Comprehensive Identification of Ginsenosides in the Roots and Rhizomes of Panax ginseng Based on Their Molecular Features-Oriented Precursor Ions Selection and Targeted MS/MS Analysis
Abstract
1. Introduction
2. Results
2.1. Data Mining
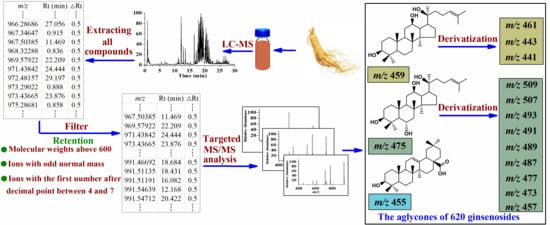
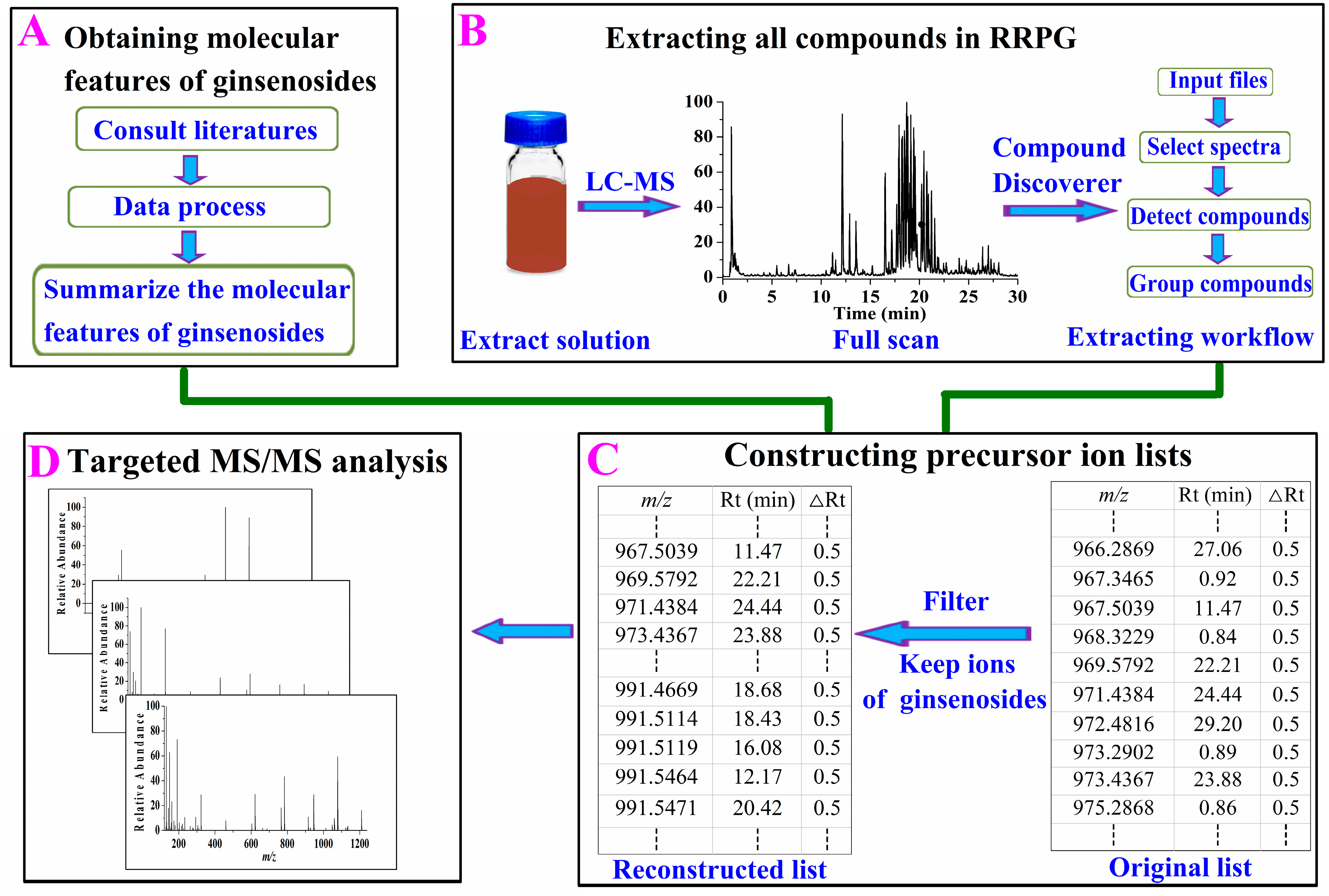
2.1.1. Obtaining Molecular Features of Ginsenosides
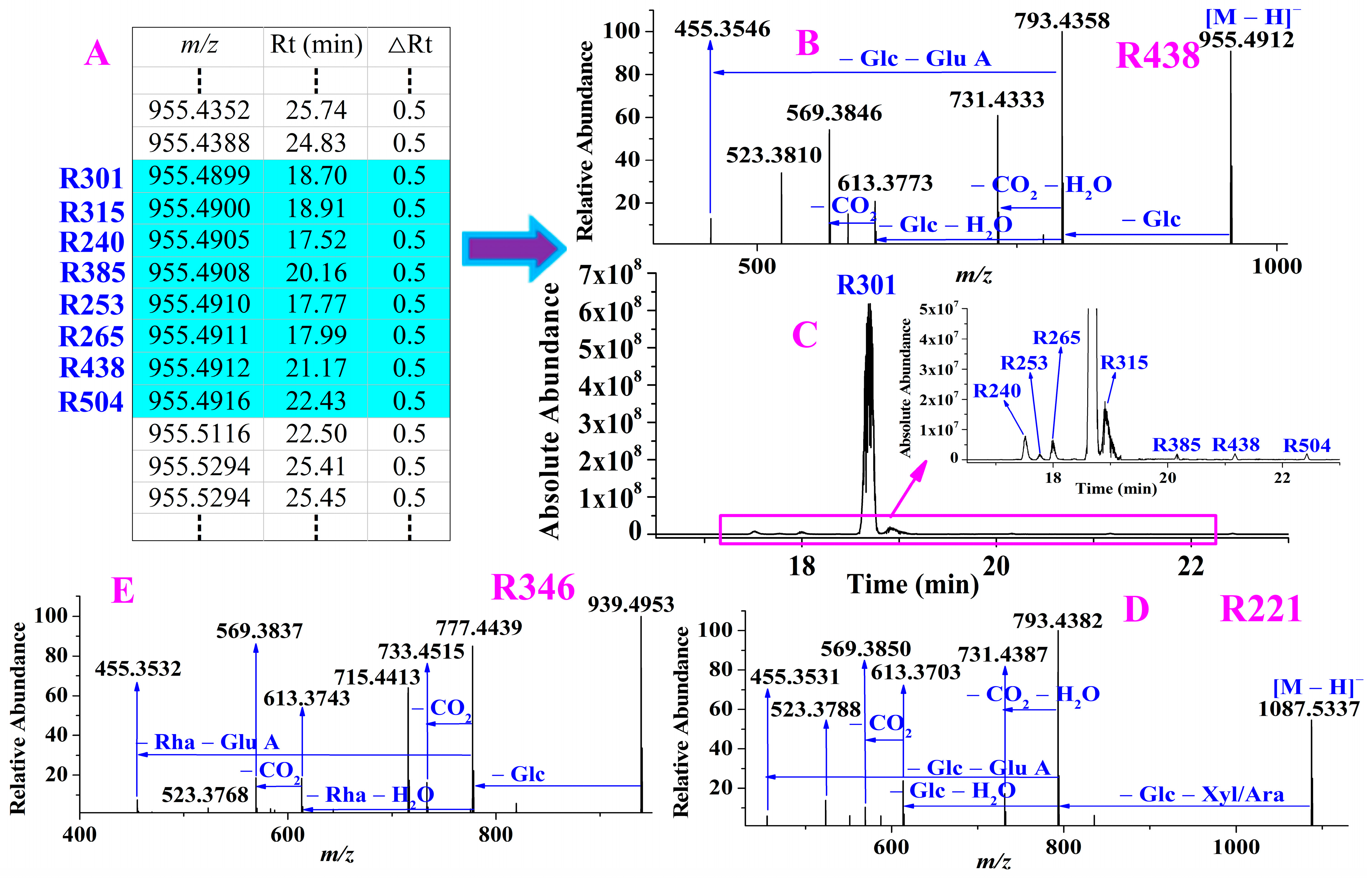
2.1.2. Extracting All Compounds in RRPG
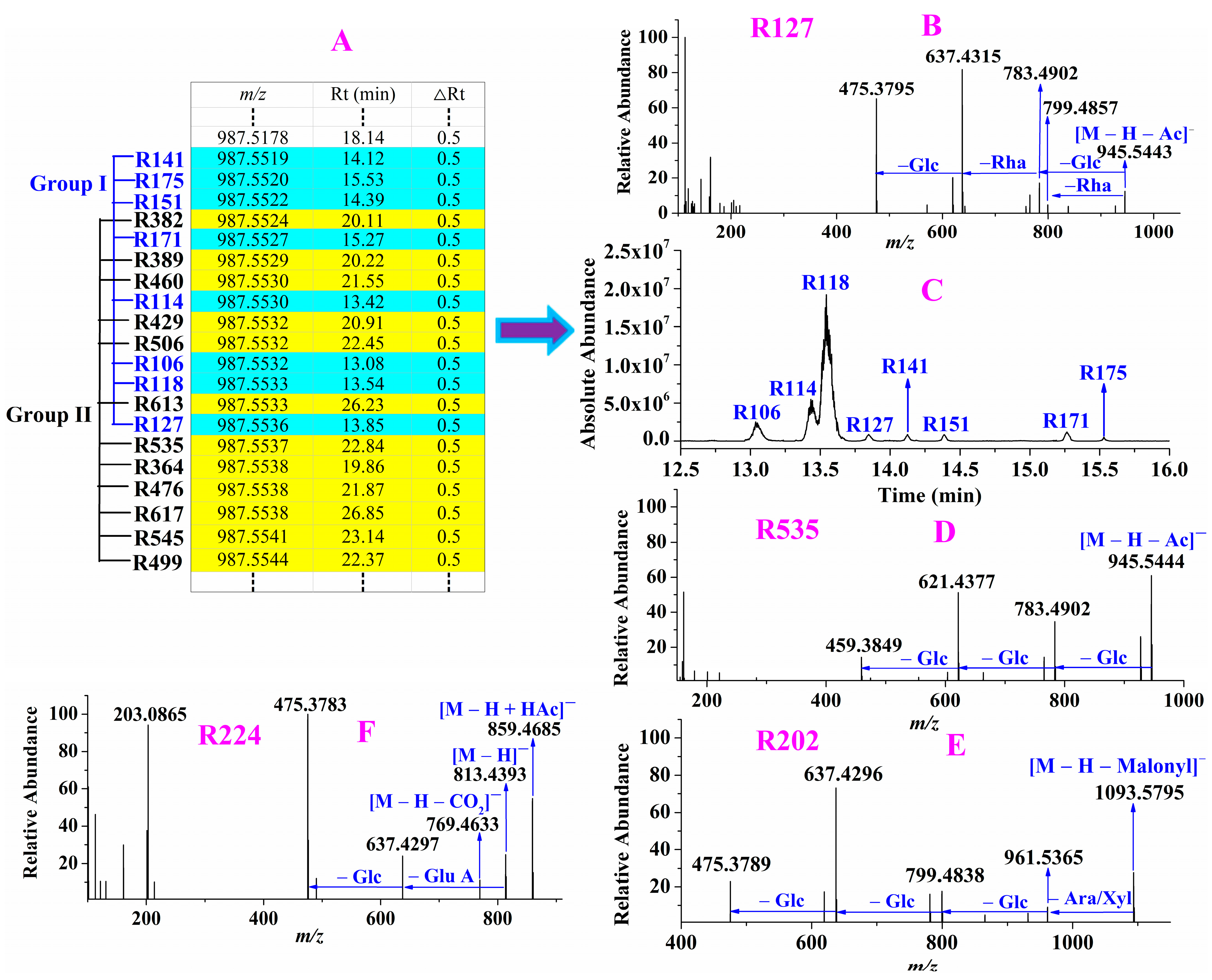
2.1.3. Constructing Precursor Ion List of Ginsenosides
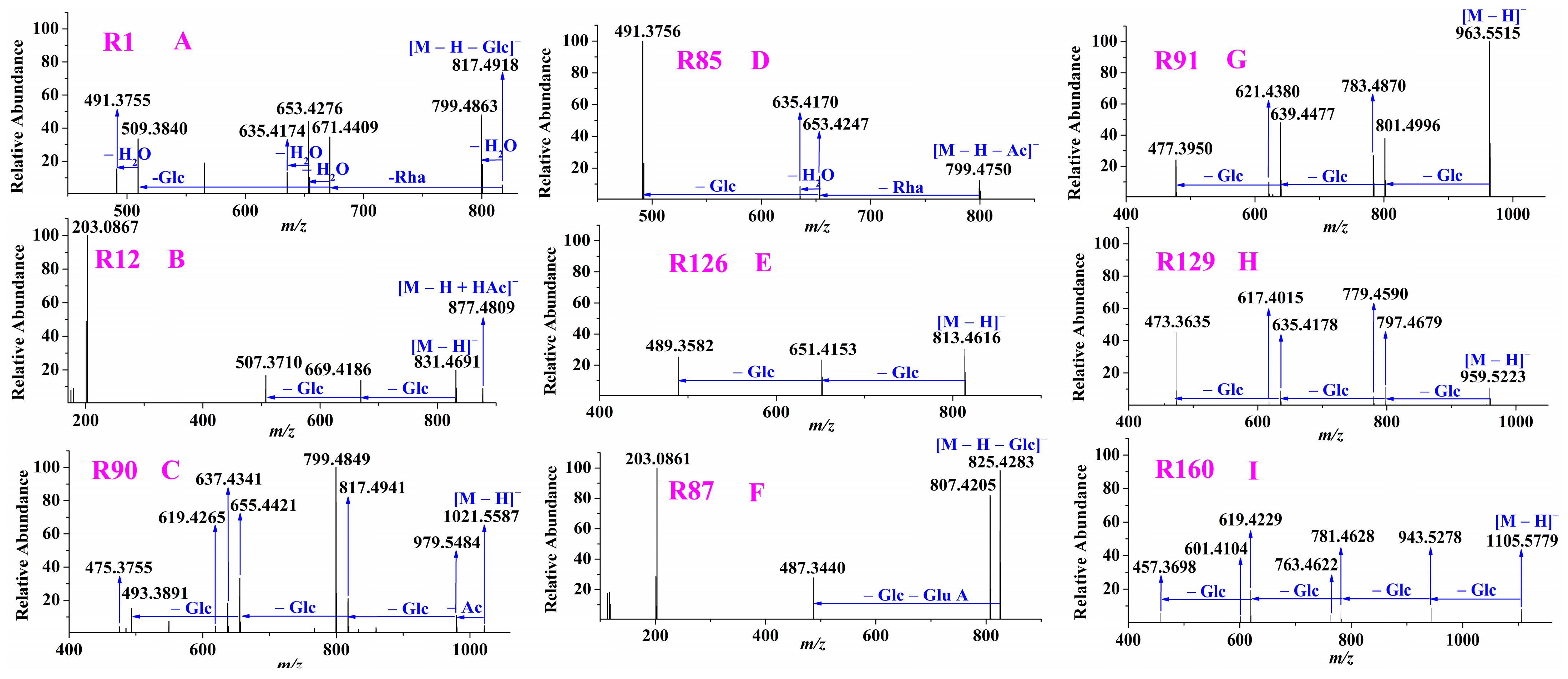
2.1.4. Targeted MS/MS Analysis and Structure Identification
2.2. Ginsenosides Identification
2.2.1. Characterization of Protopanaxadiol-Type Ginsenosides
Ginsenosides with Aglycone m/z 459
Ginsenosides with Aglycone m/z 461
Ginsenosides with Aglycone m/z 443
Ginsenosides with Aglycone m/z 441
2.2.2. Characterization of Protopanaxatriol-Type Ginsenosides
Ginsenosides with Aglycone m/z 475
Ginsenosides with Aglycone m/z 509
Ginsenosides with Aglycone m/z 507
Ginsenosides with Aglycone m/z 493
Ginsenosides with Aglycone m/z 491
Ginsenosides with Aglycone m/z 489
Ginsenosides with Aglycone m/z 487
Ginsenosides with Aglycone m/z 477
Ginsenosides with Aglycone m/z 473
Ginsenosides with Aglycone m/z 457
2.2.3. Characterization of Oleanane-Type of Ginsenosides
3. Materials and Methods
3.1. RRPG Sample, Reference Standards and Reagents
3.2. Sample and Standard Solution Preparations
3.3. Chromatographic and Mass Spectrometric Conditions
3.4. Data Acquisition and Data Processing
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Wang, Q.; Wu, M.Q.; Zhao, L.H.; Yang, H.K.; Lv, X.H. Effect of ginsenoside Rh2 on transplanted-tumor and expression of JAM in mice. China J. Chin. Mater. Med. 2008, 33, 2116–2119. [Google Scholar]
- Wang, T.F.; Meng, Z.M. Experiment for immunity effects of ginsenoside Rg3. J. China Pharmaceut. Univ. 1999, 30, 133–135. [Google Scholar]
- Pan, S.Y.; Liu, D.Y.; Yu, L.; Wang, S.P. The effect of ginsenosides on the axon growth of spinal ganglionic neurons induced by NGF. Chin. J. Neurosci. 2000, 16, 345–348. [Google Scholar]
- Chen, L.M.; Zhou, X.M.; Cao, Y.L.; Hu, W.X. Neuroprotection of ginsenoside Re in cerebral ischemia-reperfusion injury in rats. J. Asian Nat. Prod. Res. 2008, 10, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zhang, J.T. Effects of ginsenoside Rb1 and Rg1 on synaptosomal free calcium level, ATPase and calmodulin in rat hippocampus. Chin. Med. J. (Engl.) 1995, 108, 544–547. [Google Scholar]
- Wang, H.P.; Yang, X.B.; Yang, X.W.; Liu, J.X.; Wang, Y.P.; Zhang, L.X. Chemical constituents from the roots and rhizomes of panax ginseng cultivated in Jilin province. China J. Chin. Mater. Med. 2013, 38, 6–15. [Google Scholar]
- Li, H.J.; Liu, J.P.; Lu, D.; Ming, L.; Wang, P.; Li, P.Y. Studies on chemical constituents of panax ginseng. Chin. J. Exp. Tradit. Med. Formu. 2010, 16, 38–40. [Google Scholar]
- Yang, X.B.; Yang, X.W.; Liu, J.X. Study on ginsenosides in the roots and rhizomes of panax ginseng. Mod. Chin. Med. 2013, 15, 349–358. [Google Scholar]
- Zhang, X.X.; Wang, H.P.; Yang, Y.; Du, M.B.; Mao, S.; Chen, C.; Liu, Y.X.; Li, S.J. Rapid analysis of ginsenosides in dried fresh Ginseng by ultra-performance liquid chromatography with quadrupole time-of-flight mass spectrometry. Chin. Med. Heral. 2015, 12, 130–136. [Google Scholar]
- Wang, H.P.; Zhang, Y.B.; Yang, X.W.; Zhao, D.Q.; Wang, Y.P. Rapid characterization of ginsenosides in the roots and rhizomes of Panax ginseng by UPLC-DAD-QTOF-MS/MS and simultaneous determination of 19 ginsenosides by HPLC-ESI-MS. J. Ginseng Res. 2016, 40, 382–394. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.Z.; Liu, Z.; Li, X.G.; Zheng, Y.N.; Wang, J.Y. Isolation and identification of two malonyl-ginsenosides from the fresh root of panax ginseng. Chin. J. Anal. Chem. 2005, 33, 1783–1786. [Google Scholar]
- Wang, H.P.; Yang, X.B.; Yang, X.W.; Liu, J.X.; Xu, W.; Zhang, Y.B.; Zhang, L.X.; Wang, Y.P. Ginsenjilinol, a new protopanaxatrioltype saponin with inhibitory activity on LPS-activated NO production in macrophage RAW 264.7 cells from the roots and rhizomes of Panax ginseng. J. Asian Nat. Prod. Res. 2013, 15, 579–587. [Google Scholar] [CrossRef]
- Shen, D.; Tang, S.; Lu, P.; Yang, H.J. Analysis on composition principles of Chinese patent drugs containing Ginseng. China J. Chin. Mater. Med. 2013, 38, 2027–2032. [Google Scholar]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.-P.; Wang, Z.-J.; Du, J.; Lin, Z.-Z.; Zhao, C.; Zhang, R.; Yin, Q.; Fan, C.-L.; Peng, P.; Wang, Z.-B. Comprehensive Identification of Ginsenosides in the Roots and Rhizomes of Panax ginseng Based on Their Molecular Features-Oriented Precursor Ions Selection and Targeted MS/MS Analysis. Molecules 2023, 28, 941. https://doi.org/10.3390/molecules28030941
Wang H-P, Wang Z-J, Du J, Lin Z-Z, Zhao C, Zhang R, Yin Q, Fan C-L, Peng P, Wang Z-B. Comprehensive Identification of Ginsenosides in the Roots and Rhizomes of Panax ginseng Based on Their Molecular Features-Oriented Precursor Ions Selection and Targeted MS/MS Analysis. Molecules. 2023; 28(3):941. https://doi.org/10.3390/molecules28030941
Chicago/Turabian StyleWang, Hong-Ping, Zi-Jian Wang, Jing Du, Zhao-Zhou Lin, Chen Zhao, Run Zhang, Qiong Yin, Chun-Lan Fan, Ping Peng, and Zhi-Bin Wang. 2023. "Comprehensive Identification of Ginsenosides in the Roots and Rhizomes of Panax ginseng Based on Their Molecular Features-Oriented Precursor Ions Selection and Targeted MS/MS Analysis" Molecules 28, no. 3: 941. https://doi.org/10.3390/molecules28030941
APA StyleWang, H.-P., Wang, Z.-J., Du, J., Lin, Z.-Z., Zhao, C., Zhang, R., Yin, Q., Fan, C.-L., Peng, P., & Wang, Z.-B. (2023). Comprehensive Identification of Ginsenosides in the Roots and Rhizomes of Panax ginseng Based on Their Molecular Features-Oriented Precursor Ions Selection and Targeted MS/MS Analysis. Molecules, 28(3), 941. https://doi.org/10.3390/molecules28030941