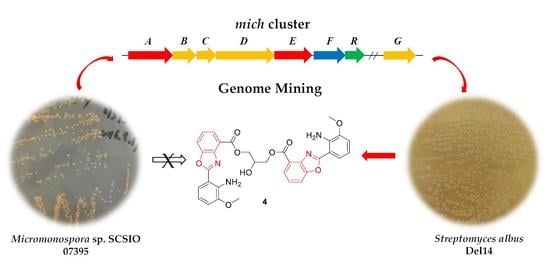
Genomics-Driven Discovery of Benzoxazole Alkaloids from the Marine-Derived Micromonospora sp. SCSIO 07395
Abstract
1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. Bacterial Strains and Reagents
3.2. General Experimental Procedures
3.3. General DNA Manipulation Techniques
3.4. Sequence Analysis
3.5. Heterologous Expression of Mich BGC
3.6. Fermentation and Metabolite Analyses by HPLC
3.7. Large-Scale Fermentation and Isolation of Compounds from Heterologous Expression Strain
3.8. Chiral HPLC Analysis
3.9. Bioactivity Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Kerru, N.; Gummidi, L.; Maddila, S.; Gangu, K.K.; Jonnalagadda, S.B. A Review on recent advances in nitrogen-containing molecules and their biological applications. Molecules 2020, 25, 1909. [Google Scholar] [CrossRef] [PubMed]
- Wong, X.K.; Yeong, K.Y. A patent review on the current developments of benzoxazoles in drug discovery. ChemMedChem 2021, 16, 3237–3262. [Google Scholar] [CrossRef] [PubMed]
- Hohmann, C.; Schneider, K.; Bruntner, C.; Irran, E.; Nicholson, G.; Bull, A.T.; Jones, A.L.; Brown, R.; Stach, J.E.; Goodfellow, M.; et al. Caboxamycin, a new antibiotic of the benzoxazole family produced by the deep-sea strain Streptomyces sp. NTK 937. J. Antibiot. 2009, 62, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Chaney, M.O.; Demarco, P.V.; Jones, N.D.; Occolowitz, J.L. Letter: The structure of A23187, a divalent cation ionophore. J. Am. Chem. Soc. 1974, 96, 1932–1933. [Google Scholar] [CrossRef] [PubMed]
- Michel, K.H.; Boeck, L.D.; Hoehn, M.M.; Jones, N.D.; Chaney, M.O. The discovery, fermentation, isolation, and structure of antibiotic A33853 and its tetraacetyl derivative. J. Antibiot. 1984, 37, 441–445. [Google Scholar] [CrossRef] [PubMed]
- Sommer, P.S.; Almeida, R.C.; Schneider, K.; Beil, W.; Sussmuth, R.D.; Fiedler, H.P. Nataxazole, a new benzoxazole derivative with antitumor activity produced by Streptomyces sp. Tu 6176. J. Antibiot. 2008, 61, 683–686. [Google Scholar] [CrossRef]
- Sun, M.; Zhang, X.; Hao, H.; Li, W.; Lu, C. Nocarbenzoxazoles A-G, benzoxazoles produced by halophilic Nocardiopsis lucentensis DSM 44048. J. Nat. Prod. 2015, 78, 2123–2127. [Google Scholar] [CrossRef]
- Rodriguez, A.D.; Ramirez, C.; Rodriguez, I.I.; Gonzalez, E. Novel antimycobacterial benzoxazole alkaloids, from the west Indian Sea whip Pseudopterogorgia elisabethae. Org. Lett. 1999, 1, 527–530. [Google Scholar] [CrossRef]
- Ovenden, S.P.; Nielson, J.L.; Liptrot, C.H.; Willis, R.H.; Tapiolas, D.M.; Wright, A.D.; Motti, C.A. Sesquiterpene benzoxazoles and sesquiterpene quinones from the marine sponge Dactylospongia elegans. J. Nat. Prod. 2011, 74, 65–68. [Google Scholar] [CrossRef]
- Don, M.J.; Shen, C.C.; Lin, Y.L.; Syu, W.J.; Ding, Y.H.; Sun, C.M. Nitrogen-containing compounds from Salvia miltiorrhiza. J. Nat. Prod. 2005, 68, 1066–1070. [Google Scholar] [CrossRef]
- Zhang, D.; Guo, J.; Zhang, M.; Liu, X.; Ba, M.; Tao, X.; Yu, L.; Guo, Y.; Dai, J. Oxazole-containing diterpenoids from cell cultures of Salvia miltiorrhiza and their anti-HIV-1 activities. J. Nat. Prod. 2017, 80, 3241–3246. [Google Scholar] [CrossRef]
- Lv, M.; Zhao, J.; Deng, Z.; Yu, Y. Characterization of the biosynthetic gene cluster for benzoxazole antibiotics A33853 reveals unusual assembly logic. Chem. Biol. 2015, 22, 1313–1324. [Google Scholar] [CrossRef]
- Cano-Prieto, C.; Garcia-Salcedo, R.; Sanchez-Hidalgo, M.; Brana, A.F.; Fiedler, H.P.; Mendez, C.; Salas, J.A.; Olano, C. Genome mining of Streptomyces sp. Tu 6176: Characterization of the nataxazole biosynthesis pathway. ChemBioChem 2015, 16, 1461–1473. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Liang, J.; Lin, S.; Zhou, X.; Bai, L.; Deng, Z.; Wang, Z. Characterization of the biosynthesis gene cluster for the pyrrole polyether antibiotic calcimycin (A23187) in Streptomyces chartreusis NRRL 3882. Antimicrob. Agents Chemother. 2011, 55, 974–982. [Google Scholar] [CrossRef] [PubMed]
- Losada, A.A.; Cano-Prieto, C.; Garcia-Salcedo, R.; Brana, A.F.; Mendez, C.; Salas, J.A.; Olano, C. Caboxamycin biosynthesis pathway and identification of novel benzoxazoles produced by cross-talk in Streptomyces sp. NTK 937. Microb. Biotechnol. 2017, 10, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Losada, A.A.; Mendez, C.; Salas, J.A.; Olano, C. Exploring the biocombinatorial potential of benzoxazoles: Generation of novel caboxamycin derivatives. Microb. Cell. Fact. 2017, 16, 93. [Google Scholar] [CrossRef]
- Song, H.; Rao, C.; Deng, Z.; Yu, Y.; Naismith, J.H. The biosynthesis of the benzoxazole in nataxazole proceeds via an unstable ester and has synthetic utility. Angew. Chem. Int. Ed. 2020, 59, 6054–6061. [Google Scholar] [CrossRef]
- Horch, T.; Molloy, E.M.; Bredy, F.; Haensch, V.G.; Scherlach, K.; Dunbar, K.L.; Franke, J.; Hertweck, C. Alternative benzoxazole assembly discovered in anaerobic bacteria provides access to privileged heterocyclic scaffold. Angew. Chem. Int. Ed. 2022, 61, e202205409. [Google Scholar] [CrossRef]
- Myronovskyi, M.; Luzhetskyy, A. Heterologous production of small molecules in the optimized Streptomyces hosts. Nat. Prod. Rep. 2019, 36, 1281–1294. [Google Scholar] [CrossRef]
- Huo, L.; Hug, J.J.; Fu, C.; Bian, X.; Zhang, Y.; Muller, R. Heterologous expression of bacterial natural product biosynthetic pathways. Nat. Prod. Rep. 2019, 36, 1412–1436. [Google Scholar] [CrossRef]
- Kang, H.S.; Kim, E.S. Recent advances in heterologous expression of natural product biosynthetic gene clusters in Streptomyces hosts. Curr. Opin. Biotechnol. 2021, 69, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Zhang, W.; Zhang, G.; Zhu, Y.; Chen, Y.; Liu, W.; Yuan, C.; Zhang, Q.; Zhang, H.; Zhang, L.; et al. Activation and characterization of a cryptic gene cluster reveals a cyclization cascade for polycyclic tetramate macrolactams. Chem. Sci. 2017, 8, 1607–1612. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhang, W.; Jin, H.; Zhang, Q.; Chen, Y.; Jiang, X.; Zhang, G.; Zhang, L.; Zhang, W.; She, Z.; et al. Genome mining of marine-derived Streptomyces sp. SCSIO 40010 leads to cytotoxic new polycyclic tetramate macrolactams. Mar. Drugs 2019, 17, 663. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Wang, L.; Zhang, H.; Zhang, L.; Tan, B.; Huang, Q.; Zhu, Y.; Zhang, C. Biosynthesis and engineered overproduction of everninomicins with promising activity against multidrug-resistant bacteria. 2022; submitted. [Google Scholar]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic. Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Huang, S.; Li, N.; Zhou, J.; He, J. Construction of a new bacterial artificial chromosome (BAC) vector for cloning of large DNA fragments and heterologous expression in Streptomyces. Wei Sheng Wu Xue Bao 2012, 52, 30–37. [Google Scholar]
- Myronovskyi, M.; Rosenkranzer, B.; Nadmid, S.; Pujic, P.; Normand, P.; Luzhetskyy, A. Generation of a cluster-free Streptomyces albus chassis strains for improved heterologous expression of secondary metabolite clusters. Metab. Eng. 2018, 49, 316–324. [Google Scholar] [CrossRef]
- Ahmed, Y.; Rebets, Y.; Estevez, M.R.; Zapp, J.; Myronovskyi, M.; Luzhetskyy, A. Engineering of Streptomyces lividans for heterologous expression of secondary metabolite gene clusters. Microb. Cell. Fact. 2020, 19, 5. [Google Scholar] [CrossRef]
- Tipparaju, S.K.; Joyasawal, S.; Pieroni, M.; Kaiser, M.; Brun, R.; Kozikowski, A.P. In pursuit of natural product leads: Synthesis and biological evaluation of 2-[3-hydroxy-2-[(3-hydroxypyridine-2-carbonyl)amino]phenyl]benzoxazole-4-carboxyl ic acid (A-33853) and its analogues: Discovery of N-(2-benzoxazol-2-ylphenyl)benzamides as novel antileishmanial chemotypes. J. Med. Chem. 2008, 51, 7344–7347. [Google Scholar]
- Mou, P.; Zhang, Q.; Peng, J.; Jiang, X.; Zhang, L.; Zhou, Z.; Zhang, C.; Zhu, Y. Antibacterial phenylspirodrimanes from the marine-derived fungus Stachybotrys sp. SCSIO 40434. Fitoterapia 2021, 152, 104937. [Google Scholar] [CrossRef]
- Zaburannyi, N.; Rabyk, M.; Ostash, B.; Fedorenko, V.; Luzhetskyy, A. Insights into naturally minimised Streptomyces albus J1074 genome. BMC Genom. 2014, 15, 97. [Google Scholar] [CrossRef]
- Flett, F.; Mersinias, V.; Smith, C.P. High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol. Lett. 1997, 155, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Tong, Z.W.; Xie, X.H.; Wang, T.T.; Lu, M.; Jiao, R.H.; Ge, H.M.; Hu, G.; Tan, R.X. Acautalides A-C, neuroprotective Diels-Alder adducts from solid-state cultivated Acaulium sp. H-JQSF. Org. Lett. 2021, 23, 5587–5591. [Google Scholar] [CrossRef] [PubMed]

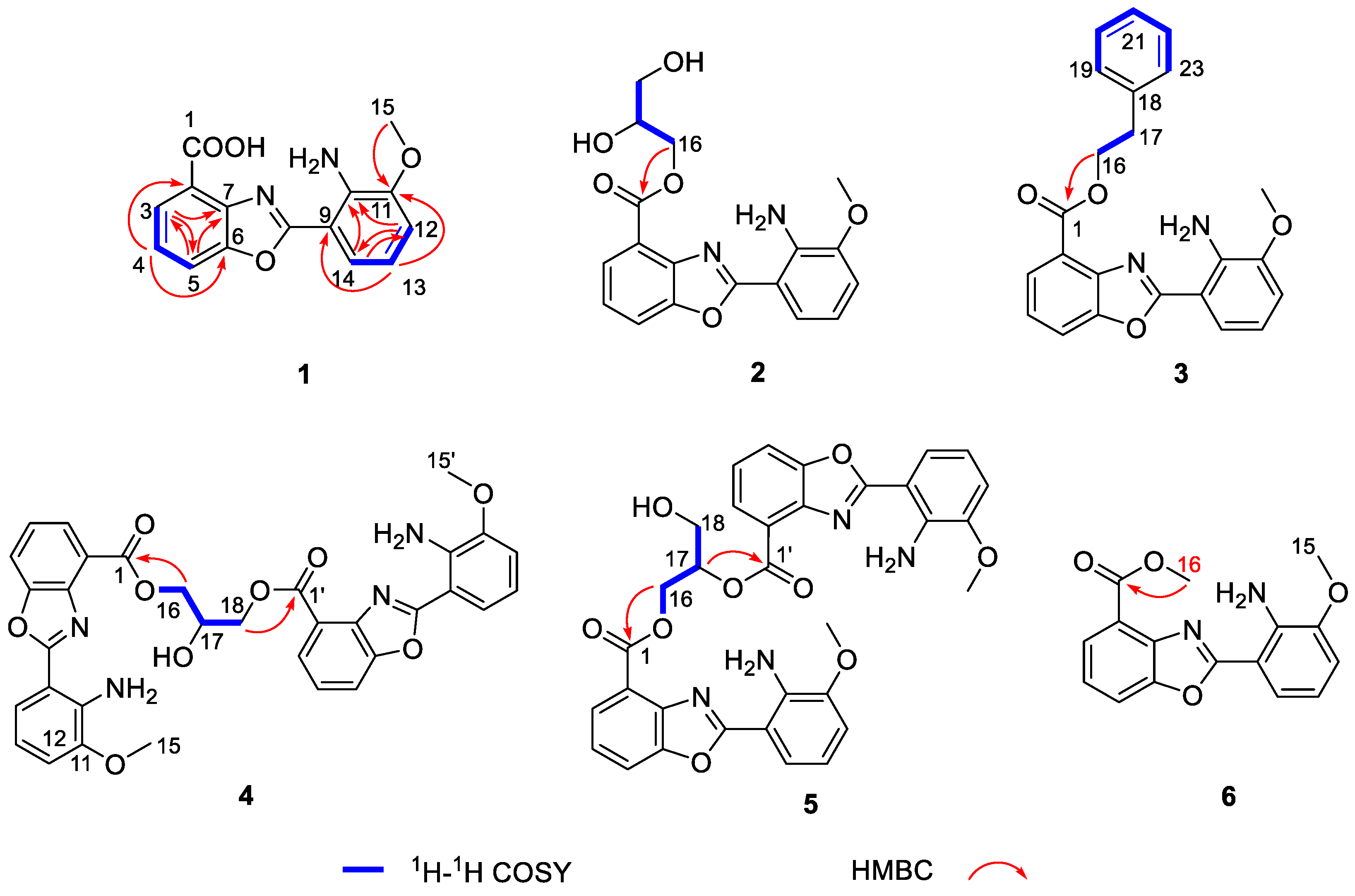
| No. | 1 a | 2 a | 3 a | 6 b | ||||
|---|---|---|---|---|---|---|---|---|
| δC | δH | δC | δH | δC | δH | δC | δH | |
| 1 | 166.9, C | 164.5, C | 164.3, C | 166.1, C | ||||
| 2 | 124.2, C | 120.7, C | 120.3, C | 121.4, C | ||||
| 3 | 126.3, CH | 7.86, d (7.7) | 126.6, CH | 7.96, dd (7.7, 0.7) | 126.3, CH | 7.88, d (7.7) | 126.8, CH | 8.00, d (7.7) |
| 4 | 124.2, CH | 7.44, dd (7.7, 7.7) | 124.4, CH | 7.51, dd (7.7, 7.7) | 124.5, CH | 7.49, dd (7.7, 7.7) | 123.9, CH | 7.37, dd (7.7, 7.7) |
| 5 | 113.7, CH | 7.93, d (7.7) | 115.1, CH | 8.03, dd (7.7, 0.7) | 114.9, CH | 8.03, d (7.7) | 114.6, CH | 7.74, d (7.7) |
| 6 | 149.2, C | 149.2, C | 149.4, C | 150.1, C | ||||
| 7 | 140.5, C | 140.8, C | 140.6, C | 141.8, C | ||||
| 8 | 163.7, C | 164.2, C | 164.3, C | 165.1, C | ||||
| 9 | 105.6, C | 113.3, C | 105.1, C | 107.1, C | ||||
| 10 | 139.7, C | 139.5, C | 139.9, C | 140.0, C | ||||
| 11 | 146.8, C | 147.0, C | 146.8, C | 147.2, C | ||||
| 12 | 112.5, CH | 7.02, dd (7.7, 0.7) | 112.4, CH | 7.03, d (7.7) | 112.6, CH | 7.03, d (7.7) | 112.1, CH | 6.88, d (7.7) |
| 13 | 114.9, CH | 6.69, dd (7.7, 8.4) | 114.9, CH | 6.69, dd (7.7, 8.4) | 115.1, CH | 6.69, dd (7.7, 7.4) | 115.4, CH | 6.72, dd (7.7, 7.7) |
| 14 | 119.5, CH | 7.57, dd (8.4, 1.4) | 119.4, CH | 7.57, dd (8.4, 1.4) | 119.4, CH | 7.57, d (7.7) | 120.3, CH | 7.68, (7.7) |
| 15 | 55.7, CH3 | 3.87, s | 55.7, CH3 | 3.90, s | 55.7, CH3 | 3.88, s | 55.9, CH3 | 3.94, s |
| 16 | 66.0, CH2 | 4.39, dd (11.2, 4.2); 4.27, dd (11.2, 5.6) | 65.3, CH2 | 4.56, dd (14.0, 7.0) | 52.4, CH3 | 4.03, s | ||
| 17 | 69.2, CH | 3.88, m | 34.4, CH2 | 3.14, m | ||||
| 18 | 62.8, CH2 | 3.53, m | 138.0, C | |||||
| 19 | 129.0, CH | 7.38, d (7.7) | ||||||
| 20 | 128.4, CH | 7.31, dd (7.7, 7.7) | ||||||
| 21 | 126.4, CH | 7.22, dd (7.7, 7.7) | ||||||
| 22 | 128.4, CH | 7.31, dd (7.7, 7.7) | ||||||
| 23 | 129.0, CH | 7.38, d (7.7) | ||||||
| No. | 4 | 5 | ||
|---|---|---|---|---|
| δC | δH | δC | δH | |
| 1 | 165.8, C | 165.8, C | ||
| 2 | 120.7, C | 120.5, C | ||
| 3 | 127.0, CH | 7.97, d (7.7) | 127.1, CH | 7.99, d (7.7) |
| 4 | 123.9, CH | 7.33, dd (7.7, 7.7) | 123.8, CH | 7.33, dd (7.7, 7.7) |
| 5 | 114.8, CH | 7.71, d (7.7) | 114.7, CH | 7.70, d (7.7) |
| 6 | 149.9, C | 150.0, C | ||
| 7 | 141.7, C | 141.8, C | ||
| 8 | 163.2, C | 164.8, C | ||
| 9 | 106.9, C | 106.9, C | ||
| 10 | 139.9, C | 139.8, C | ||
| 11 | 147.2, C | 147.3, C | ||
| 12 | 112.1, CH | 6.71, d (7.7) | 112.3, CH | 6.83, d (7.7) |
| 13 | 116.0, CH | 6.61, dd (7.7, 7.7) | 115.6, CH | 6.68, dd (7.7, 7.7) |
| 14 | 120.3, CH | 7.52, (7.7) | 120.2, CH | 7.60, (7.7) |
| 15 | 55.8, CH3 | 3.71, s | 55.8, CH3 | 3.86, s |
| 16 | 66.3, CH2 | 4.72, m | 63.4, CH2 | 4.91, m; 4.83, m |
| 17 | 68.5 CH | 4.59, m | 73.5 CH | 5.63, m |
| 18 | 66.3, CH2 | 4.72, m | 61.9 CH2 | 4.14, m |
| 1’ | 165.8, C | 165.8, C | ||
| 2’ | 120.7, C | 120.5, C | ||
| 3’ | 127.0, CH | 7.97, d (7.7) | 127.1, CH | 7.99, d (7.7) |
| 4’ | 123.9, CH | 7.33, dd (7.7, 7.7) | 123.8, CH | 7.33, dd (7.7, 7.7) |
| 5’ | 114.8, CH | 7.71, d (7.7) | 114.7, CH | 7.70, d (7.7) |
| 6’ | 149.9, C | 150.0, C | ||
| 7’ | 141.7, C | 141.8, C | ||
| 8’ | 163.2, C | 164.8, C | ||
| 9’ | 106.9, C | 106.9, C | ||
| 10’ | 149.9, C | 139.8, C | ||
| 11’ | 147.2, C | 147.3, C | ||
| 12’ | 112.1, CH | 6.71, d (7.7) | 112.3, CH | 6.83, d (7.7) |
| 13’ | 116.0, CH | 6.61, dd (7.7, 7.7) | 115.6, CH | 6.68, dd (7.7, 7.7) |
| 14’ | 120.3, CH | 7.52, (7.7) | 120.2, CH | 7.60, (7.7) |
| 15’ | 55.8, CH3 | 3.71, s | 55.8, CH3 | 3.86, s |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, Z.; Zhang, Q.; Peng, J.; Zhao, X.; Ma, L.; Zhang, C.; Zhu, Y. Genomics-Driven Discovery of Benzoxazole Alkaloids from the Marine-Derived Micromonospora sp. SCSIO 07395. Molecules 2023, 28, 821. https://doi.org/10.3390/molecules28020821
Cheng Z, Zhang Q, Peng J, Zhao X, Ma L, Zhang C, Zhu Y. Genomics-Driven Discovery of Benzoxazole Alkaloids from the Marine-Derived Micromonospora sp. SCSIO 07395. Molecules. 2023; 28(2):821. https://doi.org/10.3390/molecules28020821
Chicago/Turabian StyleCheng, Ziqian, Qingbo Zhang, Jing Peng, Xiaoyang Zhao, Liang Ma, Changsheng Zhang, and Yiguang Zhu. 2023. "Genomics-Driven Discovery of Benzoxazole Alkaloids from the Marine-Derived Micromonospora sp. SCSIO 07395" Molecules 28, no. 2: 821. https://doi.org/10.3390/molecules28020821
APA StyleCheng, Z., Zhang, Q., Peng, J., Zhao, X., Ma, L., Zhang, C., & Zhu, Y. (2023). Genomics-Driven Discovery of Benzoxazole Alkaloids from the Marine-Derived Micromonospora sp. SCSIO 07395. Molecules, 28(2), 821. https://doi.org/10.3390/molecules28020821