Abstract
Ligustrum robustum has been not only used as a heat-clearing and detoxicating functional tea (Ku-Ding-Cha) but also consumed as a hypotensive, anti-diabetic, and weight-reducing folk medicine. From the leaves of L. robustum, ten new monoterpenoid glycosides named ligurobustosides T10 (1a), T11 (1b), T12 (2a), T13 (2b), T14 (3a), T15 (3b), F1 (4b), T16 (5a), T17 (5b), and E1 (6b), together with five known ones (4a, 6a, 7, 8a, 8b), were separated and identified using the spectroscopic method and chemical method in this research. The results of biological tests exhibited that the fatty acid synthase (FAS) inhibitory action of compound 5 (IC50: 4.38 ± 0.11 μM) was as strong as orlistat (IC50: 4.46 ± 0.13 μM), a positive control; the α-glucosidase inhibitory actions of compounds 1–4 and 7–8, and the α-amylase inhibitory actions of compounds 1–8 were medium; the ABTS radical scavenging capacities of compounds 1–3 and 5–8 (IC50: 6.27 ± 0.23 ~ 8.59 ± 0.09 μM) were stronger than l-(+)-ascorbic acid (IC50: 10.06 ± 0.19 μM) served as a positive control. This research offered a theoretical foundation for the leaves of L. robustum to prevent diabetes and its complications.
1. Introduction
Diabetes, which affects about 10.5% of the population in the world, is a chronic metabolic disease with the characteristic of hyperglycemia, and its complications, such as diabetic nephropathy, neuropathy, and cardiovascular diseases, result in high morbidity and mortality [1]. Present anti-diabetic agents, including insulin, metformin, α-glucosidase inhibitors acarbose and N-substituted iminosugar C-glycoside [2], can deal with hyperglycemia, whereas their function to prevent the complications of diabetes is not ideal. Recent research [3] indicated that antioxidant natural ingredients with inhibitory actions on fatty acid synthase (FAS), α-glucosidase, and α-amylase might be a novel resource for preventing diabetes and its complications.
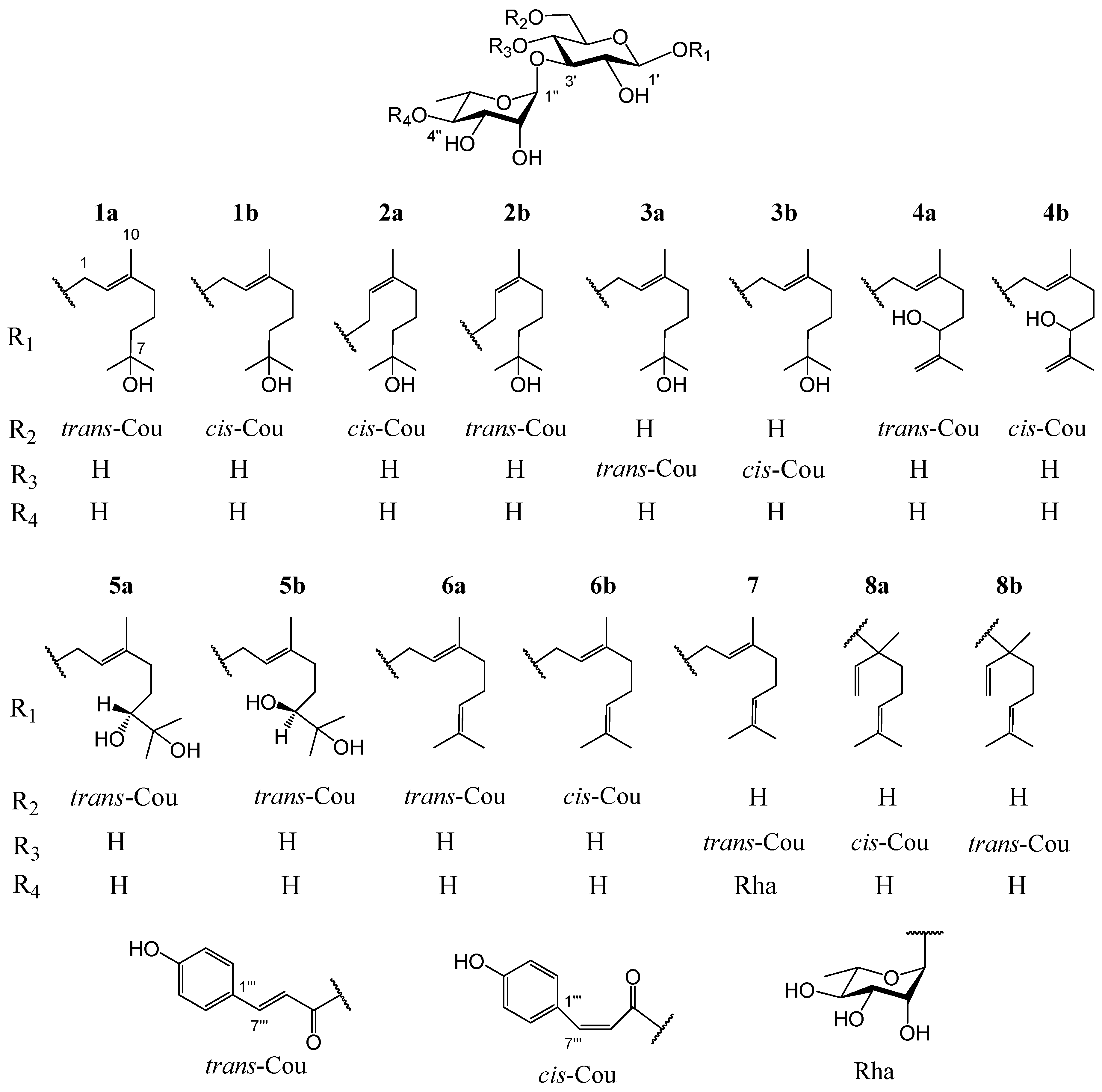
Ligustrum robustum (Roxb.) Blume, a shrub of the Oleaceae family, is widespread in Southwest China, Vietnam, Burma, Cambodia, and India [3]. The leaves of L. robustum, containing 3.96–3.99 mg·g−1 trans-p-hydroxycinnamic acid and 202.6–210.6 mg·g−1 total trans-p-hydroxycinnamic acid esters [4], have been not only used as a heat-clearing and detoxicating functional tea (Ku-Ding-Cha) but also consumed as a hypotensive, anti-diabetic, and weight-reducing folk medicine [3]. In the preceding researches on L. robustum [3,4,5,6,7,8,9,10,11,12,13,14,15], about 90 chemical ingredients (of which 34 ingredients belonged to trans-p-hydroxycinnamic acid esters) involving phenylmethanoid glycosides, phenylethanoid glycosides, monoterpenoid glycosides, iridoid glycosides, butenol glycosides, hexenol glycosides, lignan glycosides, flavonoid glycosides, triterpenoids, sugar esters, and other compounds were separated and elucidated. At the same time, the antioxidant capacities and inhibitory actions of FAS, α-glucosidase, and α-amylase of many constituents were reported. In order to elucidate more trans-p-hydroxycinnamic acid esters and discover more functional ingredients for preventing diabetes and its complications, the study on the chemical compositions from the leaves of L. robustum and their bioactivities, which had been performed preliminary [3,15], was ulteriorly carried out. In this investigation, ten new monoterpenoid glycosides named ligurobustosides T10 (1a), T11 (1b), T12 (2a), T13 (2b), T14 (3a), T15 (3b), F1 (4b), T16 (5a), T17 (5b), and E1 (6b), along with five reported ones (4a, 6a, 7, 8a, 8b) (Figure 1), were separated and elucidated from the leaves of L. robustum. This article discusses the separation and elucidation of compounds 1–8 while reporting their inhibitory actions on FAS, α-glucosidase, α-amylase, and their antioxidant capacities.
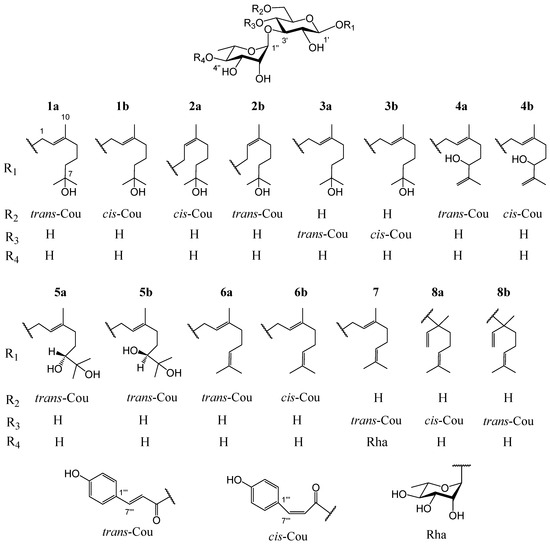
Figure 1.
Structures of compounds 1–8 from the leaves of L. robustum.
2. Results and Discussion
2.1. Identification of Compounds 1–8
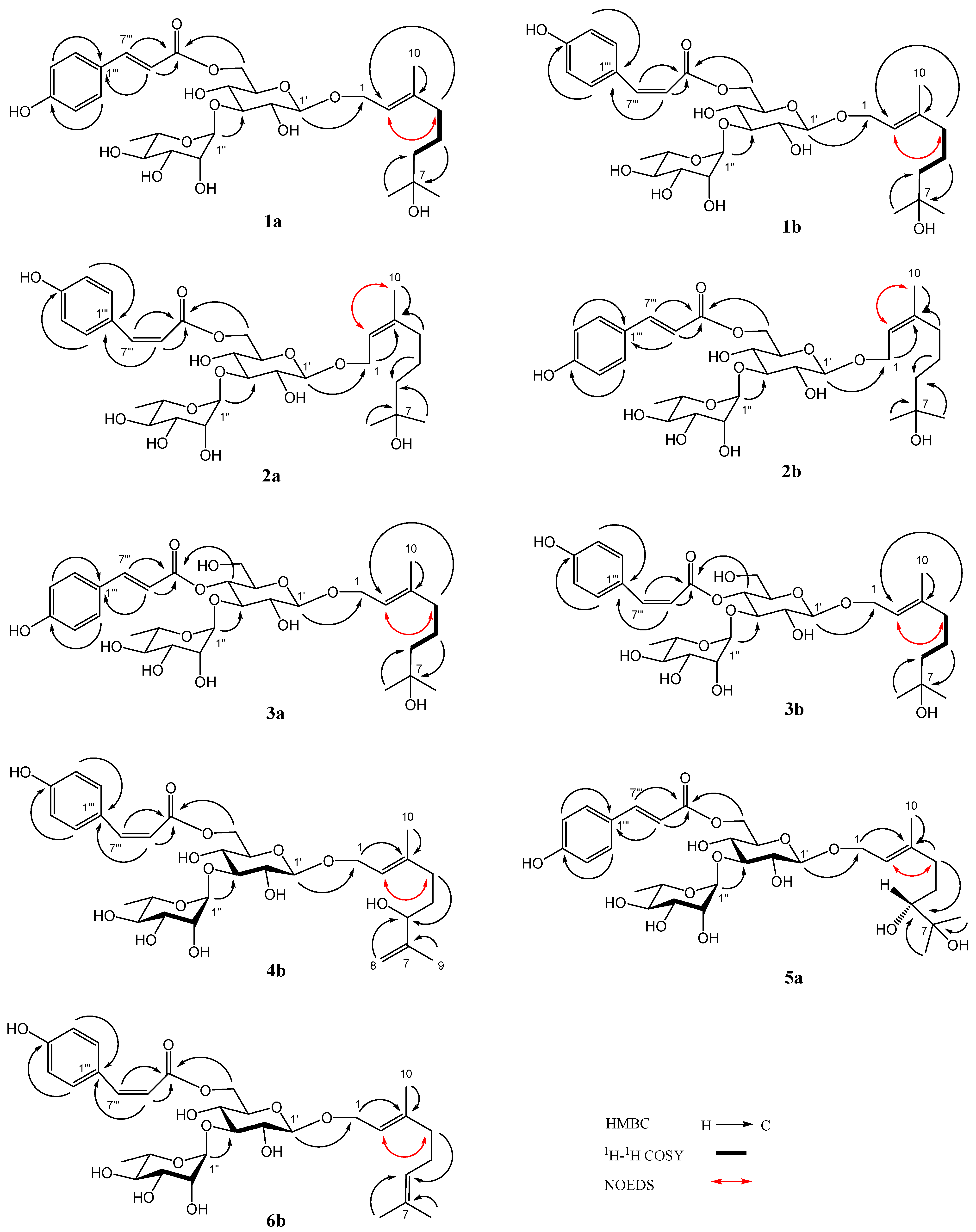
Compound 1 was a yellowish amorphous powder. The HRESIMS of compound 1 showed a base peak at m/z 649.2836 [M + Na]+ (calculated for C31H46NaO13, 649.2836), confirming that its molecular formula was C31H46O13, which contains nine degrees of unsaturation. The IR spectrum of 1 suggested the presence of hydroxy (3366 cm−1), carbonyl (1693 cm−1), double bond (1632 cm−1), and phenyl (1604, 1515, 1447 cm−1). Two stereoisomers, 1a and 1b (3:1), were observed in the NMR spectra of compound 1. The 1H NMR spectrum of 1a (Table 1) indicated the presence of a trans-p-coumaroyl [δH 6.81, 7.45 (2 H each, d, J = 8.4 Hz), 6.35, 7.64 (1 H each, d, J = 16.2 Hz)], two monosaccharide residues [δH 4.32 (1 H, d, J = 7.8 Hz), 5.18 (1 H, d, J = 1.8 Hz), 1.25 (3 H, d, J = 6.6 Hz)], and a monoterpenoid aglycone [δH 5.37 (1 H, t, J = 7.2 Hz), 4.26, 4.27 (1 H each, d, J = 7.2 Hz), 2.02 (2 H, t, J = 7.2 Hz), 1.40, 1.48 (2 H each, m), 1.15, 1.15, 1.66 (3 H each, s)]. The 13C NMR spectrum of 1a (Table 2) also confirmed the presence of a trans-p-coumaroyl (δC 114.9–169.0), two monosaccharide residues (δC 17.9–102.7), and a monoterpenoid aglycone (δC 16.4–142.4). The above 1H and 13C NMR features of 1a were closely related to those of (2E,5E)-1-(1-hydroxy-7-methoxy-3,7-dimethyl-octa-2,5-dienyl)-(3-O-α-l-rhamnopyranosyl)-(6-O-trans-p-coumaroyl)-O-β-d-glucopyranoside (ligurobustoside T8) [15], except that the C-5 double bond and C-7 methoxy in ligurobustoside T8 were replaced by a single bond and a hydroxy in 1a. The 1H-1H COSY of 1a (Figure 2) showed the correlations between H-4, 6 of aglycone (δH 2.02, 1.40), and H-5 of aglycone (δH 1.48), suggesting that there was a single bond at C-5 of 1a. The NOEDS of 1a (Figure 2) exhibited a correlation between H-2 of aglycone (δH 5.37) and H-4 of aglycone (δH 2.02), confirming that the C-2 double bond was E-configuration. Combined with the HMBC spectrum of 1a (Figure 2), the aglycone of 1a was determined to be (E)-3,7-dimethyl-2-octene-1,7-diol. In addition, d-glucose and l-rhamnose, which were identified by TLC, were obtained from the acid hydrolysis experiment of compound 1. The long-distance correlations between H-1′ of Glc (δH 4.32) and C-1 of aglycone (δC 66.2), between H-1″ of Rha (δH 5.18) and C-3′ of Glc (δC 84.0), and between H-6′ of Glc (δH 4.36, 4.51) and carbonyl of Cou (δC 169.0) were displayed in the HMBC spectrum of 1a (Figure 2). 1H-1H COSY, HSQC, HMBC, and NOEDS (Figure S1) were applied to the assignment of the 1H and 13C NMR signals of compound 1. Based on the above evidences, 1a was identified as (E)-7-hydroxy-3,7-dimethyl-2-octen-1-yl 3-O-(α-l-rhamnopyranosyl)-6-O-(trans-p-coumaroyl)-O-β-d-glucopyranoside. It is a novel monoterpenoid glycoside named ligurobustoside T10.

Table 1.
1H NMR data of compounds 1–6 from the leaves of L. robustum in CD3OD a.

Table 2.
13C NMR data of compounds 1–6 from the leaves of L. robustum in CD3OD.
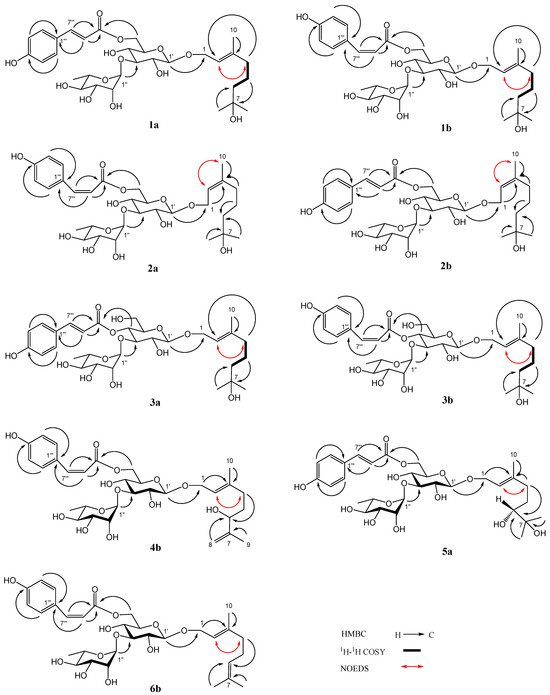
Figure 2.
Key HMBC, 1H-1H COSY, and NOEDS correlations of compounds 1–6.
The NMR characteristics of 1b (Table 1 and Table 2) were similar to those of 1a, except that the cis-p-coumaroyl [δH 6.74, 7.66 (2 H each, d, J = 8.4 Hz), 5.76, 6.87 (1 H each, d, J = 12.6 Hz)] in 1b took the place of the trans-p-coumaroyl in 1a. The long-distance correlations between H-1′ of Glc (δH 4.26) and C-1 of aglycone (δC 66.2), between H-1″ of Rha (δH 5.15) and C-3′ of Glc (δC 84.5), and between H-6′ of Glc (δH 4.29, 4.48) and carbonyl of Cou (δC 168.2) were shown in the HMBC spectrum of 1b (Figure 2). Thus, compound 1b was identified as (E)-7-hydroxy-3,7-dimethyl-2-octen-1-yl 3-O-(α-l-rhamnopyranosyl)-6-O-(cis-p-coumaroyl)-O-β-d-glucopyranoside. It is a new monoterpenoid glycoside named ligurobustoside T11. In conclusion, compound 1 is made up of ligurobustosides T10 and T11.
Compound 2 was obtained as a white amorphous powder. The molecular formula C31H46O13 containing nine degrees of unsaturation was determined by HRESIMS at m/z 649.2836 [M + Na]+ (calculated for C31H46NaO13, 649.2836). The IR spectrum of 2 indicated the existence of hydroxy (3366 cm−1), carbonyl (1693 cm−1), double bond (1632 cm−1), and phenyl (1604, 1515, 1447 cm−1). Two stereoisomers, 2a and 2b (3:1), were displayed in the NMR spectra of compound 2. The 1H and 13C NMR characteristics of 2a (Table 1 and Table 2) were very similar to those of 1b, except for the configuration of the C-2 double bond of aglycone. The correlation between H-2 of aglycone (δH 5.35) and H-10 of aglycone (δH 1.64) was revealed in the NOEDS of 2a (Figure 2), suggesting that the C-2 double bond was Z-configuration. d-glucose and l-rhamnose identified by TLC were obtained from the acid hydrolysis experiment of compound 2. Additionally, the long-distance correlations between H-1′ of Glc (δH 4.28) and C-1 of aglycone (δC 66.2), between H-1″ of Rha (δH 5.15) and C-3′ of Glc (δC 84.1), and between H-6′ of Glc (δH 4.29, 4.49) and carbonyl of Cou (δC 168.1) were displayed in the HMBC spectrum of 2a (Figure 2). HMBC and NOEDS (Figure S2) were used to assign the 1H and 13C NMR signals of compound 2. Therefore, 2a was identified as (Z)-7-hydroxy-3,7-dimethyl-2-octen-1-yl 3-O-(α-l-rhamnopyranosyl)-6-O-(cis-p-coumaroyl)-O-β-d-glucopyranoside. It is a new monoterpenoid glycoside named ligurobustoside T12.
The NMR features of 2b (Table 1 and Table 2) were closely related to those of 2a, except that the trans-p-coumaroyl [δH 6.81, 7.46 (2 H each, d, J = 8.8 Hz), 6.31, 7.64 (1 H each, d, J = 16.0 Hz)] in 2b took the place of cis-p-coumaroyl [δH 6.76, 7.65 (2 H each, d, J = 8.8 Hz), 5.80, 6.88 (1 H each, d, J = 12.8 Hz)] in 2a. From the HMBC spectrum of 2b (Figure 2), the long-distance correlations between H-1′ of Glc (δH 4.32) and C-1 of aglycone (δC 66.2), between H-1″ of Rha (δH 5.17) and C-3′ of Glc (δC 84.0), and between H-6′ of Glc (δH 4.35, 4.50) and carbonyl of Cou (δC 169.3) were observed. Thus, compound 2b was confirmed as (Z)-7-hydroxy-3,7-dimethyl-2-octen-1-yl 3-O-(α-l-rhamnopyranosyl)-6-O-(trans-p-coumaroyl)-O-β-d-glucopyranoside, which is a novel monoterpenoid glycoside named ligurobustoside T13. To sum up, compound 2 is composed of ligurobustosides T12 and T13.
Compound 3, a white amorphous powder, was determined to be C31H46O13 by HRESIMS (m/z 649.2834 [M + Na]+, calculated 649.2836 for C31H46NaO13), which contains nine degrees of unsaturation. The hydroxy (3391 cm−1), carbonyl (1698 cm−1), double bond (1632 cm−1), and phenyl (1605, 1515 cm−1) were observed in the IR spectrum of 3. Two stereoisomers, 3a and 3b (3:1), were exhibited in the NMR spectra of compound 3. The 1H and 13C NMR characteristics of 3a (Table 1 and Table 2) were closely related to those of 1a, except for a trans-p-coumaroyl linking with different positions. The correlation between H-2 of aglycone (δH 5.40) and H-4 of aglycone (δH 2.06) was shown in the NOEDS of 3a (Figure 2), affirming further that the C-2 double bond was E-configuration. d-glucose and l-rhamnose, confirmed by TLC, were offered from the acid hydrolysis experiment of compound 3. Furthermore, the long-distance correlations between H-1′ of Glc (δH 4.38) and C-1 of aglycone (δC 66.5), between H-1″ of Rha (δH 5.19) and C-3′ of Glc (δC 81.6), and between H-4′ of Glc (δH 4.92) and carbonyl of Cou (δC 168.3) were revealed in the HMBC spectrum of 3a (Figure 2). 1H-1H COSY, HSQC, HMBC, and NOEDS (Figure S3) were applied to the assignment of the 1H and 13C NMR signals of compound 3. Consequently, compound 3a was confirmed to be (E)-7-hydroxy-3,7-dimethyl-2-octen-1-yl 3-O-(α-l-rhamnopyranosyl)-4-O-(trans-p-coumaroyl)-O-β-d-glucopyranoside, which is a new monoterpenoid glycoside named ligurobustoside T14.
The NMR features of 3b (Table 1 and Table 2) were very close to those of 3a, the main difference was that the cis-p-coumaroyl [δH 6.76, 7.62 (2 H each, d, J = 8.4 Hz), 5.79, 6.95 (1 H each, d, J = 12.8 Hz)] in 3b took the place of the trans-p-coumaroyl [δH 6.81, 7.47 (2 H each, d, J = 8.4 Hz), 6.34, 7.66 (1 H each, d, J = 16.0 Hz)] in 3a. The long-distance correlations between H-1′ of Glc (δH 4.33) and C-1 of aglycone (δC 66.5), between H-1″ of Rha (δH 5.16) and C-3′ of Glc (δC 81.9), and between H-4′ of Glc (δH 4.86) and carbonyl of Cou (δC 166.9) were exhibited in the HMBC spectrum of 3b (Figure 2). Therefore, the structure of compound 3b was identified as (E)-7-hydroxy-3,7-dimethyl-2-octen-1-yl 3-O-(α-l-rhamnopyranosyl)-4-O-(cis-p-coumaroyl)-O-β-d-glucopyranoside. It is a new monoterpenoid glycoside named ligurobustoside T15. In sum, compound 3 is made up of ligurobustosides T14 and T15.
Compound 4, a white amorphous powder, was analyzed to be C31H44O13 by HRESIMS (m/z 647.2684 [M + Na]+, calculated 647.2680 for C31H44NaO13), which contains ten degrees of unsaturation. The hydroxy (3392 cm−1), carbonyl (1696 cm−1), double bond (1633 cm−1), and phenyl (1605, 1515 cm−1) were observed in the IR spectrum of 4. Two stereoisomers, 4a and 4b (5:3), were shown in the NMR spectra of compound 4. The 1H and 13C NMR data of 4a (Supplementary Material S1) were consistent with those of (2E)-6-hydroxy-3,7-dimethyl-2,7-octadien-1-yl 3-O-(α-l-rhamnopyranosyl)-6-O-(trans-p-coumaroyl)-O-β-d-glucopyranoside (ligurobustoside F) [8]. The 1H and 13C NMR characteristics of 4b (Table 1 and Table 2) were close to those of 4a, the main difference was that the cis-p-coumaroyl [δH 6.74, 7.66 (2 H each, d, J = 8.4 Hz), 5.77, 6.87 (1 H each, d, J = 12.8 Hz)] in 4b took the place of the trans-p-coumaroyl [δH 6.77, 7.44 (2 H each, d, J = 8.8 Hz), 6.32, 7.64 (1 H each, d, J = 16.0 Hz)] in 4a. The correlation between H-2 of aglycone (δH 5.38) and H-4 of aglycone (δH 2.07) was displayed in the NOEDS of 4b (Figure 2), confirming further that the C-2 double bond was E-configuration. d-glucose and l-rhamnose identified by TLC were obtained from the acid hydrolysis experiment of compound 4. Moreover, the long-distance correlations between H-1′ of Glc (δH 4.30) and C-1 of aglycone (δC 66.4), between H-1″ of Rha (δH 5.15) and C-3′ of Glc (δC 84.0), and between H-6′ of Glc (δH 4.30, 4.49) and carbonyl of Cou (δC 168.2) were exhibited in the HMBC spectrum of 4b (Figure 2). 1H-1H COSY, HSQC, HMBC, and NOEDS (Figure S4) were used to assign the 1H and 13C NMR signals of compound 4. So, 4b was elucidated to be (2E)-6-hydroxy-3,7-dimethyl-2,7-octadien-1-yl 3-O-(α-l-rhamnopyranosyl)-6-O-(cis-p-coumaroyl)-O-β-d-glucopyranoside. It is a novel monoterpenoid glycoside named ligurobustoside F1. To sum up, compound 4 is made up of ligurobustosides F and F1.
Compound 5 was acquired as a white amorphous powder. Its molecular formula C31H46O14, which contains nine degrees of unsaturation, was affirmed by HRESIMS at m/z 665.2790 [M + Na]+ (calculated for C31H46NaO14, 665.2785). The presence of hydroxy (3375 cm−1), carbonyl (1693 cm−1), double bond (1632 cm−1), and phenyl (1604, 1515, 1448 cm−1) was confirmed with the IR spectrum of 5. Two stereoisomers, 5a and 5b (6:4), were revealed in the NMR spectra of compound 5. The 1H NMR spectrum of 5a (Table 1) affirmed the existence of a trans-p-coumaroyl [δH 6.81, 7.46 (2 H each, d, J = 8.4 Hz), 6.35, 7.64 (1 H each, d, J = 16.0 Hz)], two monosaccharide residues [δH 4.32 (1 H, d, J = 7.6 Hz), 5.17 (1 H, d, J = 2.0 Hz), 1.24 (3 H, d, J = 6.4 Hz)], and a monoterpenoid aglycone [δH 5.41 (1 H, t, J = 7.2 Hz), 4.26, 4.27 (1 H each, d, J = 7.2 Hz), 3.23 (1 H, dd, J = 10.4, 2.0 Hz), 1.33, 1.76, 2.08, 2.29 (1 H each, m), 1.12, 1.16, 1.68 (3 H each, s)]. The 13C NMR spectrum of 5a (Table 2) also indicated the presence of a trans-p-coumaroyl (δC 114.9–169.1), two monosaccharide residues (δC 17.9–102.7), and a monoterpenoid aglycone (δC 16.5–142.5). The above 1H and 13C NMR characteristics of 5a were close to those of (E)-6,7-dihydroxy-3,7-dimethyl-2-octen-1-yl 3-O-(α-l-rhamnopyranosyl)-4-O-(cis-p-coumaroyl)-O-β-d-glucopyranoside (ligurobustoside L) [9], except that the cis-p-coumaroyl in ligurobustoside L was taken in place of a trans-p-coumaroyl at a different position in 5a. The correlation between H-2 of aglycone (δH 5.41) and H-4 of aglycone (δH 2.08, 2.29) was shown in the NOEDS of 5a (Figure 2), suggesting that the C-2 double bond was E-configuration. d-glucose and l-rhamnose, which were confirmed by TLC, were obtained from the acid hydrolysis experiment of compound 5. Moreover, the long-distance correlations between H-1′ of Glc (δH 4.32) and C-1 of aglycone (δC 66.2), between H-1″ of Rha (δH 5.17) and C-3′ of Glc (δC 84.0), and between H-6′ of Glc (δH 4.36, 4.51) and carbonyl of Cou (δC 169.1) were displayed in the HMBC spectrum of 5a (Figure 2). 1H-1H COSY, HSQC, HMBC, and NOEDS (Figure S5) were applied to the assignment of the 1H and 13C NMR signals of 5. Thus, the plane structure of 5a was identified as (E)-6,7-dihydroxy-3,7-dimethyl-2-octen-1-yl 3-O-(α-l-rhamnopyranosyl)-6-O-(trans-p-coumaroyl)-O-β-d-glucopyranoside.
The NMR features of 5b (Table 1 and Table 2) were very similar to those of 5a, except for the C-6 configuration of aglycone. The enzymatic hydrolysis experiment of 5 gave (E,R)-3,7-dimethyl-oct-2-ene-1,6,7-triol and (E,S)-3,7-dimethyl-oct-2-ene-1,6,7-triol (6:4). Together with 5a:5b = 6:4 observed in the 1H NMR spectrum of compound 5, the C-6 configurations of the aglycones in 5a and 5b were affirmed to be 6R and 6S, respectively. Based on the above evidences, 5a was identified as (2E,6R)-6,7-dihydroxy-3,7-dimethyl-2-octen-1-yl 3-O-(α-l-rhamnopyranosyl)-6-O-(trans-p-coumaroyl)-O-β-d-glucopyranoside, which is a new monoterpenoid glycoside named ligurobustoside T16; 5b was identified as (2E,6S)-6,7-dihydroxy-3,7-dimethyl-2-octen-1-yl 3-O-(α-l-rhamnopyranosyl)-6-O-(trans-p-coumaroyl)-O-β-d-glucopyranoside, which is a new monoterpenoid glycoside named ligurobustoside T17. In conclusion, compound 5 is composed of ligurobustosides T16 and T17.
Compound 6, a yellowish oil, was analyzed as C31H44O12 by HRESIMS (m/z 631.2728 [M + Na]+, calculated 631.2730 for C31H44NaO12), which contains ten degrees of unsaturation. The hydroxy (3391 cm−1), carbonyl (1694 cm−1), double bond (1633 cm−1), and phenyl (1605, 1515, 1444 cm−1) were observed in the IR spectrum of 6. Two stereoisomers, 6a and 6b (5:1), were displayed in the NMR spectra of compound 6. The 1H and 13C NMR data of 6a (Supplementary Material S1) were consistent with those of geraniol 3-O-(α-l-rhamnopyranosyl)-6-O-(trans-p-coumaroyl)-O-β-d-glucopyranoside (ligurobustoside E) [8]. The 1H and 13C NMR features of 6b (Table 1 and Table 2) were close to those of 6a, except that the cis-p-coumaroyl [δH 6.76, 7.64 (2 H each, d, J = 8.4 Hz), 5.79, 6.87 (1 H each, d, J = 13.2 Hz)] in 6b took the place of the trans-p-coumaroyl [δH 6.81, 7.46 (2 H each, d, J = 8.4 Hz), 6.35, 7.64 (1 H each, d, J = 16.2 Hz)] in 6a. The correlation between H-2 of aglycone (δH 5.35) and H-4 of aglycone (δH 2.03) was shown in the NOEDS of 6b (Figure 2), affirming further that the C-2 double bond was E-configuration. d-glucose and l-rhamnose identified by TLC were obtained from the acid hydrolysis experiment of compound 6. In addition, the long-distance correlations between H-1′ of Glc (δH 4.28) and C-1 of aglycone (δC 66.2), between H-1″ of Rha (δH 5.15) and C-3′ of Glc (δC 84.0), and between H-6′ of Glc (δH 4.30, 4.45) and carbonyl of Cou (δC 168.1) were revealed in the HMBC spectrum of 6b (Figure 2). 1H-1H COSY, HSQC, HMBC, and NOEDS (Figure S6) were used to assign the 1H and 13C NMR signals of compound 6. Hence, 6b was elucidated as (2E)-3,7-dimethyl-2,6-octadien-1-yl 3-O-(α-l-rhamnopyranosyl)-6-O-(cis-p-coumaroyl)-O-β-d-glucopyranoside. It is a novel monoterpenoid glycoside named ligurobustoside E1. To sum up, compound 6 is made up of ligurobustosides E and E1.
Compounds 7–8 (1H NMR, 13C NMR data see Supplementary Material S1) were confirmed to be known monoterpenoid glycosides ligurobustoside J (7) [8], lipedoside B-II (8a) [16], and lipedoside B-III (8b) [16], by comparison with the reported NMR information and 2D-NMR (1H-1H COSY, HSQC, and HMBC) experiments. Compound 8a was separated from L. robustum for the first time.
2.2. The Bioactivities of Compounds 1–8
The bioactivities of compounds 1–8 isolated from the leaves of L. robustum, including the inhibitory actions on FAS, α-glucosidase and α-amylase, and the antioxidant capacities, were measured. The results of the biological tests are listed in Table 3. As exhibited in Table 3, the FAS inhibitory action of compound 5 (IC50: 4.38 ± 0.11 μM) was as strong as orlistat (IC50: 4.46 ± 0.13 μM) applied as a positive control, whereas the FAS inhibitory actions of compounds 4, 6, and 8 (IC50: 6.78 ± 0.18~24.68 ± 0.27 μM) were weaker than orlistat; the α-glucosidase inhibitory actions of compounds 1–4 and 7–8 were medium and weaker than acarbose, a positive control; the α-amylase inhibitory actions of compounds 1–8 were medium and weaker than the positive control acarbose. Although the DPPH radical scavenging capacities of compounds 1–8 were not observed, the ABTS radical scavenging capacities of compounds 1–3 and 5–8 (IC50: 6.27 ± 0.23~8.59 ± 0.09 μM) were stronger than l-(+)-ascorbic acid (IC50: 10.06 ± 0.19 μM), which was used as a positive control.

Table 3.
Results of the biological tests of compounds 1–8 from the leaves of L. robustum a.
Because antioxidant compositions with inhibitory actions on FAS, α-glucosidase, and α-amylase might be a novel resource for preventing diabetes and its complications [3], strong antioxidant compounds 1–8, with strong or medium FAS, α-glucosidase, and α-amylase inhibitory actions, might be partial functional ingredients of L. robustum for preventing diabetes and its complications.
3. Materials and Methods
3.1. General Experimental Procedure
HRESIMS was measured using a Waters Q-TOF Premier mass spectrometer (Waters, Milford, MA, USA). The IR spectrum was recorded on a PerkinElmer Spectrum Two FT-IR spectrometer (PerkinElmer, Waltham, MA, USA). 1H NMR, 13C NMR, 1H-1H COSY, HSQC, HMBC, and NOEDS experiments were carried out on an Agilent 600/54 Premium Compact NMR spectrometer (Agilent, Santa Clara, CA, USA) or a Bruker AscendTM 400 NMR spectrometer (Bruker, Mannheim, Germany) with CD3OD as solvent at 25 °C, while tetramethylsilane (TMS) was used as an internal standard. Chemical shifts are expressed in δ (ppm), and coupling constants (J) are reported in Hz. The UV spectrum was performed with a UV2700 spectrophotometer (Shimadzu, Kyoto, Japan). The optical rotation value was examined on an AUTOPOL VI automatic polarimeter (Rudolph, Hackettstown, NJ, USA).
Column chromatography (CC) was carried out on polyamide (60–90 mesh, Jiangsu Changfeng Chemical Industry Co., Yangzhou, China), silica gel (SiO2: 200–300 mesh, Qingdao Ocean Chemical Industry Co., Qingdao, China), and MCI-gel CHP-20P (75–150 μm, Mitsubishi Chemical Co., Tokyo, Japan). Preparative HPLC was performed with a GL3000–300 mL system instrument (Chengdu Gelai Precision Instruments Co., Ltd., Chengdu, China) matching a C-18 column (5 μm, 50 × 450 mm), which was eluted with MeOH-H2O at 30 mL/min and monitored with a UV-3292 detector at 215 nm. TLC was executed on a precoated HPTLC Fertigplatten Kieselgel 60 F254 plate (Merck, Rahway, NJ, USA), which was sprayed with an α-naphthol-sulfuric acid solution or 10% sulfuric acid ethanolic solution and heated at 100–105 °C for 2–3 min. UV–vis absorbance was examined using a UV2700 spectrophotometer (Shimadzu, Kyoto, Japan) or a Spark 10M microplate reader (Tecan Trading Co. Ltd., Shanghai, China). NADPH and Ac-CoA were offered by Zeye Biochemical Co., Ltd. (Shanghai, China). Mal-CoA was obtained by Sigma-Aldrich (St. Louis, MO, USA). ABTS was supplied by Aladdin Industrial Co., Ltd. (Shanghai, China). DPPH was acquired from Macklin Biochemical Co., Ltd. (Shanghai, China).
3.2. Plant Material
The fresh leaves of L. robustum collected from Yibin City, China, were affirmed by Doctor Guo-Min Liu, Hainan University. A voucher sample (No. 201704lsh) of the leaves of L. robustum was preserved at the West China School of Pharmacy, Sichuan University.
3.3. Extraction and Separation
The fresh leaves of L. robustum were turned and heated at 115–120 °C for about 1 h, then powdered. The powdered leaves (7.0 kg) were extracted under reflux in a multi-function extractor with 70% ethanol (28 L × 1) for 2 h [15]. After filtration, the ethanol extract was condensed in vacuo to give a brownish-black paste (2.2 kg). The paste was dissolved with 95% ethanol (3 L), and then distilled water (3 L) was added to deposit the chlorophyll. After percolation, the filtrate was condensed in vacuo to acquire a brown residue (1.0 kg). The residue was chromatographed using a silica gel column, which was eluted with CH2Cl2-MeOH (10:0–0:10) to afford Fr. II (145 g) and three other fractions. Fr. II was chromatographed repeatedly on silica gel column (CH2Cl2-MeOH-H2O, 200:10:1–80:20:2; or EtOAc-MeOH-H2O, 100:4:2–100:6:2), and then separated by CC with polyamide (EtOH-H2O, 0:10–7:3) and MCI (MeOH-H2O, 1:9–7:3), and isolated finally by preparative HPLC (MeOH-H2O, 55:45–64:36) and silica gel column (EtOAc-MeOH-H2O, 100:2:1–100:6:2), to give compounds 1 (187.4 mg), 2 (17.6 mg), 3 (90.5 mg), 4 (4.2 mg), 5 (27.4 mg), 6 (169.1 mg), 7 (16.8 mg) and 8 (80.5 mg).
Compound 1: yellowish amorphous powder. UV (MeOH) λmax: (log ε) 213 (4.1), 227 (4.2), 316 (4.4) nm; IR (film) νmax: 3366, 2926, 1693, 1632, 1604, 1515, 1447, 1268, 1166, 1038, 912, 834 cm–1; 1H NMR (CD3OD, 600 MHz) data, see Table 1; 13C NMR (CD3OD, 150 MHz) data, see Table 2; HRESIMS m/z 649.2836 [M + Na]+ (calculated for C31H46NaO13, 649.2836).
Compound 2: white amorphous powder. UV (MeOH) λmax (log ε): 213 (4.1), 227 (4.2), 316 (4.4) nm; IR (film) νmax: 3366, 2926, 1693, 1632, 1604, 1515, 1447, 1268, 1166, 1038, 912, 834 cm–1; 1H NMR (CD3OD, 400 MHz) data, see Table 1; 13C NMR (CD3OD, 150 MHz) data, see Table 2; HRESIMS m/z 649.2836 [M + Na]+ (calculated for C31H46NaO13, 649.2836).
Compound 3: white amorphous powder. UV (MeOH) λmax (log ε): 214 (4.1), 228 (4.2), 316 (4.4) nm; IR (film) νmax: 3391, 2935, 1698, 1632, 1605, 1515, 1263, 1169, 1041, 835 cm–1; 1H NMR (CD3OD, 400 MHz) data, see Table 1; 13C NMR (CD3OD, 100 MHz) data, see Table 2; HRESIMS m/z 649.2834 [M + Na]+ (calculated for C31H46NaO13, 649.2836).
Compound 4: white amorphous powder. UV (MeOH) λmax (log ε): 213 (4.1), 228 (4.2), 317 (4.4) nm; IR (film) νmax: 3392, 2926, 1696, 1633, 1605, 1515, 1263, 1169, 1040, 835 cm–1; 1H NMR (CD3OD, 400 MHz) data, see Table 1; 13C NMR (CD3OD, 150 MHz) data, see Table 2; HRESIMS m/z 647.2684 [M + Na]+ (calculated for C31H44NaO13, 647.2680).
Compound 5: white amorphous powder. [α −38.7 (c 0.55, MeOH); UV (MeOH) λmax (log ε): 214 (4.1), 227 (4.2), 316 (4.4) nm; IR (film) νmax: 3375, 2925, 1693, 1632, 1604, 1515, 1448, 1262, 1170, 1070, 833 cm–1; 1H NMR (CD3OD, 400 MHz) data, see Table 1; 13C NMR (CD3OD, 150 MHz) data, see Table 2; HRESIMS m/z 665.2790 [M + Na]+ (calculated for C31H46NaO14, 665.2785).
Compound 6: yellowish oil. [α −66.2 (c 0.24, MeOH); UV (MeOH) λmax (log ε): 214 (4.1), 227 (4.2), 316 (4.4) nm; IR (film) νmax: 3391, 2925, 1694, 1633, 1605, 1515, 1444, 1264, 1170, 1037, 832 cm–1; 1H NMR (CD3OD, 600 MHz) data, see Table 1; 13C NMR (CD3OD, 100 MHz) data, see Table 2; HRESIMS m/z 631.2728 [M + Na]+ (calculated for C31H44NaO12, 631.2730).
3.4. Acid Hydrolysis of Compounds 1–6
Compounds 1–6 (2 mg, each) were hydrolyzed with 1 M H2SO4 aqueous solution at 92–95 °C for 6 h, respectively, and then neutralized with Ba(OH)2 solution. After filtration, the solvent of the hydrolyzed solution was volatilized. The monosaccharides in the concentrated solution were identified by TLC with l-rhamnose and d-glucose references, which were developed with EtOAc-MeOH-HOAc-H2O (8:1:1:0.7) [3]. The Rf values of l-rhamnose and d-glucose were 0.73 and 0.43, respectively.
3.5. Enzymatic Hydrolysis of Compound 5
Compound 5 (20 mg) and cellulase (30 mg) were added to a 12 mL HOAc-NaOAc buffer solution (pH 5.0) and incubated at 37 °C for 6 h. The mixture solution was extracted with EtOAc and isolated on a silica gel column (EtOAc) to give (E,R)-3,7-dimethyl-oct-2-ene-1,6,7-triol and (E,S)-3,7-dimethyl-oct-2-ene-1,6,7-triol (6:4), which were affirmed by [α +3.2 (c 0.19, EtOAc) [17].
3.6. Determination of Bioactivities
The bioactivities of compounds 1–8, including FAS inhibitory action, α-glucosidase inhibitory action, α-amylase inhibitory action, DPPH radical scavenging capacity, and ABTS radical scavenging capacity, were measured in this study. The published methods [3,18,19,20] were employed in the biological tests, in which orlistat, acarbose, and l-(+)-ascorbic acid served as positive controls, respectively (Supplementary Materials S2).
3.7. Statistical Analyses
GraphPad Prism 5.01 was applied in statistical analyses. All compounds were measured in triplicate, and the results are reported as mean ± SD. The difference in means was determined by ANOVA with the statistical package SPSS 25.0. The significant difference between groups was confirmed at p < 0.05.
4. Conclusions
In conclusion, ten new monoterpenoid glycosides named ligurobustosides T10 (1a), T11 (1b), T12 (2a), T13 (2b), T14 (3a), T15 (3b), F1 (4b), T16 (5a), T17 (5b), and E1 (6b), together with five known ones (4a, 6a, 7, 8a, 8b), in which nine ingredients belonged to trans-p-hydroxycinnamic acid esters, were separated from the leaves of L. robustum and identified using the spectroscopic method (1H NMR, 13C NMR, 1H-1H COSY, HSQC, HMBC, NOEDS, HRESIMS, IR) and chemical method. The results of biological tests exhibited that the FAS inhibitory action of compound 5 (IC50: 4.38 ± 0.11 μM) was as strong as asorlistat (IC50: 4.46 ± 0.13 μM) used as a positive control; the α-glucosidase inhibitory actions of compounds 1–4 and 7–8, and the α-amylase inhibitory actions of compounds 1–8 were medium and weaker than acarbose; the ABTS radical scavenging capacities of compounds 1–3 and 5–8 (IC50: 6.27 ± 0.23 ~ 8.59 ± 0.09 μM) were stronger than l-(+)-ascorbic acid (IC50: 10.06 ± 0.19 μM), a positive control. Together with our previous studies [3,15], monoterpenoid glycosides, phenylmethanoid glycosides, phenylethanoid glycosides, butenol and hexenol glycosides, sugar esters, and flavonoid glycosides, along with trans-p-hydroxycinnamic acid, were believed to be the main functional constituents of L. robustum to prevent diabetes and its complications. This research provided a theoretical foundation for the leaves of L. robustum to prevent diabetes and its complications. Nevertheless, the activities of the stereoisomer mixture might be different from those of the relevant pure compound, and the activities in vitro might be different from those in vivo. Thus, further research should be carried out to evaluate the activities of pure compounds in vivo in the future.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules28217274/s1, Figures S1–S6: 1H NMR, 13C NMR, 1H-1H COSY, HSQC, HMBC, NOEDS, HRESIMS, and IR spectra of compounds 1 (Figure S1), 2 (Figure S2), 3 (Figure S3), 4 (Figure S4), 5 (Figure S5), and 6 (Figure S6); S1: 1H NMR and 13C NMR data of compounds 4a, 6a, 7, 8a, and 8b; S2: determination of bioactivities.
Author Contributions
Conceptualization, S.-H.L. and J.H.; methodology, S.-H.L.; formal analysis, S.-H.L. and W.-N.L.; investigation, S.-H.L., X.-X.L., H.-J.Z., W.-N.L. and J.-P.P.; data curation, J.H.; writing—original draft preparation, S.-H.L.; writing—review and editing, J.H. and X.-X.L.; supervision, J.H.; funding acquisition, S.-H.L. All authors have read and agreed to the published version of the manuscript.
Funding
The research was subsidized by the Guangxi Natural Science Foundation Project (No. 2020GXNSFAA297129), the Guangxi Science and Technology Base and Talents Special Project (No. Guike AD21075006), and the Youjiang Medical University for Nationalities Science Research Project (No. yy2021sk004).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in the Supplementary Materials.
Acknowledgments
The authors sincerely thank Fu Su and You Zhou, West China School of Pharmacy, Sichuan University, for measuring the NMR spectra. The authors are obliged to Ming-Hai Tang, State Key Laboratory of Biotherapy, West China Hospital, Sichuan University, for measuring the HRESIMS.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Sample Availability
Samples of the compounds are not available from the authors.
Abbreviations
| ABTS | 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulphonic acid) ammonium salt |
| Ac-CoA | acetyl-coenzyme A |
| ANOVA | one-way analysis of variance |
| CC | column chromatography |
| Cou | coumaroyl |
| DPPH | 2,2-diphenyl-1-picrylhydrazyl |
| EtOAc | ethyl acetate |
| FAS | fatty acid synthase |
| Glc | glucosyl |
| 1H-1H COSY | 1H-1H homonuclear chemical shift correlation spectroscopy |
| HMBC | heteronuclear multiple bond coherence spectroscopy |
| HPLC | high-performance liquid chromatography |
| HRESIMS | high-resolution electrospray ionization mass spectroscopy |
| HSQC | heteronuclear single quantum coherence spectroscopy |
| IC50 | half inhibitory concentration |
| IR | infrared absorption spectrum |
| Mal-CoA | methylmalonyl coenzyme A |
| NMR | nuclear magnetic resonance |
| NOEDS | nuclear Overhauser effect difference spectrum |
| Rha | rhamnosyl |
| SD | standard deviation |
| TLC | thin-layer chromatography |
| UV | ultraviolet–visible absorption spectrum |
References
- Ansari, P.; Akther, S.; Hannan, J.M.A.; Seidel, V.; Nujat, N.J.; Abdel-Wahab, Y.H.A. Pharmacologically active phytomolecules isolated from traditional antidiabetica plants and their therapeutic role for the management of diabetes mellitus. Molecules 2022, 27, 4278. [Google Scholar] [PubMed]
- Wang, H.; Tang, S.; Zhang, G.; Pan, Y.; Jiao, W.; Shao, H. Synthesis of N-substituted iminosugar C-glycosides and evaluation as promising α-glucosidase inhibitors. Molecules 2022, 27, 5517. [Google Scholar] [PubMed]
- Lu, S.-H.; Zuo, H.-J.; Huang, J.; Li, W.-N.; Huang, J.-L.; Li, X.-X. Chemical constituents from the leaves of Ligustrum robustum and their bioactivities. Molecules 2023, 28, 362. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.-H.; Liang, X.-N.; Nong, X.-J.; Chen, R.; Li, X.-X. A new HPLC-UV method using hydrolyzation with sodium hydroxide for quantitation of trans-p-hydroxycinnamic acid and total trans-p-hydroxycinnamic acid esters in the leaves of Ligustrum robustum. Molecules 2023, 28, 5309. [Google Scholar] [CrossRef] [PubMed]
- He, Z.D.; Lau, K.M.; But, P.P.-H.; Jiang, R.W.; Dong, H.; Ma, S.C.; Fung, K.P.; Ye, W.C.; Sun, H.D. Antioxidative glycosides from the leaves of Ligustrum robustum. J. Nat. Prod. 2003, 66, 851–854. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Peng, Y.; Xu, L.J.; Wu-Lan, T.N.; Shi, R.B.; Xiao, P.G. Chemical constituents from Ligustrum robustum Bl. Biochem. Syst. Ecol. 2010, 38, 398–401. [Google Scholar] [CrossRef]
- Li, L.; Peng, Y.; Liu, Y.; Xu, L.J.; Guo, N.; Shi, R.B.; Xiao, P.G. Two new phenethanol glycosides from Ligustrum robustum. Chin. Chem. Lett. 2011, 22, 326–329. [Google Scholar] [CrossRef]
- Tian, J.; Zhang, H.J.; Sun, H.D.; Pan, L.T.; Yao, P.; Chen, D.Y. Monoterpenoid glycosides from Ligustrum robustum. Phytochemistry 1998, 48, 1013–1018. [Google Scholar] [CrossRef]
- Tian, J.; Sun, H.D. New monoterpenoid glycosides from Ligustrum robustum. Chin. J. Appl. Environ. Biol. 1999, 5, 501–506. [Google Scholar]
- Yu, Z.L.; Zeng, W.C. Antioxidant, antibrowning, and cytoprotective activities of Ligustrum robustum (Roxb.) Blume extract. J. Food Sci. 2013, 78, 1354–1362. [Google Scholar]
- Yu, Z.L.; Gao, H.X.; Zhang, Z.; He, Z.; He, Q.; Jia, L.R.; Zeng, W.C. Inhibitory effects of Ligustrum robustum (Roxb.) Blume extract on α-amylase and α-glucosidase. J. Funct. Foods 2015, 19, 204–213. [Google Scholar] [CrossRef]
- Ito, H.; Otsuki, A.; Mori, H.; Li, P.; Kinoshita, M.; Kawakami, Y.; Tsuji, H.; Fang, D.Z.; Takahashi, Y. Two new monoterpene glycosides from Qing Shan Lu Shui tea with inhibitory effects on leukocyte-type 12-lipoxygenase activity. Molecules 2013, 18, 4257–4266. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Zhang, Z.-W.; Li, C.-C.; Yang, R.-M.; Pan, R.-L. Chemical constituents of alcoholic extract from Ligustrum robustum (Roxb.) Blume. Mod. Chin. Med. 2018, 20, 540–544. [Google Scholar]
- Kawakami, Y.; Otsuki, A.; Mori, Y.; Kanzaki, K.; Suzuki-Yamamoto, T.; Fang, D.Z.; Ito, H.; Takahashi, Y. Involvement of the hydroperoxy group in the irreversible inhibition of leukocyte-type 12-lipoxygenase by monoterpene glycosides contained in the Qing Shan Lu Shui tea. Molecules 2019, 24, 304. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.-H.; Huang, J.; Zuo, H.-J.; Zhou, Z.-B.; Yang, C.-Y.; Huang, Z.-L. Monoterpenoid glycosides from the leaves of Ligustrum robustum and their bioactivities. Molecules 2022, 27, 3709. [Google Scholar] [CrossRef] [PubMed]
- He, Z.D.; Ueda, S.; Akaji, M.; Fujita, T.; Inoue, K.; Yang, C.R. Monoterpenoid and phenylethanoid glycosides from Ligustrum pedunculare. Phytochemistry 1994, 36, 709–716. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.W.; Zhang, X.T.; Zhao, Y.; Liu, X.L.; Zhang, Z.H.; Wang, M.A. Divergent synthesis of four isomers of 6,7-dihydroxy-3,7-dimethyloct-2-enoic acid, esters and evaluation for the antifungal activity. Chin. Chem. Lett. 2018, 29, 1872–1874. [Google Scholar] [CrossRef]
- Fan, H.J.; Wu, D.; Tian, W.X.; Ma, X.F. Inhibitory effects of tannic acid on fatty acid synthase and 3T3-L1 preadipocyte. Biochim. Biophys. Acta 2013, 1831, 1260–1266. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Ma, X.F.; Tian, W.X. Pomegranate husk extract, punicalagin and ellagic acid inhibit fatty acid synthase and adipogenesis of 3T3-L1 adipocyte. J. Func. Foods 2013, 5, 633–641. [Google Scholar] [CrossRef]
- Mudgil, P.; Kamal, H.; Yuen, G.C.; Maqsood, S. Characterization and identification of novel antidiabetic and anti-obesity peptides from camel milk protein hydrolysates. Food Chem. 2018, 259, 46–54. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).