Liquid–Liquid Phase Separation and Protective Protein Aggregates in Bacteria
Abstract
:1. Introduction
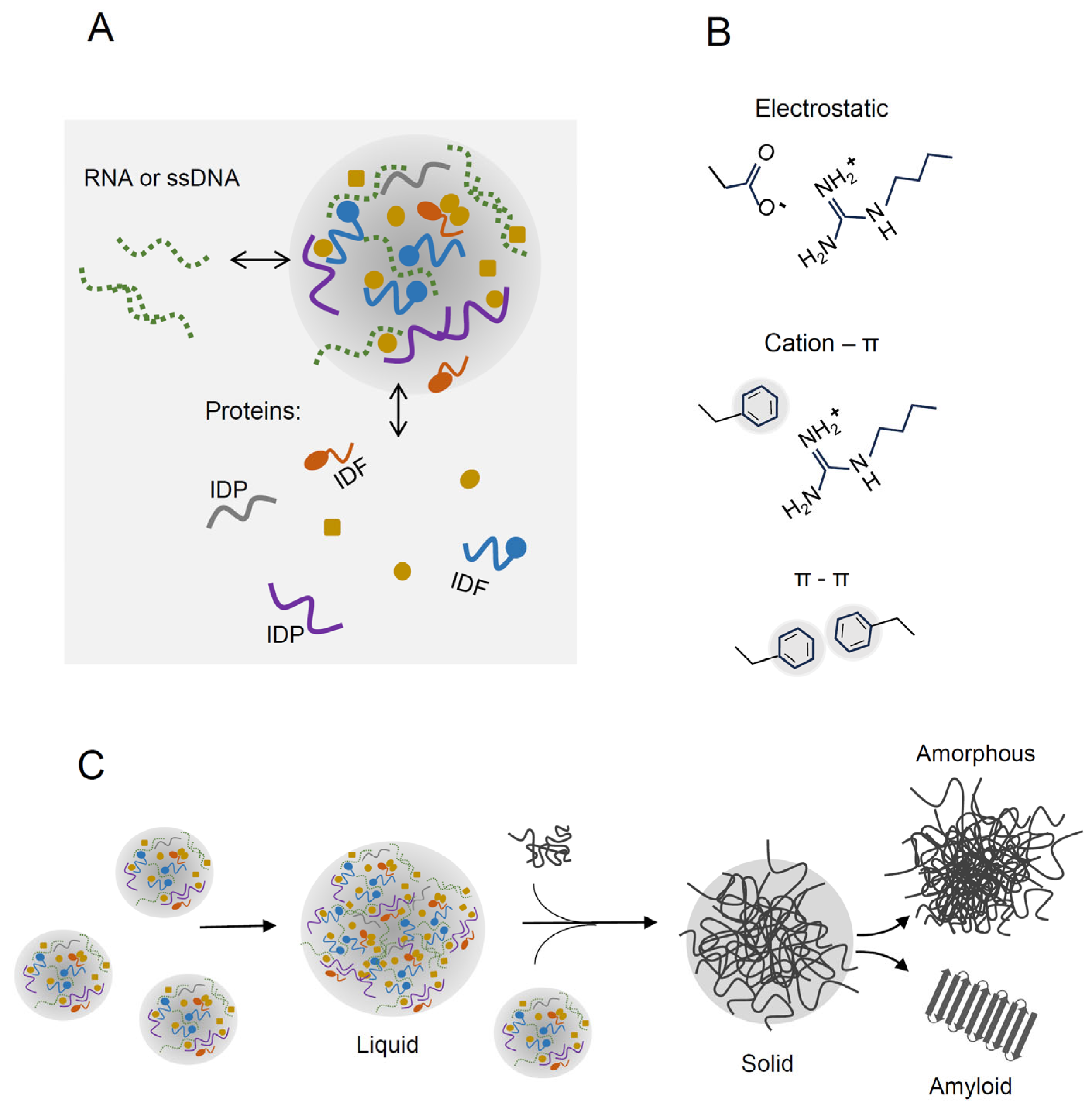
2. Liquid–Liquid Phase Separation of Proteins
2.1. Intrinsically Disordered Proteins
2.2. Post-Translational Modification of Proteins in LLPS
2.3. Transition of Liquid Droplets into Solid Aggregates
3. LLPS-Prone Proteins Participating in Physiological and Stress-Protecting Processes in Bacteria
4. Protein Aggregation in Bacteria as a Consequence of Proteostasis Disruption
4.1. Protective Aggregates and LLPS
4.1.1. Analysis of E. coli Protein Aggregates Formed during Desiccation–Rehydration Stress
5. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Shin, Y.; Brangwynne, C.P. Liquid Phase Condensation in Cell Physiology and Disease. Science 2017, 357, eaaf4382. [Google Scholar] [CrossRef]
- Franzmann, T.M.; Alberti, S. Protein Phase Separation as a Stress Survival Strategy. Cold Spring Harb. Perspect. Med. 2019, 11, a034058. [Google Scholar] [CrossRef]
- Parra, G.L.; Libich, D.S. Major Structural Features of Membrane-Less Organelles. In Droplets of Life Membrane-Less Organelles, Biomolecular Condensates, and Biological Liquid-Liquid Phase Separation; Uversky, V.N., Ed.; Elsevier: Amsterdam, The Netherlands, 2023; pp. 83–99. [Google Scholar]
- Hirose, T.; Ninomiya, K.; Nakagawa, S.; Yamazaki, T. A Guide to Membraneless Organelles and Their Various Roles in Gene Regulation. Nat. Rev. Mol. Cell Biol. 2023, 24, 288–304. [Google Scholar] [CrossRef]
- Orti, F.; Navarro, A.M.; Rabinovich, A.; Wodak, S.J.; Marino-Buslje, C. Insight into Membraneless Organelles and Their Associated Proteins: Drivers, Clients and Regulators. Comput. Struct. Biotechnol. J. 2021, 19, 3964–3977. [Google Scholar] [CrossRef]
- Darling, A.L.; Uversky Vladimir, N. Known Types of Membrane-Less Organelles and Biomolecular Condensates. In Droplets of Life: Membrane-Less Organelles, Biomolecular Condensates, and Biological Liquid-Liquid Phase Separation; Uversky, V.N., Ed.; Elsevier: Amsterdam, The Netherlands, 2023; pp. 271–335. [Google Scholar]
- Hardenberg, M.; Horvath, A.; Ambrus, V.; Fuxreiter, M.; Vendruscolo, M. Widespread Occurrence of the Droplet State of Proteins in the Human Proteome. Proc. Natl. Acad. Sci. USA 2020, 117, 33254–33262. [Google Scholar] [CrossRef]
- Saar, K.L.; Morgunov, A.S.; Qi, R.; Arter, W.E.; Krainer, G.; Lee, A.A.; Knowles, T.P.J. Learning the Molecular Grammar of Protein Condensates from Sequence Determinants and Embeddings. Proc. Natl. Acad. Sci. USA 2021, 118, e2019053118. [Google Scholar] [CrossRef]
- Nakashima, K.K.; Vibhute, M.A.; Spruijt, E. Biomolecular Chemistry in Liquid Phase Separated Compartments. Front. Mol. Biosci. 2019, 6, 21. [Google Scholar] [CrossRef]
- Gao, Z.; Zhang, W.; Chang, R.; Zhang, S.; Yang, G.; Zhao, G. Liquid-Liquid Phase Separation: Unraveling the Enigma of Biomolecular Condensates in Microbial Cells. Front. Microbiol. 2021, 12. [Google Scholar] [CrossRef]
- Azaldegui, C.A.; Vecchiarelli, A.G.; Biteen, J.S. The Emergence of Phase Separation as an Organizing Principle in Bacteria. Biophys. J. 2021, 120, 1123–1138. [Google Scholar] [CrossRef]
- Wei, S.P.; Qian, Z.G.; Hu, C.F.; Pan, F.; Chen, M.T.; Lee, S.Y.; Xia, X.X. Formation and Functionalization of Membraneless Compartments in Escherichia coli. Nat. Chem. Biol. 2020, 16, 1143–1148. [Google Scholar] [CrossRef]
- Xue, B.; Dunker, A.K.; Uversky, V.N. Orderly Order in Protein Intrinsic Disorder Distribution: Disorder in 3500 Proteomes from Viruses and the Three Domains of Life. J. Biomol. Struct. Dyn. 2012, 30, 137–149. [Google Scholar] [CrossRef]
- Zhang, H.; Shao, S.; Sun, Y. Characterization of Liquid–Liquid Phase Separation Using Super-Resolution and Single-Molecule Imaging. Biophys. Rep. 2022, 8, 2–13. [Google Scholar] [PubMed]
- Govers, S.K.; Mortier, J.; Adam, A.; Aertsen, A. Protein Aggregates Encode Epigenetic Memory of Stressful Encounters in Individual Escherichia coli Cells. PLoS Biol. 2018, 16, e2003853. [Google Scholar] [CrossRef]
- Jin, X.; Lee, J.-E.; Schaefer, C.; Luo, X.; M Wollman, A.J.; Payne-Dwyer, A.L.; Tian, T.; Zhang, X.; Chen, X.; Li, Y.; et al. Membraneless Organelles Formed by Liquid-Liquid Phase Separation Increase Bacterial Fitness. Sci. Adv. 2021, 7, eabh2929. [Google Scholar] [CrossRef]
- Wang, X.; Cole, C.G.; Dupai, C.D.; Davies, B.W. Protein Aggregation Is Associated with Acinetobacter baumannii Desiccation Tolerance. Microorganisms 2020, 8, 343. [Google Scholar] [CrossRef]
- Pu, Y.; Li, Y.; Jin, X.; Tian, T.; Ma, Q.; Zhao, Z.; Lin, S.Y.; Chen, Z.; Li, B.; Yao, G.; et al. ATP-Dependent Dynamic Protein Aggregation Regulates Bacterial Dormancy Depth Critical for Antibiotic Tolerance. Mol. Cell 2019, 73, 143–156. [Google Scholar] [CrossRef]
- Łupkowska, A.; Monem, S.; Dębski, J.; Stojowska-Swędrzyńska, K.; Kuczyńska-Wiśnik, D.; Laskowska, E. Protein Aggregation and Glycation in Escherichia coli Exposed to Desiccation-Rehydration Stress. Microbiol. Res. 2023, 270, 127335. [Google Scholar] [CrossRef]
- Sołtys, K.; Tarczewska, A.; Bystranowska, D.; Sozańska, N. Getting Closer to Decrypting the Phase Transitions of Bacterial Biomolecules. Biomolecules 2022, 12, 907. [Google Scholar] [CrossRef]
- Romero-Perez, P.S.; Dorone, Y.; Flores, E.; Sukenik, S.; Boeynaems, S. When Phased without Water: Biophysics of Cellular Desiccation, from Biomolecules to Condensates. Chem. Rev. 2022, 123, 9010–9035. [Google Scholar] [CrossRef]
- Janis, B.; Belott, C.; Brockman, T.; Menze, M.A. Functional and Conformational Plasticity of an Animal Group 1 LEA Protein. Biomolecules 2022, 12, 425. [Google Scholar] [CrossRef]
- Belott, C.; Janis, B.; Menze, M.A. Liquid-Liquid Phase Separation Promotes Animal Desiccation Tolerance. Proc. Natl. Acad. Sci. USA 2020, 117, 27676–27684. [Google Scholar] [CrossRef]
- Boothby, T.C.; Tapia, H.; Brozena, A.H.; Piszkiewicz, S.; Smith, A.E.; Giovannini, I.; Rebecchi, L.; Pielak, G.J.; Koshland, D.; Goldstein, B. Tardigrades Use Intrinsically Disordered Proteins to Survive Desiccation. Mol. Cell 2017, 65, 975–984. [Google Scholar] [CrossRef]
- Hincha, D.K.; Thalhammer, A. LEA Proteins: IDPs with Versatile Functions in Cellular Dehydration Tolerance. Biochem. Soc. Trans. 2012, 5, 1000–1003. [Google Scholar] [CrossRef]
- Green, E.R.; Fakhoury, J.N.; Monteith, A.J.; Pi, H.; Giedroc, D.P.; Skaar, E.P. Bacterial Hydrophilins Promote Pathogen Desiccation Tolerance. Cell Host Microbe 2022, 30, 975–987. [Google Scholar] [CrossRef]
- Oda, Y.; Shapiro, M.M.; Lewis, N.M.; Zhong, X.; Huse, H.K.; Zhong, W.; Bruce, J.E.; Manoil, C.; Harwood, C.S. CsrA-Controlled Proteins Reveal New Dimensions of Acinetobacter Baumannii Desiccation Tolerance. J. Bacteriol. 2022, 204, e00479-21. [Google Scholar] [CrossRef]
- Burgess, C.M.; Gianotti, A.; Gruzdev, N.; Holah, J.; Knøchel, S.; Lehner, A.; Margas, E.; Esser, S.S.; Sela Saldinger, S.; Tresse, O. The Response of Foodborne Pathogens to Osmotic and Desiccation Stresses in the Food Chain. Int. J. Food Microbiol. 2016, 221, 37–53. [Google Scholar] [CrossRef]
- Lebre, P.H.; De Maayer, P.; Cowan, D.A. Xerotolerant Bacteria: Surviving through a Dry Spell. Nat. Rev. Microbiol. 2017, 15, 285–296. [Google Scholar] [CrossRef]
- McCoy Vernon, R.; Andrew Chong, P.; Tsang, B.; Hun Kim, T.; Bah, A.; Farber, P.; Lin, H.; Deborah Forman-Kay, J. Pi-Pi Contacts Are an Overlooked Protein Feature Relevant to Phase Separation. Elife 2018, 7, e31486. [Google Scholar] [CrossRef]
- Yeong, V.; Werth, E.G.; Brown, L.M.; Obermeyer, A.C. Formation of Biomolecular Condensates in Bacteria by Tuning Protein Electrostatics. ACS Cent. Sci. 2020, 6, 2301–2310. [Google Scholar] [CrossRef]
- Dignon, G.L.; Best, R.B.; Mittal, J. Annual Review of Physical Chemistry Biomolecular Phase Separation: From Molecular Driving Forces to Macroscopic Properties. Annu. Rev. Phys. Chem. 2020, 71, 53–75. [Google Scholar] [CrossRef]
- Murthy, A.C.; Dignon, G.L.; Kan, Y.; Zerze, G.H.; Parekh, S.H.; Mittal, J.; Fawzi, N.L. Molecular Interactions Underlying Liquid−liquid Phase Separation of the FUS Low-Complexity Domain. Nat. Struct. Mol. Biol. 2019, 26, 637–648. [Google Scholar] [CrossRef]
- André, A.A.M.; Spruijt, E. Liquid–Liquid Phase Separation in Crowded Environments. Int. J. Mol. Sci. 2020, 21, 5908. [Google Scholar] [CrossRef]
- Li, J.; Zhang, M.; Ma, W.; Yang, B.; Lu, H.; Zhou, F.; Zhang, L. Post-Translational Modifications in Liquid-Liquid Phase Separation: A Comprehensive Review. Mol. Biomed. 2022, 3, 13. [Google Scholar] [CrossRef]
- Owen, I.; Shewmaker, F. The Role of Post-Translational Modifications in the Phase Transitions of Intrinsically Disordered Proteins. Int. J. Mol. Sci. 2019, 20, 5501. [Google Scholar] [CrossRef]
- Tompa, P.; Davey, N.E.; Gibson, T.J.; Babu, M.M. A Million Peptide Motifs for the Molecular Biologist. Mol. Cell 2014, 55, 161–169. [Google Scholar] [CrossRef]
- Brocca, S.; Grandori, R.; Longhi, S.; Uversky, V. Liquid–Liquid Phase Separation by Intrinsically Disordered Protein Regions of Viruses: Roles in Viral Life Cycle and Control of Virus–Host Interactions. Int. J. Mol. Sci. 2020, 21, 9045. [Google Scholar] [CrossRef]
- Crabtree, M.D.; Mendonça, C.A.T.F.; Bubb, Q.R.; Clarke, J. Folding and Binding Pathways of BH3-Only Proteins Are Encoded within Their Intrinsically Disordered Sequence, Not Templated by Partner Proteins. J. Biol. Chem. 2018, 293, 9718–9723. [Google Scholar] [CrossRef]
- Dosnon, M.; Bonetti, D.; Morrone, A.; Erales, J.; Di Silvio, E.; Longhi, S.; Gianni, S. Demonstration of a Folding after Binding Mechanism in the Recognition between the Measles Virus NTAIL and X Domains. ACS Chem. Biol. 2015, 10, 795–802. [Google Scholar] [CrossRef]
- Malagrinò, F.; Visconti, L.; Pagano, L.; Toto, A.; Troilo, F.; Gianni, S. Understanding the Binding Induced Folding of Intrinsically Disordered Proteins by Protein Engineering: Caveats and Pitfalls. Int. J. Mol. Sci. 2020, 21, 3484. [Google Scholar] [CrossRef]
- Chakrabortee, S.; Boschetti, C.; Walton, L.J.; Sarkar, S.; Rubinsztein, D.C.; Tunnacliffe, A. Hydrophilic Protein Associated with Desiccation Tolerance Exhibits Broad Protein Stabilization Function. Proc. Natl. Acad. Sci. USA 2007, 104, 18073–18078. [Google Scholar] [CrossRef]
- Schaffert, L.N.; Carter, W.G. Do Post-Translational Modifications Influence Protein Aggregation in Neurodegenerative Diseases: A Systematic Review. Brain Sci. 2020, 10, 232. [Google Scholar] [CrossRef]
- Turner, A.L.; Watson, M.; Wilkins, O.G.; Cato, L.; Travers, A.; Thomas, J.O.; Stott, K. Highly Disordered Histone H1−DNA Model Complexes and Their Condensates. Proc. Natl. Acad. Sci. USA 2018, 115, 11964–11969. [Google Scholar] [CrossRef]
- Wang, Q.; Li, Z.; Zhang, S.; Li, Y.; Wang, Y.; Fang, Z.; Ma, Y.; Liu, Z.; Zhang, W.; Li, D.; et al. Global Profiling of Arginine Dimethylation in Regulating Protein Phase Separation by a Steric Effect-Based Chemical-Enrichment Method. Proc. Natl. Acad. Sci. USA 2022, 119, e2205255119. [Google Scholar] [CrossRef]
- Macek, B.; Forchhammer, K.; Hardouin, J.; Weber-Ban, E.; Grangeasse, C.; Mijakovic, I. Protein Post-Translational Modifications in Bacteria. Nat. Rev. Microbiol. 2019, 17, 651–664. [Google Scholar] [CrossRef]
- Mijakovic, I.; Petranovic, D.; Macek, B.; Cepo, T.; Mann, M.; Davies, J.; Jensen, P.R.; Vujaklija, D. Bacterial Single-Stranded DNA-Binding Proteins Are Phosphorylated on Tyrosine. Nucleic Acids Res. 2006, 34, 1588–1596. [Google Scholar] [CrossRef]
- Szoke, T.; Albocher, N.; Govindarajan, S.; Nussbaum-Shochat, A.; Amster-Choder, O. Tyrosine Phosphorylation-Dependent Localization of TmaR That Controls Activity of a Major Bacterial Sugar Regulator by Polar Sequestration. Proc. Natl. Acad. Sci. USA 2021, 118, e2016017118. [Google Scholar] [CrossRef]
- Lima, B.P.; Antelmann, H.; Gronau, K.; Chi, B.K.; Becher, D.; Brinsmade, S.R.; Wolfe, A.J. Involvement of Protein Acetylation in Glucose-Induced Transcription of a Stress-Responsive Promoter. Mol. Microbiol. 2011, 81, 1190–1204. [Google Scholar] [CrossRef]
- Zhang, J.; Sprung, R.; Pei, J.; Tan, X.; Kim, S.; Zhu, H.; Liu, C.F.; Grishin, N.V.; Zhao, Y. Lysine Acetylation Is a Highly Abundant and Evolutionarily Conserved Modification in Escherichia coli. Mol. Cell. Proteom. 2009, 8, 215–225. [Google Scholar] [CrossRef]
- Kuczyńska-Wiśnik, D.; Moruno-Algara, M.; Stojowska-Swȩdrzyńska, K.; Laskowska, E. The Effect of Protein Acetylation on the Formation and Processing of Inclusion Bodies and Endogenous Protein Aggregates in Escherichia Coli Cells. Microb. Cell Fact. 2016, 15, 189. [Google Scholar] [CrossRef]
- Patel, A.; Lee, H.O.; Jawerth, L.; Maharana, S.; Jahnel, M.; Hein, M.Y.; Stoynov, S.; Mahamid, J.; Saha, S.; Franzmann, T.M.; et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 2015, 162, 1066–1077. [Google Scholar] [CrossRef]
- Ray, S.; Singh, N.; Kumar, R.; Patel, K.; Pandey, S.; Datta, D.; Mahato, J.; Panigrahi, R.; Navalkar, A.; Mehra, S.; et al. α-Synuclein Aggregation Nucleates through Liquid–Liquid Phase Separation. Nat. Chem. 2020, 12, 705–716. [Google Scholar] [CrossRef]
- Gui, X.; Feng, S.; Li, Z.; Li, Y.; Reif, B.; Shi, B.; Niu, Z. Liquid–Liquid Phase Separation of Amyloid-β Oligomers Modulates Amyloid Fibrils Formation. J. Biol. Chem. 2023, 299, 102926. [Google Scholar] [CrossRef]
- Pytowski, L.; Fan Lee, C.; Foley, A.C.; Vaux, D.J.; Jean, L. Liquid-Liquid Phase Separation of Type II Diabetes-Associated IAPP Initiates Hydrogelation and Aggregation. Proc. Natl. Acad. Sci. USA 2020, 11, 12050–12061. [Google Scholar] [CrossRef]
- Sutter, M.; Greber, B.; Aussignargues, C.; Kerfeld, C.A. Assembly Principles and Structure of a 6.5-MDa Bacterial Microcompartment Shell. Science 2017, 356, 1293–1297. [Google Scholar] [CrossRef]
- Bresan, S.; Sznajder, A.; Hauf, W.; Forchhammer, K.; Pfeiffer, D.; Jendrossek, D. Polyhydroxyalkanoate (PHA) Granules Have No Phospholipids. Sci. Rep. 2016, 6, 26612. [Google Scholar] [CrossRef]
- Renner, L.D.; Weibel, D.B. Cardiolipin Microdomains Localize to Negatively Curved Regions of Escherichia Coli Membranes. Proc. Natl. Acad. Sci. USA 2011, 108, 6264–6269. [Google Scholar] [CrossRef]
- Ladouceur, A.-M.; Singh Parmar, B.; Biedzinski, S.; Wall, J.; Graydon Tope, S.; Cohn, D.; Kim, A.; Soubry, N.; Reyes-Lamothe, R.; Weber, S.C.; et al. Clusters of Bacterial RNA Polymerase Are Biomolecular Condensates That Assemble through Liquid-Liquid Phase Separation Proc. Natl. Acad. Sci. USA 2020, 117, 18540–18549. [Google Scholar] [CrossRef]
- Guilhas, B.; Walter, J.C.; Rech, J.; David, G.; Walliser, N.O.; Palmeri, J.; Mathieu-Demaziere, C.; Parmeggiani, A.; Bouet, J.Y.; Le Gall, A.; et al. ATP-Driven Separation of Liquid Phase Condensates in Bacteria. Mol. Cell 2020, 79, 293–303. [Google Scholar] [CrossRef]
- Monterroso, B.; Zorrilla, S.; Sobrinos-Sanguino, M.; Robles-Ramos, M.A.; López-Álvarez, M.; Margolin, W.; Keating, C.D.; Rivas, G. Bacterial FtsZ Protein Forms Phase-separated Condensates with Its Nucleoid-associated Inhibitor SlmA. EMBO Rep. 2019, 20, e45946. [Google Scholar] [CrossRef]
- Gupta, A.; Joshi, A.; Arora, K.; Mukhopadhyay, S.; Guptasarma, P. The Bacterial Nucleoid-Associated Proteins, HU and Dps, Condense DNA into Context-Dependent Biphasic or Multiphasic Complex Coacervates. J. Biol. Chem. 2023, 299, 104637. [Google Scholar] [CrossRef]
- Harami, G.M.; Kovács, Z.J.; Pancsa, R.; Pálinkás, J.; Baráth, V.; Tárnok, K.; Málnási-Csizmadia, A.; Kovács, M. Phase Separation by SsDNA Binding Protein Controlled via Protein−protein and Protein−DNA Interactions. Proc. Natl. Acad. Sci. USA 2020, 117, 26206–26217. [Google Scholar] [CrossRef]
- Antony, E.; Lohman, T.M. Dynamics of E. Coli Single Stranded DNA Binding (SSB) Protein-DNA Complexes. Semin. Cell Dev. Biol. 2019, 86, 102–111. [Google Scholar] [CrossRef]
- Orban, K.; Finkel, S.E. Dps Is a Universally Conserved Dual-Action DNA-Binding and Ferritin Protein. J. Bacteriol. 2022, 204, e0003622. [Google Scholar] [CrossRef]
- Maisonneuve, E.; Ezraty, B.; Dukan, S. Protein Aggregates: An Aging Factor Involved in Cell Death. J. Bacteriol. 2008, 190, 6070–6075. [Google Scholar] [CrossRef]
- Kucharczyk, K.; Laskowska, E.; Taylor, A. Response of Escherichia coli Cell Membranes to Induction of X D857 Prophage by Heat Shock. Mol. Microbiol. 1991, 5, 2935–2945. [Google Scholar] [CrossRef]
- Kwiatkowska, J.; Matuszewska, E.; Kuczyńska-Wiśnik, D.; Laskowska, E. Aggregation of Escherichia coli Proteins during Stationary Phase Depends on Glucose and Oxygen Availability. Res. Microbiol. 2008, 159, 651–657. [Google Scholar] [CrossRef]
- Schramm, F.D.; Schroeder, K.; Jonas, K. Protein Aggregation in Bacteria. FEMS Microbiol. Rev. 2019, 44, 54–72. [Google Scholar] [CrossRef]
- Fernandez-Escamilla, A.M.; Rousseau, F.; Schymkowitz, J.; Serrano, L. Prediction of Sequence-Dependent and Mutational Effects on the Aggregation of Peptides and Proteins. Nat. Biotechnol. 2004, 22, 1302–1306. [Google Scholar] [CrossRef]
- Singh, A.; Upadhyay, V.; Singh, A.; Panda, A.K. Structure-Function Relationship of Inclusion Bodies of a Multimeric Protein. Front. Microbiol. 2020, 11, 876. [Google Scholar] [CrossRef]
- García-Fruitõs, E.; Sabate, R.; De Groot, N.S.; Villaverde, A.; Ventura, S. Biological Role of Bacterial Inclusion Bodies: A Model for Amyloid Aggregation. FEBS J. 2011, 278, 2419–2427. [Google Scholar] [CrossRef]
- Ganesan, A.; Debulpaep, M.; Wilkinson, H.; Van Durme, J.; De Baets, G.; Jonckheere, W.; Ramakers, M.; Ivarsson, Y.; Zimmermann, P.; Van Eldere, J.; et al. Selectivity of Aggregation-Determining Interactions. J. Mol. Biol. 2015, 427, 236–247. [Google Scholar] [CrossRef]
- Bednarska, N.G.; van Eldere, J.; Gallardo, R.; Ganesan, A.; Ramakers, M.; Vogel, I.; Baatsen, P.; Staes, A.; Goethals, M.; Hammarström, P.; et al. Protein Aggregation as an Antibiotic Design Strategy. Mol. Microbiol. 2016, 99, 849–865. [Google Scholar] [CrossRef]
- Khodaparast, L.; Khodaparast, L.; Gallardo, R.; Louros, N.N.; Michiels, E.; Ramakrishnan, R.; Ramakers, M.; Claes, F.; Young, L.; Shahrooei, M.; et al. Aggregating Sequences That Occur in Many Proteins Constitute Weak Spots of Bacterial Proteostasis. Nat. Commun. 2018, 9, 1–15. [Google Scholar] [CrossRef]
- Hipp, M.S.; Kasturi, P.; Hartl, F.U. The Proteostasis Network and Its Decline in Ageing. Nat. Rev. Mol. Cell Biol. 2019, 20, 421–435. [Google Scholar] [CrossRef]
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular Chaperones in Protein Folding and Proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef]
- Winkler, J.; Seybert, A.; König, L.; Pruggnaller, S.; Haselmann, U.; Sourjik, V.; Weiss, M.; Frangakis, A.S.; Mogk, A.; Bukau, B. Quantitative and Spatio-Temporal Features of Protein Aggregation in Escherichia coli and Consequences on Protein Quality Control and Cellular Ageing. EMBO J. 2010, 29, 910–923. [Google Scholar] [CrossRef]
- Lindner, A.B.; Madden, R.; Demarez, A.; Stewart, E.J.; Taddei, F. Asymmetric Segregation of Protein Aggregates Is Associated with Cellular Aging and Rejuvenation. Proc. Natl. Acad. Sci. USA 2008, 26, 3076–3081. [Google Scholar] [CrossRef]
- Rokney, A.; Shagan, M.; Kessel, M.; Smith, Y.; Rosenshine, I.; Oppenheim, A.B. E. Coli Transports Aggregated Proteins to the Poles by a Specific and Energy-Dependent Process. J. Mol. Biol. 2009, 392, 589–601. [Google Scholar] [CrossRef]
- Kuczyńska-Wiśnik, D.; Żurawa-Janicka, D.; Narkiewicz, J.; Kwiatkowska, J.; Lipińska, B.; Laskowska, E. Escherichia coli Small Heat Shock Proteins IbpA/B Enhance Activity of Enzymes Sequestered in Inclusion Bodies. Acta Biochim. Pol. 2004, 51, 925–931. [Google Scholar]
- García-Fruitós, E.; González-Montalbán, N.; Morell, M.; Vera, A.; Ferraz, R.M.; Arís, A.; Ventura, S.; Villaverde, A. Aggregation as Bacterial Inclusion Bodies Does Not Imply Inactivation of Enzymes and Fluorescent Proteins. Microb. Cell Fact. 2005, 4, 27. [Google Scholar] [CrossRef]
- García-Fruitós, E.; Arís, A.; Villaverde, A. Localization of Functional Polypeptides in Bacterial Inclusion Bodies. Appl. Environ. Microbiol. 2007, 73, 289–294. [Google Scholar] [CrossRef]
- Leszczynska, D.; Matuszewska, E.; Kuczynska-Wisnik, D.; Furmanek-Blaszk, B.; Laskowska, E. The Formation of Persister Cells in Stationary-Phase Cultures of Escherichia coli Is Associated with the Aggregation of Endogenous Proteins. PLoS ONE 2013, 8, e54737. [Google Scholar] [CrossRef]
- Gollan, B.; Grabe, G.; Michaux, C.; Helaine, S.; ARjatscls, H. Annual Review of Microbiology Bacterial Persisters and Infection: Past, Present, and Progressing. Annu. Rev. Microbiol. 2019, 73, 359–385. [Google Scholar] [CrossRef]
- Bollen, C.; Dewachter, L.; Michiels, J. Protein Aggregation as a Bacterial Strategy to Survive Antibiotic Treatment. Front. Mol. Biosci. 2021, 8. [Google Scholar] [CrossRef]
- Balaban, N.Q.; Helaine, S.; Lewis, K.; Ackermann, M.; Aldridge, B.; Andersson, D.I.; Brynildsen, M.P.; Bumann, D.; Camilli, A.; Collins, J.J.; et al. Definitions and Guidelines for Research on Antibiotic Persistence. Nat. Rev. Microbiol. 2019, 17, 441–448. [Google Scholar] [CrossRef]
- Bollen, C.; Louwagie, E.; Verstraeten, N.; Michiels, J.; Ruelens, P. Environmental, Mechanistic and Evolutionary Landscape of Antibiotic Persistence. EMBO Rep. 2023, 23, e57309. [Google Scholar] [CrossRef]
- Patel, A.; Malinovska, L.; Saha, S.; Wang, J.; Alberti, S.; Krishnan, Y.; Hyman, A.A. ATP as a Biological Hydrotrope. Science 2017, 19, 753–756. [Google Scholar] [CrossRef]
- Oates, M.E.; Romero, P.; Ishida, T.; Ghalwash, M.; Mizianty, M.J.; Xue, B.; Dosztányi, Z.; Uversky, V.N.; Obradovic, Z.; Kurgan, L.; et al. D2P2: Database of Disordered Protein Predictions. Nucleic Acids Res. 2013, 41, D508–D516. [Google Scholar] [CrossRef]
- Iannuzzi, C.; Irace, G.; Sirangelo, I. Differential Effects of Glycation on Protein Aggregation and Amyloid Formation. Front. Mol. Biosci. 2014, 1, 9. [Google Scholar] [CrossRef]
- Fournet, M.; Bonté, F.; Desmoulière, A. Glycation Damage: A Possible Hub for Major Pathophysiological Disorders and Aging. Aging Dis. 2018, 9, 880–900. [Google Scholar] [CrossRef]
- Rabbani, N.; Thornalley, P.J. Protein Glycation—Biomarkers of Metabolic Dysfunction and Early-Stage Decline in Health in the Era of Precision Medicine. Redox Biol. 2021, 42, 101920. [Google Scholar] [CrossRef]
- Shiraki, K.; Mimura, M.; Nishinami, S.; Ura, T. Effect of Additives on Liquid Droplets and Aggregates of Proteins. Biophys. Rev. 2020, 12, 587–592. [Google Scholar] [CrossRef]
- Hatos, A.; Tosatto, S.C.E.; Vendruscolo, M.; Fuxreiter, M. FuzDrop on AlphaFold: Visualizing the Sequence-Dependent Propensity of Liquid-Liquid Phase Separation and Aggregation of Proteins. Nucleic Acids Res. 2022, 50, W337–W344. [Google Scholar] [CrossRef]
- Chu, X.; Sun, T.; Li, Q.; Xu, Y.; Zhang, Z.; Lai, L.; Pei, J. Prediction of Liquid–Liquid Phase Separating Proteins Using Machine Learning. BMC Bioinform. 2022, 23, 1–13. [Google Scholar] [CrossRef]
- Moruno Algara, M.; Kuczyńska-Wiśnik, D.; Dębski, J.; Stojowska-Swędrzyńska, K.; Sominka, H.; Bukrejewska, M.; Laskowska, E. Trehalose Protects Escherichia coli against Carbon Stress Manifested by Protein Acetylation and Aggregation. Mol. Microbiol. 2019, 112, 866–880. [Google Scholar] [CrossRef]
- Lancaster, A.K.; Nutter-Upham, A.; Lindquist, S.; King, O.D. PLAAC: A Web and Command-Line Application to Identify Proteins with Prion-like Amino Acid Composition. Bioinformatics 2014, 30, 2501–2502. [Google Scholar] [CrossRef]
- Uversky, V.N.; Kurgan, L. Overview Update: Computational Prediction of Intrinsic Disorder in Proteins. Curr. Protoc. 2023, 3, e802. [Google Scholar] [CrossRef]
- Fonseca, F.; Meneghel, J.; Cenard, S.; Passot, S.; Morris, G.J. Determination of Intracellular Vitrification Temperatures for Unicellular Micro Organisms under Conditions Relevant for Cryopreservation. PLoS ONE 2016, 11, e0152939. [Google Scholar] [CrossRef]
- Parry, B.R.; Surovtsev, I.V.; Cabeen, M.T.; O’Hern, C.S.; Dufresne, E.R.; Jacobs-Wagner, C. The Bacterial Cytoplasm Has Glass-like Properties and Is Fluidized by Metabolic Activity. Cell 2014, 156, 183–194. [Google Scholar] [CrossRef]
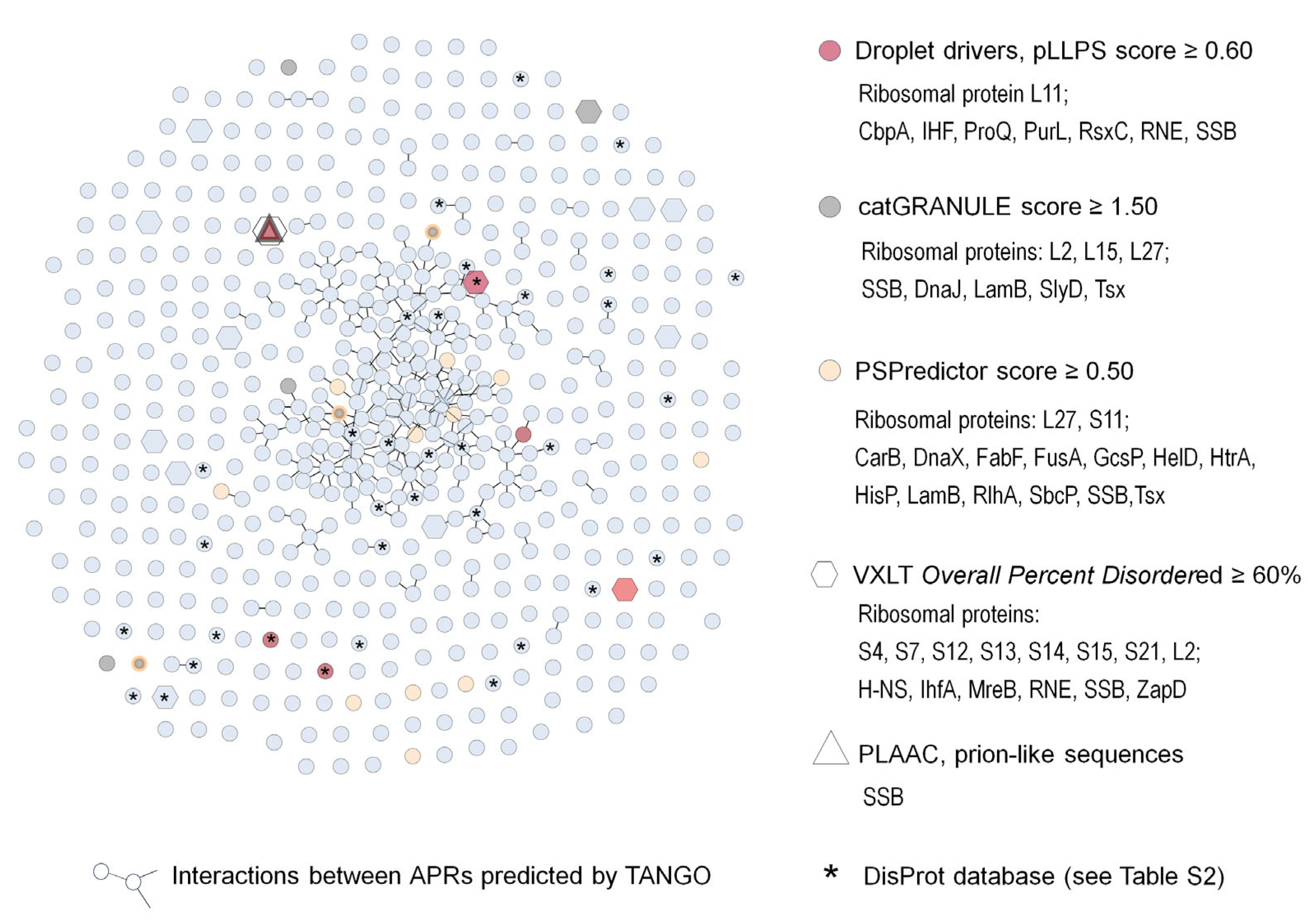
| Species | Conditions Inducing Aggregation | Protection Against | Proposed Mechanism of Protection | Comments | Ref. |
|---|---|---|---|---|---|
| E. coli | Sublethal heat stress, hydrogen peroxide, streptomycin | More severe heat shock | Induction of protein quality control components | “Memory” aggregates | [15] |
| A. baumannii | Desiccation, streptomycin, Δlon mutation | Desiccation | Protection of sequestered proteins | Preserved activity of a model enzyme | [17] |
| E. coli | Stationary phase | Antibiotics | Dormancy | [84] | |
| E. coli and other Gram-negative species | Stationary phase, heat shock, streptomycin, hydrogen peroxide | Antibiotics | Dormancy | MLOs | [16,18] |
| E. coli | Desiccation–rehydration | Desiccation–rehydration stress | Protection of sequestered proteins | Contain LLPS-prone proteins and IDPs | [19] |
| ID | Protein Names | emPAI % | Abundance PaxDb (ppm) | catGranule | FuzDrop pLLPS | PSPredictor | |
|---|---|---|---|---|---|---|---|
| 1 | P0AG51 | 50S ribosomal protein L30 | 2.88 | 6056 | −0.970 | 0.117 | 0.005 |
| 2 | P02413 | 50S ribosomal protein L15 | 2.82 | 3541 | 2.075 | 0.514 | 0.285 |
| 3 | P62399 | 50S ribosomal protein L5 | 2.79 | 5965 | 0.283 | 0.114 | 0.002 |
| 4 | P0CE47 | Elongation factor Tu 1 | 2.25 | 27,871 | 0.850 | 0.147 | 0.043 |
| 5 | P60422 | 50S ribosomal protein L2 | 1.60 | 1852 | 1.887 | 0.421 | 0.245 |
| 6 | P61175 | 50S ribosomal protein L22 | 1.55 | 6098 | −0.529 | 0.237 | 0.023 |
| 7 | P02359 | 30S ribosomal protein S7 | 1.32 | 10,524 | −0.038 | 0.310 | 0.004 |
| 8 | P0AG55 | 50S ribosomal protein L6 | 1.25 | 3285 | 0.829 | 0.179 | 0.063 |
| 9 | P0A7 × 3 | 30S ribosomal protein S9 | 1.23 | 1374 | 0.811 | 0.262 | 0.119 |
| 10 | P0ADY3 | 50S ribosomal protein L14 | 1.19 | 4074 | 0.585 | 0.139 | 0.094 |
| 11 | P0A7V0 | 30S ribosomal protein S2 | 1.18 | 3366 | 0.411 | 0.255 | 0.051 |
| 12 | P0A7W7 | 30S ribosomal protein S8 | 1.09 | 2813 | 0.038 | 0.123 | 0.003 |
| 13 | P0A7M2 | 50S ribosomal protein L28 | 1.00 | 4610 | −0.283 | 0.142 | 0.013 |
| 14 | P0A7S9 | 30S ribosomal protein S13 | 1.00 | 6249 | 0.322 | 0.245 | 0.014 |
| 15 | P0A7V8 | 30S ribosomal protein S4 | 0.96 | 2549 | 0.690 | 0.178 | 0.003 |
| 16 | P0A7R9 | 30S ribosomal protein S11 | 0.90 | 735 | 0.545 | 0.170 | 0.595 |
| 17 | P68919 | 50S ribosomal protein L25 | 0.89 | 18,593 | −0.224 | 0.144 | 0.006 |
| 18 | P0A853 | Tryptophanase, TnaA | 0.89 | 830 | 0.538 | 0.141 | 0.005 |
| 19 | P0ABT2 | Dps | 0.89 | 7698 | −0.527 | 0.126 | 0.012 |
| 20 | P60438 | 50S ribosomal protein L3 | 0.88 | 5500 | 1.392 | 0.235 | 0.103 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuczyńska-Wiśnik, D.; Stojowska-Swędrzyńska, K.; Laskowska, E. Liquid–Liquid Phase Separation and Protective Protein Aggregates in Bacteria. Molecules 2023, 28, 6582. https://doi.org/10.3390/molecules28186582
Kuczyńska-Wiśnik D, Stojowska-Swędrzyńska K, Laskowska E. Liquid–Liquid Phase Separation and Protective Protein Aggregates in Bacteria. Molecules. 2023; 28(18):6582. https://doi.org/10.3390/molecules28186582
Chicago/Turabian StyleKuczyńska-Wiśnik, Dorota, Karolina Stojowska-Swędrzyńska, and Ewa Laskowska. 2023. "Liquid–Liquid Phase Separation and Protective Protein Aggregates in Bacteria" Molecules 28, no. 18: 6582. https://doi.org/10.3390/molecules28186582
APA StyleKuczyńska-Wiśnik, D., Stojowska-Swędrzyńska, K., & Laskowska, E. (2023). Liquid–Liquid Phase Separation and Protective Protein Aggregates in Bacteria. Molecules, 28(18), 6582. https://doi.org/10.3390/molecules28186582






