.; O’Shea, J.J. The JAK-STAT pathway at 30: Much learned, much more to do. Cell 2022, 185, 3857–3876. [Google Scholar] [CrossRef]
- Deisseroth, A.; Kaminskas, E.; Grillo, J.; Chen, W.; Saber, H.; Lu, H.L.; Rothmann, M.D.; Brar, S.; Wang, J.; Garnett, C.; et al. Food and Drug Administration approval: Ruxolitinib for the treatment of patients with intermediate and high-risk myelofibrosis. Clin. Cancer Res. 2012, 18, 3212–3217. [Google Scholar] [CrossRef]
- Becker, H.; Engelhardt, M.; von Bubnoff, N.; Wäsch, R. Ruxolitinib. Recent Results Cancer Res. 2014, 201, 249–257. [Google Scholar]
- Markham, A. Baricitinib: First Global Approval. Drugs 2017, 77, 697–704. [Google Scholar] [CrossRef]
- King, B.; Ko, J.; Forman, S.; Ohyama, M.; Mesinkovska, N.; Yu, G.; McCollam, J.; Gamalo, M.; Janes, J.; Edson-Heredia, E.; et al. Efficacy and safety of the oral Janus kinase inhibitor baricitinib in the treatment of adults with alopecia areata: Phase 2 results from a randomized controlled study. J. Am. Acad. Dermatol. 2021, 85, 847–853. [Google Scholar] [CrossRef]
- Flanagan, M.E.; Blumenkopf, T.A.; Brissette, W.H.; Brown, M.F.; Casavant, J.M.; Shang-Poa, C.; Doty, J.L.; Elliott, E.A.; Fisher, M.B.; Hines, M.; et al. Discovery of CP-690,550: A potent and selective Janus kinase (JAK) inhibitor for the treatment of autoimmune diseases and organ transplant rejection. J. Med. Chem. 2010, 53, 8468–8484. [Google Scholar] [CrossRef]
- van Vollenhoven, R.F. Rheumatoid arthritis in 2012: Progress in RA genetics, pathology and therapy. Nat. Rev. Rheumatol. 2013, 9, 70–72. [Google Scholar] [CrossRef]
- López-Sanromán, A.; Esplugues, J.V.; Domènech, E. Pharmacology and safety of tofacitinib in ulcerative colitis. Gastroenterol. Hepatol. 2021, 44, 39–48. [Google Scholar] [CrossRef]
- Noji, S.; Hara, Y.; Miura, T.; Yamanaka, H.; Maeda, K.; Hori, A.; Yamamoto, H.; Obika, S.; Inoue, M.; Hase, Y.; et al. Discovery of a Janus Kinase Inhibitor Bearing a Highly Three-Dimensional Spiro Scaffold: JTE-052 (Delgocitinib) as a New Dermatological Agent to Treat Inflammatory Skin Disorders. J. Med. Chem. 2020, 63, 7163–7185. [Google Scholar] [CrossRef]
- Farmer, L.J.; Ledeboer, M.W.; Hoock, T.; Arnost, M.J.; Bethiel, R.S.; Bennani, Y.L.; Black, J.J.; Brummel, C.L.; Chakilam, A.; Dorsch, W.A.; et al. Discovery of VX-509 (Decernotinib): A Potent and Selective Janus Kinase 3 Inhibitor for the Treatment of Autoimmune Diseases. J. Med. Chem. 2015, 58, 7195–7216. [Google Scholar] [CrossRef] [PubMed]
- Ho Lee, Y.; Gyu Song, G. Comparative efficacy and safety of tofacitinib, baricitinib, upadacitinib, filgotinib and peficitinib as monotherapy for active rheumatoid arthritis. J. Clin. Pharm. Ther. 2020, 45, 674–681. [Google Scholar] [CrossRef] [PubMed]
- Menet, C.J.; Fletcher, S.R.; Van Lommen, G.; Geney, R.; Blanc, J.; Smits, K.; Jouannigot, N.; Deprez, P.; van der Aar, E.M.; Clement-Lacroix, P.; et al. Triazolopyridines as selective JAK1 inhibitors: From hit identification to GLPG0634. J. Med. Chem. 2014, 57, 9323–9342. [Google Scholar] [CrossRef] [PubMed]
- Parmentier, J.M.; Voss, J.; Graff, C.; Schwartz, A.; Argiriadi, M.; Friedman, M.; Camp, H.S.; Padley, R.J.; George, J.S.; Hyland, D.; et al. In vitro and in vivo characterization of the JAK1 selectivity of upadacitinib (ABT-494). BMC Rheumatol. 2018, 2, 23. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.A. Filgotinib, a JAK1 Inhibitor, for Treatment-Resistant Rheumatoid Arthritis. JAMA 2019, 322, 309–311. [Google Scholar] [CrossRef]
- Tanaka, Y.; Kavanaugh, A.; Wicklund, J.; McInnes, I.B. Filgotinib, a novel JAK1-preferential inhibitor for the treatment of rheumatoid arthritis: An overview from clinical trials. Mod. Rheumatol. 2022, 32, 1–11. [Google Scholar] [CrossRef]
- Degirmenci, U.; Wang, M.; Hu, J. Targeting Aberrant RAS/RAF/MEK/ERK Signaling for Cancer Therapy. Cells 2020, 9, 198. [Google Scholar] [CrossRef]
- Ullah, R.; Yin, Q.; Snell, A.H.; Wan, L. RAF-MEK-ERK pathway in cancer evolution and treatment. Semin. Cancer Biol. 2022, 85, 123–154. [Google Scholar] [CrossRef]
- Hatzivassiliou, G.; Song, K.; Yen, I.; Brandhuber, B.J.; Anderson, D.J.; Alvarado, R.; Ludlam, M.J.; Stokoe, D.; Gloor, S.L.; Vigers, G.; et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature 2010, 464, 431–435. [Google Scholar] [CrossRef]
- Karoulia, Z.; Gavathiotis, E.; Poulikakos, P.I. New perspectives for targeting RAF kinase in human cancer. Nat. Rev. Cancer 2017, 17, 676–691. [Google Scholar] [CrossRef]
- Ritterhouse, L.L.; Barletta, J.A. BRAF V600E mutation-specific antibody: A review. Semin. Diagn Pathol. 2015, 32, 400–408. [Google Scholar] [CrossRef]
- Gunderwala, A.; Cope, N.; Wang, Z. Mechanism and inhibition of BRAF kinase. Curr. Opin. Chem. Biol. 2022, 71, 102205. [Google Scholar] [CrossRef]
- Poulikakos, P.I.; Sullivan, R.J.; Yaeger, R. Molecular Pathways and Mechanisms of BRAF in Cancer Therapy. Clin. Cancer Res. 2022, 28, 4618–4628. [Google Scholar] [CrossRef]
- Man, R.J.; Zhang, Y.L.; Jiang, A.Q.; Zhu, H.L. A patent review of RAF kinase inhibitors (2010–2018). Expert Opin. Ther. Pat. 2019, 29, 675–688. [Google Scholar] [CrossRef]
- Garbe, C.; Eigentler, T.K. Vemurafenib. Recent Results Cancer Res. 2018, 211, 77–89. [Google Scholar]
- Agianian, B.; Gavathiotis, E. Current Insights of BRAF Inhibitors in Cancer. J. Med. Chem. 2018, 61, 5775–5793. [Google Scholar] [CrossRef]
- Bollag, G.; Hirth, P.; Tsai, J.; Zhang, J.Z.; Ibrahim, P.N.; Cho, H.N.; Spevak, W.; Zhang, C.; Zhang, Y.; Habets, G.; et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature 2010, 467, 596–599. [Google Scholar] [CrossRef]
- Livingstone, E.; Zimmer, L.; Piel, S.; Schadendorf, D. PLX4032: Does it keep its promise for metastatic melanoma treatment? Expert Opin. Investig. Drugs 2010, 19, 1439–1449. [Google Scholar] [CrossRef]
- Karoulia, Z.; Wu, Y.; Ahmed, T.A.; Xin, Q.; Bollard, J.; Krepler, C.; Wu, X.; Zhang, C.; Bollag, G.; Herlyn, M.; et al. An Integrated Model of RAF Inhibitor Action Predicts Inhibitor Activity against Oncogenic BRAF Signaling. Cancer Cell 2016, 30, 485–498. [Google Scholar] [CrossRef]
- Poulikakos, P.I.; Persaud, Y.; Janakiraman, M.; Kong, X.; Ng, C.; Moriceau, G.; Shi, H.; Atefi, M.; Titz, B.; Gabay, M.T.; et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature 2011, 480, 387–390. [Google Scholar] [CrossRef]
- Shi, H.; Moriceau, G.; Kong, X.; Lee, M.K.; Lee, H.; Koya, R.C.; Ng, C.; Chodon, T.; Scolyer, R.A.; Dahlman, K.B.; et al. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat. Commun. 2012, 3, 724. [Google Scholar] [CrossRef] [PubMed]
- Long, G.V.; Stroyakovskiy, D.; Gogas, H.; Levchenko, E.; de Braud, F.; Larkin, J.; Garbe, C.; Jouary, T.; Hauschild, A.; Grob, J.J.; et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N. Engl. J. Med. 2014, 371, 1877–1888. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Yang, X.; Liu, J.; Zhao, B.; Cai, W.; Li, Y.; Hu, D. Efficacy and safety of BRAF inhibition alone versus combined BRAF and MEK inhibition in melanoma: A meta-analysis of randomized controlled trials. Oncotarget 2017, 8, 32258–32269. [Google Scholar] [CrossRef] [PubMed]
- Fruman, D.A.; Chiu, H.; Hopkins, B.D.; Bagrodia, S.; Cantley, L.C.; Abraham, R.T. The PI3K Pathway in Human Disease. Cell 2017, 170, 605–635. [Google Scholar] [CrossRef] [PubMed]
- Rommel, C.; Camps, M.; Ji, H. PI3K delta and PI3K gamma: Partners in crime in inflammation in rheumatoid arthritis and beyond? Nat. Rev. Immunol. 2007, 7, 191–201. [Google Scholar] [CrossRef]
- Ma, X.D.; Wei, J.; Wang, C.; Gu, D.Y.; Hu, Y.Z.; Sheng, R. Design, synthesis and biological evaluation of novel benzothiadiazine derivatives as potent PI3K delta-selective inhibitors for treating B-cell-mediated malignancies. Eur. J. Med. Chem. 2019, 170, 112–125. [Google Scholar] [CrossRef]
- Meng, D.D.; He, W.; Zhang, Y.; Liang, Z.G.; Zheng, J.L.; Zhang, X.; Zheng, X.; Zhan, P.; Chen, H.F.; Li, W.J.; et al. Development of PI3K inhibitors: Advances in clinical trials and new strategies (Review). Pharmacol. Res. 2021, 173, 105900. [Google Scholar] [CrossRef]
- He, Y.; Sun, M.M.; Zhang, G.G.; Yang, J.; Chen, K.S.; Xu, W.W.; Li, B. Targeting PI3K/Akt signal transduction for cancer therapy. Signal Transduct. Tar. 2021, 6, 425. [Google Scholar] [CrossRef]
- Markham, A. Idelalisib: First Global Approval. Drugs 2014, 74, 1701–1707, Erratum in Drugs 2014, 74, 1839. [Google Scholar] [CrossRef]
- Dhillon, S.; Keam, S.J. Umbralisib: First Approval. Drugs 2021, 81, 857–866. [Google Scholar] [CrossRef]
- Blair, H.A. Duvelisib: First Global Approval. Drugs 2018, 78, 1847–1853. [Google Scholar] [CrossRef]
- Knight, Z.A. Small Molecule Inhibitors of the PI3-Kinase Family. Curr. Top. Microbiol. 2011, 347, 263–278. [Google Scholar]
- Knight, Z.A.; Gonzalez, B.; Feldman, M.E.; Zunder, E.R.; Goldenberg, D.D.; Williams, O.; Loewith, R.; Stokoe, D.; Balla, A.; Toth, B.; et al. A pharmacological map of the PI3-K family defines a role for p110alpha in insulin signaling. Cell 2006, 125, 733–747. [Google Scholar] [CrossRef]
- Garces, A.E.; Stocks, M.J. Class 1 PI3K Clinical Candidates and Recent Inhibitor Design Strategies: A Medicinal Chemistry Perspective. J. Med. Chem. 2019, 62, 4815–4850. [Google Scholar] [CrossRef]
- Yang, C.; Xu, C.; Li, Z.; Chen, Y.; Wu, T.; Hong, H.; Lu, M.; Jia, Y.; Yang, Y.; Liu, X.; et al. Bioisosteric replacements of the indole moiety for the development of a potent and selective PI3Kδ inhibitor: Design, synthesis and biological evaluation. Eur. J. Med. Chem. 2021, 223, 113661. [Google Scholar] [CrossRef]
- Saha, D.; Kharbanda, A.; Yan, W.; Lakkaniga, N.R.; Frett, B.; Li, H.Y. The Exploration of Chirality for Improved Druggability within the Human Kinome. J. Med. Chem. 2020, 63, 441–469. [Google Scholar] [CrossRef]
- Lannutti, B.J.; Meadows, S.A.; Herman, S.E.; Kashishian, A.; Steiner, B.; Johnson, A.J.; Byrd, J.C.; Tyner, J.W.; Loriaux, M.M.; Deininger, M.; et al. CAL-101, a p110delta selective phosphatidylinositol-3-kinase inhibitor for the treatment of B-cell malignancies, inhibits PI3K signaling and cellular viability. Blood 2011, 117, 591–594. [Google Scholar] [CrossRef]
- Lampson, B.L.; Brown, J.R. PI3Kdelta-selective and PI3Kalpha/delta-combinatorial inhibitors in clinical development for B-cell non-Hodgkin lymphoma. Expert Opin. Investig. Drugs 2017, 26, 1267–1279. [Google Scholar] [CrossRef]
- Winkler, D.G.; Faia, K.L.; DiNitto, J.P.; Ali, J.A.; White, K.F.; Brophy, E.E.; Pink, M.M.; Proctor, J.L.; Lussier, J.; Martin, C.M.; et al. PI3K-delta and PI3K-gamma inhibition by IPI-145 abrogates immune responses and suppresses activity in autoimmune and inflammatory disease models. Chem. Biol. 2013, 20, 1364–1374. [Google Scholar] [CrossRef]
- Methot, J.L.; Zhou, H.; McGowan, M.A.; Anthony, N.J.; Christopher, M.; Garcia, Y.; Achab, A.; Lipford, K.; Trotter, B.W.; Altman, M.D.; et al. Projected Dose Optimization of Amino- and Hydroxypyrrolidine Purine PI3Kdelta Immunomodulators. J. Med. Chem. 2021, 64, 5137–5156. [Google Scholar] [CrossRef]
- Gockeritz, E.; Kerwien, S.; Baumann, M.; Wigger, M.; Vondey, V.; Neumann, L.; Landwehr, T.; Wendtner, C.M.; Klein, C.; Liu, N.S.; et al. Efficacy of phosphatidylinositol-3 kinase inhibitors with diverse isoform selectivity profiles for inhibiting the survival of chronic lymphocytic leukemia cells. Int. J. Cancer 2015, 137, 2234–2242. [Google Scholar] [CrossRef]
- Rodrigues, D.A.; Sagrillo, F.S.; Fraga, C.A.M. Duvelisib: A 2018 Novel FDA-Approved Small Molecule Inhibiting Phosphoinositide 3-Kinases. Pharmaceuticals 2019, 12, 69. [Google Scholar] [CrossRef] [PubMed]
- Burris, H.A.; Flinn, I.W.; Patel, M.R.; Fenske, T.S.; Deng, C.C.; Brander, D.M.; Gutierrez, M.; Essell, J.H.; Kuhn, J.G.; Miskin, H.P.; et al. Umbralisib, a novel PI3K delta and casein kinase-1 epsilon inhibitor, in relapsed or refractory chronic lymphocytic leukaemia and lymphoma: An open-label, phase 1, dose-escalation, first-in-human study. Lancet Oncol. 2018, 19, 486–496. [Google Scholar] [CrossRef] [PubMed]
- Elmenier, F.M.; Lasheen, D.S.; Abouzid, K.A.M. Phosphatidylinositol 3 kinase (PI3K) inhibitors as new weapon to combat cancer. Eur. J. Med. Chem. 2019, 183, 111718. [Google Scholar] [CrossRef] [PubMed]
- Khotskaya, Y.B.; Holla, V.R.; Farago, A.F.; Mills Shaw, K.R.; Meric-Bernstam, F.; Hong, D.S. Targeting TRK family proteins in cancer. Pharmacol. Ther. 2017, 173, 58–66. [Google Scholar] [CrossRef]
- Huang, E.J.; Reichardt, L.F. Trk receptors: Roles in neuronal signal transduction. Annu. Rev. Biochem. 2003, 72, 609–642. [Google Scholar] [CrossRef]
- Cocco, E.; Scaltriti, M.; Drilon, A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat. Rev. Clin. Oncol. 2018, 15, 731–747. [Google Scholar] [CrossRef]
- Drilon, A. TRK inhibitors in TRK fusion-positive cancers. Ann. Oncol. 2019, 30, viii23–viii30. [Google Scholar] [CrossRef]
- Scott, L.J. Larotrectinib: First Global Approval. Drugs 2019, 79, 201–206. [Google Scholar] [CrossRef]
- Al-Salama, Z.T.; Keam, S.J. Entrectinib: First Global Approval. Drugs 2019, 79, 1477–1483. [Google Scholar] [CrossRef]
- Harada, G.; Santini, F.C.; Wilhelm, C.; Drilon, A. NTRK fusions in lung cancer: From biology to therapy. Lung Cancer-J. Iaslc. 2021, 161, 108–113. [Google Scholar] [CrossRef]
- Choi, H.S.; Rucker, P.V.; Wang, Z.; Fan, Y.; Albaugh, P.; Chopiuk, G.; Gessier, F.; Sun, F.; Adrian, F.; Liu, G.; et al. (R)-2-Phenylpyrrolidine Substituted Imidazopyridazines: A New Class of Potent and Selective Pan-TRK Inhibitors. ACS Med. Chem. Lett. 2015, 6, 562–567. [Google Scholar] [CrossRef]
- Doebele, R.C.; Davis, L.E.; Vaishnavi, A.; Le, A.T.; Estrada-Bernal, A.; Keysar, S.; Jimeno, A.; Varella-Garcia, M.; Aisner, D.L.; Li, Y.; et al. An Oncogenic NTRK Fusion in a Patient with Soft-Tissue Sarcoma with Response to the Tropomyosin-Related Kinase Inhibitor LOXO-101. Cancer Discov. 2015, 5, 1049–1057. [Google Scholar] [CrossRef]
- Liu, Z.; Yu, P.; Dong, L.; Wang, W.; Duan, S.; Wang, B.; Gong, X.; Ye, L.; Wang, H.; Tian, J. Discovery of the Next-Generation Pan-TRK Kinase Inhibitors for the Treatment of Cancer. J. Med. Chem. 2021, 64, 10286–10296. [Google Scholar] [CrossRef]
- Rolfo, C.; Ruiz, R.; Giovannetti, E.; Gil-Bazo, I.; Russo, A.; Passiglia, F.; Giallombardo, M.; Peeters, M.; Raez, L. Entrectinib: A potent new TRK, ROS1, and ALK inhibitor. Expert Opin. Inv. Drug 2015, 24, 1493–1500. [Google Scholar] [CrossRef]
- Ardini, E.; Menichincheri, M.; Banfi, P.; Bosotti, R.; De Ponti, C.; Pulci, R.; Ballinari, D.; Ciomei, M.; Texido, G.; Degrassi, A.; et al. Entrectinib, a Pan-TRK, ROS1, and ALK Inhibitor with Activity in Multiple Molecularly Defined Cancer Indications. Mol. Cancer Ther. 2016, 15, 628–639. [Google Scholar] [CrossRef]
- Menichincheri, M.; Ardini, E.; Magnaghi, P.; Avanzi, N.; Banfi, P.; Bossi, R.; Buffa, L.; Canevari, G.; Ceriani, L.; Colombo, M.; et al. Discovery of Entrectinib: A New 3-Aminoindazole As a Potent Anaplastic Lymphoma Kinase (ALK), c-ros Oncogene 1 Kinase (ROS1), and Pan-Tropomyosin Receptor Kinases (Pan-TRKs) inhibitor. J. Med. Chem. 2016, 59, 3392–3408. [Google Scholar] [CrossRef]
- Roskoski, R. Anaplastic lymphoma kinase (ALK): Structure, oncogenic activation, and pharmacological inhibition. Pharmacol. Res. 2013, 68, 68–94. [Google Scholar] [CrossRef]
- Folkman, J. Role of angiogenesis in tumor growth and metastasis. Semin. Oncol. 2002, 29, 15–18. [Google Scholar] [CrossRef]
- Hoeben, A.; Landuyt, B.; Highley, M.S.; Wildiers, H.; Van Oosterom, A.T.; De Bruijn, E.A. Vascular endothelial growth factor and angiogenesis. Pharmacol. Rev. 2004, 56, 549–580. [Google Scholar] [CrossRef]
- Schenone, S.; Bondavalli, F.; Botta, M. Antiangiogenic agents: An update on small molecule VEGFR inhibitors. Curr. Med. Chem. 2007, 14, 2495–2516. [Google Scholar] [CrossRef] [PubMed]
- Modi, S.J.; Kulkarni, V.M. Vascular Endothelial Growth Factor Receptor (VEGFR-2)/KDR Inhibitors: Medicinal Chemistry Perspective. Med. Drug Discov. 2019, 2, 100009. [Google Scholar] [CrossRef]
- Shibuya, M. Vascular Endothelial Growth Factor (VEGF) and Its Receptor (VEGFR) Signaling in Angiogenesis: A Crucial Target for Anti- and Pro-Angiogenic Therapies. Genes Cancer 2011, 2, 1097–1105. [Google Scholar] [CrossRef] [PubMed]
- Miettinen, M.; Rikala, M.-S.; Rys, J.; Lasota, J.; Wang, Z.-F. Vascular Endothelial Growth Factor Receptor 2 as a Marker for Malignant Vascular Tumors and Mesothelioma: An Immunohistochemical Study of 262 Vascular Endothelial and 1640 Nonvascular Tumors. Am. J. Surg. Pathol. 2012, 36, 629. [Google Scholar] [CrossRef] [PubMed]
- Cortes, F.; Debacker, C.; Peault, B.; Labastie, M.C. Differential expression of KDR/VEGFR-2 and CD34 during mesoderm development of the early human embryo. Mech. Develop. 1999, 83, 161–164. [Google Scholar] [CrossRef]
- Cheng, K.; Liu, C.F.; Rao, G.W. Anti-angiogenic Agents: A Review on Vascular Endothelial Growth Factor Receptor-2 (VEGFR-2) Inhibitors. Curr. Med. Chem. 2021, 28, 2540–2564. [Google Scholar] [CrossRef]
- Zhang, C.L.; Tan, C.Y.; Ding, H.W.; Xin, T.; Jiang, Y.Y. Selective VEGFR Inhibitors for Anticancer Therapeutics in Clinical Use and Clinical Trials. Curr. Pharm. Design. 2012, 18, 2921–2935. [Google Scholar] [CrossRef]
- Li, H.T.; Zhu, X. Quinoline-based Compounds with Potential Activity against Drugresistant Cancers. Curr. Top. Med. Chem. 2021, 21, 426–437. [Google Scholar] [CrossRef]
- Syed, Y.Y. Anlotinib: First Global Approval. Drugs 2018, 78, 1057–1062. [Google Scholar] [CrossRef]
- Zhou, M.; Chen, X.; Zhang, H.; Xia, L.; Tong, X.; Zou, L.; Hao, R.; Pan, J.; Zhao, X.; Chen, D.; et al. China National Medical Products Administration approval summary: Anlotinib for the treatment of advanced non-small cell lung cancer after two lines of chemotherapy. Cancer Commun. 2019, 39, 36. [Google Scholar] [CrossRef]
- Lin, B.Y.; Song, X.M.; Yang, D.W.; Bai, D.S.; Yao, Y.Y.; Lu, N. Anlotinib inhibits angiogenesis via suppressing the activation of VEGFR2, PDGFR beta and FGFR1 (vol 654, pg 77, 2018). Gene 2020, 723, 77–86. [Google Scholar]
- Lu, C.; Zhang, Q.Y.; Zhang, H.Y.; Li, X.M.; Jiang, Q.; Yao, J. A small molecular multi-targeting tyrosine kinase inhibitor, anlotinib, inhibits pathological ocular neovascularization. Biomed Pharm. 2021, 138, 111493. [Google Scholar] [CrossRef]
- Xie, C.Y.; Wan, X.Z.; Quan, H.T.; Zheng, M.Y.; Fu, L.; Li, Y.; Lou, L.G. Preclinical characterization of anlotinib, a highly potent and selective vascular endothelial growth factor receptor-2 inhibitor. Cancer Sci. 2018, 109, 1207–1219. [Google Scholar] [CrossRef]
- Parkin, D.M.; Bray, F.; Ferlay, J.; Pisani, P. Global cancer statistics, 2002. Ca-Cancer J. Clin. 2005, 55, 74–108. [Google Scholar] [CrossRef]
- Ferlay, J.; Shin, H.R.; Bray, F.; Forman, D.; Mathers, C.; Parkin, D.M. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer. 2010, 127, 2893–2917. [Google Scholar] [CrossRef]
- Kania, R.S. Structure-Based Design and Characterization of Axitinib. In Kinase Inhibitor Drugs; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2009; pp. 167–201. [Google Scholar]
- Hu-Lowe, D.D.; Zou, H.Y.; Grazzini, M.L.; Hallin, M.E.; Wickman, G.R.; Amundson, K.; Chen, J.H.; Rewolinski, D.A.; Yamazaki, S.; Wu, E.Y.; et al. Nonclinical antiangiogenesis and antitumor activities of axitinib (AG-013736), an oral, potent, and selective inhibitor of vascular endothelial growth factor receptor tyrosine kinases 1, 2, 3. Clin. Cancer Res. 2008, 14, 7272–7283. [Google Scholar] [CrossRef]
- Fenton, B.M.; Paoni, S.F. The addition of AG-013736 to fractionated radiation improves tumor response without functionally normalizing the tumor vasculature. Cancer Res. 2007, 67, 9921–9928. [Google Scholar] [CrossRef]
- Larkin, J.; Fishman, M.; Wood, L.; Negrier, S.; Olivier, K.; Pyle, L.; Gorbunovn, V.; Jonasch, E.; Andrews, L.; Staehler, M. Axitinib for the Treatment of Metastatic Renal Cell Carcinoma Recommendations for Therapy Management to Optimize Outcomes. Am. J. Clin. Oncol.-Canc. 2014, 37, 397–403. [Google Scholar] [CrossRef]
- Matiadis, D.; Sagnou, M. Pyrazoline Hybrids as Promising Anticancer Agents: An Up-to-Date Overview. Int. J. Mol. Sci. 2020, 21, 5507. [Google Scholar] [CrossRef]
- Gajiwala, K.S.; Wu, J.C.; Christensen, J.; Deshmukh, G.D.; Diehl, W.; DiNitto, J.P.; English, J.M.; Greig, M.J.; He, Y.A.; Jacques, S.L.; et al. KIT kinase mutants show unique mechanisms of drug resistance to imatinib and sunitinib in gastrointestinal stromal tumor patients. Proc. Natl. Acad. Sci. USA 2009, 106, 1542–1547. [Google Scholar] [CrossRef]
- Roskoski, R. Classification of small molecule protein kinase inhibitors based upon the structures of their drug-enzyme complexes. Pharmacol. Res. 2016, 103, 26–48. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Liang, C.; Shirazian, S.; Zhou, Y.; Miller, T.; Cui, J.; Fukuda, J.Y.; Chu, J.Y.; Nematalla, A.; Wang, X.Y.; et al. Discovery of 5-[5-Fluoro-2-oxo-1,2-dihydroindol-(3Z)-ylidenemethyl]-2,4-dimethyl-1H-pyrrole-3-carboxylic acid (2-diethylaminoethyl)amide, a novel tyrosine kinase inhibitor targeting vascular endothelial and platelet-derived growth factor receptor tyrosine kinase. J. Med. Chem. 2003, 46, 1116–1119. [Google Scholar] [PubMed]
- Saha, D.; Ryan, K.R.; Lakkaniga, N.R.; Acharya, B.; Garcia, N.G.; Smith, E.L.; Frett, B. Targeting Rearranged during Transfection in Cancer: A Perspective on Small-Molecule Inhibitors and Their Clinical Development. J. Med. Chem. 2021, 64, 11747–11773. [Google Scholar] [CrossRef] [PubMed]
- Roth, G.J.; Heckel, A.; Colbatzky, F.; Handschuh, S.; Kley, J.; Lehmann-Lintz, T.; Lotz, R.; Tontsch-Grunt, U.; Walter, R.; Hilberg, F. Design, Synthesis, and Evaluation of Indolinones as Triple Angiokinase Inhibitors and the Discovery of a Highly Specific 6-Methoxycarbonyl-Substituted Indolinone (BIBF 1120). J. Med. Chem. 2009, 52, 4466–4480. [Google Scholar] [CrossRef] [PubMed]
- Hilberg, F.; Roth, G.J.; Krssak, M.; Kautschitsch, S.; Sommergruber, W.; Tontsch-Grunt, U.; Garin-Chesa, P.; Bader, G.; Zoephel, A.; Quant, J.; et al. BIBF 1120: Triple angiokinase inhibitor with sustained receptor blockade and good antitumor efficacy. Cancer Res. 2008, 68, 4774–4782. [Google Scholar] [CrossRef]
- Roth, G.J.; Binder, R.; Colbatzky, F.; Dallinger, C.; Schlenker-Herceg, R.; Hilberg, F.; Wollin, S.-L.; Kaiser, R. Nintedanib: From Discovery to the Clinic. J. Med. Chem. 2015, 58, 1053–1063. [Google Scholar] [CrossRef]
- Miyamoto, S.; Kakutani, S.; Sato, Y.; Hanashi, A.; Kinoshita, Y.; Ishikawa, A. Drug review: Pazopanib. Jpn. J. Clin. Oncol. 2018, 48, 503–513. [Google Scholar] [CrossRef]
- Harris, P.A.; Boloor, A.; Cheung, M.; Kumar, R.; Crosby, R.M.; Davis-Ward, R.G.; Epperly, A.H.; Hinkle, K.W.; Hunter, R.N.; Johnson, J.H.; et al. Discovery of 5-[[4-[(2,3-dimethyl-2H-indazol-6-yl)methylamino]-2-pyrimidinyl]amino]-2-methylbenzenesulfonamide (pazopanib), a novel and potent vascular endothelial growth factor receptor inhibitor. J. Med. Chem. 2008, 51, 4632–4640. [Google Scholar] [CrossRef]
- Markham, A. Selpercatinib: First Approval. Drugs 2020, 80, 1119–1124. [Google Scholar] [CrossRef]
- Subbiah, V.; Yang, D.; Velcheti, V.; Drilon, A.; Meric-Bernstam, F. State-of-the-Art Strategies for Targeting RET-Dependent Cancers. J. Clin. Oncol. 2020, 38, 1209–1221. [Google Scholar] [CrossRef]
- Evans, E.K.; Hodous, B.L.; Gardino, A.K.; Davis, A.; Zhu, J.; Shutes, A.; Kim, J.L.; Wilson, K.J.; Wilson, D.; Zhang, Y.; et al. Abstract 791: BLU-285, the first selective inhibitor of PDGFRα D842V and KIT Exon 17 mutants. Cancer Res. 2015, 75, 791. [Google Scholar] [CrossRef]
- Dhillon, S. Avapritinib: First Approval. Drugs 2020, 80, 433–439. [Google Scholar] [CrossRef]
- Winger, B.A.; Cortopassi, W.A.; Ruiz, D.G.; Ding, L.; Jang, K.; Leyte-Vidal, A.; Zhang, N.; Esteve-Puig, R.; Jacobson, M.P.; Shah, N.P. ATP-Competitive Inhibitors Midostaurin and Avapritinib Have Distinct Resistance Profiles in Exon 17-Mutant KIT. Cancer Res. 2019, 79, 4283–4292. [Google Scholar] [CrossRef]
- Liu, X.D.; Wang, Q.; Yang, G.J.; Marando, C.; Koblish, H.K.; Hall, L.M.; Fridman, J.S.; Behshad, E.; Wynn, R.; Li, Y.; et al. A Novel Kinase Inhibitor, INCB28060, Blocks c-MET-Dependent Signaling, Neoplastic Activities, and Cross-Talk with EGFR and HER-3. Clin. Cancer Res. 2011, 17, 7127–7138. [Google Scholar] [CrossRef]
- Dhillon, S. Capmatinib: First Approval. Drugs 2020, 80, 1125–1131. [Google Scholar] [CrossRef]
- Alzofon, N.; Jimeno, A. Capmatinib for non-small cell lung cancer. Drugs Today 2021, 57, 17–25. [Google Scholar] [CrossRef]
- Mathieu, L.N.; Larkins, E.; Akinboro, O.; Roy, P.; Amatya, A.K.; Fiero, M.H.; Mishra-Kalyani, P.S.; Helms, W.S.; Myers, C.E.; Skinner, A.M.; et al. FDA Approval Summary: Capmatinib and Tepotinib for the Treatment of Metastatic NSCLC Harboring MET Exon 14 Skipping Mutations or Alterations. Clin. Cancer. Res. 2022, 28, 249–254. [Google Scholar] [CrossRef]
- Wu, Y.L.; Smit, E.F.; Bauer, T.M. Capmatinib for patients with non-small cell lung cancer with MET exon 14 skipping mutations: A review of preclinical and clinical studies. Cancer Treat. Rev. 2021, 95, 102173. [Google Scholar] [CrossRef]
- Gainor, J.F.; Chabner, B.A. Ponatinib: Accelerated Disapproval. Oncologist 2015, 20, 847–848. [Google Scholar] [CrossRef]
- Cameron, F.; Sanford, M. Ibrutinib: First global approval. Drugs 2014, 74, 263–271. [Google Scholar] [CrossRef]
- Markham, A.; Dhillon, S. Acalabrutinib: First Global Approval. Drugs 2018, 78, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, S. Tirabrutinib: First Approval. Drugs 2020, 80, 835–840. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.S. Abemaciclib: First Global Approval. Drugs 2017, 77, 2063–2070. [Google Scholar] [CrossRef] [PubMed]
- Syed, Y.Y. Ribociclib: First Global Approval. Drugs 2017, 77, 799–807. [Google Scholar] [CrossRef] [PubMed]
- Lamb, Y.N. Pexidartinib: First Approval. Drugs 2019, 79, 1805–1812. [Google Scholar] [CrossRef]
- Lee, A. Tucatinib: First Approval. Drugs 2020, 80, 1033–1038. [Google Scholar] [CrossRef]
- Jiang, T.; Luo, Y.; Wang, B. Almonertinib-induced interstitial lung disease: A case report. Medicine (Baltimore) 2021, 100, e24393. [Google Scholar] [CrossRef]
- Ajayi, S.; Becker, H.; Reinhardt, H.; Engelhardt, M.; Zeiser, R.; von Bubnoff, N.; Wasch, R. Ruxolitinib. Recent Results Cancer Res. 2018, 212, 119–132. [Google Scholar]
- Padda, I.S.; Bhatt, R.; Parmar, M. Tofacitinib; StatPearls: Treasure Island, FL, USA, 2022. [Google Scholar]
- Dhillon, S. Delgocitinib: First Approval. Drugs 2020, 80, 609–615. [Google Scholar] [CrossRef]
- Markham, A.; Keam, S.J. Peficitinib: First Global Approval. Drugs 2019, 79, 887–891. [Google Scholar] [CrossRef]
- Dhillon, S.; Keam, S.J. Filgotinib: First Approval. Drugs 2020, 80, 1987–1997. [Google Scholar] [CrossRef]
- Khaddour, K.; Kurn, H.; Zito, P.M. Vemurafenib; StatPearls: Treasure Island, FL, USA, 2022. [Google Scholar]
- Akaza, H.; Fukuyama, T. Axitinib for the treatment of advanced renal cell carcinoma. Expert Opin. Pharm. 2014, 15, 283–297. [Google Scholar] [CrossRef]
- Tzogani, K.; Skibeli, V.; Westgaard, I.; Dalhus, M.; Thoresen, H.; Slot, K.B.; Damkier, P.; Hofland, K.; Borregaard, J.; Ersboll, J.; et al. The European Medicines Agency approval of axitinib (Inlyta) for the treatment of advanced renal cell carcinoma after failure of prior treatment with sunitinib or a cytokine: Summary of the scientific assessment of the committee for medicinal products for human use. Oncologist 2015, 20, 196–201. [Google Scholar]
- Rini, B.I. Sunitinib. Expert Opin. Pharmacother. 2007, 8, 2359–2369. [Google Scholar] [CrossRef]
- McCormack, P.L. Nintedanib: First global approval. Drugs 2015, 75, 129–139. [Google Scholar] [CrossRef]
- Nguyen, D.T.; Shayahi, S. Pazopanib: Approval for soft-tissue sarcoma. J. Adv. Pr. Oncol. 2013, 4, 53–57. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).