Tolyporphins–Exotic Tetrapyrrole Pigments in a Cyanobacterium—A Review
Abstract
“To know well the full biodiversity of Earth is not important simply to add figures to textbooks. The real purpose of science must be the original Linnaean goal: to find and take full account of each and every species of organism on Earth”.E. O. Wilson [1]
1. Introduction to a Singular Discovery
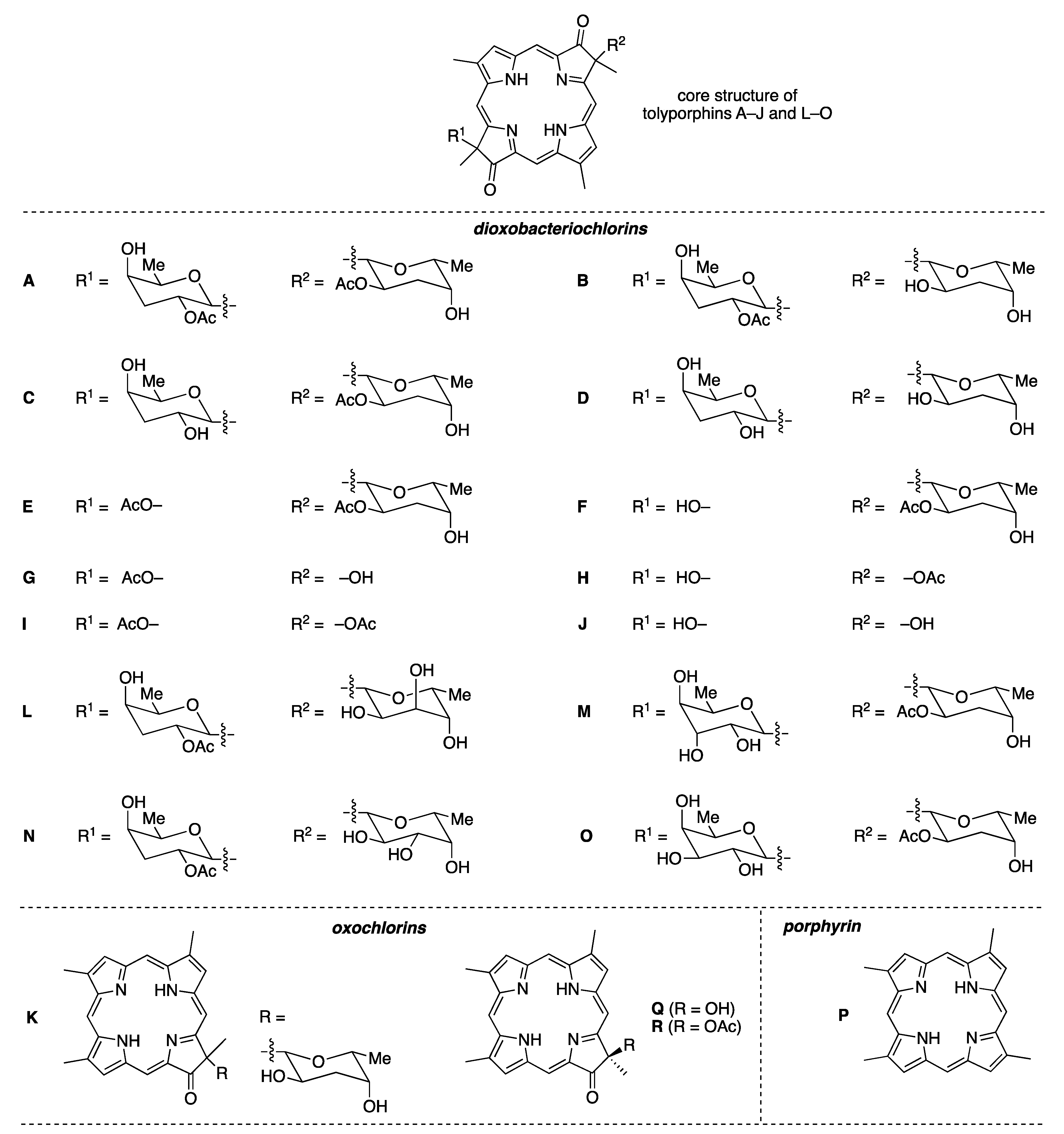
2. Structure and Diversity of Tolyporphins
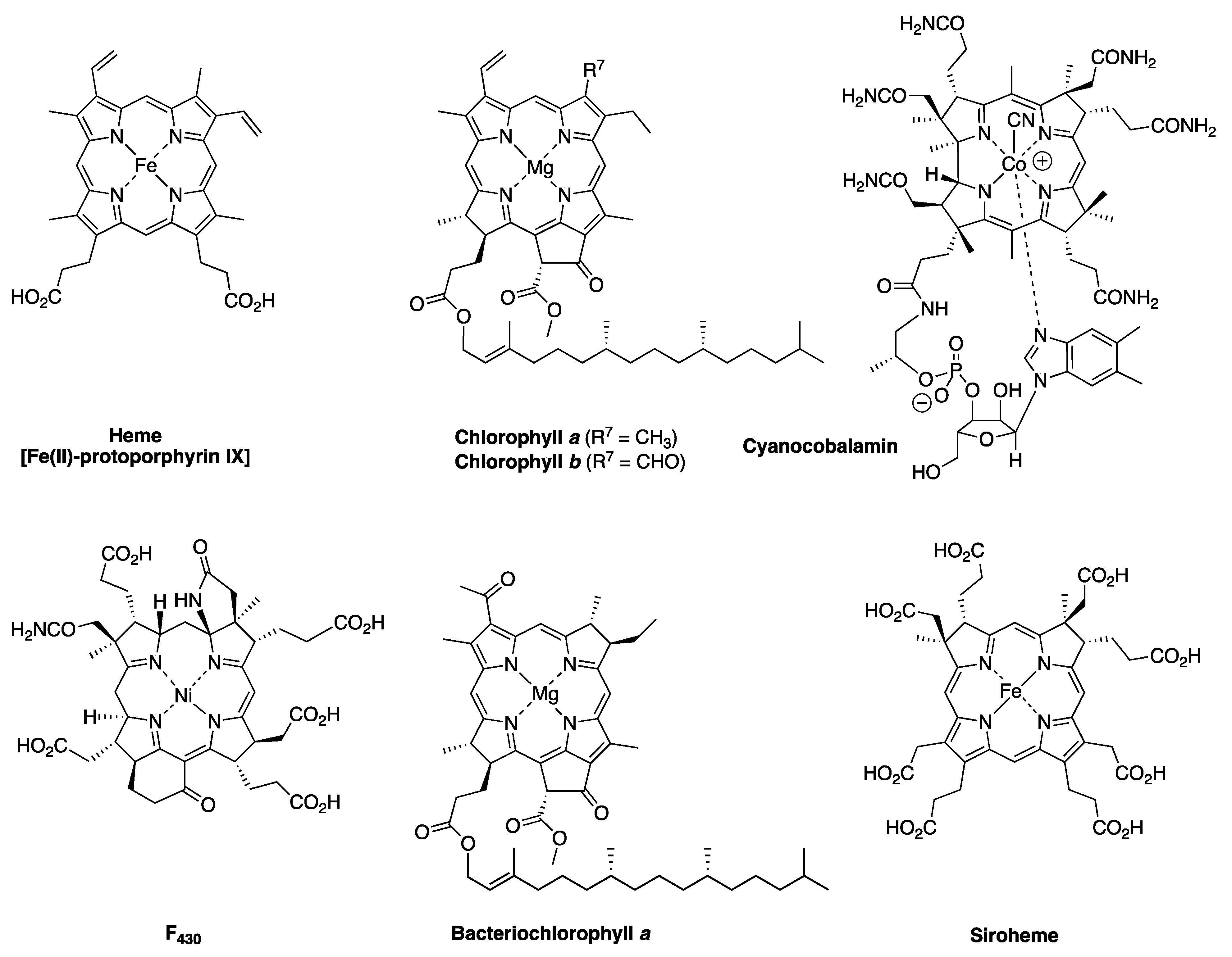
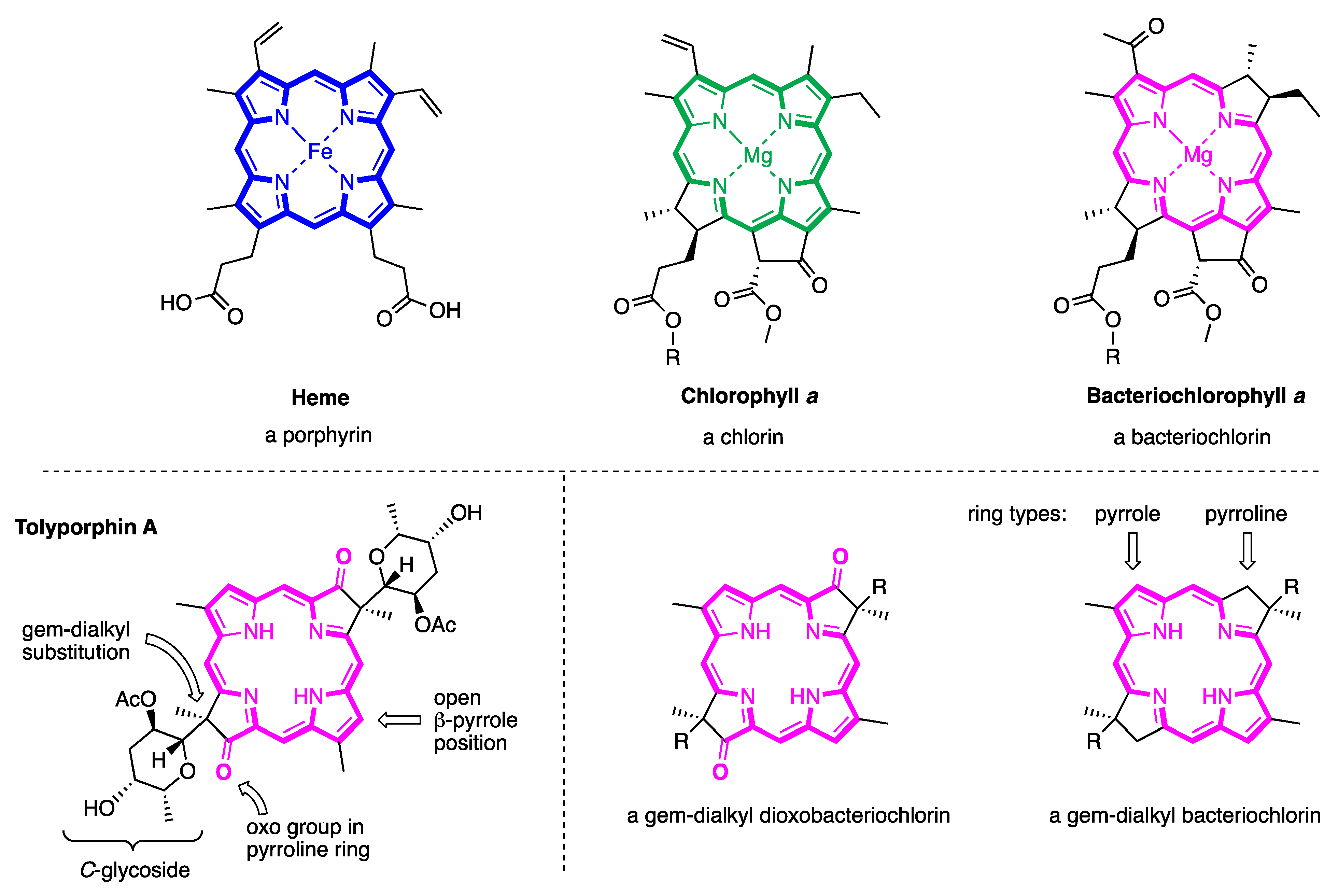
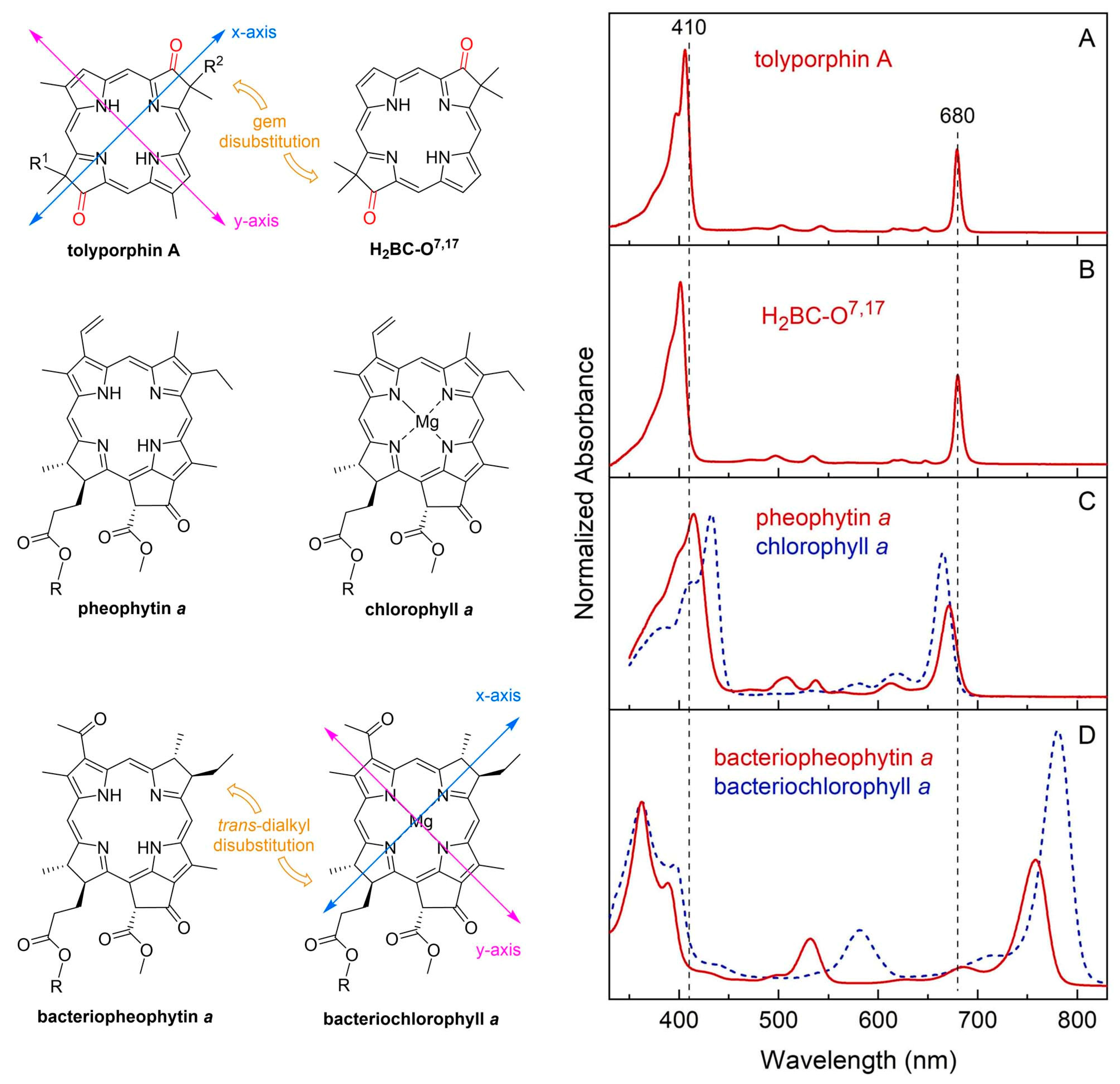
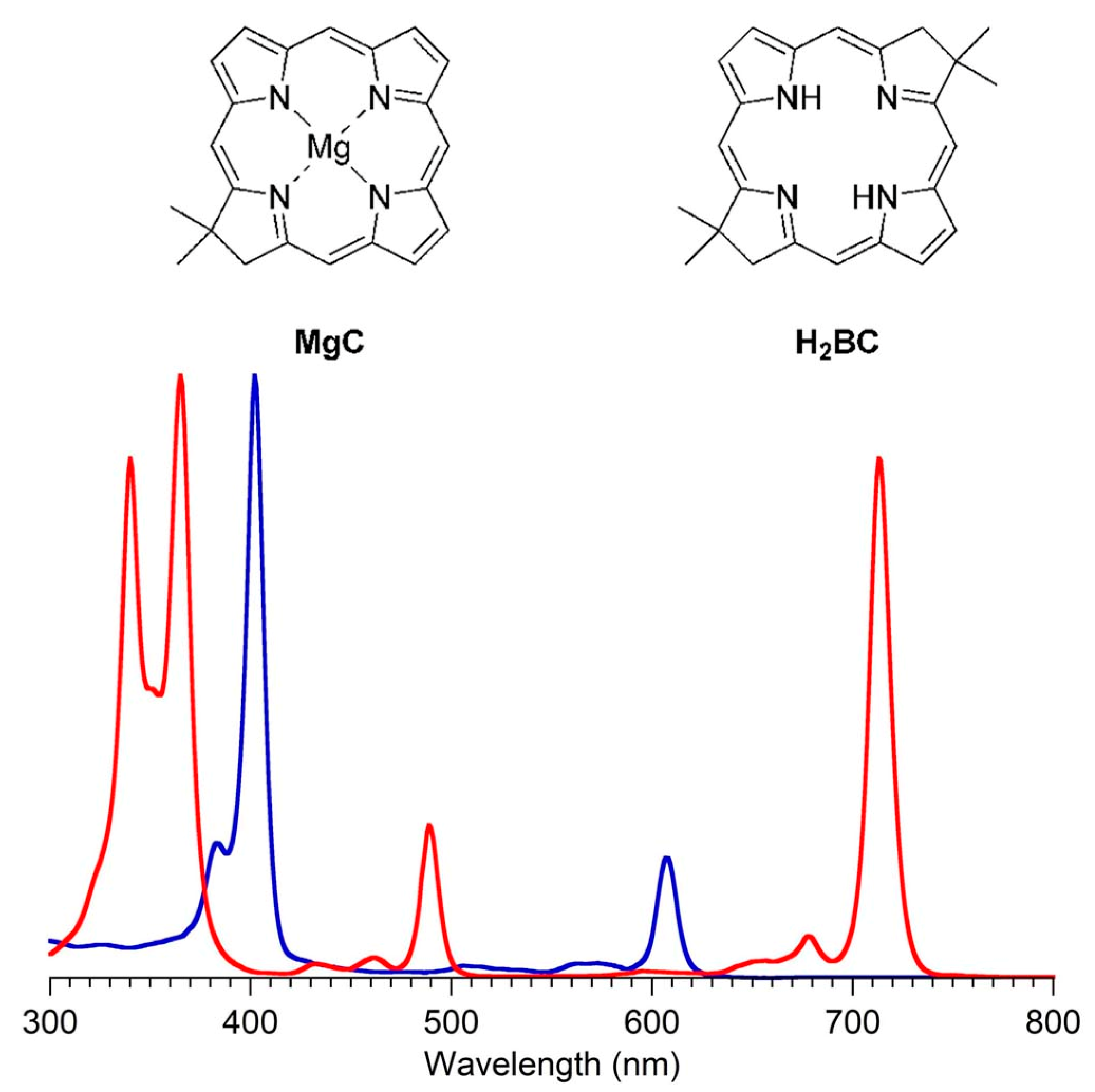
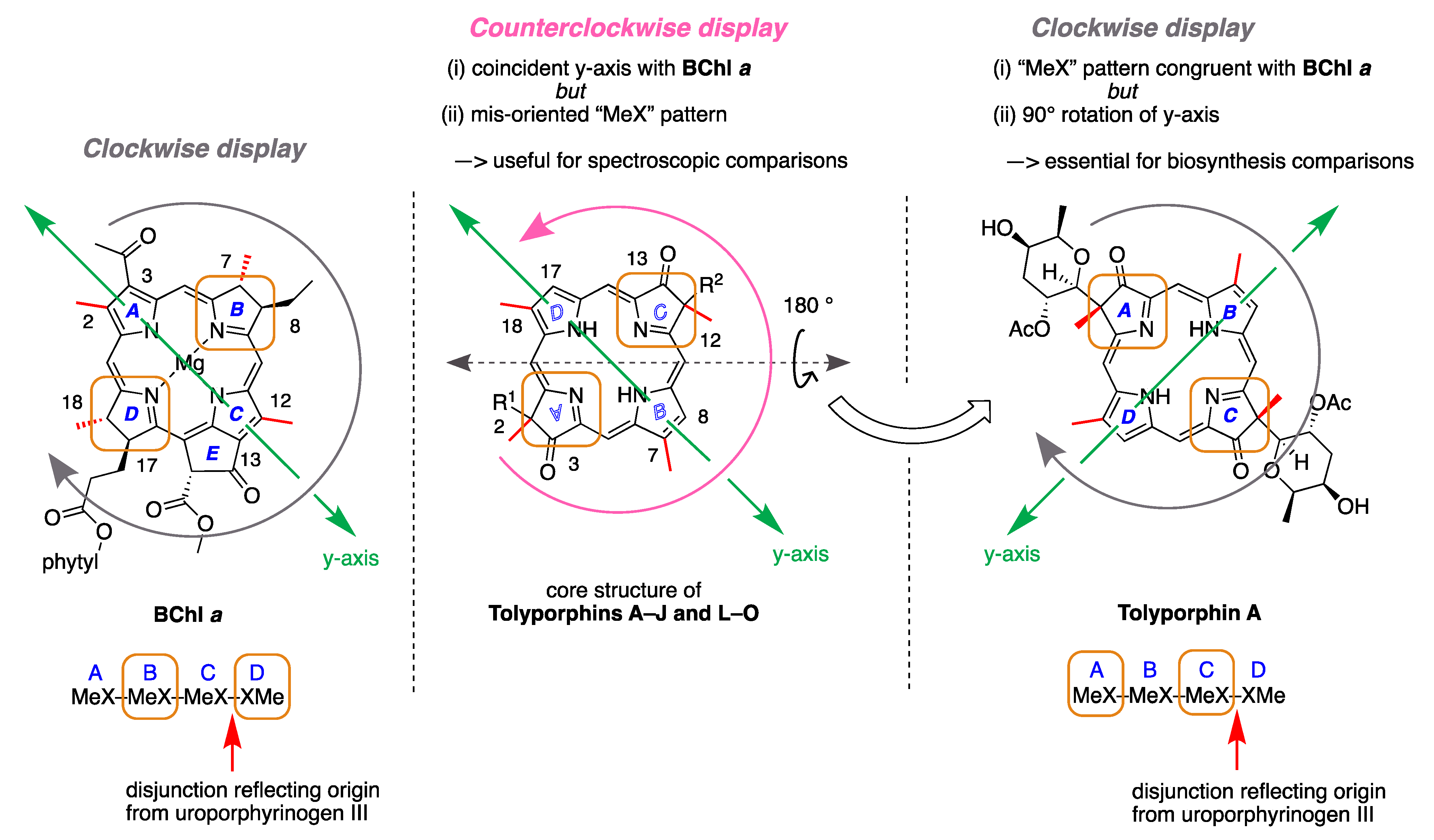
- First, tolyporphins A–J and L–O are bacteriochlorins; tolyporphins K, Q, and R are chlorins; and tolyporphin P is a porphyrin. The porphyrin, chlorin, and bacteriochlorin chromophores are highlighted using heme, chlorophyll a, and bacteriochlorophyll a as archetypes in Figure 7. The distinction between porphyrin, chlorin, and bacteriochlorin may seem simple, but the consequences are profound in terms of absorption spectra, biosynthesis, and physiological function.
- Second, the pyrroline ring (i.e., the reduced ring) in all tolyporphins (except P, which has no reduced ring) bears an oxo group and a flanking geminal disubstituted motif. Accordingly, tolyporphins A–J and L–O are dioxobacteriochlorins, and tolyporphins K, Q, and R are oxochlorins. The gem-disubstitution motif is distinct from the trans-dialkyl substitution motif of native photosynthetic tetrapyrroles—the members of the chlorophyll and bacteriochlorophyll family. The respective structural motifs in the pyrroline rings—gem-disubstitution versus trans-dialkyl—underpin a fundamental dichotomy between the two families of macrocycles.
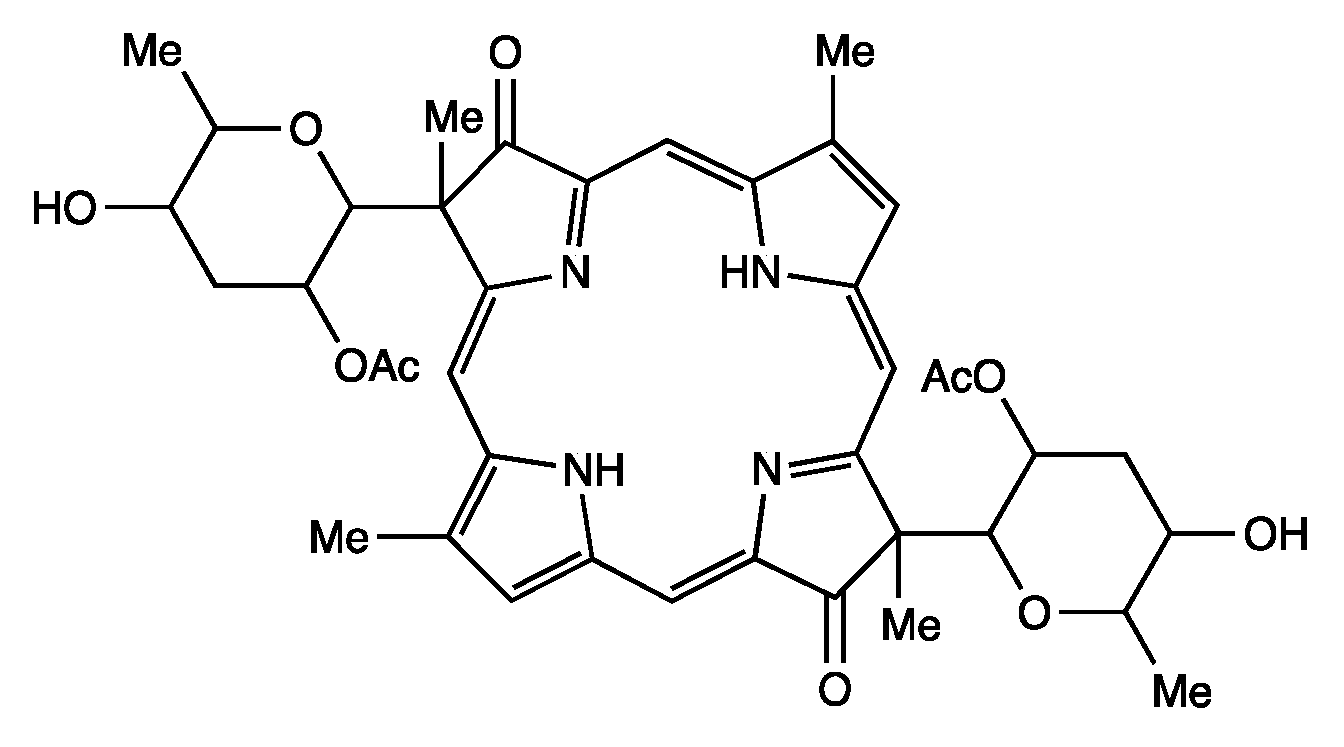
- Third, each geminal disubstituted motif contains one methyl group and one variable entity composed of a C-glycoside, O-acetyl, or OH group. The two C-glycosides, at least for tolyporphins A, E, and R, are known to be positioned on the same face of the macrocycle (Figure 8) [49], affording a Janus-like structure (methyl groups on one face, C-glycosides on the other). A similar diagram, illustrating the location of both C-glycosides on the same face of the tolyporphin A macrocycle, is shown by Smith et al. [21].
- Fourth, each pyrrole ring bears one unsubstituted (i.e., open) β-position, which is exceptionally rare among natural tetrapyrroles. By contrast, heme contains vinyl, methyl, or propionic acid groups at all eight β-pyrrolic positions. Chlorophylls at the six β-pyrrolic positions contain methyl, ethyl, β-ketoester, vinyl, acetyl, and formyl substituents.
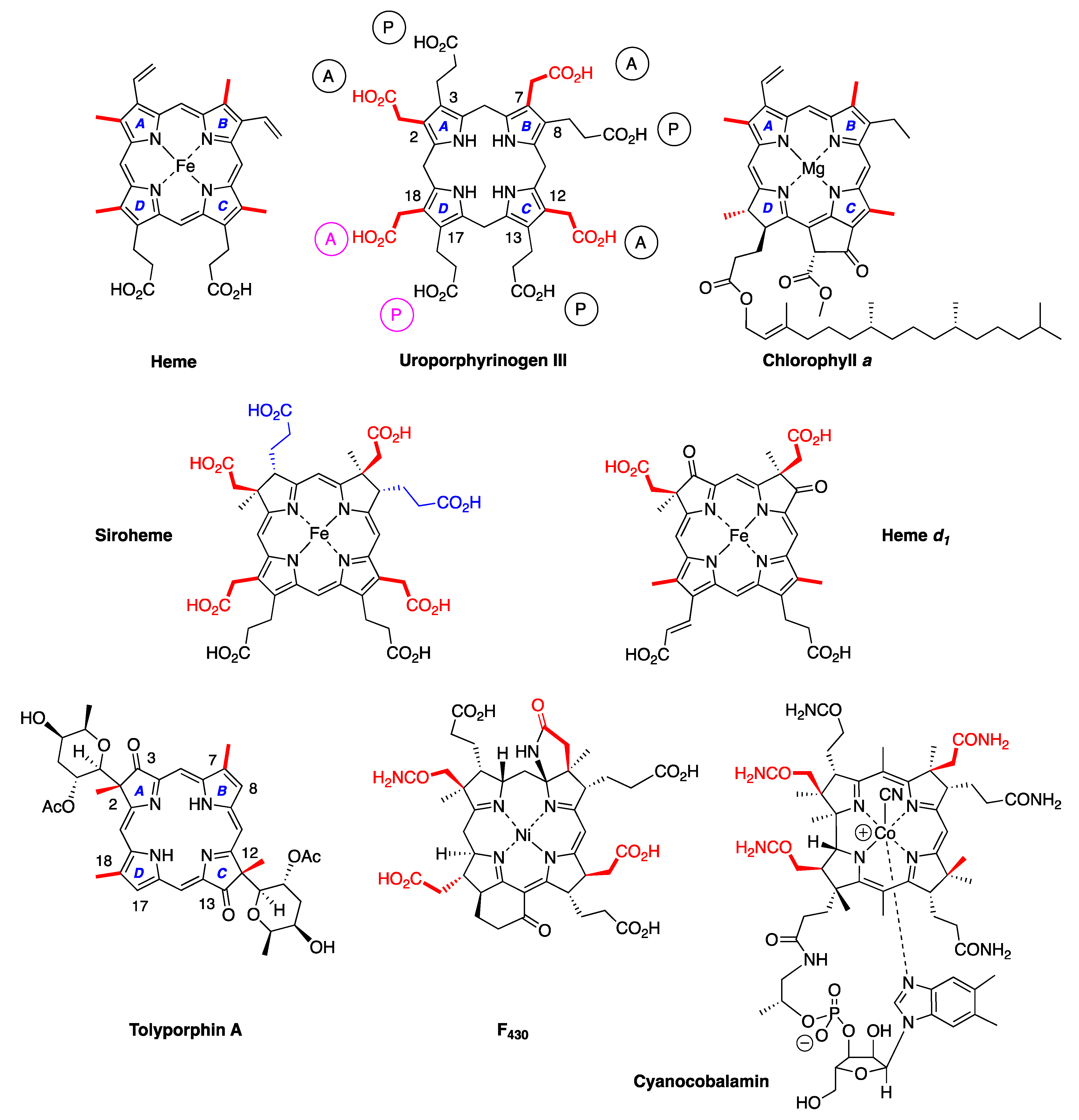
- Fifth, a metal is absent, whereas the vast majority of natural tetrapyrroles occur as the metal chelate: iron in heme, magnesium in the (bacterio)chlorophylls, cobalt in cobalamin, and nickel in F430.
3. Resuscitation of HT-58-2
4. Fascination with Tolyporphins
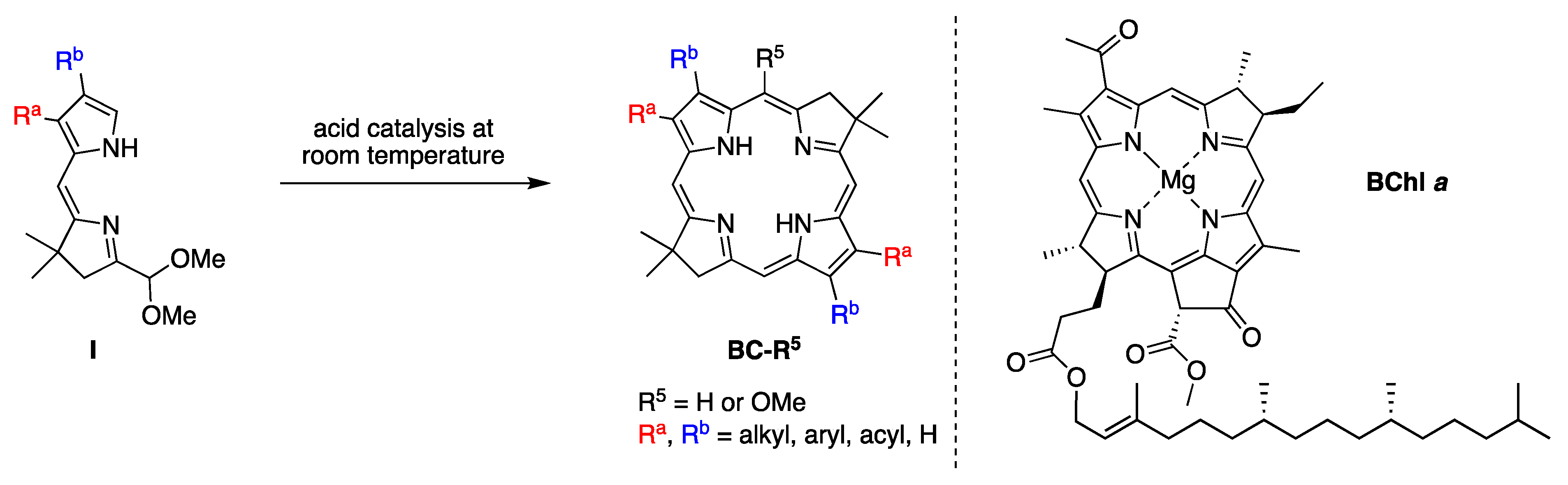
4.1. Gem-Dialkyl Motif in Hydroporphyrins
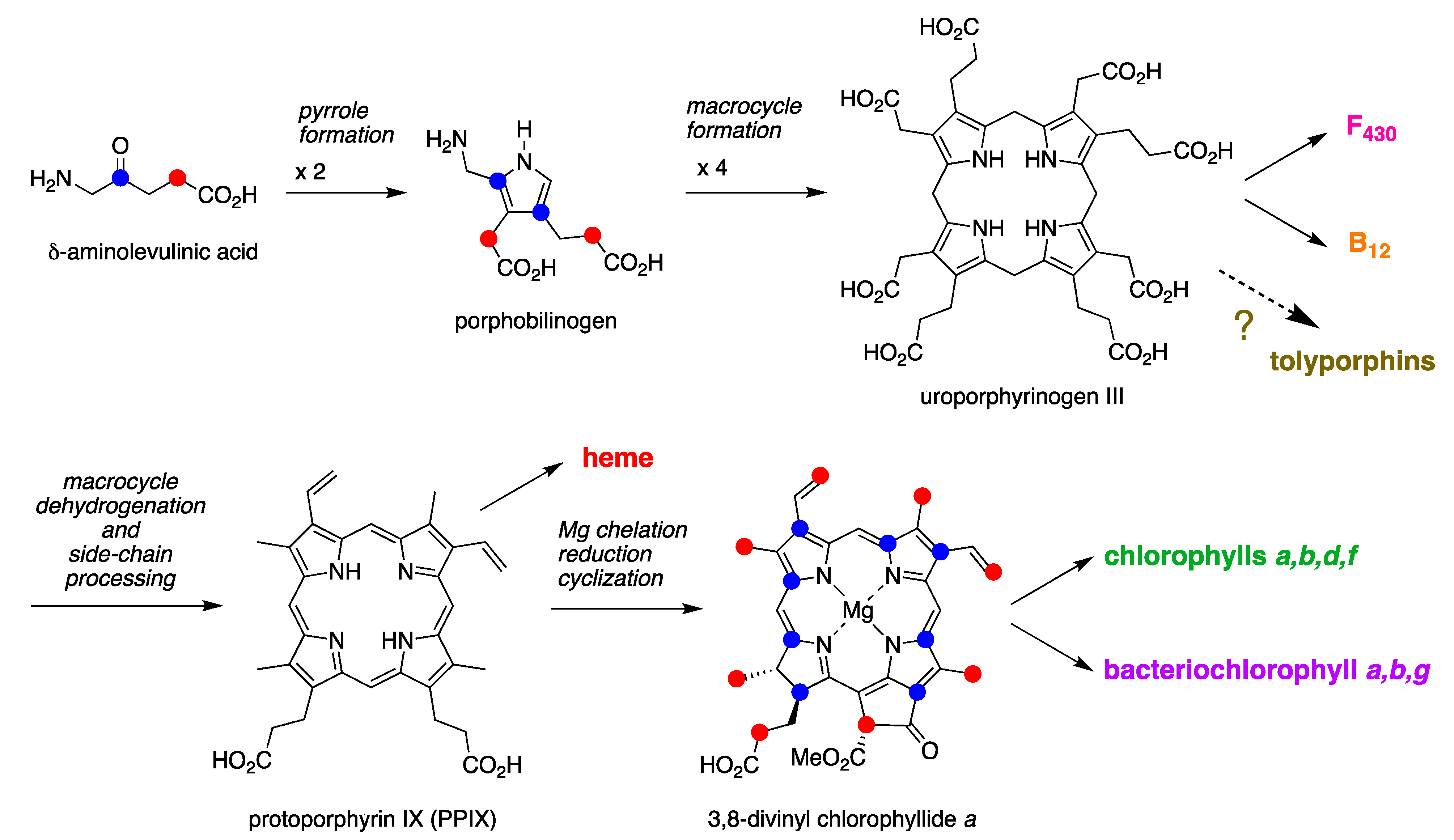
4.2. Uroporphyrinogen III and Tolyporphins
4.3. Questions
- (1)
- All tolyporphins display at least two open β-pyrrole positions, and most contain at least one gem-dialkyl motif. Could enzymes from the tolyporphins biosynthetic pathway be exploited in chemoenzymatic syntheses of sparsely substituted, gem-dialkyl-equipped tetrapyrrole macrocycles to create tailored target compounds [66]?
- (2)
- Tolyporphins collectively certainly might be regarded as un mélange bizarre—all like chlorophyll per provenance in a cyanobacterium, many part bacteriochlorophyll by virtue of the π-chromophore, many part chlorophyll on the basis of the absorption spectral properties (see Section 5), almost all part heme d1 given the gem-dialkyl groups, and many like other natural products given the appended C-glycosides with structural diversity therein. Could knowledge of the biosynthesis of tolyporphins deepen understanding of tetrapyrrole biosynthesis broadly, as well as perhaps shed light on the (evolutionary) dichotomy between gem-dialkylated hydroporphyrins (cobalamin, F430, siroheme, and heme d1) and trans-dialkylated hydroporphyrins (chlorophylls and bacteriochlorophylls) [66]? What can be said or learned about the origin and distribution of putative enzymes for removing propionic acid groups from tetrapyrrole macrocycles? If such questions can be addressed, the richness of the repertoire of tolyporphins may prove to be an invaluable scientific portal.
- (3)
- Cyanobacteria are known to produce abundant quantities of chlorophyll a, as required for photosynthesis, yet it is unprecedented for cyanobacteria to produce bacteriochlorin macrocycles of any type (14 of the 18 known tolyporphins are dioxobacteriochlorins). What are dioxobacteriochlorins doing, after all, in a photosynthetic bacterium [49]? An evolutionary model of the origin of natural molecular diversity [50,67,68] posits that [68] “most natural products will possess no biological activity of value to the producer and any biological activity found could well be fortuitous”. An ultimate finding that the existence of tolyporphins is nugatory with regards to physiological activity, for example, would not detract from the two aforementioned opportunities.
5. Photochemical Activity of Tolyporphin A
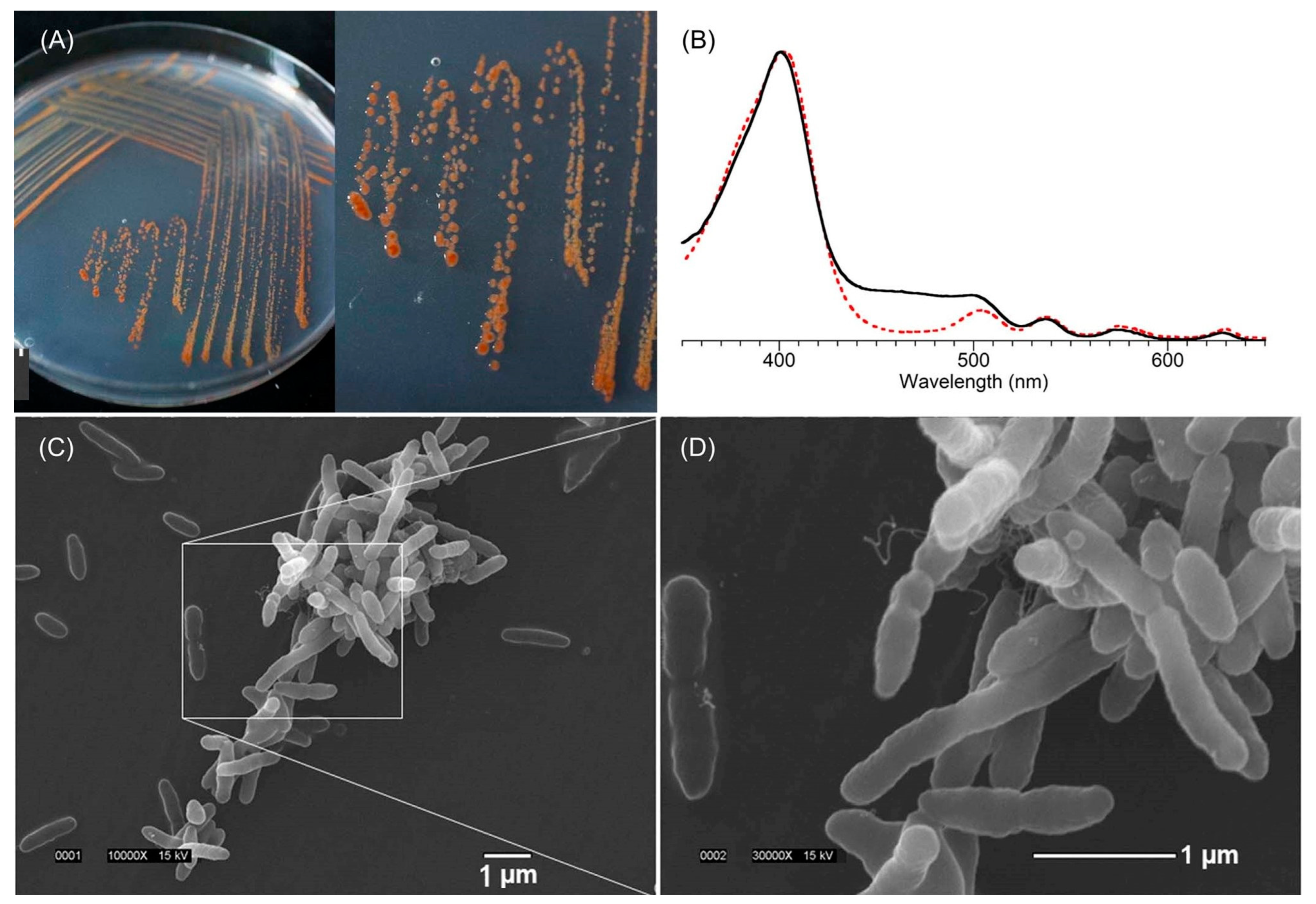
6. Biology of the Tolyporphins-Producing Culture
6.1. The Culture
- (1)
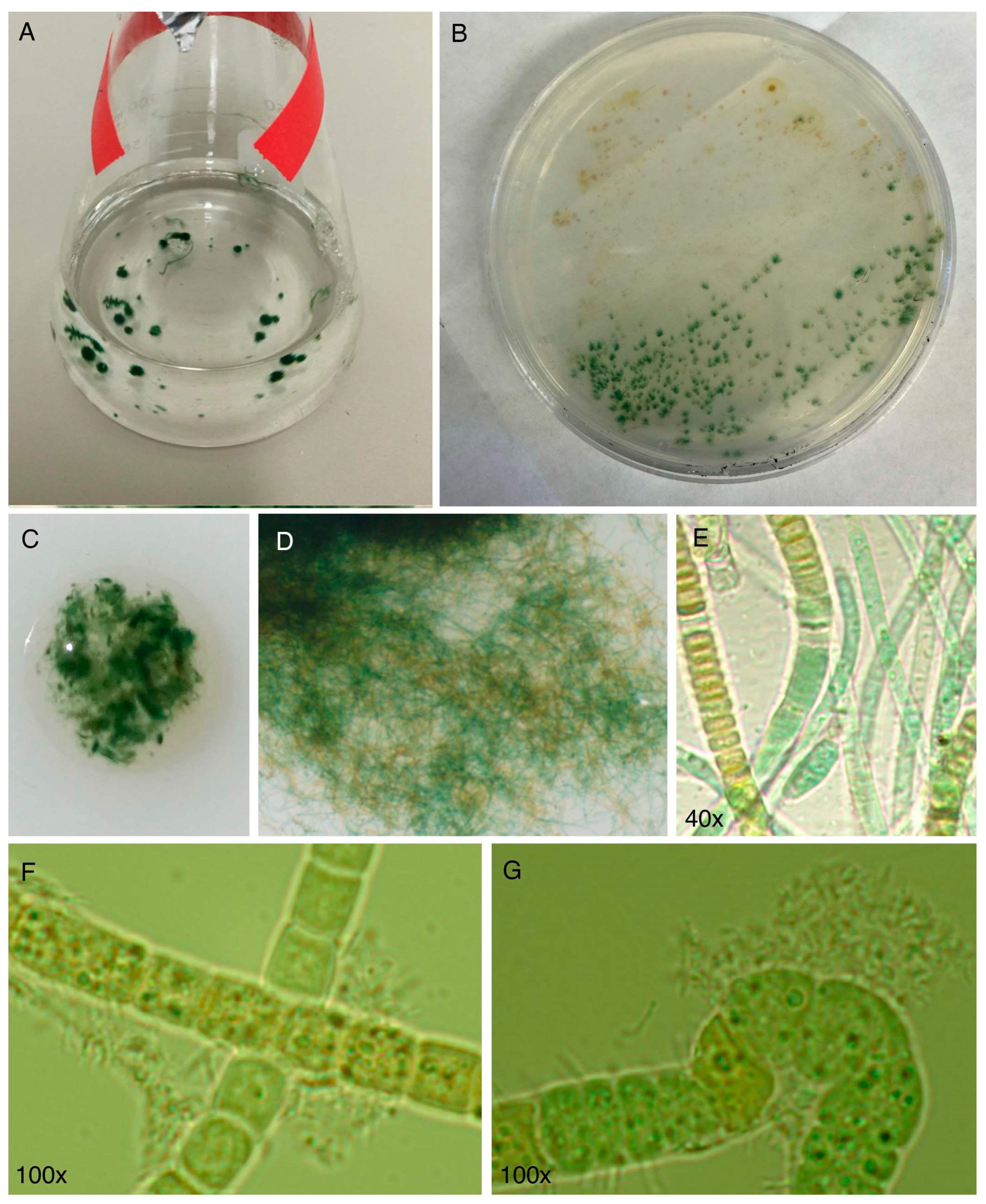
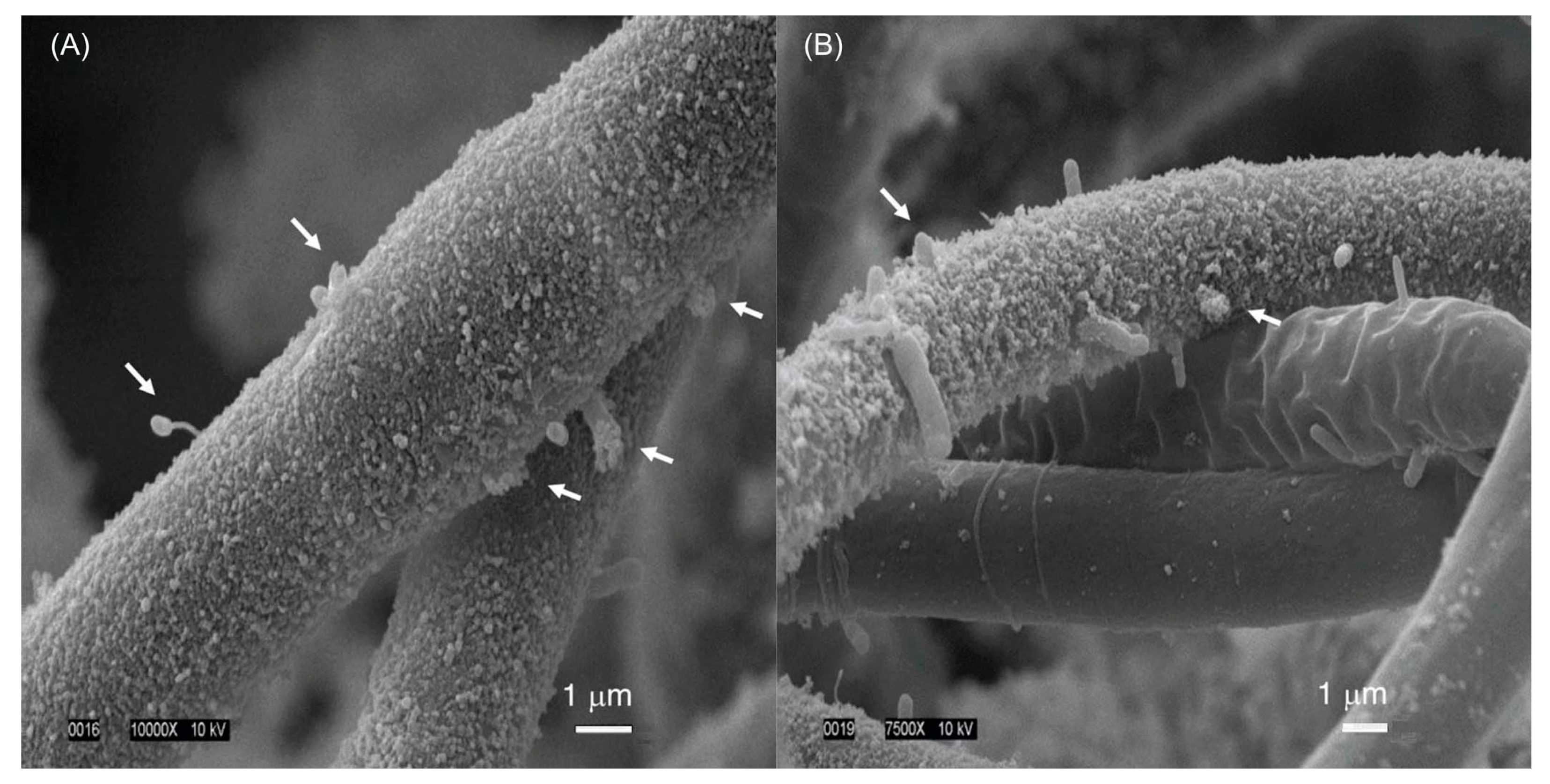
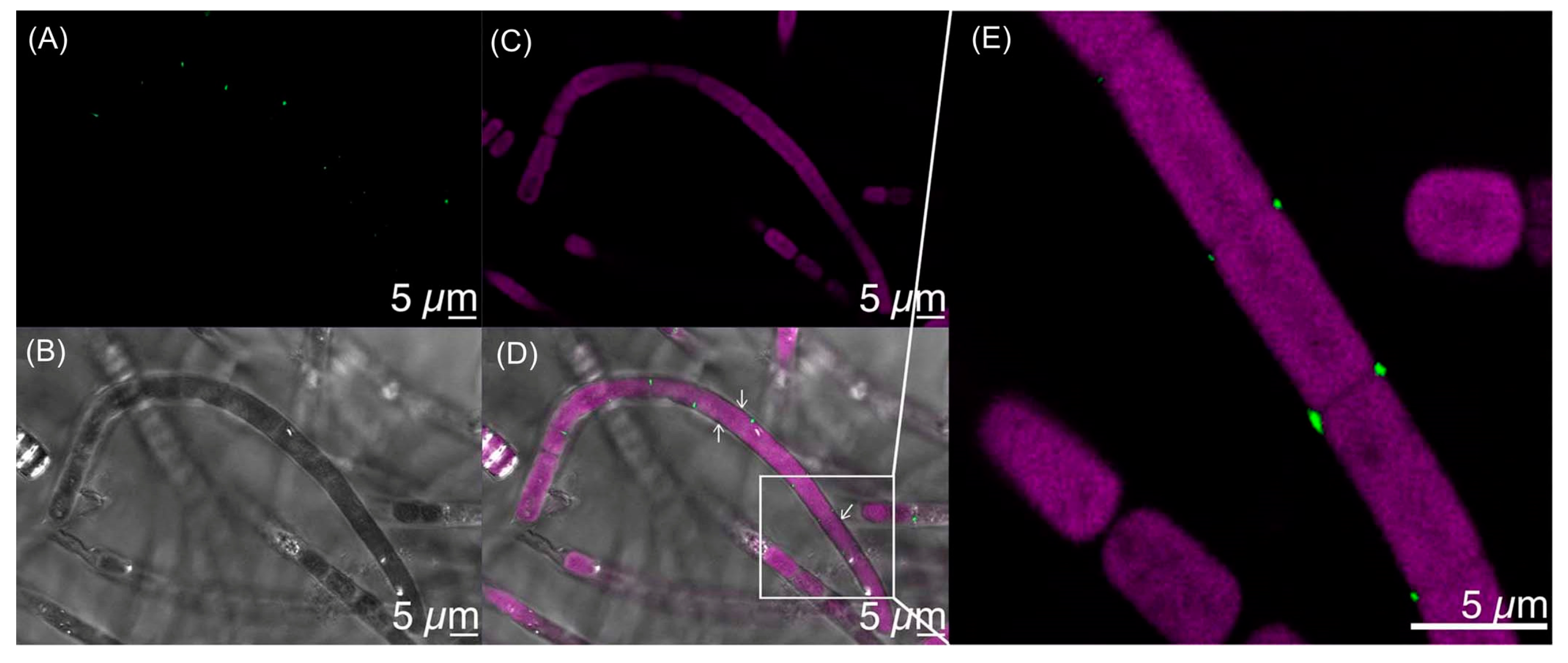
- The culture is not axenic. Examination by light microscopy, fluorescence microscopy, and scanning electron microscopy revealed cyanobacterial filaments, not single cells, with filaments decorated with attached bacteria and the presence of free bacteria (Figure 14). In other words, HT-58-2 is not a pure culture; indeed, HT-58-2 could not be made axenic, regardless of treatment applied including organic solvents (dimethyl sulfoxide), biofilm inhibitors, antibiotics, cycloserine/cycloheximide, arsenite, and anaerobiosis [79]. The non-axenic nature of the culture has significant consequences for numerous studies. The filamentous nature of the cyanobacterium also presents challenges to investigation.
- (2)
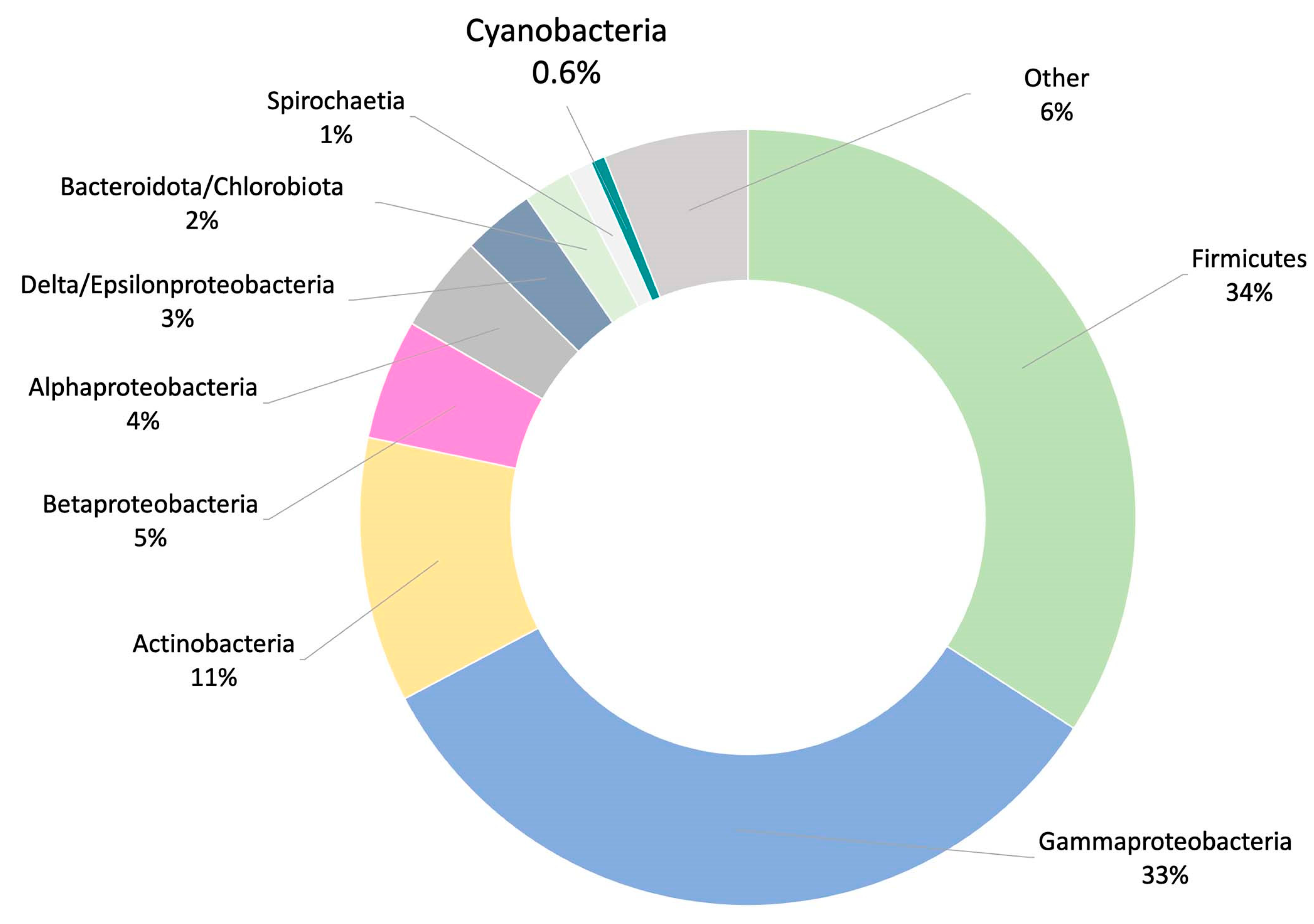
- Metagenomic surveys of the HT-58-2 culture in either BG-11 or BG-110 media revealed a single cyanobacterium accompanied by diverse bacteria. Other operational taxonomic units (OTUs) in the cultures were dominated by the bacterial family Erythrobacteraceae, representing 35% and 24% of all OTUs in BG-11 and BG-110, respectively. The dominant OTU within the Erythrobacteraceae family aligned with the 16S rRNA of Porphyrobacter sp [80], which accounted for 97% of the reads under both growth conditions. The remaining OTUs represented Sphingomonadacea, Proteobacteria, alphaproteobacteria, and unknown bacteria (Figure 15). Further studies of the effects of nutrients on the community bacterial population would be required if the production of tolyporphins requires participation by community bacteria.
- (3)
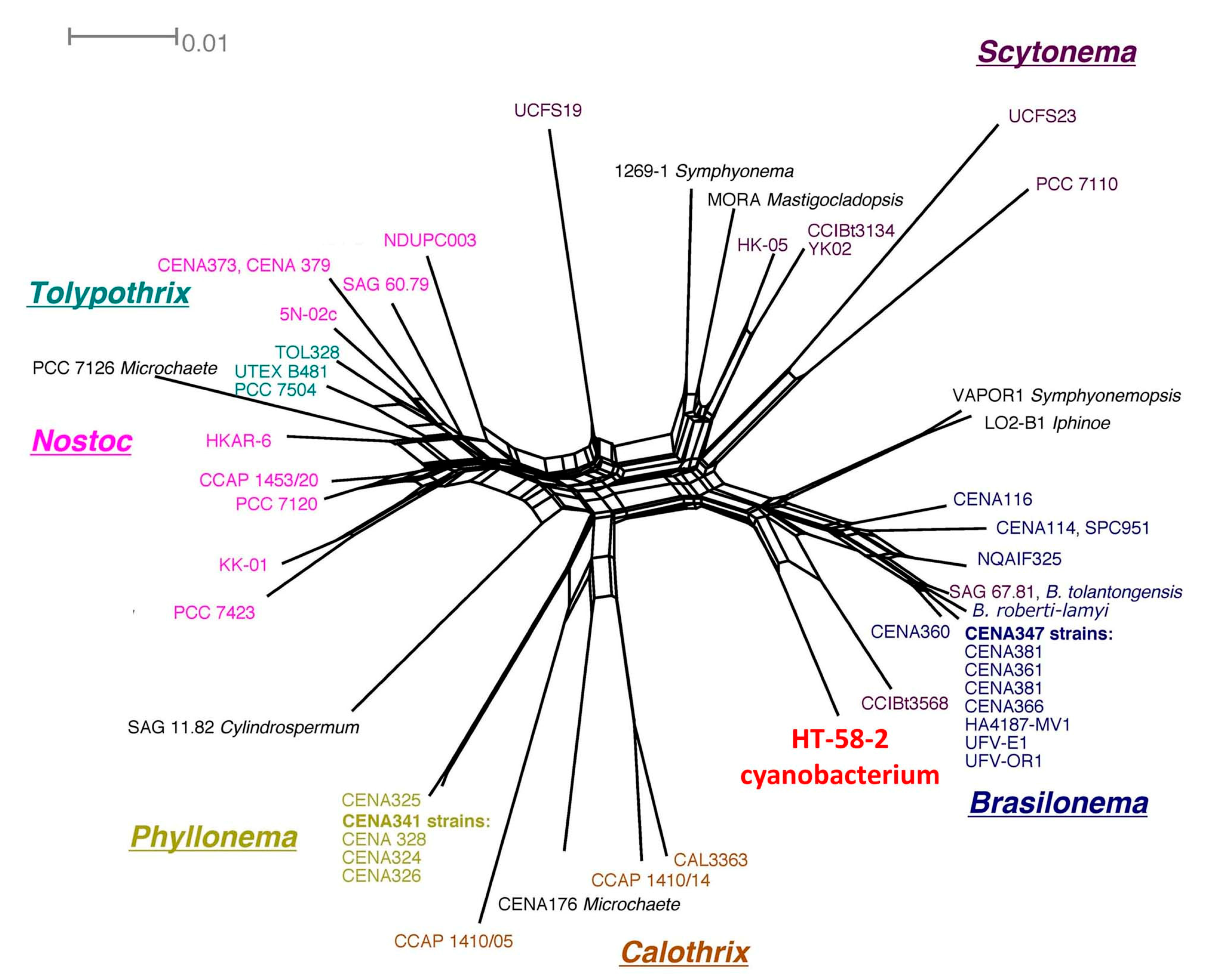
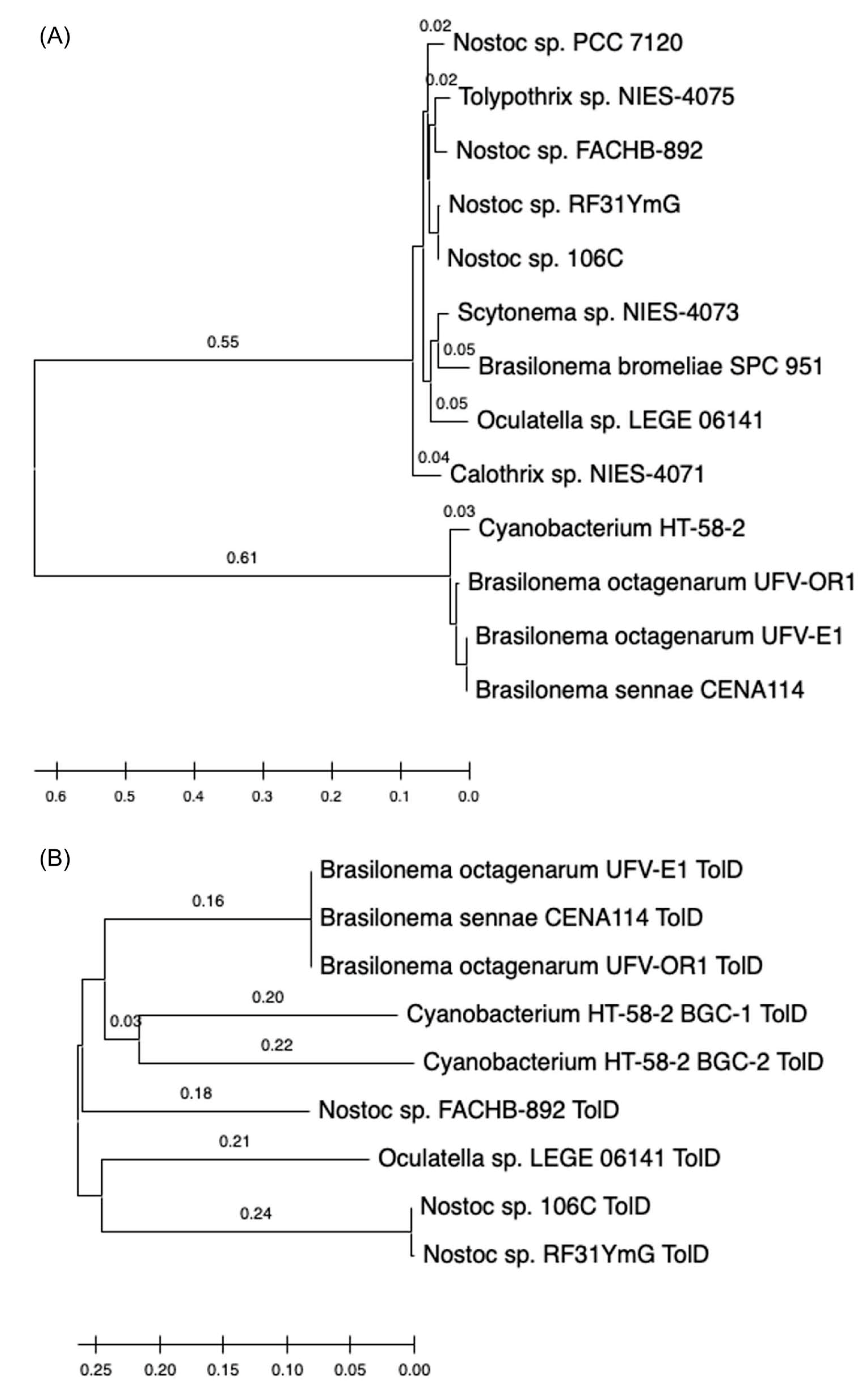
- The 16S rRNA sequence of the HT-58-2 cyanobacterium showed close alignment with the proposed Brasilonema clade and not with the Tolypothrix clade. The 16S rRNA phylogenetic map is shown in Figure 16. The closest strain to the HT-58-2 cyanobacterium is assigned Scytonema CCIBt3568 [81]. The genus Brasilonema was first described only 16 years ago by Fiore as a genus apart from that of Scytonema [82]. Since then, other Brasilonema species have been identified [83,84]; indeed, at the time of this writing, a Web of Science search (all databases) for “cyanobacteria and brasilonema” resulted in 62 hits.
6.2. The Cyanobacterium
6.3. Community Bacteria
6.4. Cyanobacterial Genomics Broadly
7. The Locale of Tolyporphins
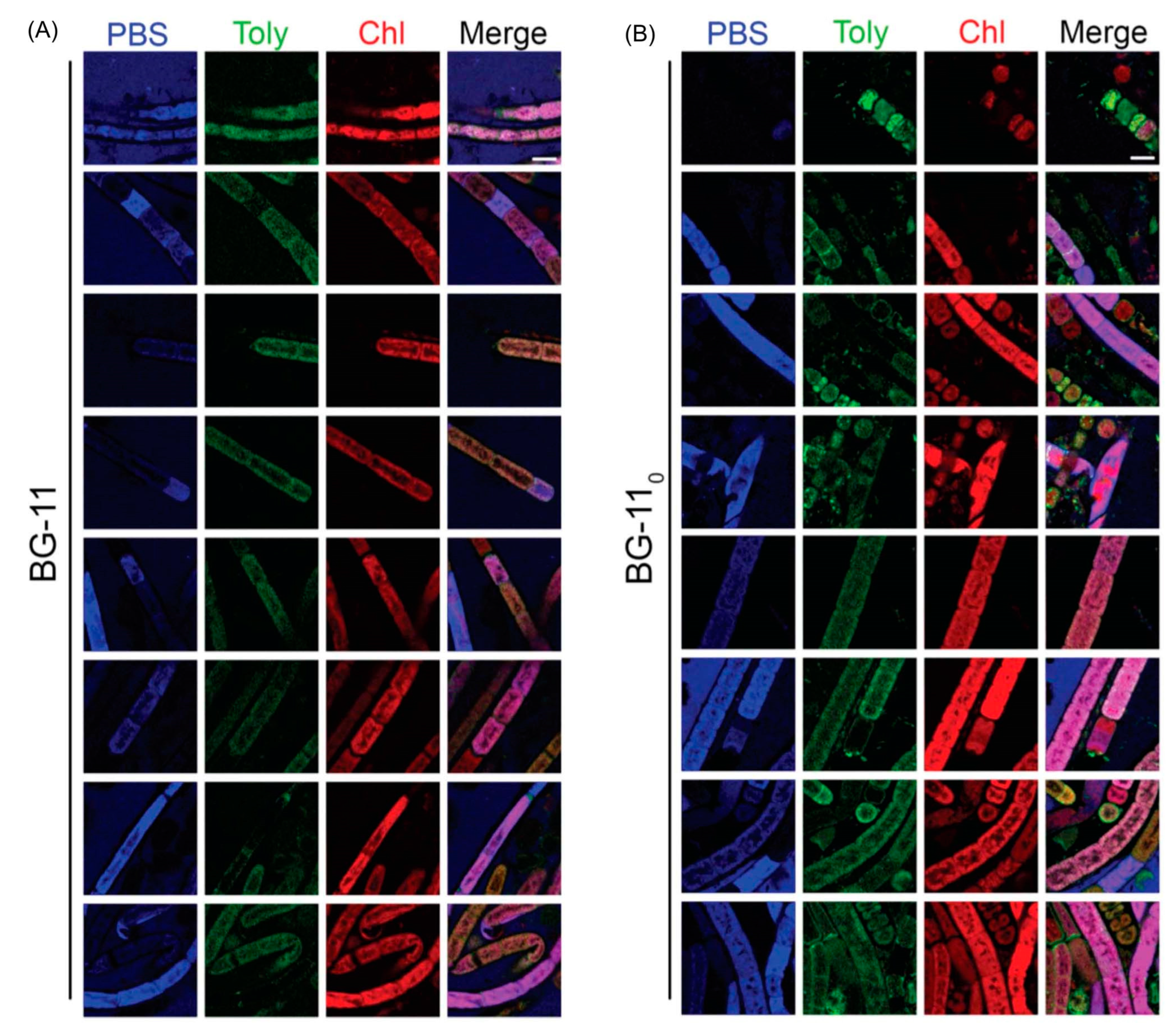
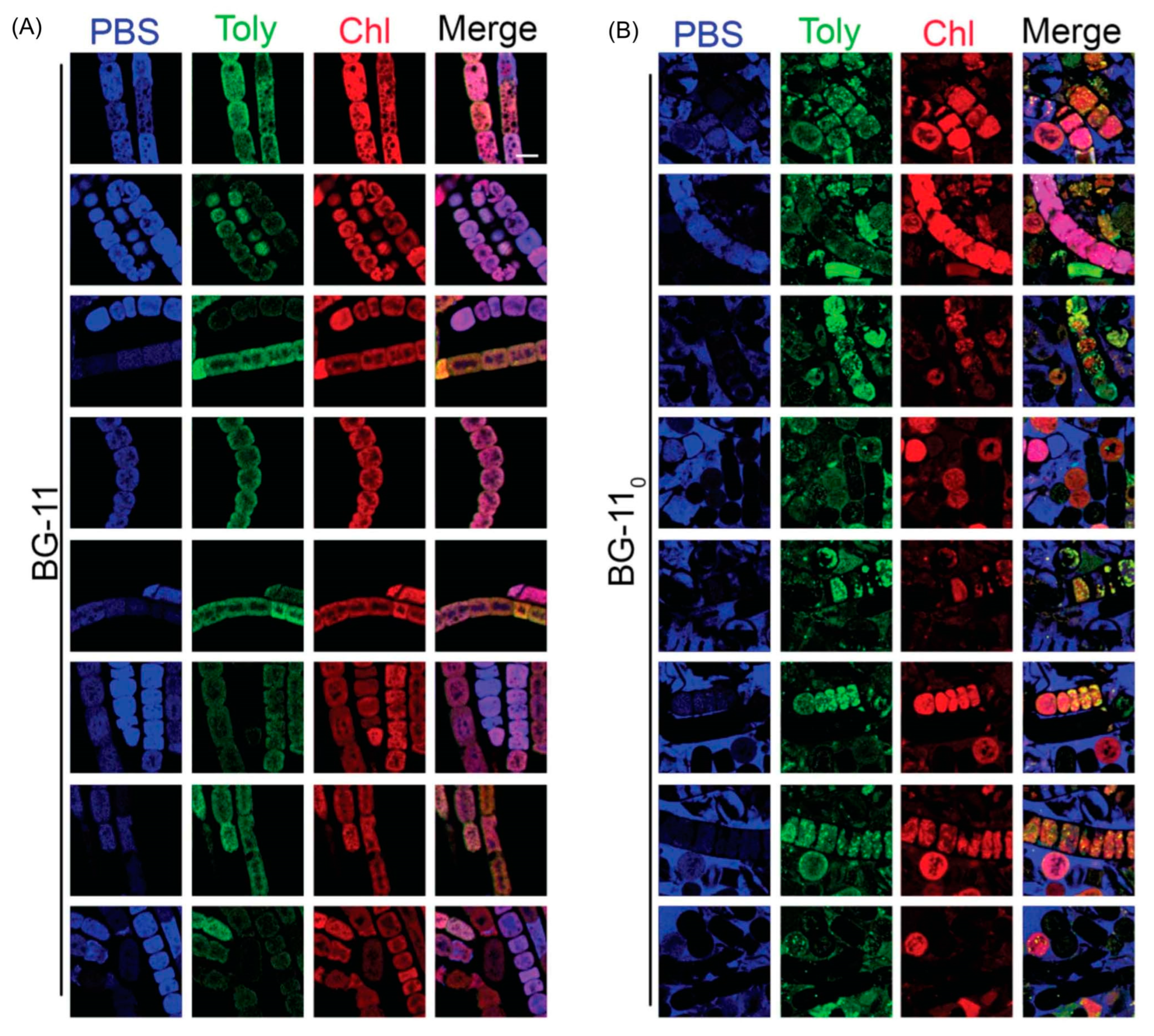
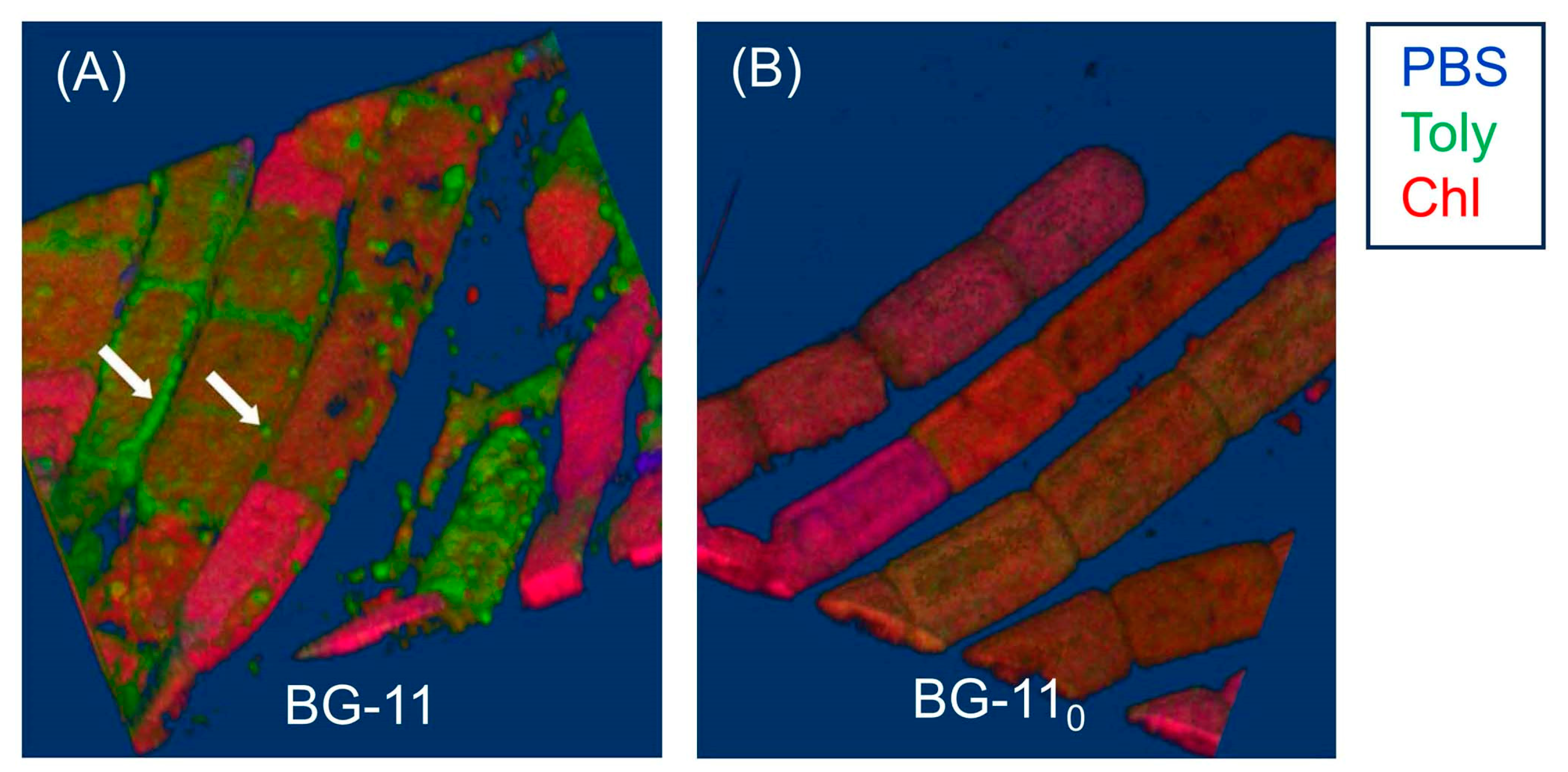
7.1. Imaging Studies
7.2. Estimates of Polarity
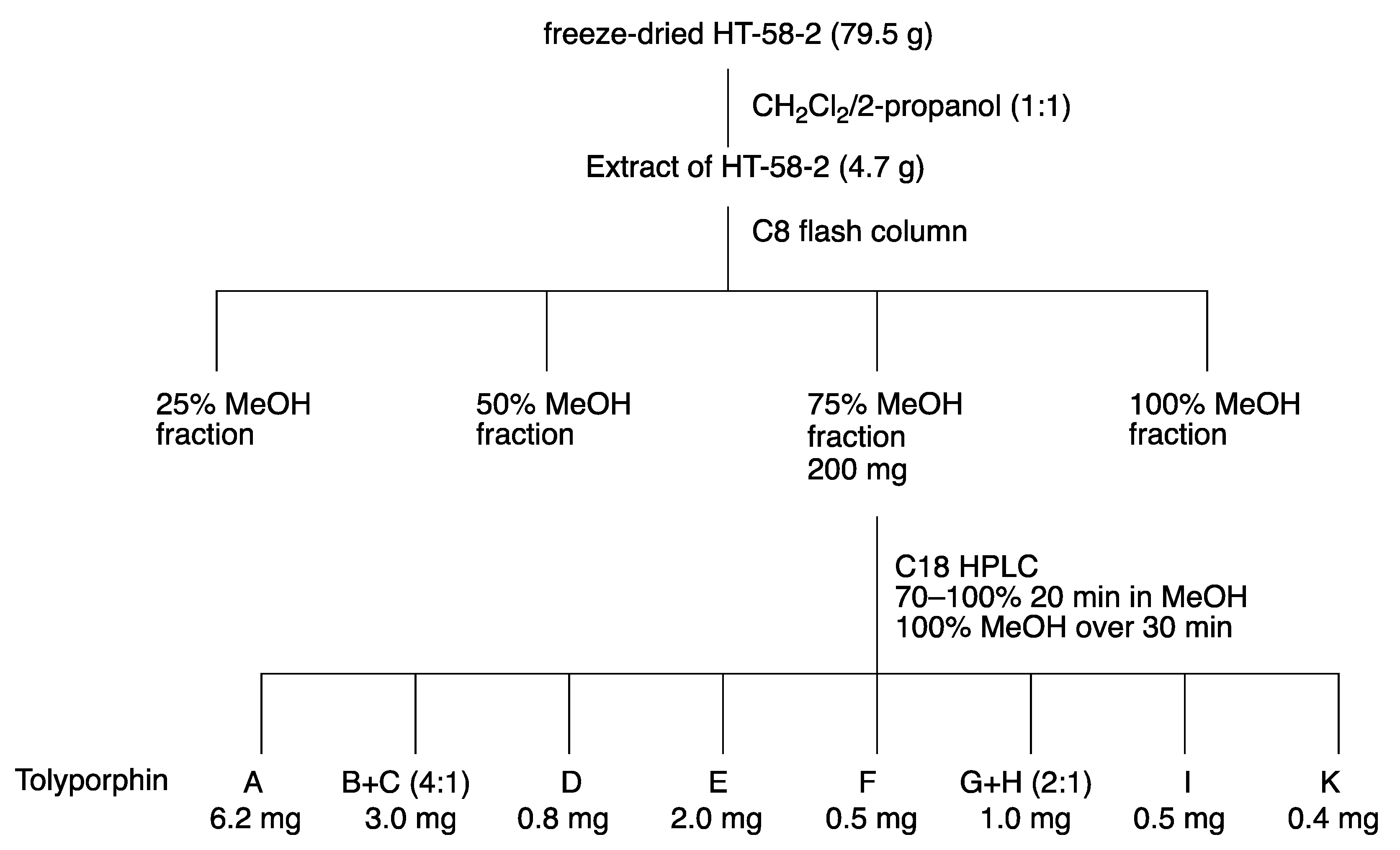
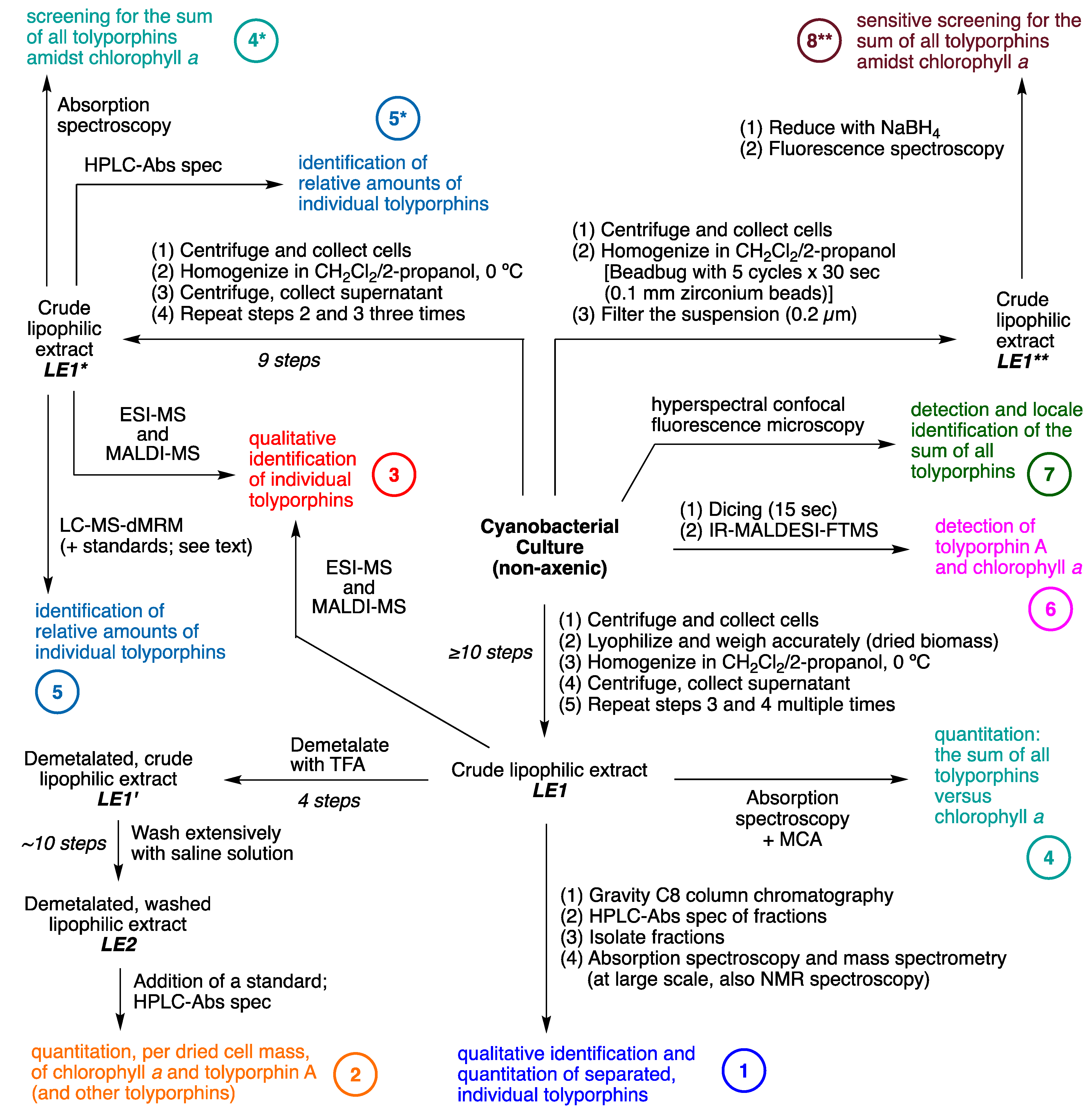
8. Isolation of Tolyporphins
9. Analytical Features of Tolyporphins
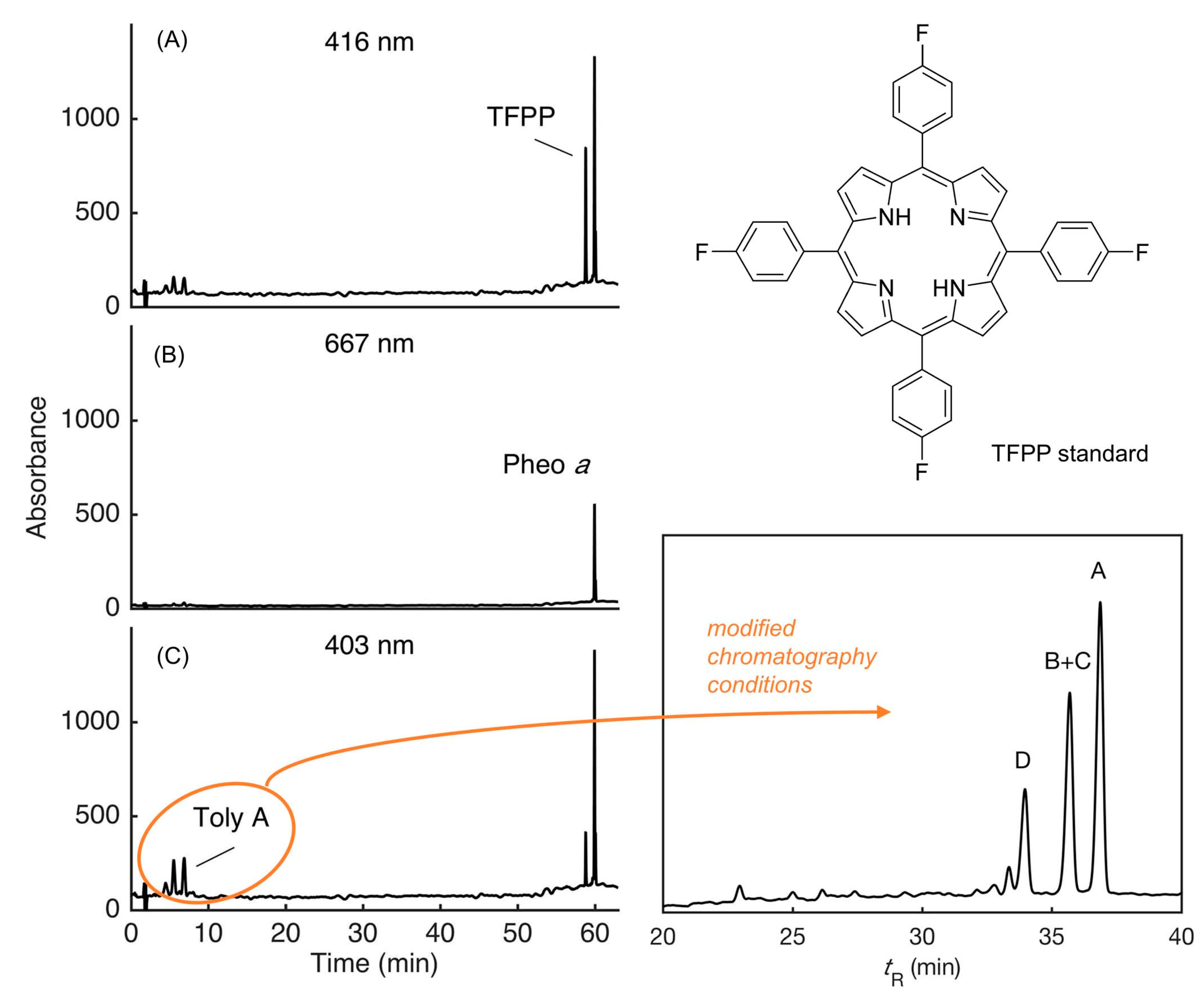
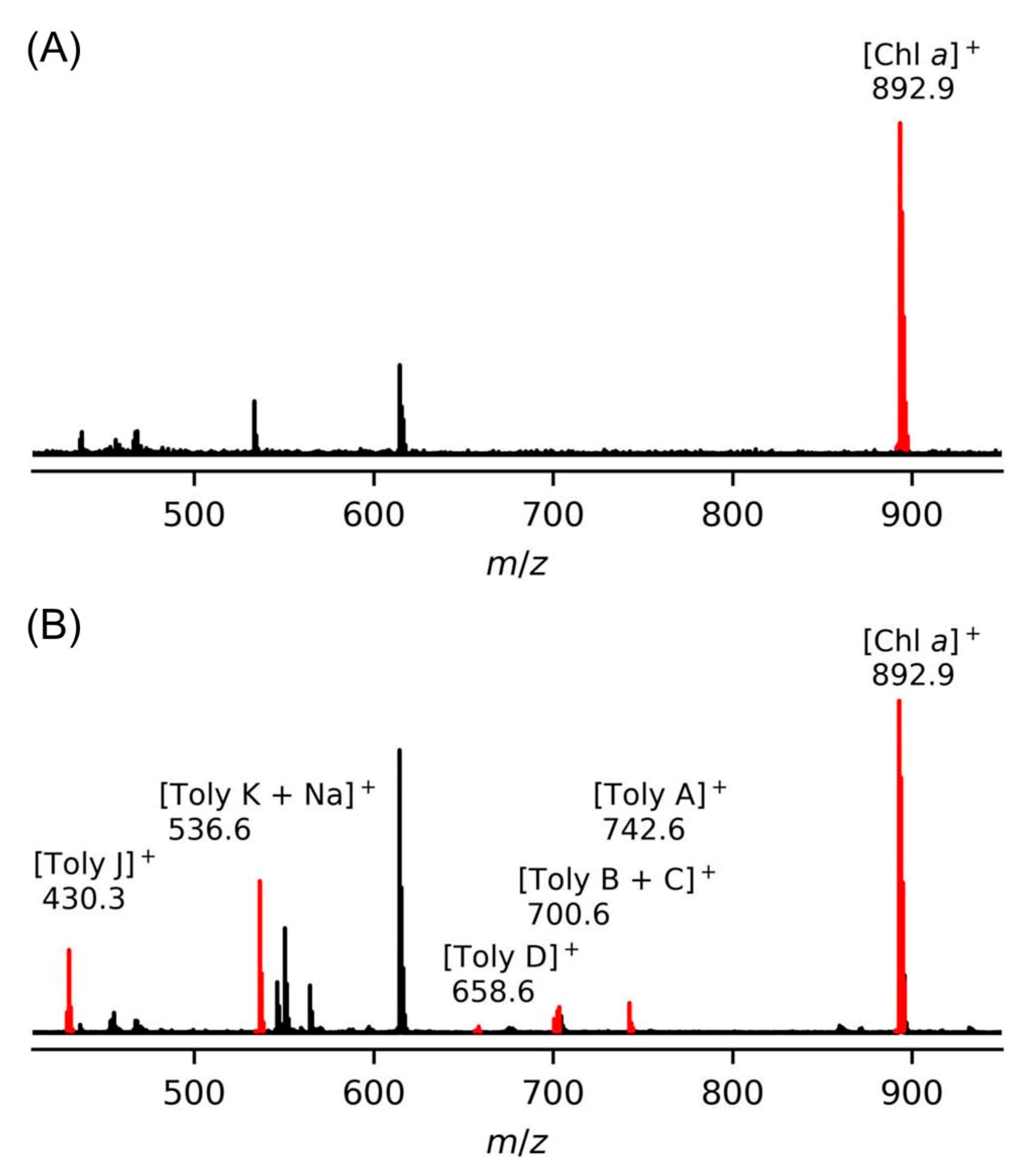
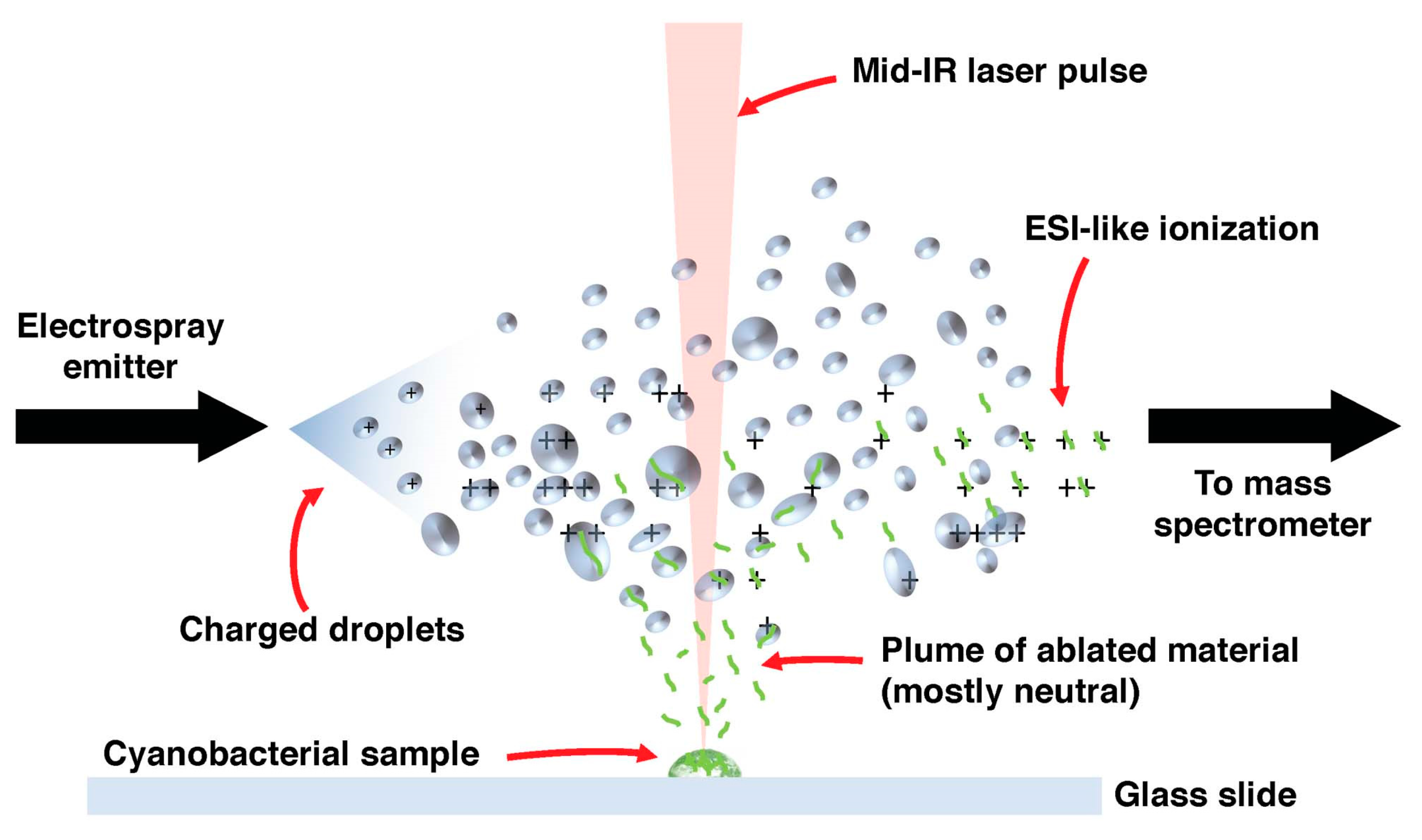
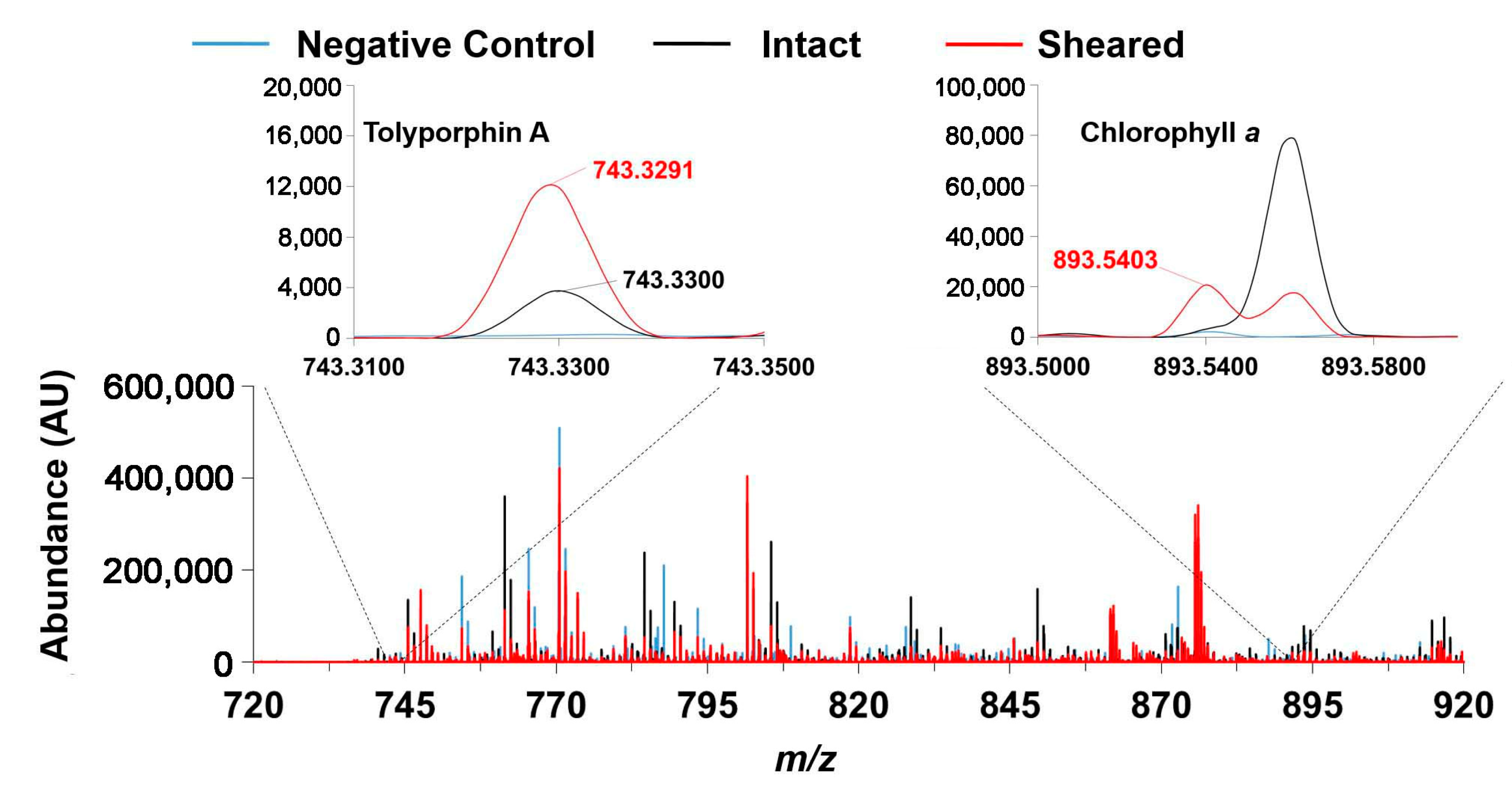
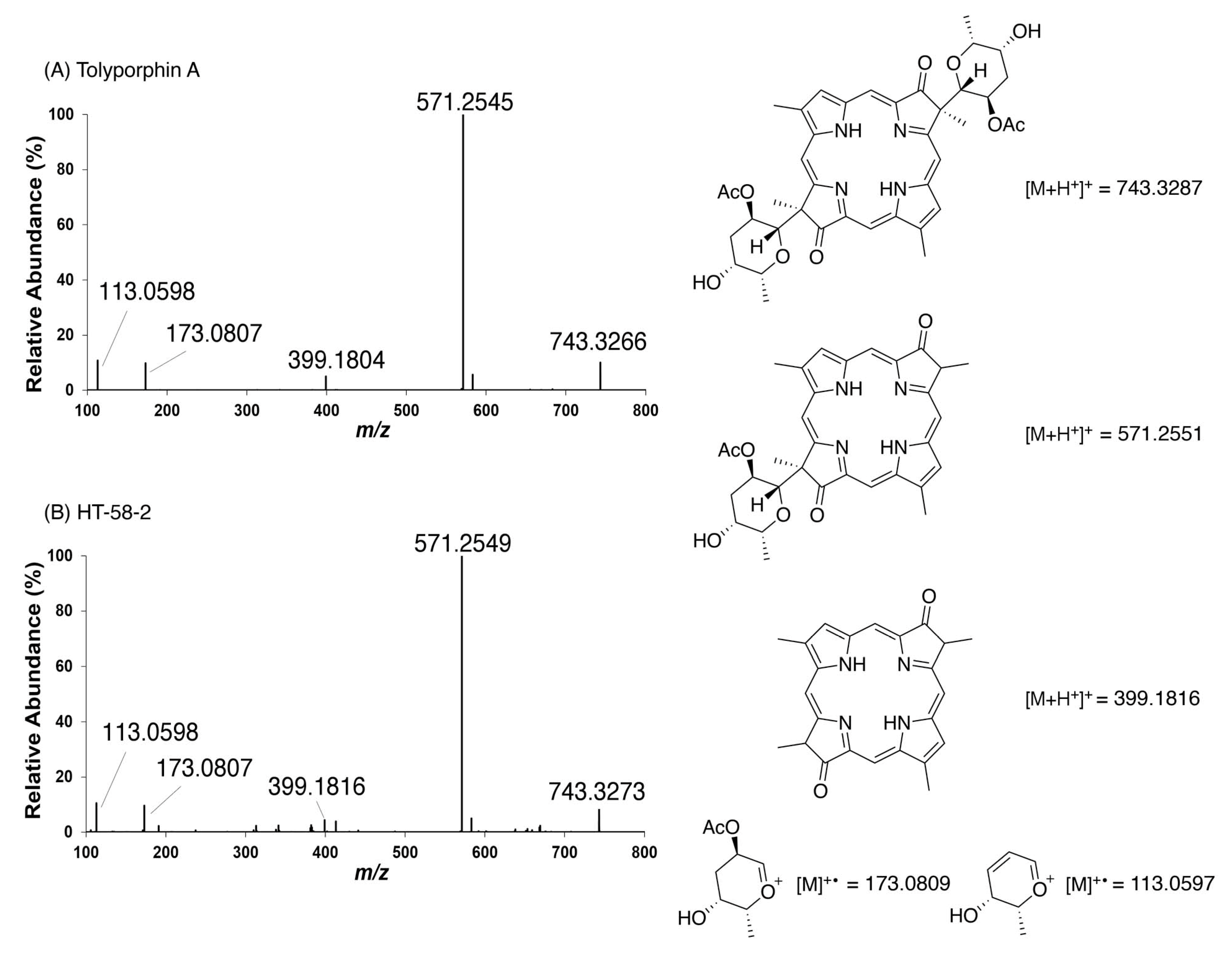
9.1. Mass Spectrometry
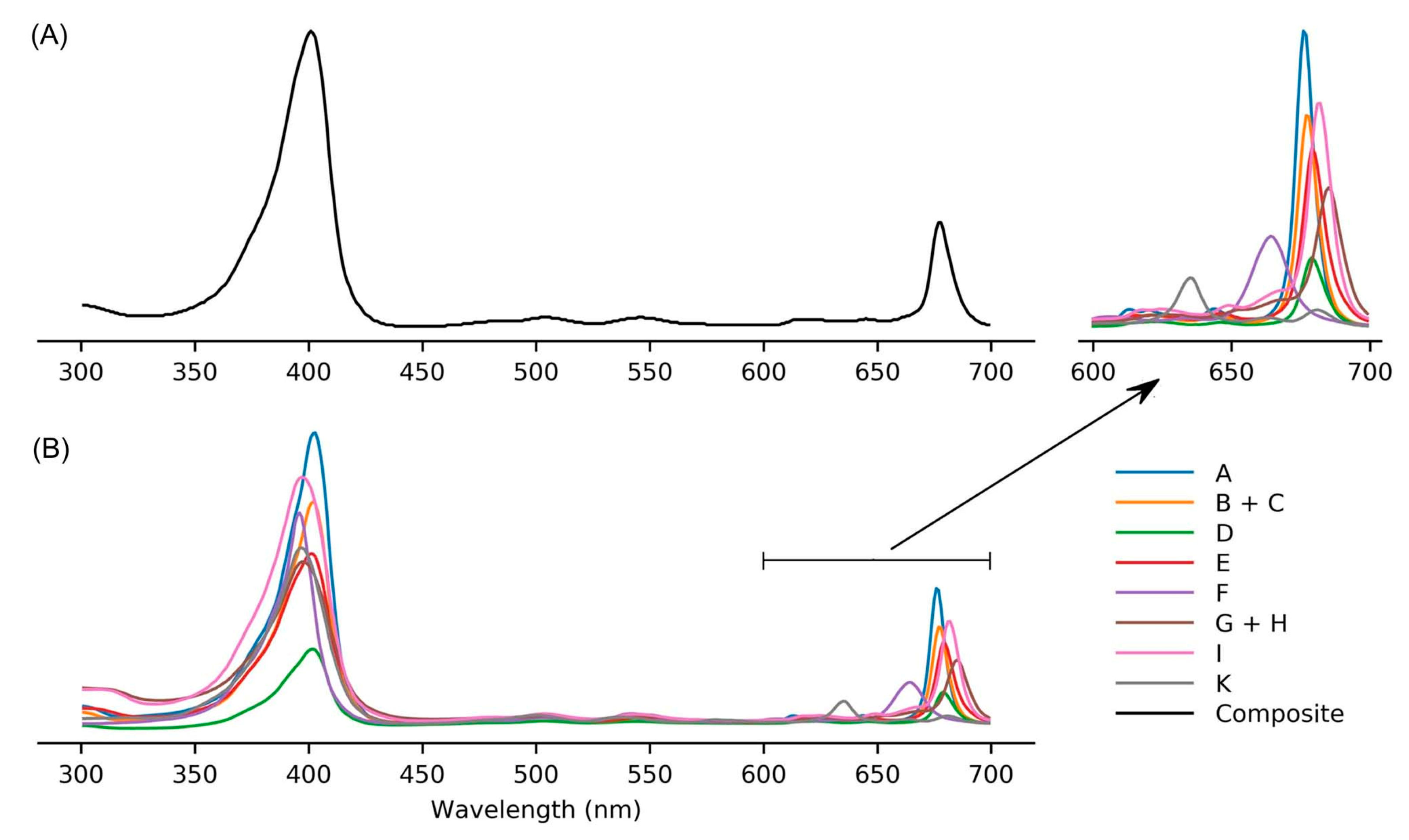
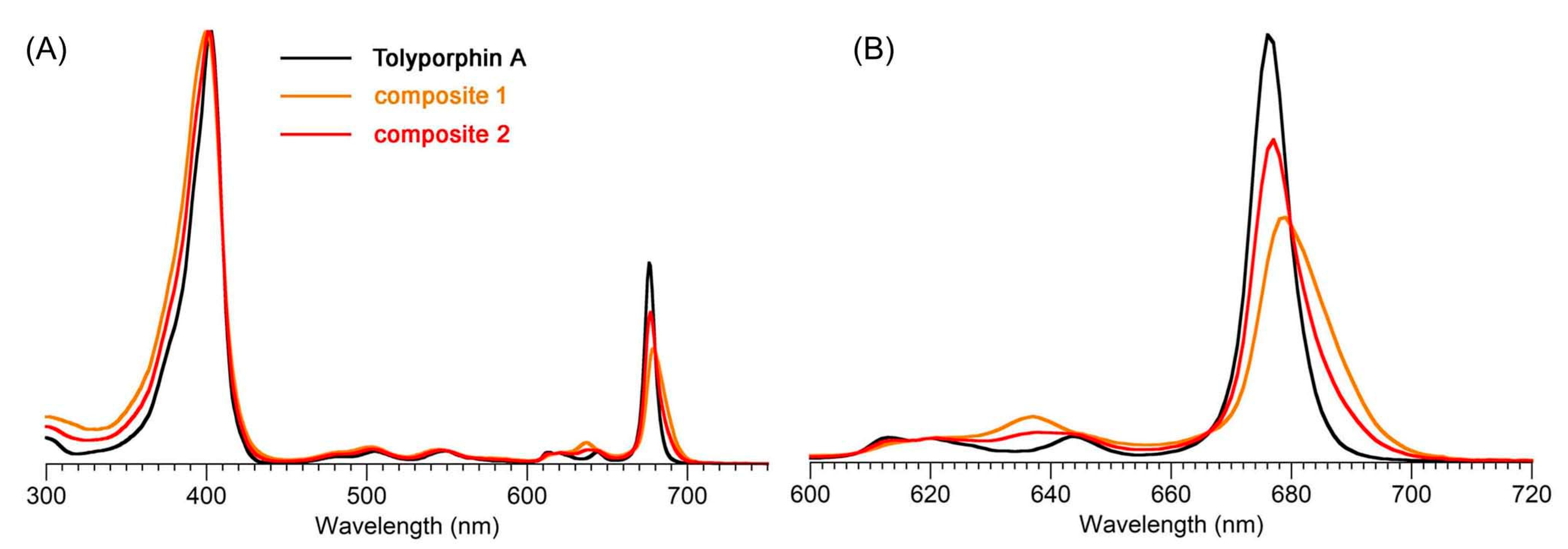
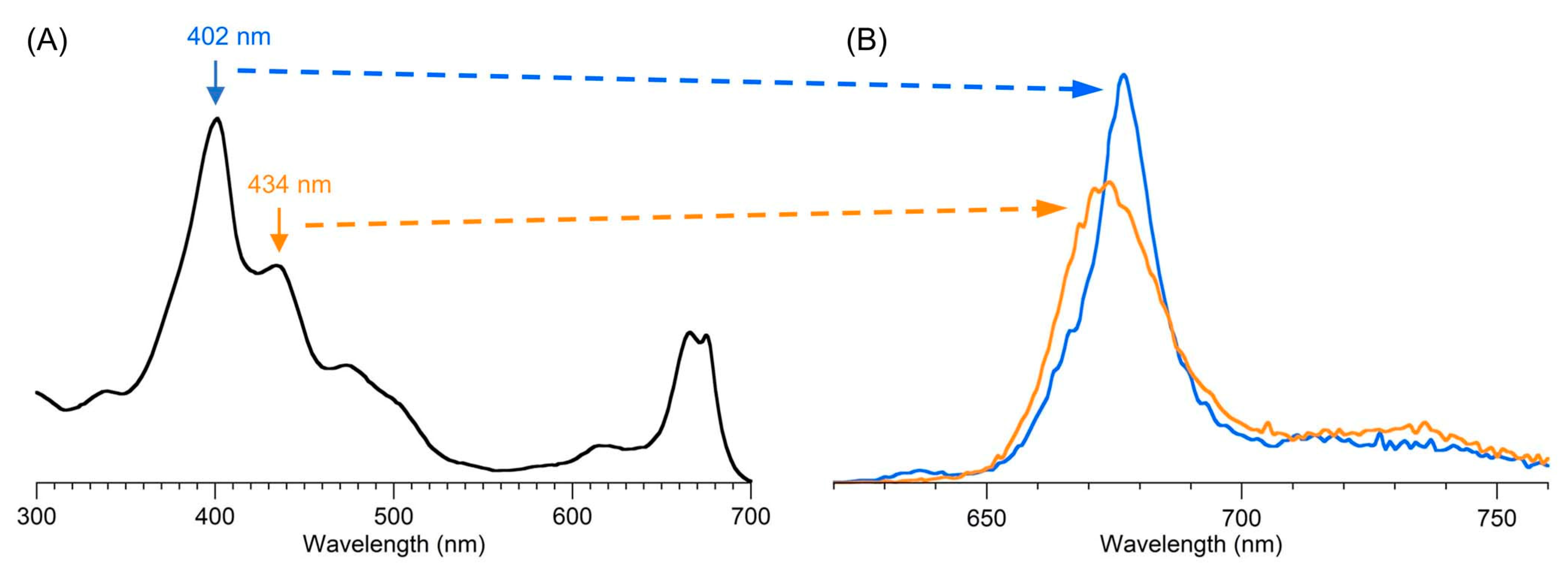
9.2. Absorption Spectroscopy
10. Quantitative Analysis of Tolyporphins
- What quantities of tolyporphins are biosynthesized?
- How does the production change with growth conditions?
- How can tolyporphins be detected easily in the search for new producers?
10.1. Process 1
10.2. Process 2
- (1)
- Regardless of conditions or the growth period, the HPLC peak due to tolyporphin A was the highest among all discernible tolyporphins. The percentage of tolyporphin A among all tolyporphins in the small-scale culture was ~40%, to be compared with 55% in large-scale growth experiments [45].
- (2)
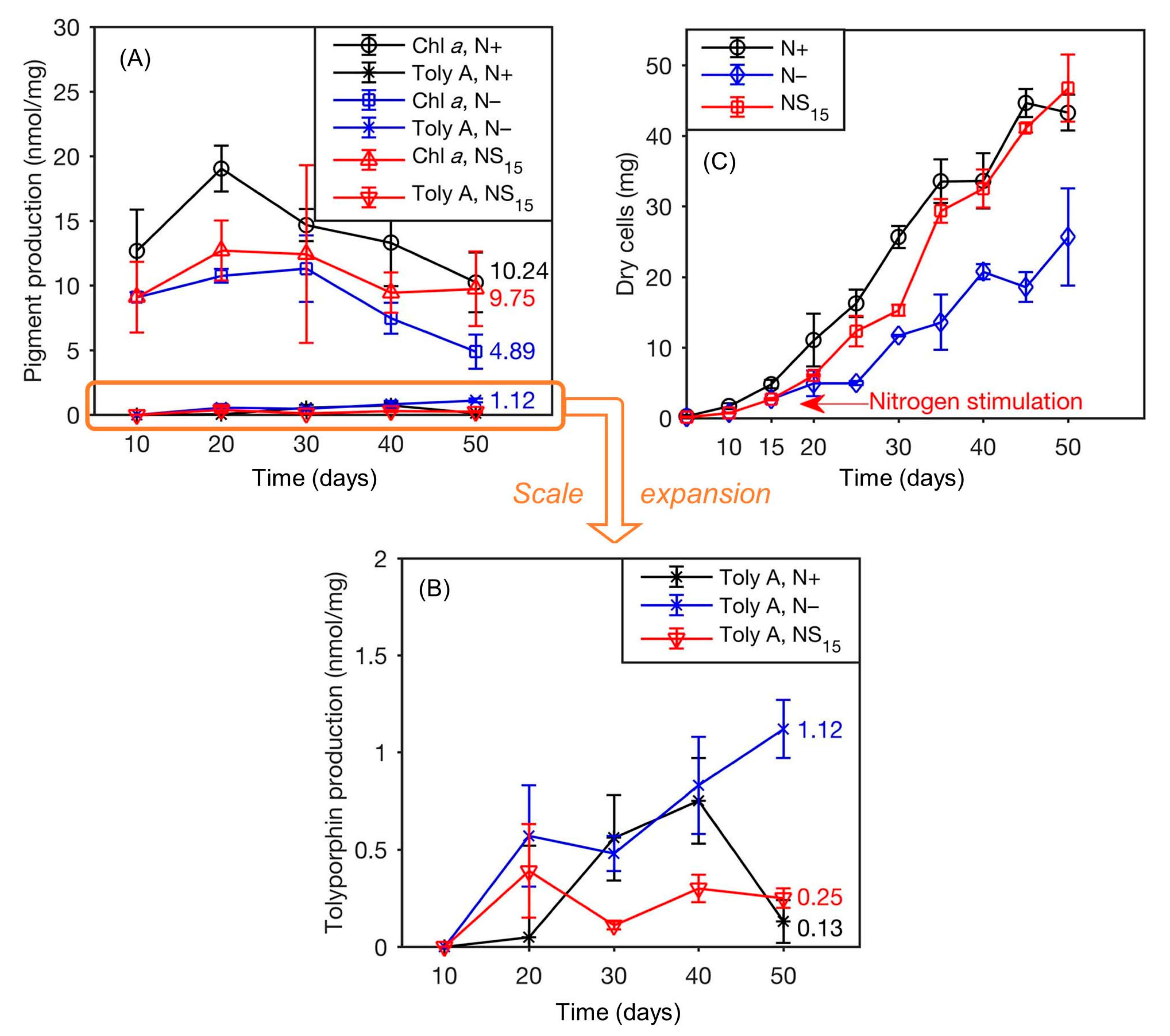
- The yield of tolyporphin A was lower than that of chlorophyll a under all growth conditions (Figure 32, panels A and B). Note that panel B is an expansion of the tolyporphin A data in panel A.
- (3)
- More chlorophyll a was produced in N+ than in N− or NS15 during the 50-day period. Upon nitrogen stimulation at day 15, the production of chlorophyll a increased to 9.75 nmol/mg dry cells at day 50, which is comparable to that in N+ (10.2 nmol/mg) and greater than that in N− (4.89 nmol/mg). For units conversion, given the molecular weight of chlorophyll a of 893.5 Da, 10 nmol/mg corresponds to 8.95 mg/g or 0.895%, which is in accord with values given in Table 8.
- (4)
- The yield of tolyporphin A was higher in conditions of N− than in N+ or NS15. The maximum yield of tolyporphin A was 1.12 nmol/mg, which was reached on day 50 in N−; the yield was approximately 10 times greater than for the sample in N+ (0.13 nmol/mg) or NS15 (0.25 nmol/mg), but still less than the yield of chlorophyll a in N− (4.89 nmol/mg) (Figure 32, panels A and B).
10.3. Process 3
10.4. Processes 4 and 4*
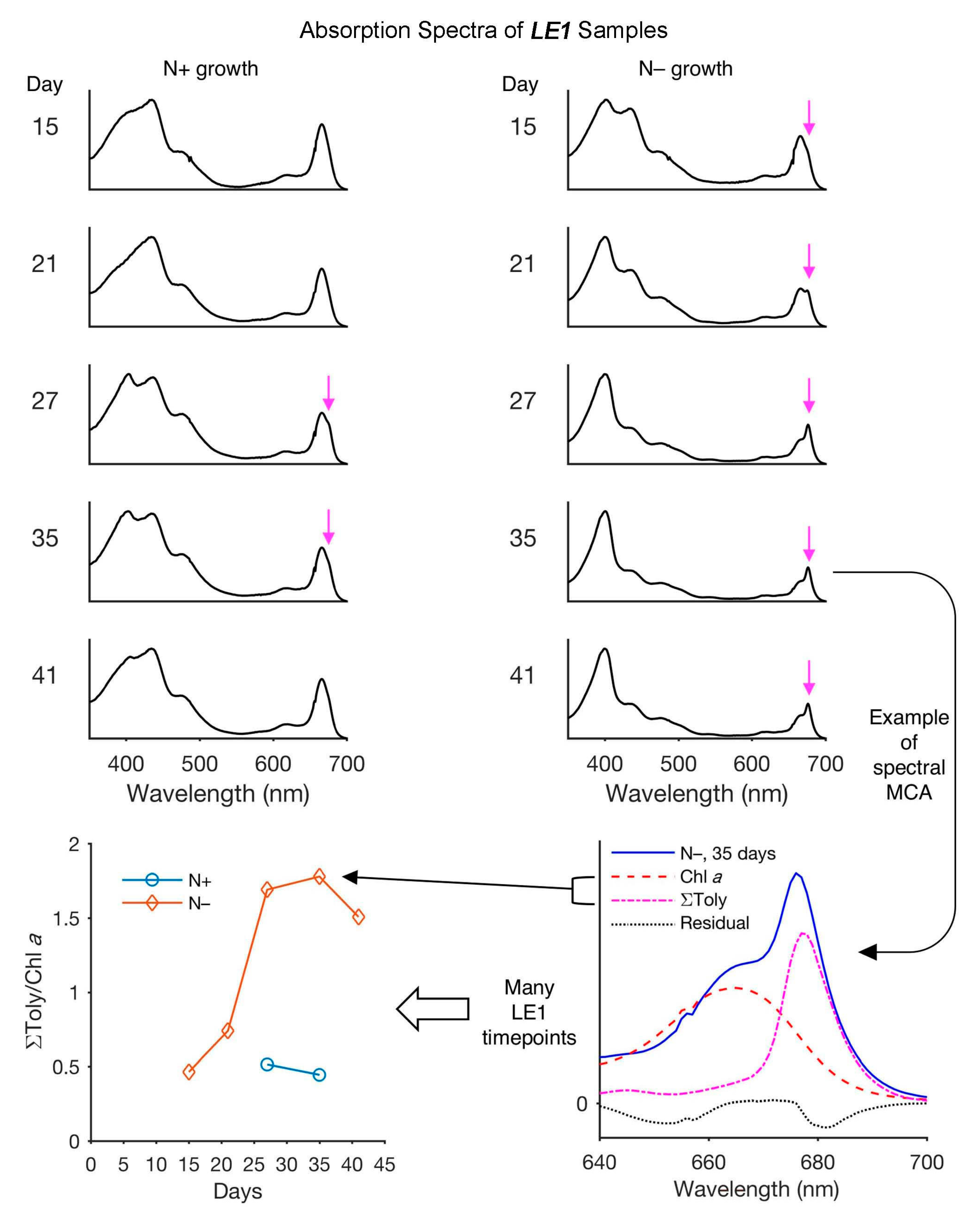
- (1)
- The results obtained by HPLC fractionation were obtained with 25 mL cultures, whereas those by absorption spectroscopy were with 2 mL cultures. Different scales of growth may alter the inherent production of chlorophyll a and tolyporphins.
- (2)
- The HPLC fractionation requires more extensive sample processing of LE1 (Process 2, Figure 30), whereas absorption spectroscopy directly interrogates LE1 (Process 4, Figure 30). Components may be lost upon performing HPLC. Any yield (of minor components) determined by gravimetry of minute samples may be fraught, just as determination of molar absorption coefficients from minute samples of tetrapyrroles has proven historically to be a precarious endeavor.
- (3)
- There may be compounds other than tolyporphins and chlorophyll a that absorb in the Qy region and make MCA inaccurate.
- (4)
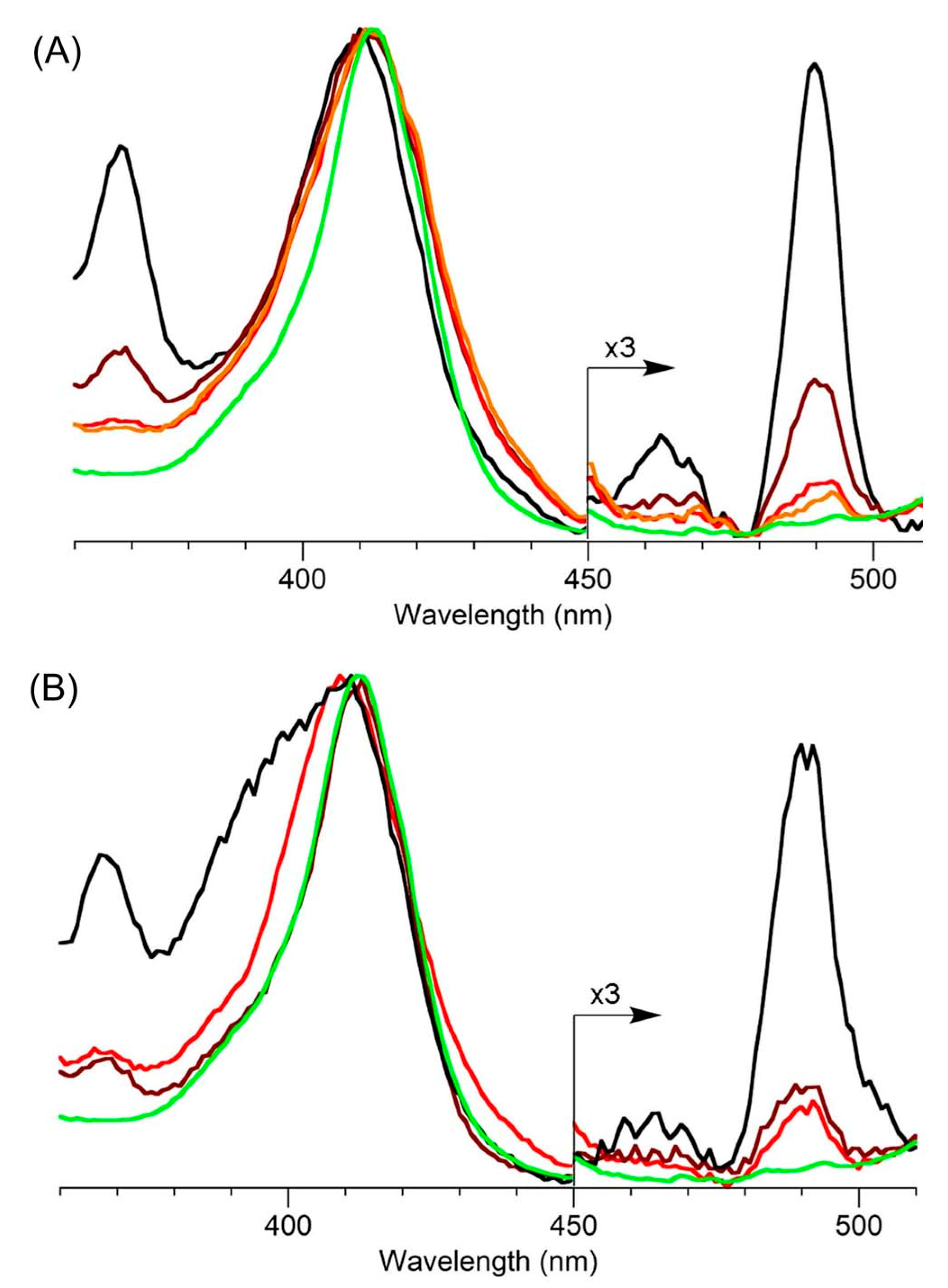
- The mole fraction-weighted basis of isolated samples to generate the composite spectrum depends on the uniform representation of tolyporphins in LE1 from the cyanobacterial sample. The extraction may not be uniform or degradation could occur (particularly of the acetates and the C-glycosides). The incomplete or skewed extraction may not be a significant concern for examination of LE1 by absorption spectroscopy, but the generation of the composite absorption spectrum relies not only on extraction, but also accurate fractionation to determine the mole fraction. While a somewhat circular process, possible pitfalls are likely mitigated by the similar distribution of tolyporphin members under various culture conditions, and the close similarity of the absorption spectra among all dioxobacteriochlorin-type tolyporphins, as described in ensuing paragraphs.
- (5)
10.5. Processes 5 and 5*
10.6. Process 6
10.7. Process 7
10.8. Process 8**
11. HT-58-2 Genome—Genes for Tolyporphins
12. HT-58-2 Genome—Genes for Other Natural Products
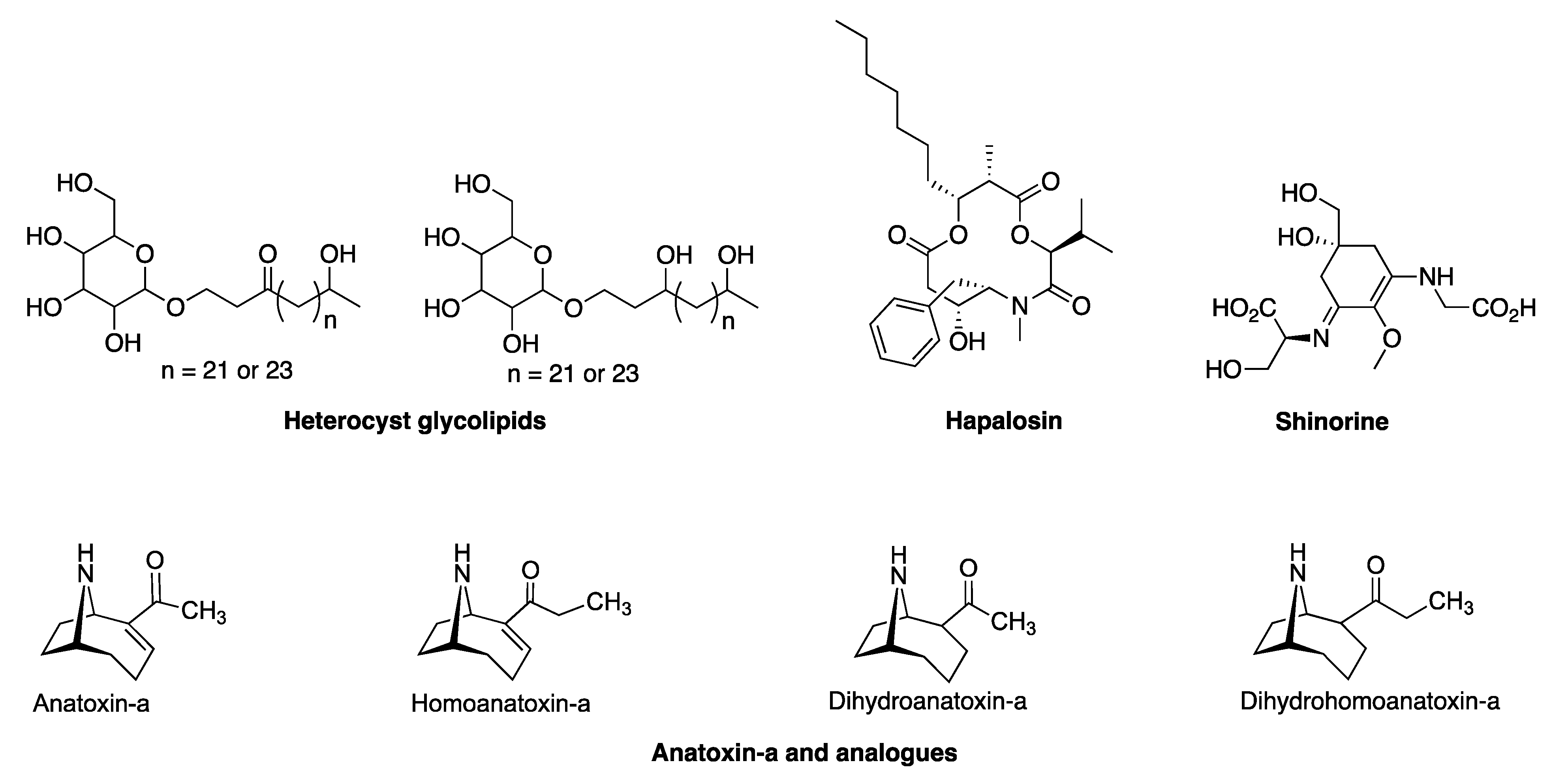
- Heterocyst glycolipids (HGs) are known to form a protective layer for oxygen-sensitive nitrogenase enzymes [169] in the envelope of heterocystous nitrogen-fixing cyanobacteria.
- Hapalosin is a cyclodepsipeptide that has been reported to reverse multidrug resistance in tumor cell lines [170].
- Shinorine is one example of a mycosporine-like amino acid that functions as an ultraviolet sunscreen in cyanobacteria [173].
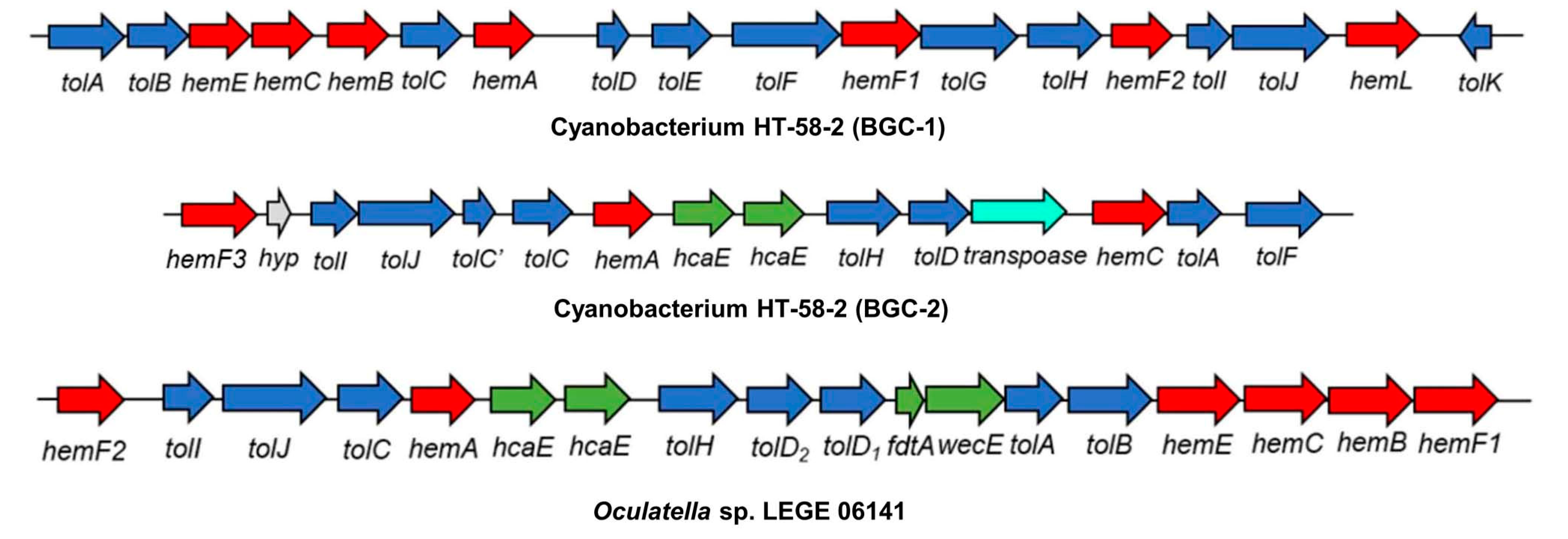
13. Searching for New Producers of Tolyporphins—Informed by Bioinformatics
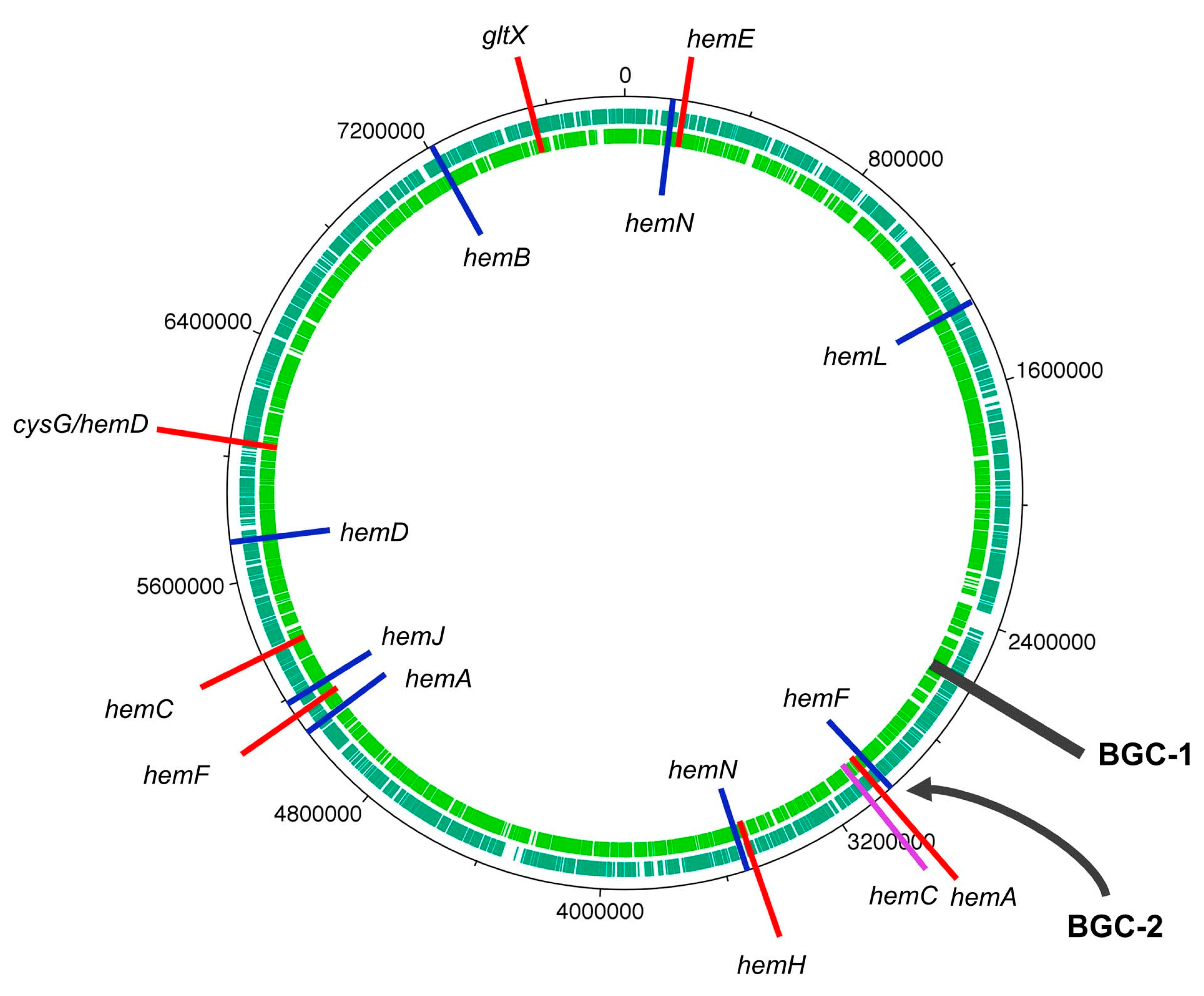
- BGC-2 contains coding sequences pertaining to seven putative proteins that correspond with >50% identity to Tol proteins from BGC-1 (TolACDHIJ), and all are arranged in the same orientation.
- There is only one cytochrome P450 in BGC-2, which shares higher identity to TolH (CYP88A) than TolG, versus two in BGC-1. However, two other P450 genes (yellow arrows in Figure 54) are adjacent to BGC-2.
- Duplicate hcaE genes encoding aromatic ring-hydroxylating dioxygenases [179] are present within BGC-2, but not within BGC-1. The relevance of such genes is unknown.
- Three transport-related protein genes (DUF3102 domain-containing proteins DevB and DevC) are present at the leftmost end of BGC-2, similar to that of BGC-1.
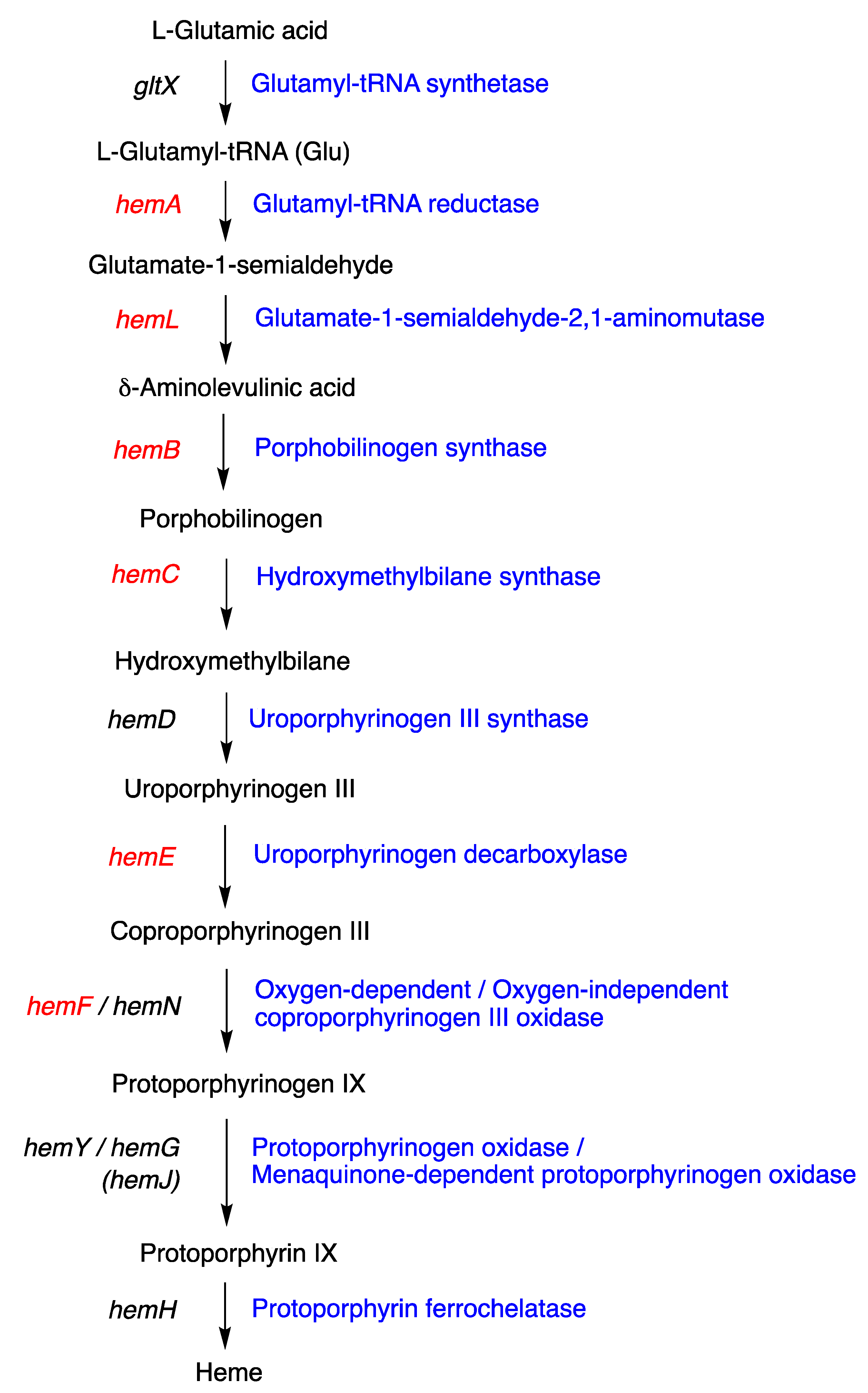
- Each BGC contains the unusual clustering of multiple hem genes, including hemABCEF, that are adjacent to several tol-like genes found in BGC-1 of HT-58-2.
- None of the BGCs includes hemD (UroS).
- Each Nostoc spp. and Brasilonema spp. contains two hemF genes (the aerobic coproporphyrinogen decarboxylase).
- Each BGC contains tolB (the RfbA orthologue, glucose-1-phosphate thymidylyltransferase) and tolD (the glycosyltransferase).
- Each BGC has nearby genes for secretory and transport proteins (DevB and DevC families).
14. Searching for New Producers of Tolyporphins—Classical Approach
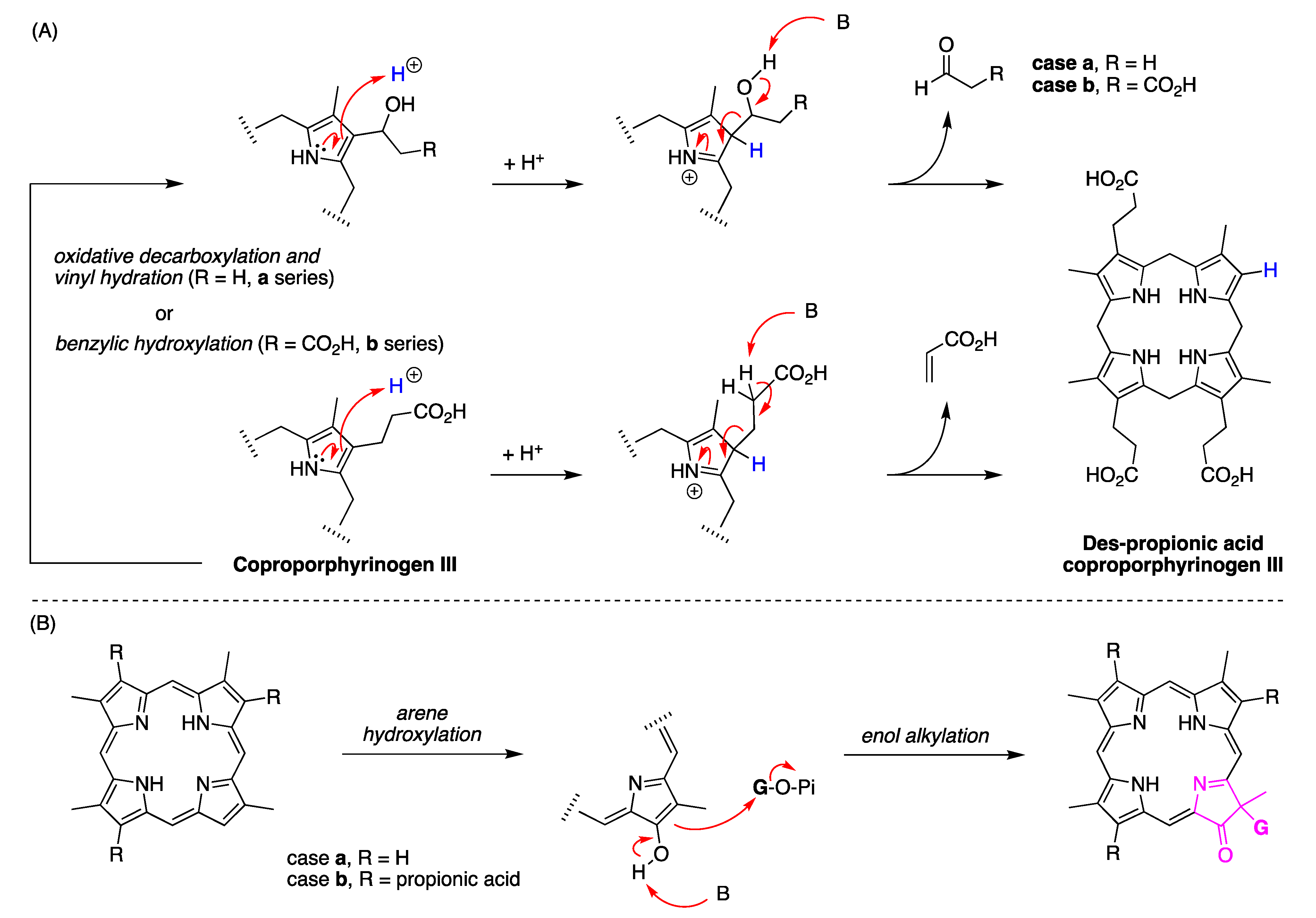
15. Biosynthesis Considerations
16. Abe’s Studies of Tolyporphins Biosynthesis
17. Potential Pharmacological Properties of Tolyporphins
17.1. Reversal of Multidrug Resistance
17.2. Photodynamic Inactivation
17.3. Studies with Metal Chelates of Tolyporphins
17.4. Computational Studies
- (1)
- As a photoprotective agent by absorption of light, given that the absorption spectrum of a dioxobacteriochlorin-type tolyporphin resembles that of chlorophyll [49]; indeed, cyanobactera are known to make scytonemins, which provide a sunscreen-like function to protect the organisms from the adverse effects of ultraviolet light [212,213].
- (2)
- (3)
18. Perspective
- (1)
- What?—the molecular structures of tolyporphins have been fairly established, although stereochemical configuration remains to be elucidated for a number of members. The molecular diversity suggests that tolyporphins are secondary metabolites (natural products), not the singular end product of core metabolism. While the molecular structures of tolyporphins have largely been elucidated, studies of electronic and tautomeric features remain to be carried out (for components other than tolyporphin A [49]); results in this regard can be compared with those for a variety of synthetic lactone-containing tetrapyrrole macrocycles [216].
- (2)
- Why?—the microbial–physiological rationale for the biosynthesis of tolyporphins is unclear, although the increase in production upon nitrogen stress is a hallmark of a microbial defensive function. Suggestions for physiological function include an efflux pump inhibitor in the cyanobacterium; a photodynamic agent; or a photoprotective agent. The existential question concerning dioxobacteriochlorins in an organism that ordinarily produces a chlorin (i.e., chlorophyll a) in ample quantity remains perhaps almost koanistic. Could it be that tolyporphins have hardly any native physiological function, and the biosynthetic machinery is merely a vestigial rarity of no significance other than an intriguing scientific curiosity?
- (3)
- Where?—imaging experiments have shown tolyporphins in the cyanobacterial envelope and septa between cyanobacterial cells, yet the questions of how tolyporphins arrived there, and where they arrived from, are unknown. The extent to which one or more community bacteria may participate in the biosynthesis remains undetermined.
- (4)
- When?—the evolutionary origin of tolyporphins is unknown. The finding of additional native producers beyond that of the now sole producer HT-58-2 (among the universe of cyanobacteria and other organisms) would help to inform evolutionary considerations.
- (5)
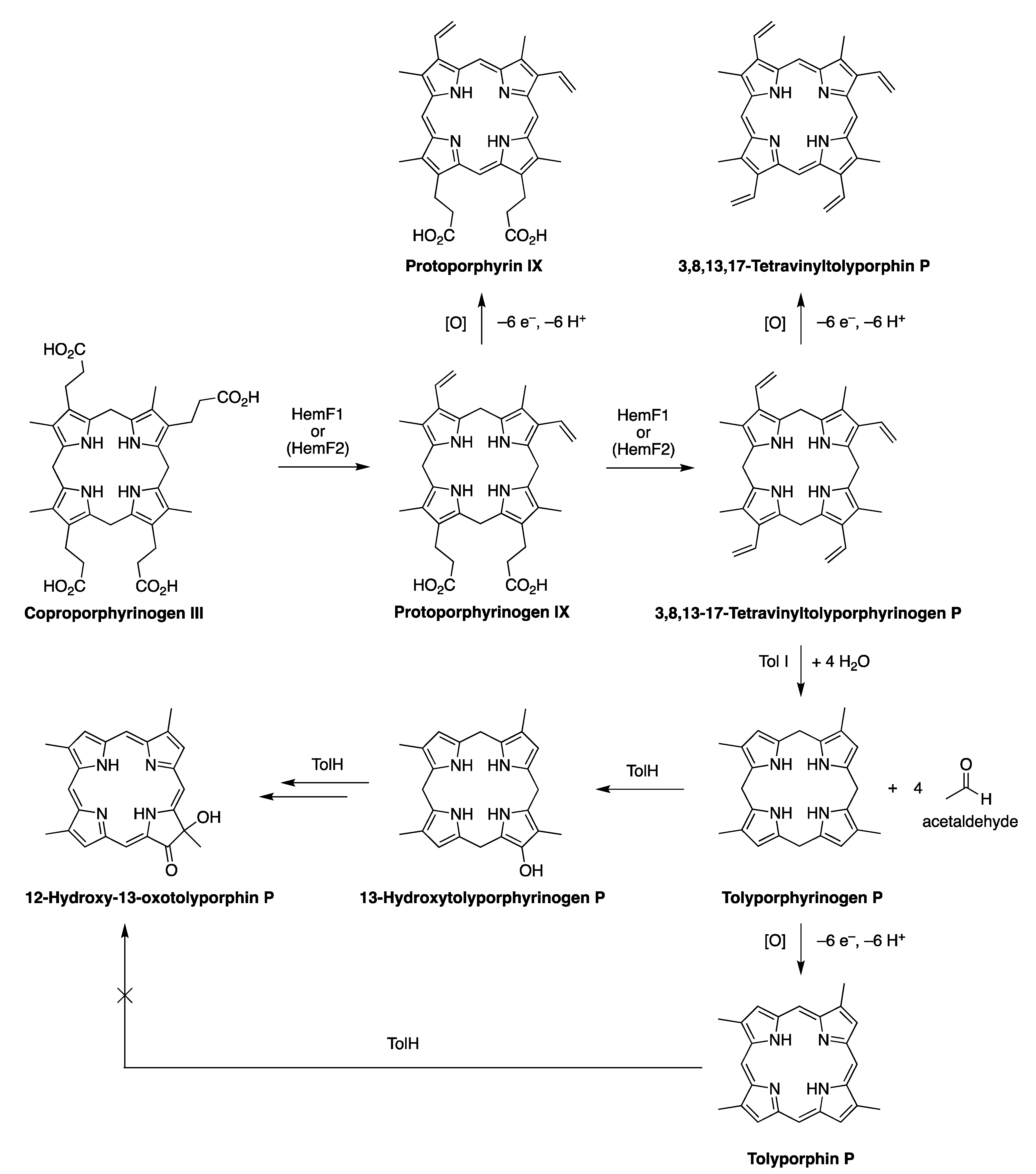
- How?—knowledge of the biosynthesis is rapidly advancing given heterologous expression of genes from the putative BGCs of the HT-58-2 cyanobacterium, as well as Oculatella sp. LEGE 06141. Understanding how hydrocarbon (vinyl) groups are removed from the β-pyrrolic positions downstream from uroporphyrinogen III, presumably by the TolI enzyme, is of utmost interest. Research on this topic is under active investigation [217]. One pathway entails formation of tetravinylporphyrinogen P, followed by hydration and loss of acetaldehyde from each position, thereby forming tolyporphyrinogen P. The role of tolyporphyrinogen P as a possible intermediate for late-stage diversification to form the entire repertoire of tolyporphins is a recent and intriguing hypothesis. The presence of the gem-dialkyl motifs in tolyporphins (all except P, which is a porphyrin) has structural relationship with the hydroporphyrins siroheme, heme d1, cobalamin and F430; but such may be purely coincidental—the results of Abe and coworkers indicate that tolyporphins emanate from a new biosynthetic branch in the tree of the pigments of life. The question of whether there are protein hosts or chaperones for tolyporphins (and tolyporphyrinogens), which are relatively hydrophobic structures, is unknown.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Wilson, E.O. Exploring a Little-Known Planet. Scientist 2011, 25, 64. [Google Scholar]
- Battersby, A.R.; Fookes, C.J.R.; Matcham, G.W.J.; McDonald, E. Biosynthesis of the pigments of life: Formation of the macrocycle. Nature 1980, 285, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Battersby, A.R. Biosynthesis of the Pigments of Life. Proc. R. Soc. Lond. B 1985, 225, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Battersby, A.R. Tetrapyrroles: The pigments of life. Nat. Prod. Rep. 2000, 17, 507–526. [Google Scholar] [CrossRef] [PubMed]
- Montforts, F.-P.; Gerlach, B.; Höper, F. Discovery and Synthesis of Less Common Natural Hydroporphyrins. Chem. Rev. 1994, 94, 327–347. [Google Scholar] [CrossRef]
- Manning, W.M.; Strain, H.H. Chlorophyll d, a green pigment of red algae. J. Biol. Chem. 1943, 151, 1–19. [Google Scholar] [CrossRef]
- Miyashita, H.; Ikemoto, H.; Kurano, N.; Adachi, K.; Chihara, M.; Miyachi, S. Chlorophyll d as a major pigment. Nature 1996, 383, 402. [Google Scholar] [CrossRef]
- Chen, M.; Schliep, M.; Willows, R.D.; Cai, Z.-L.; Neilan, B.A.; Scheer, H. A Red-Shifted Chlorophyll. Science 2010, 329, 1318–1319. [Google Scholar] [CrossRef]
- Ohashi, S.; Miyashita, H.; Okada, N.; Iemura, T.; Watanabe, T.; Kobayashi, M. Unique photosystems in Acaryochloris marina. Photosynth. Res. 2008, 98, 141–149. [Google Scholar] [CrossRef]
- Li, Y.; Lin, Y.; Loughlin, P.C.; Chen, M. Optimization and effects of different culture conditions on growth of Halomicronema hongdechloris—A filamentous cyanobacterium containing chlorophyll f. Front. Plant Sci. 2014, 5, 67. [Google Scholar] [CrossRef]
- Stirbet, A.; Lazár, D.; Papageorgiou, G.C.; Govindjee. Chlorophyll a Fluorescence in Cyanobacteria: Relation to Photosynthesis. In Cyanobacteria—From Basic Science to Applications; Mishra, A.K., Tiwari, D.N., Rai, A.N., Eds.; Academic Press: London, UK, 2019; pp. 79–130. [Google Scholar]
- Gaysina, L.A.; Saraf, A.; Singh, P. Cyanobacteria in Diverse Habitats. In Cyanobacteria—From Basic Science to Applications; Mishra, A.K., Tiwari, D.N., Rai, A.N., Eds.; Academic Press: London, UK, 2019; pp. 1–28. [Google Scholar]
- Gerwick, W.H.; Tan, L.T.; Sitachitta, N. Nitrogen-containing Metabolites From Marine Cyanobacteria. In The Alkaloids: Chemistry and Biology; Cordell, G.A., Ed.; Academic Press: San Diego, CA, USA, 2001; Volume 57, pp. 75–184. [Google Scholar]
- Macedo, M.F.; Miller, A.Z.; Dionísio, A.; Saiz-Jimenez, C. Biodiversity of cyanobacteria and green algae on monuments in the Mediterranean Basin: An overview. Microbiology 2009, 155, 3476–3490. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.; Rath, J. Extremophilic Cyanobacteria For Novel Drug Development; Springer: Cham, Switzerland, 2015; Volume 7. [Google Scholar] [CrossRef]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2016, 33, 382–431. [Google Scholar] [CrossRef] [PubMed]
- Allen, M.B. The Cultivation of Myxophyceae. Arch. Mikrobiol. 1952, 17, 34–53. [Google Scholar] [CrossRef]
- Patterson, G.M.L.; Larsen, L.K.; Moore, R.E. Bioactive natural products from blue-green algae. J. Appl. Phycol. 1994, 6, 151–157. [Google Scholar] [CrossRef]
- Patterson, G.M.L.; Baldwin, C.L.; Bolis, C.M.; Caplan, F.R.; Karuso, H.; Larsen, L.K.; Levine, I.A.; Moore, R.E.; Nelson, C.S.; Tschappat, K.D.; et al. Antineoplastic Activity of Cultured Blue-Green Algae (Cyanophyta). J. Phycol. 1991, 27, 530–536. [Google Scholar] [CrossRef]
- Prinsep, M.R.; Caplan, F.R.; Moore, R.E.; Patterson, G.M.L.; Smith, C.D. Tolyporphin, a Novel Multidrug Resistance Reversing Agent from the Blue-Green Alga Tolypothrix nodosa. J. Am. Chem. Soc. 1992, 114, 385–387. [Google Scholar] [CrossRef]
- Smith, C.D.; Prinsep, M.R.; Caplan, F.R.; Moore, R.E.; Patterson, G.M.L. Reversal of Multiple Drug Resistance by Tolyporphin, a Novel Cyanobacterial Natural Product. Oncol. Res. 1994, 6, 211–218. [Google Scholar]
- Trimurtulu, G.; Ohtani, I.; Patterson, G.M.L.; Moore, R.E.; Corbett, T.H.; Valeriote, F.A.; Demchik, L. Total Structures of Cryptophycins, Potent Antitumor Depsipeptides from the Blue-Green Alga Nostoc sp. Strain GSV 224. J. Am. Chem. Soc. 1994, 116, 4729–4737. [Google Scholar] [CrossRef]
- Magarvey, N.A.; Beck, Z.Q.; Golakoti, T.; Ding, Y.; Huber, U.; Hemscheidt, T.K.; Abelson, D.; Moore, R.E.; Sherman, D.H. Biosynthetic Characterization and Chemoenzymatic Assembly of the Cryptophycins. Potent Anticancer Agents from Nostoc Cyanobionts. ACS Chem. Biol. 2006, 1, 766–779. [Google Scholar] [CrossRef]
- Dai, J.; Philbin, C.S.; Wakano, C.; Yoshida, W.Y.; Williams, P.G. New Nostocyclophanes from Nostoc linckia. Mar. Drugs 2023, 21, 101. [Google Scholar] [CrossRef]
- Karolle, B.G. Atlas of Micronesia, 2nd ed.; The Bess Press, Inc.: Honolulu, HI, USA, 1993. [Google Scholar]
- Carucci, L.M.; Poyer, L. The West Central Pacific. In Oceania—An Introduction to the Cultures and Identities of Pacific Islanders; Strathern, A., Stewart, P.J., Carucci, L.M., Poyer, L., Feinberg, R., Macpherson, C., Eds.; Carolina Academic Press: Durham, NC, USA, 2002; pp. 183–249. [Google Scholar]
- Childress, D.H. Ancient Micronesia & The Lost City of Nan Madol; Adventures Unlimited Press: Kempton, IL, USA, 1998. [Google Scholar]
- Dextro, R.B.; Delbaje, E.; Cotta, S.R.; Zehr, J.P.; Fiore, M.F. Trends in Free-access Genomic Data Accelerate Advances in Cyanobacteria Taxonomy. J. Phycol. 2021, 57, 1392–1402. [Google Scholar] [CrossRef] [PubMed]
- Cavaleiro, J.A.S.; Tomé, J.P.C.; Faustino, M.A.F. Synthesis of Glycoporphyrins. Top. Heterocycl. Chem. 2007, 7, 179–248. [Google Scholar]
- Puddick, J.; Prinsep, M.R. MALDI-TOF Mass Spectrometry of Cyanobacteria: A Global Approach to the Discovery of Novel Secondary Metabolites. Chem. NZ 2008, 72, 68–71. [Google Scholar]
- Tidgewell, K.; Clark, B.R.; Gerwick, W.H. The Natural Products Chemistry of Cyanobacteria. In Comprehensive Natural Products II: Chemistry and Biology; Mander, L., Lui, H.-W., Eds.; Elsevier: Oxford, UK, 2010; Volume 2, pp. 141–188. [Google Scholar]
- Singh, R.K.; Tiwari, S.P.; Rai, A.K.; Mohapatra, T.M. Cyanobacteria: An emerging source for drug discovery. J. Antibiot. 2011, 64, 401–412. [Google Scholar] [CrossRef]
- Elshahawi, S.I.; Shaaban, K.A.; Kharel, M.K.; Thorson, J.S. A comprehensive review of glycosylated bacterial natural products. Chem. Soc. Rev. 2015, 44, 7591–7697. [Google Scholar] [CrossRef]
- Brückner, C. Tolyporphin–An Unusual Green Chlorin-like Dioxobacteriochlorin. Photochem. Photobiol. 2017, 93, 1320–1325. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.M. Honoring the 70th birthday of Professor Yoshito Kishi. Heterocycles 2007, 72, 1–3. [Google Scholar] [CrossRef]
- Wang, A.H. Yoshito Kishi, Organic Chemist Who Climbed ‘Mount Everest’ of Synthesis, Dies at 85. Harv. Crimson 2023, 21, 1–9. [Google Scholar]
- Lowe, D. Prof. Yoshito Kishi, 1937–2023. Science 2023. Available online: https://www.science.org/content/blog-post/prof-yoshito-kishi-1937-2023 (accessed on 12 July 2023).
- Minehan, T.G.; Kishi, Y. Extension of the Eschenmoser Sulfide Contraction/Iminoester Cyclization Method to the Synthesis of Tolyporphin Chromophore. Tetrahedron Lett. 1997, 38, 6811–6814. [Google Scholar] [CrossRef]
- Minehan, T.G.; Kishi, Y. β-Selective C-Glycosidations: Lewis-Acid Mediated Reactions of Carbohydrates with Silyl Ketene Acetals. Tetrahedron Lett. 1997, 38, 6815–6818. [Google Scholar] [CrossRef]
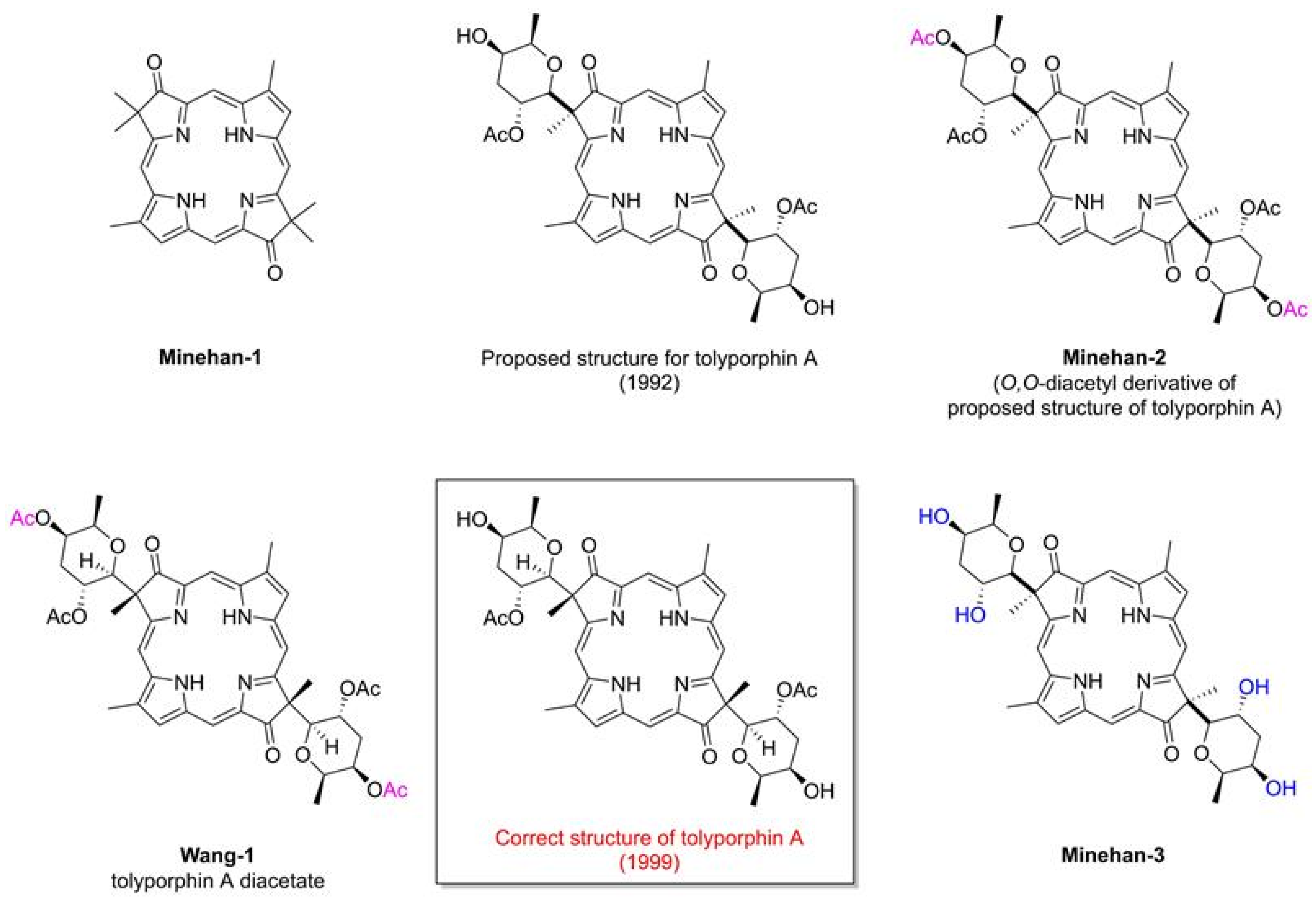
- Minehan, T.G.; Kishi, Y. Total Synthesis of the Proposed Structure of (+)-Tolyporphin A O,O-Diacetate. Angew. Chem. Int. Ed. 1999, 38, 923–925. [Google Scholar] [CrossRef]
- Minehan, T.G.; Cook-Blumberg, L.; Kishi, Y.; Prinsep, M.R.; Moore, R.E. Revised Structure of Tolyporphin A. Angew. Chem. Int. Ed. 1999, 38, 926–928. [Google Scholar] [CrossRef]
- Wang, W.; Kishi, Y. Synthesis and Structure of Tolyporphin A O,O-Diacetate. Org. Lett. 1999, 1, 1129–1132. [Google Scholar] [CrossRef] [PubMed]
- Eliel, E.L.; Wilen, S.H. Stereochemistry of Organic Compounds; John Wiley & Sons, Inc.: New York, NY, USA, 1994. [Google Scholar]
- Liu, Y.; Zhang, S.; Lindsey, J.S. Total synthesis campaigns toward chlorophylls and related natural hydroporphyrins—Diverse macrocycles, unrealized opportunities. Nat. Prod. Rep. 2018, 35, 879–901. [Google Scholar] [CrossRef]
- Prinsep, M.R.; Patterson, G.M.L.; Larsen, L.K.; Smith, C.D. Further Tolyporphins from the Blue-Green Alga Tolypothrix nodosa. Tetrahedron 1995, 51, 10523–10530. [Google Scholar] [CrossRef]
- Prinsep, M.R.; Patterson, G.M.L.; Larsen, L.K.; Smith, C.D. Tolyporphins J and K, Two Further Porphinoid Metabolites from the Cyanobacterium Tolypothrix nodosa. J. Nat. Prod. 1998, 61, 1133–1136. [Google Scholar] [CrossRef]
- Prinsep, M.R.; Puddick, J. Laser Desorption Ionisation-Time of Flight Mass Spectrometry of the Tolyporphins, Bioactive Metabolites from the Cyanobacterium Tolypothrix nodosa. Phytochem. Anal. 2011, 22, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Gurr, J.R.; Dai, J.; Philbin, C.S.; Sartain, H.T.; O’Donnell, T.J.; Yoshida, W.Y.; Rheingold, A.L.; Williams, P.G. Tolyporphins L–R: Unusual Tetrapyrroles from a Brasilonema sp. of Cyanobacterium. J. Org. Chem. 2020, 85, 318–326. [Google Scholar] [CrossRef]
- Hood, D.; Niedzwiedzki, D.M.; Zhang, R.; Zhang, Y.; Dai, J.; Miller, E.S.; Bocian, D.F.; Williams, P.G.; Lindsey, J.S.; Holten, D. Photophysical Characterization of the Naturally Occurring Dioxobacteriochlorin Tolyporphin A and Synthetic Oxobacteriochlorin Analogues. Photochem. Photobiol. 2017, 93, 1204–1215. [Google Scholar] [CrossRef]
- Firn, R.D.; Jones, C.G. A Darwinian view of metabolism: Molecular properties determine fitness. J. Exp. Bot. 2009, 60, 719–726. [Google Scholar] [CrossRef]
- Prinsep, M.R.; Thomson, R.A.; West, M.L.; Wylie, B.L. Tolypodiol, an Antiinflammatory Diterpenoid from the Cyanobacterium Tolypothrix nodosa. J. Nat. Prod. 1996, 59, 786–788. [Google Scholar] [CrossRef] [PubMed]
- Morlière, P.; Mazière, J.-C.; Santus, R.; Smith, C.D.; Prinsep, M.R.; Stobbe, C.C.; Fenning, M.C.; Golberg, J.L.; Chapman, J.D. Tolyporphin: A Natural Product from Cyanobacteria with Potent Photosensitizing Activity against Tumor Cells in Vitro and in Vivo. Cancer Res. 1998, 58, 3571–3578. [Google Scholar] [PubMed]
- Cardellina, J.H.; Moore, B.S. Richard E. Moore (1933–2007). J. Nat. Prod. 2010, 73, 301–302. [Google Scholar] [CrossRef]
- Williams, P.G.; (Department of Chemistry, University of Hawaii, Honolulu, HI, USA). Personal communication, 2015.
- Kim, H.-J.; Lindsey, J.S. De Novo Synthesis of Stable Tetrahydroporphyrinic Macrocycles: Bacteriochlorins and a Tetradehydrocorrin. J. Org. Chem. 2005, 70, 5475–5486. [Google Scholar] [CrossRef]
- Krayer, M.; Ptaszek, M.; Kim, H.-J.; Meneely, K.R.; Fan, D.; Secor, K.; Lindsey, J.S. Expanded Scope of Synthetic Bacteriochlorins via Improved Acid Catalysis Conditions and Diverse Dihydrodipyrrin-Acetals. J. Org. Chem. 2010, 75, 1016–1039. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, J.S. De Novo Synthesis of Gem-Dialkyl Chlorophyll Analogues for Probing and Emulating Our Green World. Chem. Rev. 2015, 115, 6534–6620. [Google Scholar] [CrossRef]
- Bryant, D.A.; Hunter, C.N.; Warren, M.J. Biosynthesis of the modified tetrapyrroles—The pigments of life. J. Biol. Chem. 2020, 295, 6888–6925. [Google Scholar] [CrossRef]
- Taniguchi, M.; Ptaszek, M.; Chandrashaker, V.; Lindsey, J.S. The Porphobilinogen Conundrum in Prebiotic Routes to Tetrapyrrole Macrocycles. Orig. Life Evol. Biosph. 2017, 47, 93–119. [Google Scholar] [CrossRef]
- Mauzerall, D. The Condensation of Porphobilinogen to Uroporphyrinogen. J. Am. Chem. Soc. 1960, 82, 2605–2609. [Google Scholar] [CrossRef]
- Mauzerall, D. The Thermodynamic Stability of Porphyrinogens. J. Am. Chem. Soc. 1960, 82, 2601–2605. [Google Scholar] [CrossRef]
- Battersby, A.R.; McDonald, E. Biosynthesis of Porphyrins, Chlorins and Corrins. In Porphyrins and Metalloporphyrins; Smith, K.M., Ed.; Elsevier: New York, NY, USA, 1975; pp. 61–122. [Google Scholar]
- Mauzerall, D.C. Evolution of Porphyrins. Clin. Dermatol. 1998, 16, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Soares, A.R.M.; Anderson, D.R.; Chandrashaker, V.; Lindsey, J.S. Catalytic diversification upon metal scavenging in a prebiotic model for formation of tetrapyrrole macrocycles. New J. Chem. 2013, 37, 2716–2732. [Google Scholar] [CrossRef]
- Moss, G.P. Nomenclature of Tetrapyrroles. Pure Appl. Chem. 1987, 59, 779–832. [Google Scholar] [CrossRef]
- Lindsey, J.S. Considerations of the biosynthesis and molecular diversity of tolyporphins. New J. Chem. 2021, 45, 12097–12107. [Google Scholar] [CrossRef]
- Firn, R.D.; Jones, C.G. The evolution of secondary metabolism—A unifying model. Mol. Microbiol. 2000, 37, 989–994. [Google Scholar] [CrossRef] [PubMed]
- Firn, R.D.; Jones, C.G. Natural products—A simple model to explain chemical diversity. Nat. Prod. Rep. 2003, 20, 382–391. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, R.; Hughes, R.-A.; Dai, J.; Gurr, J.R.; Williams, P.G.; Miller, E.S.; Lindsey, J.S. Quantitation of Tolyporphins, Diverse Tetrapyrrole Secondary Metabolites with Chlorophyll-Like Absorption, from a Filamentous Cyanobacterium–Microbial Community. Phytochem. Anal. 2018, 29, 205–216. [Google Scholar] [CrossRef]
- Taniguchi, M.; Lindsey, J.S. Absorption and Fluorescence Spectral Database of Chlorophylls and Analogues. Photochem. Photobiol. 2021, 97, 136–165. [Google Scholar] [CrossRef]
- Yang, E.; Kirmaier, C.; Krayer, M.; Taniguchi, M.; Kim, H.-J.; Diers, J.R.; Bocian, D.F.; Lindsey, J.S.; Holten, D. Photophysical Properties and Electronic Structure of Stable, Tunable Synthetic Bacteriochlorins: Extending the Features of Native Photosynthetic Pigments. J. Phys. Chem. B 2011, 115, 10801–10816. [Google Scholar] [CrossRef]
- Chen, C.-Y.; Sun, E.; Fan, D.; Taniguchi, M.; McDowell, B.E.; Yang, E.; Diers, J.R.; Bocian, D.F.; Holten, D.; Lindsey, J.S. Synthesis and Physicochemical Properties of Metallobacteriochlorins. Inorg. Chem. 2012, 51, 9443–9464. [Google Scholar] [CrossRef]
- Springer, J.W.; Faries, K.M.; Diers, J.R.; Muthiah, C.; Mass, O.; Kee, H.L.; Kirmaier, C.; Lindsey, J.S.; Bocian, D.F.; Holten, D. Effects of Substituents on Synthetic Analogs of Chlorophylls. Part 3: The Distinctive Impact of Auxochromes at the 7- versus 3-Positions. Photochem. Photobiol. 2012, 88, 651–674. [Google Scholar] [CrossRef]
- Lindsey, J.S.; Taniguchi, M.; Bocian, D.F.; Holten, D. The fluorescence quantum yield parameter in Förster resonance energy transfer (FRET)—Meaning, misperception, and molecular design. Chem. Phys. Rev. 2021, 2, 011302. [Google Scholar] [CrossRef]
- Taniguchi, M.; Lindsey, J.S. Synthetic Chlorins, Possible Surrogates for Chlorophylls, Prepared by Derivatization of Porphyrins. Chem. Rev. 2017, 117, 344–535. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, M.; Lindsey, J.S. Absorption and fluorescence spectra of organic compounds from 40 sources—Archives, repositories, databases, and literature search engines. Proc. SPIE 2020, 11256, 112560J. [Google Scholar] [CrossRef]
- Castenholz, R.W. Culturing Methods for Cyanobacteria. Methods Enzymol. 1988, 167, 68–93. [Google Scholar] [CrossRef]
- Moss, N.A.; Leao, T.; Glukhov, E.; Gerwick, L.; Gerwick, W.H. Collection, Culturing, and Genome Analyses of Tropical Marine Filamentous Benthic Cyanobacteria. Methods Enzymol. 2018, 604, 3–43. [Google Scholar] [CrossRef]
- Hughes, R.-A.; Zhang, Y.; Zhang, R.; Williams, P.G.; Lindsey, J.S.; Miller, E.S. Genome Sequence and Composition of a Tolyporphin-Producing Cyanobacterium-Microbial Community. Appl. Environ. Microbiol. 2017, 83, e01068-17. [Google Scholar] [CrossRef]
- Lee, K.-B.; Liu, C.-T.; Anzai, Y.; Kim, H.; Aono, T.; Oyaizu, H. The hierarchical system of the ‘Alphaproteobacteria’: Description of Hyphomonadaceae fam. nov., Xanthobacteraceae fam. nov. and Erythrobacteraceae fam. nov. Int. J. Syst. Evol. Microbiol. 2005, 55, 1907–1919. [Google Scholar] [CrossRef]
- Komárek, J.; Sant’Anna, C.L.; Bohunická, M.; Mareš, J.; Hentschke, G.S.; Rigonato, J.; Fiore, M.F. Phenotype diversity and phylogeny of selected Scytonema-species (Cyanoprokaryota) from SE Brazil. Fottea 2013, 13, 173–200. [Google Scholar] [CrossRef]
- Fiore, M.F.; Sant’Anna, C.L.; Azevedo, M.T.d.P.; Komárek, J.; Kaštovský, J.; Sulek, J.; Lorenzi, A.S. The Cyanobacterial Genus Brasilonema, Gen. Nov., a Molecular and Phenotypic Evaluation. J. Phycol. 2007, 43, 789–798. [Google Scholar] [CrossRef]
- Vaccarino, M.A.; Johansen, J.R. Brasilonema Angustatum Sp. Nov. (Nostocales), a New Filamentous Cyanobacterial Species from the Hawaiian Islands. J. Phycol. 2012, 48, 1178–1186. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, M.; Berthold, D.E.; Lefler, F.W.; Laughinghouse IV, H.D. Diversity of the genus Brasilonema (Nostocales, Cyanobacteria) in plant nurseries of central Florida (USA) with the description of three new species: B. fioreae sp. nov., B. santannae sp. nov. and B. wernerae sp. nov. Fottea 2021, 21, 82–99. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Bryant, D. Application of Phylogenetic Networks in Evolutionary Studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Alvarenga, D.O.; Fiore, M.F.; Varani, A.M. A Metagenomic Approach to Cyanobacterial Genomics. Front. Microbiol. 2017, 8, 809. [Google Scholar] [CrossRef]
- Jin, X.; Miller, E.S.; Lindsey, J.S. Natural Product Gene Clusters in the Filamentous Nostocales Cyanobacterium HT-58-2. Life 2021, 11, 356. [Google Scholar] [CrossRef]
- Pfeffer, S.; Brown, R.M., Jr. Complete Genome Sequence of the Cyanobacterium Anabaena sp. 33047. Genome Announc. 2016, 4, e00809-16. [Google Scholar] [CrossRef]
- Kaneko, T.; Sato, S.; Kotani, H.; Tanaka, A.; Asamizu, E.; Nakamura, Y.; Miyajima, N.; Hirosawa, M.; Sugiura, M.; Sasamoto, S.; et al. Sequence Analysis of the Genome of the Unicellular Cyanobacterium Synechocystis sp. Strain PCC6803. II. Sequence Determination of the Entire Genome and Assignment of Potential Protein-coding Regions. DNA Res. 1996, 3, 109–136. [Google Scholar] [CrossRef]
- Partensky, F.; Garczarek, L. Prochlorococcus: Advantages and Limits of Minimalism. Ann. Rev. Mar. Sci. 2010, 2, 305–331. [Google Scholar] [CrossRef]
- Hughes, R.-A.; Jin, X.; Zhang, Y.; Zhang, R.; Tran, S.; Williams, P.G.; Lindsey, J.S.; Miller, E.S. Genome sequence, metabolic properties and cyanobacterial attachment of Porphyrobacter sp. HT-58-2 isolated from a filamentous cyanobacterium–microbial consortium. Microbiology 2018, 164, 1229–1239. [Google Scholar] [CrossRef]
- Hanada, S.; Kawase, Y.; Hiraishi, A.; Takaichi, S.; Matsuura, K.; Shimada, K.; Nagashima, K.V.P. Porphyrobacter tepidarius sp. nov., a Moderately Thermophilic Aerobic Photosynthetic Bacterium Isolated from a Hot Spring. Int. J. Syst. Bacteriol. 1997, 47, 408–413. [Google Scholar] [CrossRef] [PubMed]
- Sieber, J.R.; McInerney, M.J.; Gunsalus, R.P. Genomic Insights into Syntrophy: The Paradigm for Anaerobic Metabolic Cooperation. Annu. Rev. Microbiol. 2012, 66, 429–452. [Google Scholar] [CrossRef] [PubMed]
- Kouzuma, A.; Kato, S.; Watanabe, K. Microbial interspecies interactions: Recent findings in syntrophic consortia. Front. Microbiol. 2015, 6, 477. [Google Scholar] [CrossRef]
- Romine, M.F.; Rodionov, D.A.; Maezato, Y.; Osterman, A.L.; Nelson, W.C. Underlying mechanisms for syntrophic metabolism of essential enzyme cofactors in microbial communities. ISME J. 2017, 11, 1434–1446. [Google Scholar] [CrossRef]
- Flores, E.; López-Lozano, A.; Herrero, A. Nitrogen Fixation in the Oxygenic Phototrophic Prokaryotes (Cyanobacteria): The Fight Against Oxygen. In Biological Nitrogen Fixation; de Bruijn, F.J., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015; Volume 2, pp. 879–890. [Google Scholar]
- Leao, T.; Castelão, G.; Korobeynikov, A.; Monroe, E.A.; Podell, S.; Glukhov, E.; Allen, E.E.; Gerwick, W.H.; Gerwick, L. Comparative genomics uncovers the prolific and distinctive metabolic potential of the cyanobacterial genus Moorea. Proc. Natl. Acad. Sci. USA 2017, 114, 3198–3203. [Google Scholar] [CrossRef] [PubMed]
- Dvořák, P.; Hašler, P.; Casamatta, D.A.; Poulíčková, A. Underestimated cyanobacterial diversity: Trends and perspectives of research in tropical environments. Fottea 2021, 21, 110–127. [Google Scholar] [CrossRef]
- Rigonato, J.; Alvarenga, D.O.; Fiore, M.F. Tropical Cyanobacteria and their Biotechnological Applications. In Diversity and Benefits of Microorganisms from the Tropics; de Azevedo, J.L., Quecine, M.C., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 139–167. [Google Scholar]
- Genuário, D.B.; Vaz, M.G.M.V.; de Melo, I.S. Phylogenetic insights into the diversity of homocytous cyanobacteria from Amazonian rivers. Mol. Phylogenet. Evol. 2017, 116, 120–135. [Google Scholar] [CrossRef]
- de Oliveira, E.D.C.; Da Cunha, A.C.; Da Silva, N.B.; Castelo-Branco, R.; Morais, J.; Schneider, M.P.C.; Faustino, S.M.M.; Ramos, V.; Vasconcelos, V. Morphological and molecular characterization of cyanobacterial isolates from the mouth of the Amazon River. Phytotaxa 2019, 387, 269–288. [Google Scholar] [CrossRef]
- Leão, T.; Wang, M.; Moss, N.; da Silva, R.; Sanders, J.; Nurk, S.; Gurevich, A.; Humphrey, G.; Reher, R.; Zhu, Q.; et al. A Multi-Omics Characterization of the Natural Product Potential of Tropical Filamentous Marine Cyanobacteria. Mar. Drugs 2021, 19, 20. [Google Scholar] [CrossRef]
- O’Donnell, T.J.; Gurr, J.R.; Dai, J.; Taniguchi, M.; Williams, P.G.; Lindsey, J.S. Tolyporphins A–R, unusual tetrapyrrole macrocycles in a cyanobacterium from Micronesia, assessed quantitatively from the culture HT-58-2. New J. Chem. 2021, 45, 11481–11494. [Google Scholar] [CrossRef]
- Majumder, E.L.-W.; Wolf, B.M.; Liu, H.; Berg, R.H.; Timlin, J.A.; Chen, M.; Blankenship, R.E. Subcellular pigment distribution is altered under far-red light acclimation in cyanobacteria that contain chlorophyll f. Photosynth. Res. 2017, 134, 183–192. [Google Scholar] [CrossRef]
- Barnhart-Dailey, M.; Zhang, Y.; Zhang, R.; Anthony, S.M.; Aaron, J.S.; Miller, E.S.; Lindsey, J.S.; Timlin, J.A. Cellular localization of tolyporphins, unusual tetrapyrroles, in a microbial photosynthetic community determined using hyperspectral confocal fluorescence microscopy. Photosynth. Res. 2019, 141, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Soares, A.R.M.; Thanaiah, Y.; Taniguchi, M.; Lindsey, J.S. Aqueous–membrane partitioning of β-substituted porphyrins encompassing diverse polarity. New J. Chem. 2013, 37, 1087–1097. [Google Scholar] [CrossRef]
- Gurnani, M.; Chauhan, A.; Ranjan, A.; Gopi, P.; Ghosh, A.; Tuli, H.S.; Haque, S.; Pandya, P.; Lal, R.; Jindal, T. Cyanobacterial compound Tolyporphine K as an inhibitor of Apo-PBP (penicillin-binding protein) in A. baumannii and its ADME assessment. J. Biomol. Struct. Dyn. 2023, 41, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Karg, C.A.; Taniguchi, M.; Lindsey, J.S.; Moser, S. Phyllobilins—Bioactive Natural Products Derived from Chlorophyll—Plant Origins, Structures, Absorption Spectra, and Biomedical Properties. Planta Med. 2023, 89, 637–662. [Google Scholar] [CrossRef]
- Taniguchi, M.; LaRocca, C.A.; Bernat, J.D.; Lindsey, J.S. Digital Database of Absorption Spectra of Diverse Flavonoids Enables Structural Comparisons and Quantitative Evaluations. J. Nat. Prod. 2023, 86, 1087–1119. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, M.; Lindsey, J.S. Absorption and fluorescence spectra of open-chain tetrapyrrole pigments–bilirubins, biliverdins, phycobilins, and synthetic analogues. J. Photochem. Photobiol. C Photochem. Rev. 2023, 55, 100585. [Google Scholar] [CrossRef]
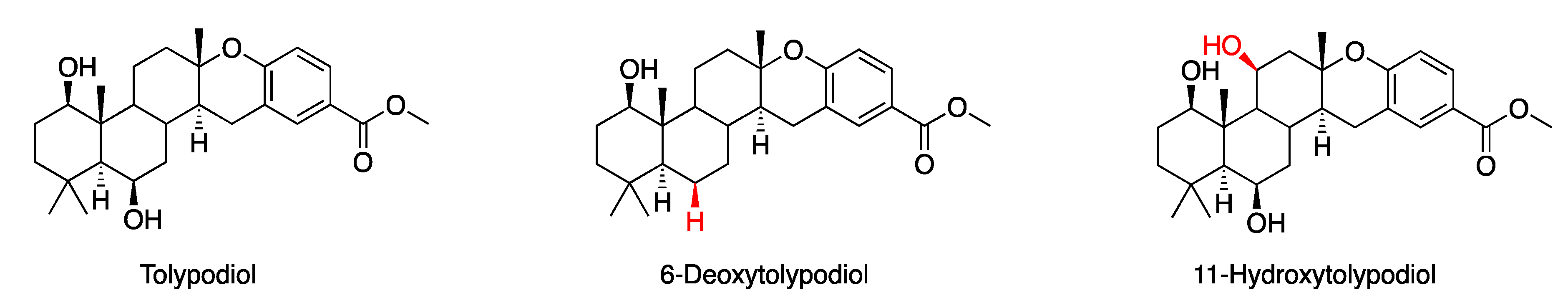
- Gurr, J.R.; O’Donnell, T.J.; Luo, Y.; Yoshida, W.Y.; Hall, M.L.; Mayer, A.M.S.; Sun, R.; Williams, P.G. 6-Deoxy- and 11-Hydroxytolypodiols: Meroterpenoids from the Cyanobacterium HT-58-2. J. Nat. Prod. 2020, 83, 1691–1695. [Google Scholar] [CrossRef]
- Prinsep, M.; (Department of Chemistry and Applied Physics, University of Waikato, Hamilton, NZ, USA). Personal communication, 2023.
- Rippka, R.; Deruelles, J.; Waterbury, J.B.; Herdman, M.; Stanier, R.Y. Generic Assignments, Strain Histories and Properties of Pure Cultures of Cyanobacteria. J. Gen. Microbiol. 1979, 111, 1–61. [Google Scholar] [CrossRef]
- Allen, M.M. Simple Conditions for Growth of Unicellular Blue-green Algae on Plates. J. Phycol. 1968, 4, 1–4. [Google Scholar] [CrossRef]
- Esteves-Ferreira, A.A.; Inaba, M.; Fort, A.; Araújo, W.L.; Sulpice, R. Nitrogen metabolism in cyanobacteria: Metabolic and molecular control, growth consequences and biotechnological applications. Crit. Rev. Microbiol. 2018, 44, 541–560. [Google Scholar] [CrossRef] [PubMed]
- Soret, J.-L. Analyse spectrale: Sur le spectre d’absorption du sang dans la partie violette et ultra-violette. Compt. Rend. Acad. Sci. 1883, 97, 1269–1270. [Google Scholar]
- Taniguchi, M.; Lindsey, J.S.; Bocian, D.F.; Holten, D. Comprehensive review of photophysical parameters (ε, Φf, τs) of tetraphenylporphyrin (H2TPP) and zinc tetraphenylporphyrin (ZnTPP)—Critical benchmark molecules in photochemistry and photosynthesis. J. Photochem. Photobiol. C Photochem. Rev. 2021, 46, 100401. [Google Scholar] [CrossRef]
- Gouterman, M. Optical Spectra and Electronic Structure of Porphyrins and Related Rings. In The Porphyrins, Dolphin, D., Ed.; Academic Press: New York, NY, USA, 1978; Volume 3, pp. 1–165. [Google Scholar]
- Minehan, T.G. Total Synthesis of the Proposed Structure of Tolyporphin A O,O-Diacetate and Structural Revision of the Tolyporphin Family of Natural Products. Ph.D. Thesis, Harvard University, Cambridge, MA, USA, 1998. [Google Scholar]
- Taniguchi, M.; Wu, Z.; Sterling, C.D.; Lindsey, J.S. Digitization of print-based absorption and fluorescence spectra—Extracting clarity from clutter. Proc. SPIE 2023, 12398, 1239806. [Google Scholar] [CrossRef]
- Magdaong, N.C.M.; Taniguchi, M.; Diers, J.R.; Niedzwiedzki, D.M.; Kirmaier, C.; Lindsey, J.S.; Bocian, D.F.; Holten, D. Photophysical Properties and Electronic Structure of Zinc(II) Porphyrins Bearing 0–4 meso-Phenyl Substituents: Zinc Porphine to Zinc Tetraphenylporphyrin (ZnTPP). J. Phys. Chem. A 2020, 124, 7776–7794. [Google Scholar] [CrossRef]
- Taniguchi, M.; Lindsey, J.S. Database of Absorption and Fluorescence Spectra of >300 Common Compounds for use in PhotochemCAD. Photochem. Photobiol. 2018, 94, 290–327. [Google Scholar] [CrossRef]
- Naes, H.; Aarnes, H.; Utkilen, H.C.; Nilsen, S.; Skulberg, O.M. Effect of Photon Fluence Rate and Specific Growth Rate on Geosmin Production of the Cyanobacterium Oscillatoria brevis (Kütz.) Gom. Appl. Environ. Microbiol. 1985, 49, 1538–1540. [Google Scholar] [CrossRef]
- Vendruscolo, R.G.; Fernandes, A.S.; Fagundes, M.B.; Zepka, L.Q.; de Menezes, C.R.; Jacob–Lopes, E.; Wagner, R. Development of a new method for simultaneous extraction of chlorophylls and carotenoids from microalgal biomass. J. Appl. Phycol. 2021, 33, 1987–1997. [Google Scholar] [CrossRef]
- Kuhne, S.; Lakatos, M.; Foltz, S.; Muffler, K.; Ulber, R. Characterization of terrestrial cyanobacteria to increase process efficiency in low energy consuming production processes. Sust. Chem. Proc. 2013, 1, 6. [Google Scholar] [CrossRef]
- Demain, A.L. Induction of microbial secondary metabolism. Int. Microbiol. 1998, 1, 259–264. [Google Scholar]
- Martín, J.F.; Sola-Landa, A.; Santos-Beneit, F.; Fernández-Martínez, L.T.; Prieto, C.; Rodríguez-García, A. Cross-talk of global nutritional regulators in the control of primary and secondary metabolism in Streptomyces. Microb. Biotechnol. 2011, 4, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Tudzynski, B. Nitrogen regulation of fungal secondary metabolism in fungi. Front. Microbiol. 2014, 5, 656. [Google Scholar] [CrossRef] [PubMed]
- Fenyo, D.; Chait, B.T.; Johnson, T.E.; Lindsey, J.S. Laser Desorption Mass Spectrometry of Synthetic Multiporphyrin Arrays. J. Porphyr. Phthalocyanines 1997, 1, 93–99. [Google Scholar] [CrossRef]
- Srinivasan, N.; Haney, C.A.; Lindsey, J.S.; Zhang, W.; Chait, B.T. Investigation of MALDI-TOF Mass Spectrometry of Diverse Synthetic Metalloporphyrins, Phthalocyanines and Multiporphyrin Arrays. J. Porphyr. Phthalocyanines 1999, 3, 283–291. [Google Scholar] [CrossRef]
- Calvano, C.D.; Ventura, G.; Cataldi, T.R.I.; Palmisano, F. Improvement of chlorophyll identification in foodstuffs by MALDI ToF/ToF mass spectrometry using 1,5-diaminonaphthalene electron transfer secondary reaction matrix. Anal. Bioanal. Chem. 2015, 407, 6369–6379. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, R.; Nazari, M.; Bagley, M.C.; Miller, E.S.; Williams, P.G.; Muddiman, D.C.; Lindsey, J.S. Mass spectrometric detection of chlorophyll a and the tetrapyrrole secondary metabolite tolyporphin A in the filamentous cyanobacterium HT-58-2. Approaches to high-throughput screening of intact cyanobacteria. J. Porphyr. Phthalocyanines 2017, 21, 759–768. [Google Scholar] [CrossRef]
- Parsons, T.R. On the Pigment Composition of Eleven Species of Marine Phytoplankters. J. Fish. Res. Board Can. 1961, 18, 1017–1025. [Google Scholar] [CrossRef]
- Merzlyak, M.N.; Chivkunova, O.B.; Maslova, I.P.; Naqvi, K.R.; Solovchenko, A.E.; Klyachko-Gurvich, G.L. Light Absorption and Scattering by Cell Suspensions of Some Cyanobacteria and Microalgae. Russ. J. Plant Physiol. 2008, 55, 420–425. [Google Scholar] [CrossRef]
- Thrane, J.-E.; Kyle, M.; Striebel, M.; Haande, S.; Grung, M.; Rohrlack, T.; Andersen, T. Spectrophotometric Analysis of Pigments: A Critical Assessment of a High-Throughput Method for Analysis of Algal Pigment Mixtures by Spectral Deconvolution. PLoS ONE 2015, 10, e0137645. [Google Scholar] [CrossRef]
- Watanabe, T.; Hongu, A.; Honda, K.; Nakazato, M.; Konno, M.; Saitoh, S. Preparation of Chlorophylls and Pheophytins by Isocratic Liquid Chromatography. Anal. Chem. 1984, 56, 251–256. [Google Scholar] [CrossRef]
- Taniguchi, M.; Du, H.; Lindsey, J.S. PhotochemCAD 3: Diverse Modules for Photophysical Calculations with Multiple Spectral Databases. Photochem. Photobiol. 2018, 94, 277–289. [Google Scholar] [CrossRef] [PubMed]
- Strain, H.H.; Svec, W.A. Extraction, Separation, Estimation, and Isolation of the Chlorophylls. In The Chlorophylls; Vernon, L.P., Seely, G.R., Eds.; Academic Press: New York, NY, USA, 1966; pp. 21–66. [Google Scholar]
- Simon, D.; Helliwell, S. Extraction and quantification of chlorophyll a from freshwater green algae. Wat. Res. 1998, 32, 2220–2223. [Google Scholar] [CrossRef]
- Chen, Y.; Li, G.; Pandey, R.K. Synthesis of Bacteriochlorins and Their Potential Utility in Photodynamic Therapy (PDT). Curr. Org. Chem. 2004, 8, 1105–1134. [Google Scholar] [CrossRef]
- Sasaki, S.; Tamiaki, H. Synthesis and Optical Properties of Bacteriochlorophyll-a Derivatives Having Various C3 Substituents on the Bacteriochlorin π-System. J. Org. Chem. 2006, 71, 2648–2654. [Google Scholar] [CrossRef]
- Grin, M.A.; Mironov, A.F.; Shtil, A.A. Bacteriochlorophyll a and Its Derivatives: Chemistry and Perspectives for Cancer Therapy. Anti-Cancer Agents Med. Chem. 2008, 8, 683–697. [Google Scholar] [CrossRef]
- Grin, M.A.; Mironov, A.F. Chemical transformations of bacteriochlorophyll a and its medical applications. Russ. Chem. Bull. Int. Ed. 2016, 65, 333–349. [Google Scholar] [CrossRef]
- Robichaud, G.; Barry, J.A.; Muddiman, D.C. IR-MALDESI Mass Spectrometry Imaging of Biological Tissue Sections Using Ice as a Matrix. J. Am. Soc. Mass Spectrom. 2014, 25, 319–328. [Google Scholar] [CrossRef]
- Bagley, M.C.; Muddiman, D.C. Investigations of β-carotene radical cation formation in infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI). Rapid Commun. Mass Spectrom. 2021, 35, e9133. [Google Scholar] [CrossRef]
- Judd, R.; Bagley, M.C.; Li, M.; Zhu, Y.; Lei, C.; Yuzuak, S.; Ekelöf, M.; Pu, G.; Zhao, X.; Muddiman, D.C.; et al. Artemisinin Biosynthesis in Non-glandular Trichome Cells of Artemisia annua. Mol. Plant 2019, 12, 704–714. [Google Scholar] [CrossRef]
- Bagley, M.C.; Stepanova, A.N.; Ekelöf, M.; Alonso, J.M.; Muddiman, D.C. Development of a relative quantification method for infrared matrix-assisted laser desorption electrospray ionization mass spectrometry imaging of Arabidopsis seedlings. Rapid Commun. Mass Spectrom. 2020, 34, e8616. [Google Scholar] [CrossRef]
- Kalmar, J.G.; Oh, Y.; Dean, R.A.; Muddiman, D.C. Investigating host-pathogen meta-metabolic interactions of Magnaporthe oryzae infected barley using infrared matrix-assisted laser desorption electrospray ionization mass spectrometry. Anal. Bioanal. Chem. 2020, 412, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Barnes, A.C.; Rodríguez-Zapata, F.; Juárez-Núñez, K.A.; Gates, D.J.; Janzen, G.M.; Kur, A.; Wang, L.; Jensen, S.E.; Estévez-Palmas, J.M.; Crow, T.M.; et al. An adaptive teosinte mexicana introgression modulates phosphatidylcholine levels and is associated with maize flowering time. Proc. Natl. Acad. Sci. USA 2022, 119, e2100036119. [Google Scholar] [CrossRef] [PubMed]
- Ekelöf, M.; McMurtrie, E.K.; Nazari, M.; Johanningsmeier, S.D.; Muddiman, D.C. Direct Analysis of Triterpenes from High-Salt Fermented Cucumbers Using Infrared Matrix-Assisted Laser Desorption Electrospray Ionization (IR-MALDESI). J. Am. Soc. Mass Spectrom. 2017, 28, 370–375. [Google Scholar] [CrossRef] [PubMed]
- Bagley, M.C.; Pace, C.L.; Ekelöf, M.; Muddiman, D.C. Infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) mass spectrometry imaging analysis of endogenous metabolites in cherry tomatoes. Analyst 2020, 145, 5516–5523. [Google Scholar] [CrossRef]
- Kasha, M. Characterization of Electronic Transitions in Complex Molecules. Disc. Faraday Soc. 1950, 9, 14–19. [Google Scholar] [CrossRef]
- Taniguchi, M.; Hu, G.; Liu, R.; Du, H.; Lindsey, J.S. Red and near-infrared fluorophores inspired by chlorophylls. Consideration of practical brightness in multicolor flow cytometry and biomedical sciences. Proc. SPIE 2018, 10508, 1050806. [Google Scholar] [CrossRef]
- Taniguchi, M.; Ptaszek, M.; McDowell, B.E.; Lindsey, J.S. Sparsely substituted chlorins as core constructs in chlorophyll analogue chemistry. Part 2: Derivatization. Tetrahedron 2007, 63, 3840–3849. [Google Scholar] [CrossRef]
- Taniguchi, M.; Ptaszek, M.; McDowell, B.E.; Boyle, P.D.; Lindsey, J.S. Sparsely substituted chlorins as core constructs in chlorophyll analogue chemistry. Part 3: Spectral and structural properties. Tetrahedron 2007, 63, 3850–3863. [Google Scholar] [CrossRef]
- Taniguchi, M.; Cramer, D.L.; Bhise, A.D.; Kee, H.L.; Bocian, D.F.; Holten, D.; Lindsey, J.S. Accessing the near-infrared spectral region with stable, synthetic, wavelength-tunable bacteriochlorins. New J. Chem. 2008, 32, 947–958. [Google Scholar] [CrossRef]
- Nguyen, K.-U.; Zhang, R.; Taniguchi, M.; Lindsey, J.S. Fluorescence Assay for Tolyporphins Amidst Abundant Chlorophyll in Crude Cyanobacterial Extracts. Photochem. Photobiol. 2021, 97, 1507–1515. [Google Scholar] [CrossRef]
- Hansson, M.; Rutberg, L.; Schröder, I.; Hederstedt, L. The Bacillus subtilis hemAXCDBL Gene Cluster, Which Encodes Enzymes of the Biosynthetic Pathway from Glutamate to Uroporphyrinogen III. J. Bacteriol. 1991, 173, 2590–2599. [Google Scholar] [CrossRef] [PubMed]
- Morita, H.; Toh, H.; Fukuda, S.; Horikawa, H.; Oshima, K.; Suzuki, T.; Murakami, M.; Hisamatsu, S.; Kato, Y.; Takizawa, T.; et al. Comparative Genome Analysis of Lactobacillus reuteri and Lactobacillus fermentum Reveal a Genomic Island for Reuterin and Cobalamin Production. DNA Res. 2008, 15, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Kafala, B.; Sasarman, A. Isolation of the Staphylococcus aureus hemCDBL gene cluster coding for early steps in heme biosynthesis. Gene 1997, 199, 231–239. [Google Scholar] [CrossRef]
- Echelard, Y.; Dymetryszyn, J.; Drolet, M.; Sasarman, A. Nucleotide Sequence of the hemB Gene of Escherichia coli K12. Mol. Gen. Genet. 1988, 214, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Elliott, T. Cloning, Genetic Characterization, and Nucleotide Sequence of the hemA-prfA Operon of Salmonella typhimurium. J. Bacteriol. 1989, 171, 3948–3960. [Google Scholar] [CrossRef]
- Jin, X.; Zhang, Y.; Zhang, R.; Nguyen, K.-U.; Lindsey, J.S.; Miller, E.S. Identification of Putative Biosynthetic Gene Clusters for Tolyporphins in Multiple Filamentous Cyanobacteria. Life 2021, 11, 758. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, J.S.; Chandrashaker, V.; Taniguchi, M.; Ptaszek, M. Abiotic formation of uroporphyrinogen and coproporphyrinogen from acyclic reactants. New J. Chem. 2011, 35, 65–75. [Google Scholar] [CrossRef]
- Taniguchi, M.; Soares, A.R.M.; Chandrashaker, V.; Lindsey, J.S. A tandem combinatorial model for the prebiogenesis of diverse tetrapyrrole macrocycles. New J. Chem. 2012, 36, 1057–1069. [Google Scholar] [CrossRef]
- Dittmann, E.; Gugger, M.; Sivonen, K.; Fewer, D.P. Natural Product Biosynthetic Diversity and Comparative Genomics of the Cyanobacteria. Trends Microbiol. 2015, 23, 642–652. [Google Scholar] [CrossRef]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Müller, R.; Wohlleben, W.; et al. antiSMASH 3.0—A comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef]
- Wörmer, L.; Cirés, S.; Velázquez, D.; Quesada, A.; Hinrichs, K.-U. Cyanobacterial heterocyst glycolipids in cultures and environmental samples: Diversity and biomarker potential. Limnol. Oceanogr. 2012, 57, 1775–1788. [Google Scholar] [CrossRef]
- Stratmann, K.; Burgoyne, D.L.; Moore, R.E.; Patterson, G.M.L.; Smith, C.D. Hapalosin, a Cyanobacterial Cyclic Depsipeptide with Multidrug-Resistance Reversing Activity. J. Org. Chem. 1994, 59, 7219–7226. [Google Scholar] [CrossRef]
- Rogers, E.H.; Hunter III, E.S.; Moser, V.C.; Phillips, P.M.; Herkovits, J.; Muñoz, L.; Hall, L.L.; Chernoff, N. Potential developmental toxicity of anatoxin-a, a cyanobacterial toxin. J. Appl. Toxicol. 2005, 25, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Carmichael, W.W.; Gorham, P.R.; Biggs, D.F. Two laboratory case studies on the oral toxicity to calves of the freshwater cyanophyte (blue-green alga) Anabaena flos-aquae NRC-44-1. Can. Vet. J. 1977, 18, 71–75. [Google Scholar] [PubMed]
- Rastogi, R.P.; Incharoensakdi, A. Characterization of UV-screening compounds, mycosporine-like amino acids, and scytonemin in the cyanobacterium Lyngbya sp. CU2555. FEMS Microbiol. Ecol. 2014, 87, 244–256. [Google Scholar] [CrossRef]
- Back, D.; O’Donnell, T.J.; Axt, K.K.; Gurr, J.R.; Vanegas, J.M.; Williams, P.G.; Philmus, B. Identification, Heterologous Expression, and Characterization of the Tolypodiol Biosynthetic Gene Cluster through an Integrated Approach. ACS Chem. Biol. 2023. [Google Scholar] [CrossRef]
- Welker, M.; von Döhren, H. Cyanobacterial peptides—Nature’s own combinatorial biosynthesis. FEMS Microbiol. Rev. 2006, 30, 530–563. [Google Scholar] [CrossRef]
- Gilchrist, C.L.M.; Booth, T.J.; van Wersch, B.; van Grieken, L.; Medema, M.H.; Chooi, Y.-H. cblaster: A remote search tool for rapid identification and visualization of homologous gene clusters. Bioinform. Adv. 2021, 1, vbab016. [Google Scholar] [CrossRef]
- van den Belt, M.; Gilchrist, C.; Booth, T.J.; Chooi, Y.-H.; Medema, M.H.; Alanjary, M. CAGECAT: The CompArative GEne Cluster Analysis Toolbox for rapid search and visualisation of homologous gene clusters. BMC Bioinform. 2023, 24, 181. [Google Scholar] [CrossRef]
- Jensen, P.R. Natural Products and the Gene Cluster Revolution. Trends Microbiol. 2016, 24, 968–977. [Google Scholar] [CrossRef]
- Díaz, E.; Ferrández, A.; García, J.L. Characterization of the hca Cluster Encoding the Dioxygenolytic Pathway for Initial Catabolism of 3-Phenylpropionic Acid in Escherichia coli K-12. J. Bacteriol. 1998, 180, 2915–2923. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-García, K.; Bustos-Díaz, E.D.; Corona-Gómez, J.A.; Ramos-Aboites, H.E.; Sélem-Mojica, N.; Cruz-Morales, P.; Pérez-Farrera, M.A.; Barona-Gómez, F.; Cibrían-Jaramillo, A. Cycad Coralloid Roots Contain Bacterial Communities Including Cyanobacteria and Caulobacter spp. That Encode Niche-Specific Biosynthetic Gene Clusters. Genome Biol. Evol. 2019, 11, 319–334. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Zhang, D.; Huang, Z.; Liu, Y. The vertical microdistribution of cyanobacteria and green algae within desert crusts and the development of the algal crusts. Plant Soil 2003, 257, 97–111. [Google Scholar] [CrossRef]
- Sokolovskaya, O.M.; Shelton, A.N.; Taga, M.E. Sharing vitamins: Cobamides unveil microbial interactions. Science 2020, 369, eaba0165. [Google Scholar] [CrossRef] [PubMed]
- Beck, C.; Knoop, H.; Axmann, I.M.; Steuer, R. The diversity of cyanobacterial metabolism: Genome analysis of multiple phototrophic microorganisms. BMC Genom. 2012, 13, 56. [Google Scholar] [CrossRef]
- Guiry, M.D. How Many Species of Algae Are There? J. Phycol. 2012, 48, 1057–1063. [Google Scholar] [CrossRef]
- Nabout, J.C.; da Silva Rocha, B.; Carneiro, F.M.; Sant’Anna, C.L. How many species of Cyanobacteria are there? Using a discovery curve to predict the species number. Biodivers. Conserv. 2013, 22, 2907–2918. [Google Scholar] [CrossRef]
- Natural History Museum of London. Available online: http://www.nhm.ac.uk/our-science/collections/botany-collections/algae-collections.html (accessed on 22 July 2017).
- Pasteur Culture Collection. Available online: http://www.cyanodb.cz (accessed on 14 October 2017).
- Hallé, F. Atlas of Poetic Botany; The MIT Press: Cambridge, MA, USA, 2018. [Google Scholar]
- Lincoln, N.K. Amy Greenwell Garden Ethnobotanical Guide to Native Hawaiian Plants & Polynesian-Introduced Plants; Bishop Museum Press: Honolulu, HI, USA, 2009. [Google Scholar]
- Lutkenhouse, D.K. Hawaii Tropical Botanical Garden; Editorial Services: Santa Barbara, CA, USA, 1994. [Google Scholar]
- Kueffer, C.; Drake, D.R.; Fernández-Palacios, J.M. Island biology: Looking towards the future. Biol. Lett. 2014, 10, 20140719. [Google Scholar] [CrossRef]
- Juvik, J.O.; Singleton, D.C.; Clarke, G.G. Climate and Water Balance on the Island of Hawaii. In Mauna Loa Observatory—A 20th Anniversary Report; Miller, J., Ed.; U.S. Department of Commerce, National Oceanic and Atmospheric Administration: Silver Spring, MD, USA, 1978; pp. 128–139. [Google Scholar]
- Giambelluca, T.W.; Chen, Q.; Frazier, A.G.; Price, J.P.; Chen, Y.-L.; Chu, P.-S.; Eischeid, J.K.; Delparte, D.M. Online Rainfall Atlas of Hawaii. Bull. Am. Meteorol. Soc. 2013, 94, 313–316. [Google Scholar] [CrossRef]
- Rainfall. Available online: http://rainfall.geography.hawaii.edu (accessed on 19 June 2023).
- Giambelluca, T.W.; Schroeder, T.A. Climate. In Atlas of Hawaii, 3rd ed.; Juvik, S.P., Juvik, J.O., Eds.; University of Hawaii Press: Honolulu, HI, USA, 1998; pp. 49–59. [Google Scholar]
- Juvik, J.O.; Perreira, D.J. Fog interception on Mauna Loa, Hawaii. Proc. Assoc. Amer. Geogr. 1974, 6, 22–25. [Google Scholar]
- Myronovskyi, M.; Luzhetskyy, A. Native and engineered promoters in natural product discovery. Nat. Prod. Rep. 2016, 33, 1006–1019. [Google Scholar] [CrossRef]
- Nigst, T.A.; Westermaier, M.; Ofial, A.R.; Mayr, H. Nucleophilic Reactivities of Pyrroles. Eur. J. Org. Chem. 2008, 2008, 2369–2374. [Google Scholar] [CrossRef]
- Mayr, H.; Lakhdar, S.; Maji, B.; Ofial, A.R. A quantitative approach to nucleophilic organocatalysis. Beilstein J. Org. Chem. 2012, 8, 1458–1478. [Google Scholar] [CrossRef]
- Samuel, G.; Reeves, P. Biosynthesis of O-antigens: Genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr. Res. 2003, 338, 2503–2519. [Google Scholar] [CrossRef] [PubMed]
- He, J.-B.; Zhao, P.; Hu, Z.-M.; Liu, S.; Kuang, Y.; Zhang, M.; Li, B.; Yun, C.-H.; Qiao, X.; Ye, M. Molecular and Structural Characterization of a Promiscuous C-Glycosyltransferase from Trollius chinensis. Angew. Chem. Int. Ed. 2019, 58, 11513–11520. [Google Scholar] [CrossRef] [PubMed]
- Ushimaru, R.; Lyu, J.; Ling, M.; Abe, I. Multiple C–C Bond Cleavage Reactions Catalyzed by Tolyporphin Tetrapyrrole Biosynthetic Enzymes. J. Am. Chem. Soc. 2023, 145, 9834–9839. [Google Scholar] [CrossRef]
- Smith, C.D.; Carmeli, S.; Moore, R.E.; Patterson, G.M.L. Scytophycins, Novel Microfilament-depolymerizing Agents Which Circumvent P-Glycoprotein-mediated Multidrug Resistance. Cancer Res. 1993, 53, 1343–1347. [Google Scholar]
- Lopez, D.; Martinez-Luis, S. Marine Natural Products with P-Glycoprotein Inhibitor Properties. Mar. Drugs 2014, 12, 525–546. [Google Scholar] [CrossRef]
- Moore, R.E. Cyclic peptides and depsipeptides from cyanobacteria: A review. J. Ind. Microbiol. 1996, 16, 134–143. [Google Scholar] [CrossRef]
- Valeriote, F.; Moore, R.E.; Patterson, G.M.L.; Paul, V.P.; Scheuer, P.J.; Corbett, T. Discovery of natural products from microalgae and marine organisms. In Anticancer Drug Discovery and Development: Natural Products and New Molecular Models; Valeriote, F.A., Corbett, T.H., Baker, L.H., Eds.; Kluwer Academic Publishers: Norwell, MA, USA, 1994; pp. 1–25. [Google Scholar]
- Ke, W.; Yu, P.; Wang, J.; Wang, R.; Guo, C.; Zhou, L.; Li, C.; Li, K. MCF-7/ADR cells (re-designated NCI/ADR-RES) are not derived from MCF-7 breast cancer cells: A loss for breast cancer multidrug-resistant research. Med. Oncol. 2011, 28, S135–S141. [Google Scholar] [CrossRef]
- Liscovitch, M.; Ravid, D. A case study in misidentification of cancer cell lines: MCF-7/AdrR cells (re-designated NCI/ADR-RES) are derived from OVCAR-8 human ovarian carcinoma cells. Cancer Lett. 2007, 245, 350–352. [Google Scholar] [CrossRef]
- Prinsep, M.R.; Appleton, T.G.; Hanson, G.R.; Lane, I.; Smith, C.D.; Puddick, J.; Fairlie, D.P. Tolyporphin Macrocycles from the Cyanobacterium Tolypothrix nodosa Selectively Bind Copper and Silver and Reverse Multidrug Resistance. Inorg. Chem. 2017, 56, 5577–5585. [Google Scholar] [CrossRef]
- de Pinillos Bayona, A.M.; Mroz, P.; Thunshelle, C.; Hamblin, M.R. Design features for optimization of tetrapyrrole macrocycles as antimicrobial and anticancer photosensitizers. Chem. Biol. Drug Des. 2017, 89, 192–206. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-Y.; Bocian, D.F.; Lindsey, J.S. Synthesis of 24 Bacteriochlorin Isotopologues, Each Containing a Symmetrical Pair of 13C or 15N Atoms in the Inner Core of the Macrocycle. J. Org. Chem. 2014, 79, 1001–1016. [Google Scholar] [CrossRef] [PubMed]
- Gröniger, A.; Sinha, R.P.; Klisch, M.; Häder, D.-P. Photoprotective compounds in cyanobacteria, phytoplankton and macroalgae—A database. J. Photochem. Photobiol. B Biol. 2000, 58, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Sinha, R.P.; Häder, D.-P. UV-protectants in cyanobacteria. Plant Sci. 2008, 174, 278–289. [Google Scholar] [CrossRef]
- Diers, J.R.; Kirmaier, C.; Taniguchi, M.; Lindsey, J.S.; Bocian, D.F.; Holten, D. A perspective on the redox properties of tetrapyrrole macrocycles. Phys. Chem. Chem. Phys. 2021, 23, 19130–19140. [Google Scholar] [CrossRef]
- Erdman, D.V. Auguries of Innocence. In The Complete Poetry & Prose of William Blake; Doubleday: New York, NY, USA, 1988. [Google Scholar]
- Guberman-Pfeffer, M.J.; Lalisse, R.F.; Hewage, N.; Brückner, C.; Gascón, J.A. Origins of the Electronic Modulations of Bacterio- and Isobacteriodilactone Regioisomers. J. Phys. Chem. A 2019, 123, 7470–7485. [Google Scholar] [CrossRef]
- Ushimaru, R.; Lyu, J.; Abe, I. Diverse enzymatic chemistry for propionate side chain cleavages in tetrapyrrole biosynthesis. J. Ind. Microbiol. Biotechnol. 2023, kuad016. [Google Scholar] [CrossRef]
- Lindsey, J.S.; Taniguchi, M. Core Chromophores of Native Photosynthetic Pigments. In Photosynthesis: From Plants to Nanomaterials; Hou, J.M.H., Allakhverdiev, S.I., Eds.; Elsevier: London, UK, 2023; Chapter 3; pp. 33–48. [Google Scholar]
- Allen, M.B.; Arnon, D.I. Studies on Nitrogen-Fixing Blue-Green Algae. I. Growth and Nitrogen Fixation by Anabaena Cylindrica Lemm. Plant Physiol. 1955, 30, 366–372. [Google Scholar] [CrossRef]
- Pukui, M.K.; Elbert, S.H.; Mookini, E.T. Place Names of Hawaii; University of Hawaii Press: Honolulu, HI, USA, 1976. [Google Scholar]
- IsotopePlotter. Available online: https://www.sisweb.com/mstools/isotope.htm (accessed on 21 June 2020).

















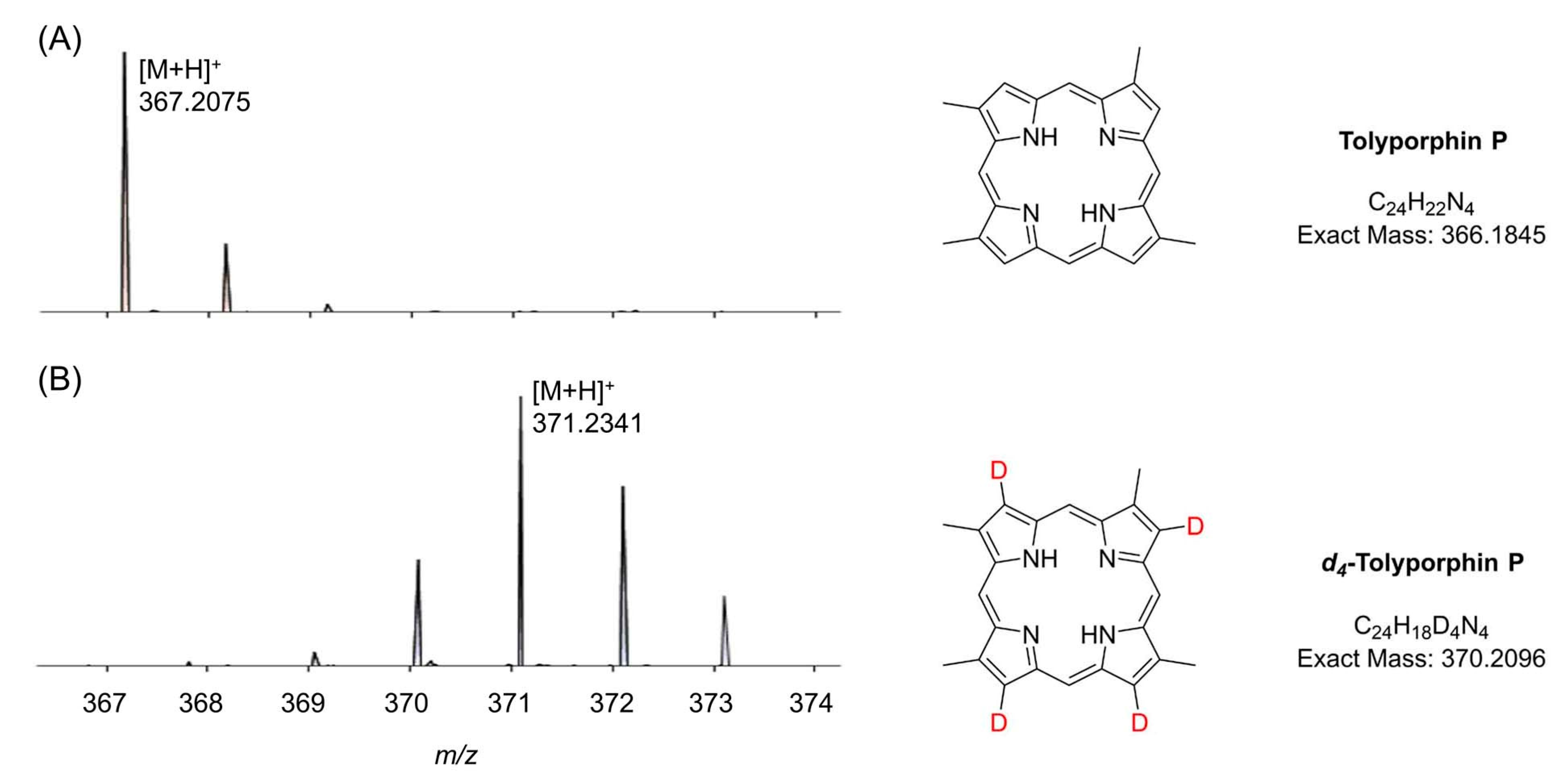
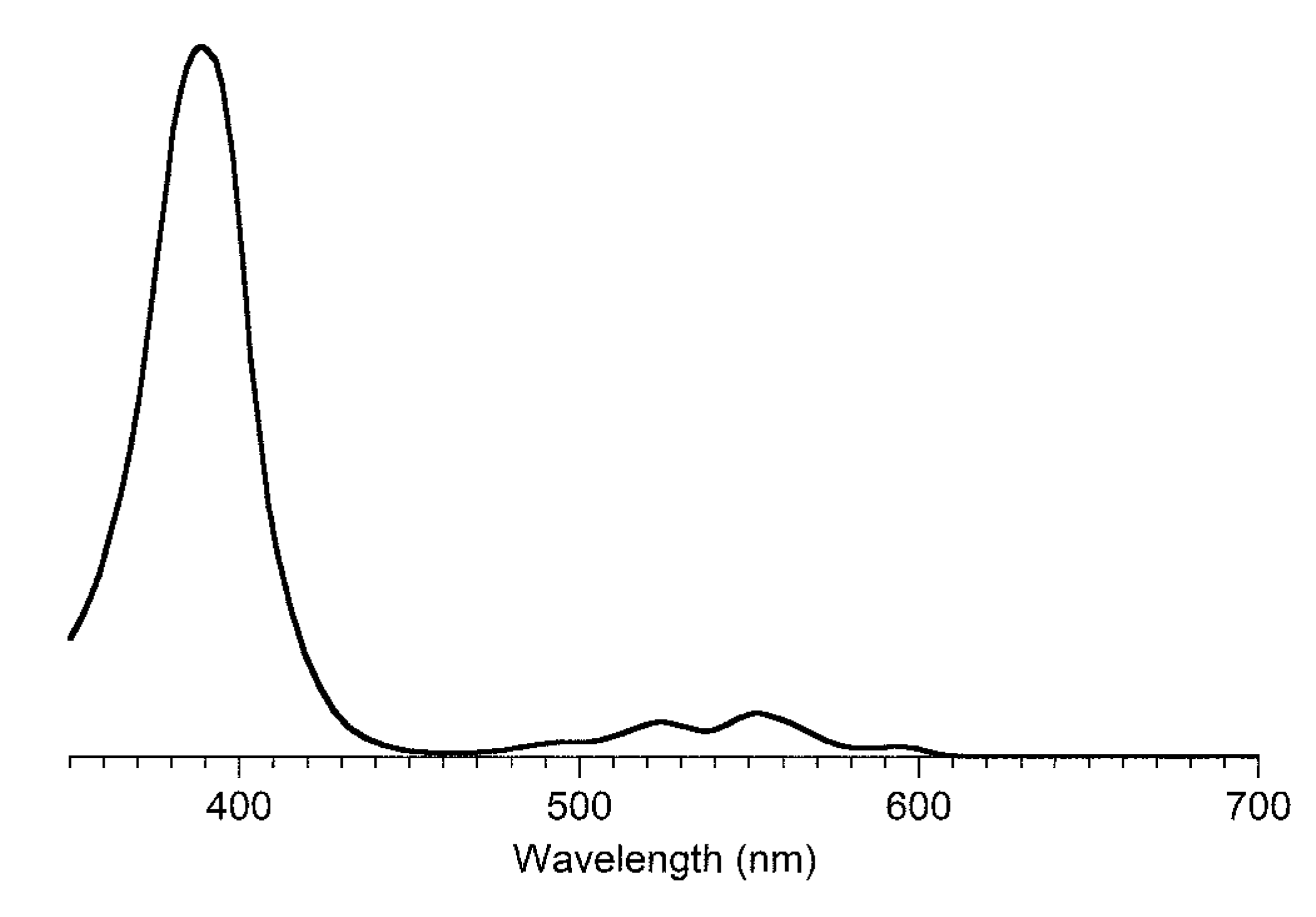
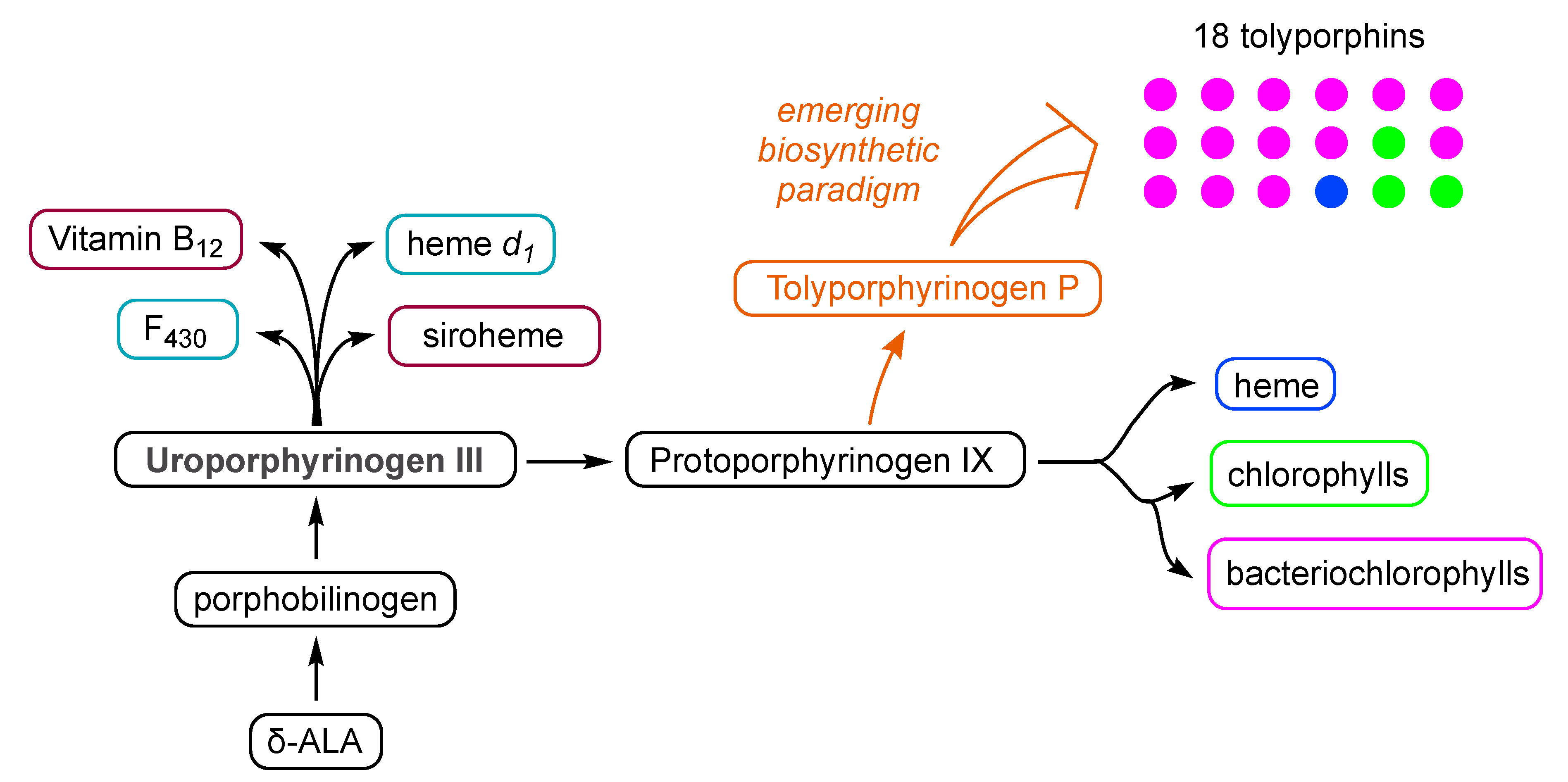
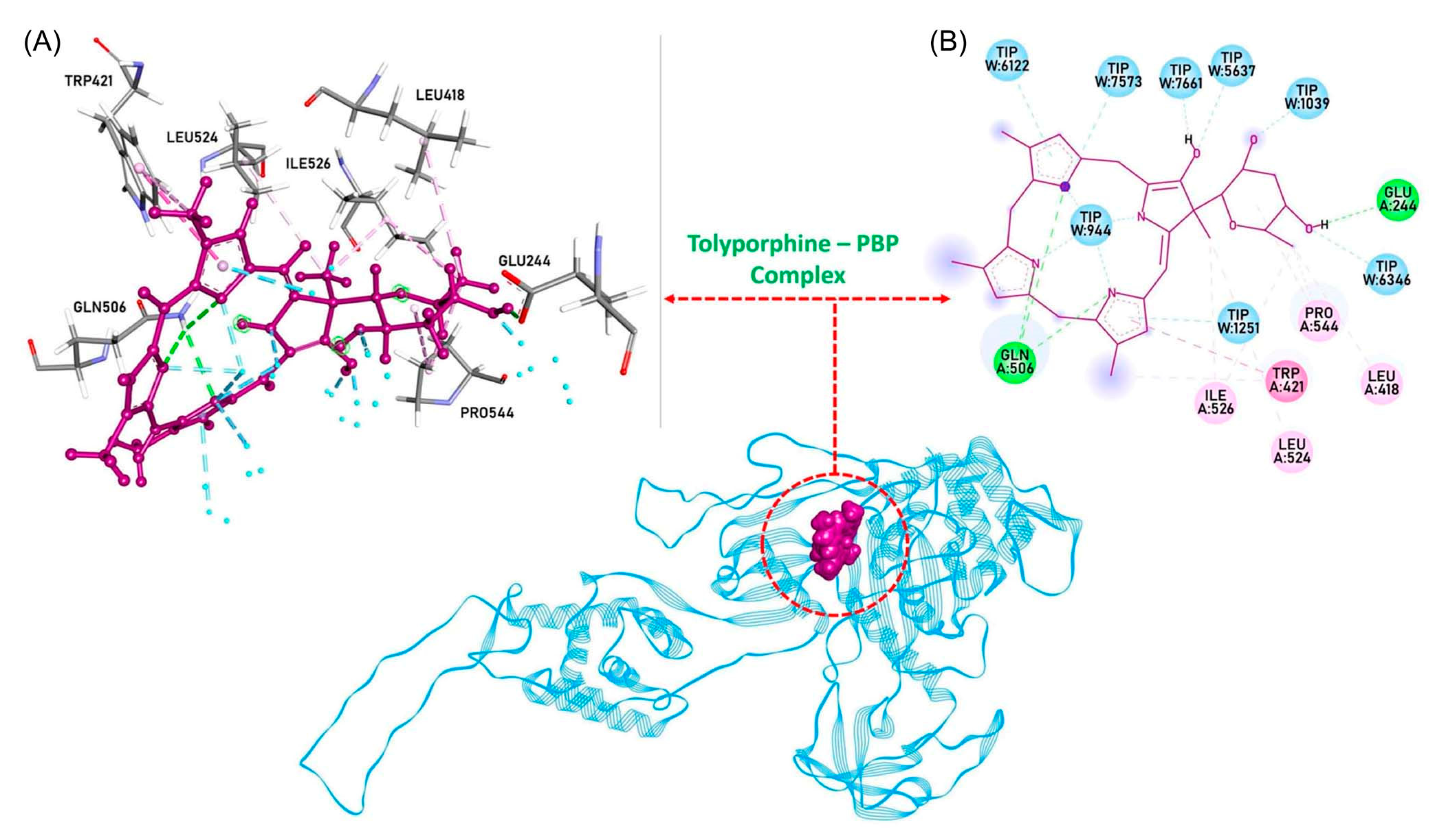
| Compound | By λabs (nm) | Bx λabs (nm) | Qy λabs (nm) | Qy λem (nm) | τs (ns) | Φf | Φisc | Φic |
|---|---|---|---|---|---|---|---|---|
| tolyporphin A | 397 | 406 | 679 | 681 | 3.9 | 0.14 | 0.77 | 0.09 |
| H2BC-O7,17 | 391 | 401 | 680 | 682 | 3.8 | 0.16 | 0.77 | 0.07 |
| pheophytin a b | 400 | 415 | 671 | 676 | 6.7 | 0.24 | 0.60 | 0.16 |
| chlorophyll a b | 413 | 432 | 665 | 671 | 6.4 | 0.33 | 0.55 | 0.12 |
| bacteriopheophytin a c | 363 | 389 | 758 | 768 | 2.7 | 0.1 | 0.57 | 0.33 |
| bacteriochlorophyll a d | 363 | 396 | 781 | 789 | 3.1 | 0.12 | 0.30 | 0.58 |
| Solvent | SDC b | Bx λabs (nm) | Qy λabs (nm) | Qy λem (nm) | τs (ns) | Φf | Φisc | Φic |
|---|---|---|---|---|---|---|---|---|
| toluene | 2.4 | 406 | 679 | 681 | 3.9 | 0.14 | 0.77 | 0.09 |
| diethyl ether | 4.3 | 401 | 678 | 679 | 4.2 | 0.11 | 0.81 | 0.08 |
| ethyl acetate | 6.1 | 402 | 677 | 678 | 3.9 | 0.12 | 0.80 | 0.08 |
| dichloromethane | 8.9 | 406 | 679 | 680 | 3.7 | 0.11 | 0.83 | 0.06 |
| pentan-1-ol | 13 | 404 | 677 | 678 | 4.4 | 0.11 | 0.82 | 0.07 |
| butan-2-one | 18.5 | 402 | 677 | 682 | 4.1 | 0.13 | 0.82 | 0.05 |
| ethanol | 24.6 | 405 | 676 | 677 | 4.1 | 0.12 | 0.81 | 0.07 |
| methanol | 32.7 | 402 | 676 | 677 | 4.3 | 0.11 | 0.85 | 0.04 |
| DMF c | 36.7 | 404 | 678 | 680 | 3.8 | 0.11 | 0.79 | 0.10 |
| dimethyl sulfoxide | 46.7 | 406 | 679 | 680 | 3.9 | 0.11 | 0.79 | 0.10 |
| Entry | Tetrapyrrole Macrocycle | cLogP Value a |
|---|---|---|
| 1 | Tolyporphin A | 3.4 |
| 2 | Tolyporphin B | 2.6 |
| 3 | Tolyporphin C | 2.6 |
| 4 | Tolyporphin D | 1.7 |
| 5 | Tolyporphin E | 2.6 |
| 6 | Tolyporphin F | 1.9 |
| 7 | Tolyporphin G | 1.7 |
| 8 | Tolyporphin H | 1.7 |
| 9 | Tolyporphin I | 2.4 |
| 10 | Tolyporphin J | 0.9 |
| 11 | Tolyporphin K | 3.1 |
| 12 | Tolyporphin L | 2.2 |
| 13 | Tolyporphin M | 2.2 |
| 14 | Tolyporphin N | 2.2 |
| 15 | Tolyporphin O | 2.2 |
| 16 | Tolyporphin P | 5.0 |
| 17 | Tolyporphin Q | 3.0 |
| 18 | Tolyporphin R | 3.7 |
| 19 | Chlorophyll a | 14.8 |
| 20 | Bacteriochlorophyll a | 13.3 |
| Component a | 1995 Culture b | 2017 Culture e | 2017 vs. 1995 f | ||
|---|---|---|---|---|---|
| Amount (mg) | % vs. Cells | Amount (mg) | % vs. Cells | ||
| cells | 93,000 | 79,500 | |||
| extract | 7500 | 4750 | |||
| A | 123 | 0.13 | 6.2 | 0.008 | –17 X |
| B + C | 38 (4:1) | 0.032, 0.008 | 3.0 | 0.0038 | –11 X |
| D | 5 | 0.005 | 0.8 | 0.001 | –5 X |
| E | 46 | 0.049 | 2.0 | 0.0025 | –19 X |
| F | 12 | 0.013 | 0.5 | 0.0006 | –21 X |
| G + H | -- (2:1) | 0.004, 0.002 | 1.0 | 0.0013 | –5 X |
| I | 1.5 | 0.002 | 0.5 | 0.0006 | –3 X |
| J | 1.5 c | 0.0016 c | -- | -- | -- |
| K | 1 c | 0.0011 c | 0.4 | 0.0005 | –2 X |
| L + M | 3.5 (2:1) d | 0.0025, 0.0013 d | -- | -- | -- |
| Component | Relative Abundance of Tolyporphins | |
|---|---|---|
| A3M7 Medium | BG-11 Medium | |
| A | 22.0% | 11.0% |
| B + C | 1.7% | 1.6% |
| D | 0.8% | 1.0% |
| E | 11.4% | 2.8% |
| F | 6.0% | 3.9% |
| G + H | 1.5% | 1.7% |
| I | 13.5% | 9.4% |
| J | 0.5% | 1.2% |
| K | 4.8% | 4.7% |
| L + M + N + O | 0.6% a | 0.9% a |
| P | 13.1% | 20.4% |
| Q | 5.2% | 2.7% |
| R | 18.8% | 38.7% |
| Tolyporphin | Formula | Mol wt (Da) | # of Sugars | Chromophore |
|---|---|---|---|---|
| A | C40H46N4O10 | 742.83 | 2 | dioxobacteriochlorin |
| L, M, N, O | C38H44N4O10 | 716.79 | 2 | dioxobacteriochlorin |
| B, C | C38H44N4O9 | 700.79 | 2 | dioxobacteriochlorin |
| D | C36H42N4O10 | 690.75 | 2 | dioxobacteriochlorin |
| E | C34H36N4O8 | 628.68 | 1 | dioxobacteriochlorin |
| F | C32H34N4O7 | 586.64 | 1 | dioxobacteriochlorin |
| I | C28H26N4O6 | 514.54 | 0 | dioxobacteriochlorin |
| G, H | C26H24N4O5 | 472.50 | 0 | dioxobacteriochlorin |
| J | C24H22N4O4 | 430.46 | 0 | dioxobacteriochlorin |
| K | C30H32N4O4 | 512.61 | 1 | oxochlorin |
| R | C26H24N4O3 | 440.50 | 0 | oxochlorin |
| Q | C24H22N4O2 | 398.47 | 0 | oxochlorin |
| P | C24H22N4 | 366.47 | 0 | porphyrin |
| Tolyporphin | Solvent | Absorption in nm (ε in M−1cm−1) | fwhm (Qy), nm | mg g | Reference | Proposed (ε in M−1cm−1) | |
|---|---|---|---|---|---|---|---|
| Soret Band | Qy Band | Qy Band | |||||
| A | MeOH | 401 (49,000) | 675 (22,000) | 7.7 | --- | [20] | 100,000 |
| A | EtOH | 402 (148,000) | 676 (68,500) | -- | --- | [52] | 100,000 |
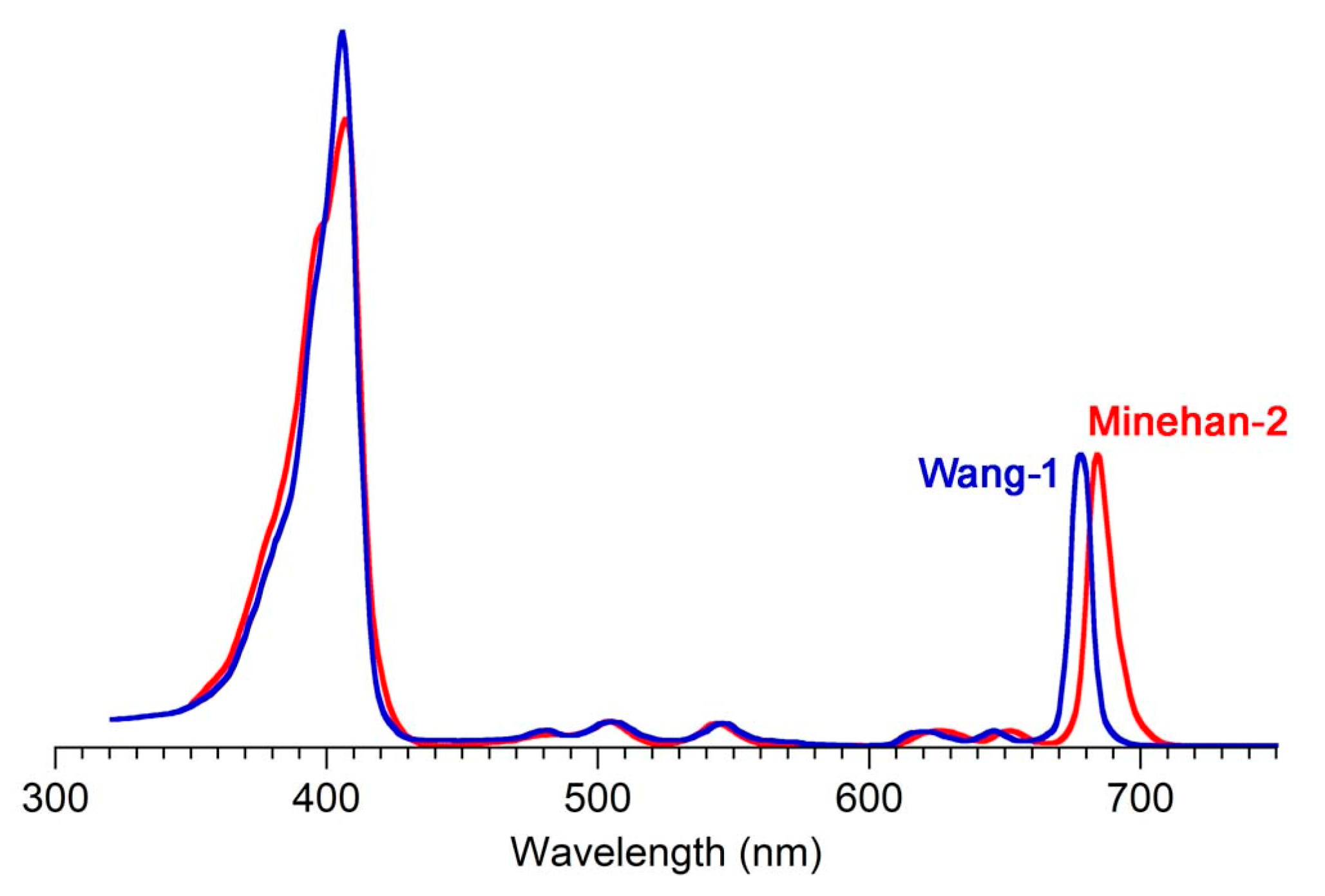
| Wang-1 | CH2Cl2 | 406 (107,463) | 678 (44,600) | 8.9 f | 0.38 | [42] | 100,000 |
| Minehan-2 | CH2Cl2 | 407 (110,500) | 684 (51,530) | 10.8 | 0.5 | [120] | 100,000 |
| B, C | MeOH | 406 (2100) c | 680 (13,000) | 9.0 | 38 | [45] | 100,000 |
| D | MeOH | 368 (24,000) | 680 (12,000) | 8.9 | 5 | [45] | 100,000 |
| E | MeOH | 388 (16,000) | 686 (7300) | 9.4 | 46 | [45] | 100,000 |
| F | MeOH | 386 (39,000) | 684 (22,000) | 15.3 | 12 | [45] | 100,000 |
| G, H | MeOH | 368 (18,000) | 686 (10,000) | 11.2 | -- | [45] | 100,000 |
| I | MeOH | 376 (14,000) | 684 (7800) | 9.6 | 1.5 | [45] | 100,000 |
| J | MeOH | 396 (7900) | 688 (1500) | -- | 1.5 | [46] | 100,000 |
| L, M | MeOH | 401 (130,000) | 677 (50,000) | -- | 0.9 | [48] | 100,000 |
| N, O | MeOH | 401 (130,000) | 677 (50,000) | -- | 2.3 | [48] | 100,000 |
| K a | MeOH | 397 (3500) | 635 d (380) | 10.6 | 1 | [46] | 35,000 |
| Q a | MeOH | 394 (63,000) | 666 (5000) | -- | 3.5 | [48] | 35,000 |
| R a | MeOH | 395 (50,000) | 636 (13,000) | -- | 1.0 | [48] | 35,000 |
| P b | CHCl3 | 398 (79,000) | 619 e (1300) | -- | 1.0 | [48] | 14,500 h |
| Organism | Chl a/Organism b | % | Reference |
|---|---|---|---|
| Oscillatoria brevis | 12–16 mg/g | 1.2–1.6 | [124] |
| Spirulina sp. | 7.2 mg/g | 0.72 | [125] |
| Nostoc muscorum | 2.61–3.80 μg/g | 0.00026–0.00038 | [126] |
| HT-58-2 (BG-11) a | 9.1–17.0 mg/g | 0.91–1.7 | [69] |
| HT-58-2 (BG-110) a | 4.4–10.1 mg/g | 0.44–1.01 | [69] |
| Tolyporphin | Mass a | LE1 b,c | HPLC fractions c | ||
|---|---|---|---|---|---|
| MALDI-MS | ESI-MS | Abs d | MALDI-MS e | ||
| A | 742.3214 | + | + | + | + |
| L, M | 716.3057 | − | + | − | − |
| B, C | 700.3108 | + | + | + | + |
| D | 658.3003 | + | − | + | + |
| E | 628.2533 | − | + | + | − |
| F | 586.2428 | − | + | + | − |
| I | 514.1852 | − | + | + | − |
| K | 512.2424 | + | + | + | − |
| G, H | 472.1747 | − | + | + | − |
| J | 430.1641 | + | + | − | − |
| Gene | Possible Function | Start | End | Size (bp) | Direction |
|---|---|---|---|---|---|
| tolA | dTDP-glucose 4,6-dehydratase | 2,586,793 | 2,587,887 | 1095 | + |
| tolB | glucose-1-phosphate thymidylyltransferase | 2,587,907 | 2,588,851 | 945 | + |
| tolC | acyltransferase | 2,592,203 | 2,593,597 | 1395 | + |
| tolD | glycosyltransferase | 2,595,115 | 2,596,395 | 1281 | + |
| tolE | UDP-glucose 4-epimerase | 2,596,678 | 2,597,817 | 1140 | + |
| tolF | aminotransferase | 2,598,123 | 2,599,451 | 1329 | + |
| tolG | cytochrome P450 | 2,600,647 | 2,602,017 | 1371 | + |
| tolH | cytochrome P450 | 2,602,062 | 2,603,444 | 1383 | + |
| tolI | L-2-amino-thiazoline-4-CO2H hydrolase | 2,604,869 | 2,605,492 | 624 | + |
| tolJ | FAD-binding protein | 2,605,501 | 2,606,913 | 1413 | + |
| tolK | aldo/keto reductase | 2,608,856 | 2,609,884 | 1029 | − |
| Number | Region (bp) | Length (nt) | Type | Similar Known Cluster |
|---|---|---|---|---|
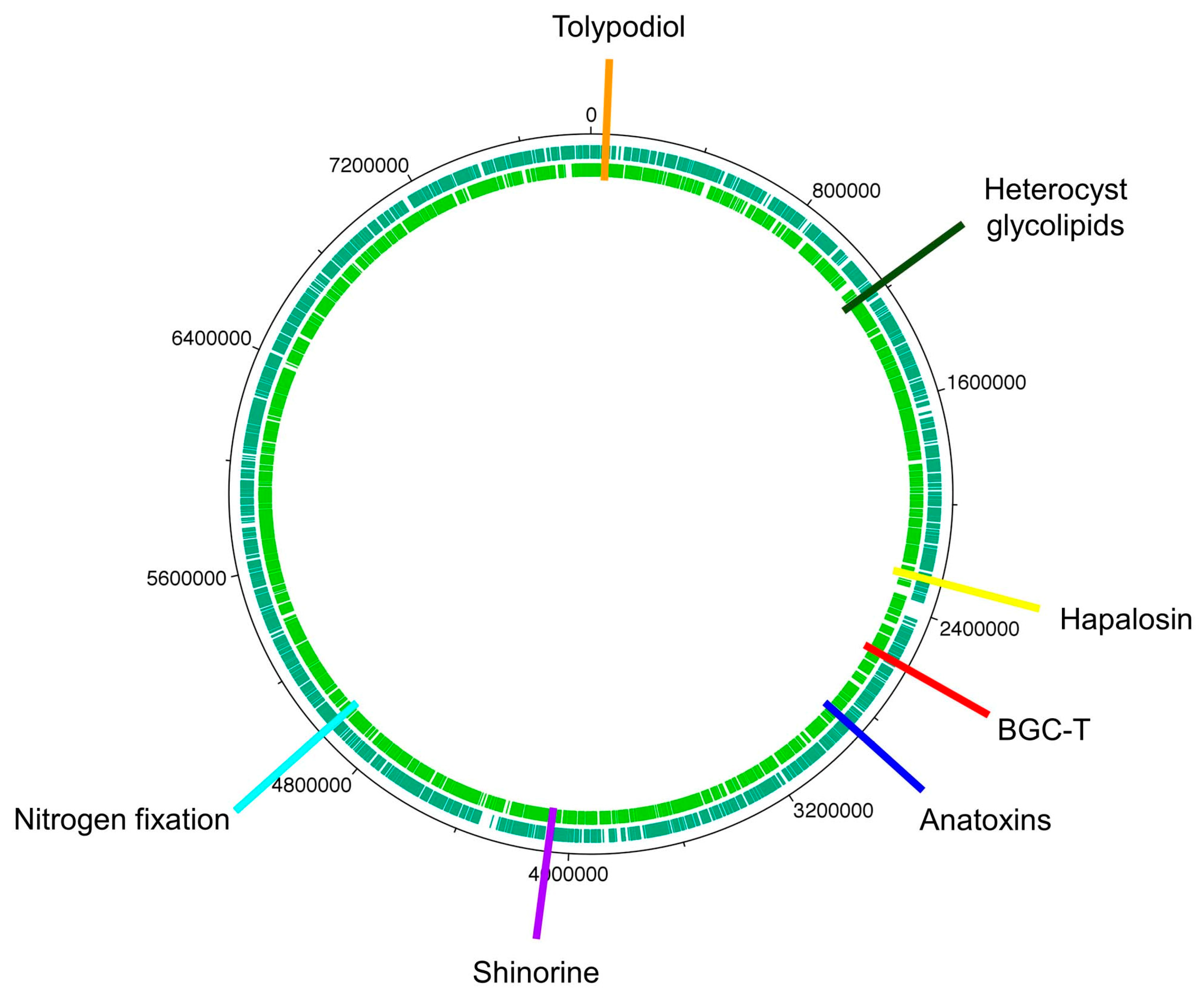
| 4 | 958,778–1,011,938 | 53,161 | hglE-KS, a T1PKS b | heterocyst glycolipids |
| 9 | 2,268,098–2,291,911 | 240,666 | NRPS, c T1PKS | hapalosin |
| 11 | 2,683,411–2,768,967 | 85,557 | T1PKS | anatoxin-a |
| 15 | 4,105,987–4,016,220 | 42,348 | NRPS | shinorine |
| Strain | Location | Origin | Number of Genes | Reference | |
|---|---|---|---|---|---|
| hem | tol | ||||
| HT-58-2 BGC-1 | Pohnpei, Micronesia | Soil | 7 | 11 | [79,164] |
| HT-58-2 BGC-2 | Pohnpei, Micronesia | Soil | 3 | 7 | [164] |
| Nostoc sp.106C | Chiapas, Mexico | Coralloid roots | 6 | 7 | [180] |
| Nostoc sp. RF31YmG | Chiapas, Mexico | Coralloid roots | 6 | 7 | [180] |
| Nostoc sp. FACHB-892 | Tengger Desert, China | Soil crusts | 6 | 6 | [181] |
| Brasilonema octagenarum UFV-OR1 | Minas Gerais, Brazil | Orchid leaves | 8 | 6 | [82] |
| Brasilonema octagenarum UFV-E1 | Minas Gerais, Brazil | Eucalyptus grandis leaves | 8 | 6 | [82] |
| Brasilonema sennae CENA114 | São Paulo, Brazil | Iron water pipe | 8 | 6 | [82] |
| Oculatella sp. LEGE 06141 | Praia de Luz, Lagos, Portugal | Green macroalgae in an intertidal zone | 6 | 9 | [164] |
| Substituent | Tolyporphin Where the Substituent Is Found | |
|---|---|---|
| Abbreviation | Common Name | |
| Ac-dd-Gal | 2′-O-acetylabequose | A, a B, C, E, F, L, M, N, O |
| dd-Gal | abequose | B, C, D, a K |
| d-Gal | D-fucose | N, O |
| d-Gul | antiarose | L, M |
| AcO | acetoxy | E, G, H, I, a R |
| HO | hydroxyl | F, G, H, J, a Q |
| Oculatella sp. LEGE 06141 | HT-58-2 BGC-1 (% identity) a | HT-58-2 BGC-2 (% identity) a |
|---|---|---|
| HemF2 | 56 | 67 |
| TolI | 59 | 66 |
| TolJ | 57 | 72 |
| TolC | 52 | 53 |
| HemA | 56 | 57 |
| HcaE | N/A | 50 |
| HcaE | N/A | 51 |
| TolH | 55 | 66 |
| TolD2 | 69 | 62 |
| TolD1 | 66 | 60 |
| FdtA | N/A | N/A |
| WecE | N/A | N/A |
| TolA | 82 | 46 |
| TolB | 83 | N/A |
| HemE | 86 | N/A |
| HemC | 76 | 80 |
| HemB | 88 | N/A |
| HemF1 | 78 | 41 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, K.-U.; Zhang, Y.; Liu, Q.; Zhang, R.; Jin, X.; Taniguchi, M.; Miller, E.S.; Lindsey, J.S. Tolyporphins–Exotic Tetrapyrrole Pigments in a Cyanobacterium—A Review. Molecules 2023, 28, 6132. https://doi.org/10.3390/molecules28166132
Nguyen K-U, Zhang Y, Liu Q, Zhang R, Jin X, Taniguchi M, Miller ES, Lindsey JS. Tolyporphins–Exotic Tetrapyrrole Pigments in a Cyanobacterium—A Review. Molecules. 2023; 28(16):6132. https://doi.org/10.3390/molecules28166132
Chicago/Turabian StyleNguyen, Kathy-Uyen, Yunlong Zhang, Qihui Liu, Ran Zhang, Xiaohe Jin, Masahiko Taniguchi, Eric S. Miller, and Jonathan S. Lindsey. 2023. "Tolyporphins–Exotic Tetrapyrrole Pigments in a Cyanobacterium—A Review" Molecules 28, no. 16: 6132. https://doi.org/10.3390/molecules28166132
APA StyleNguyen, K.-U., Zhang, Y., Liu, Q., Zhang, R., Jin, X., Taniguchi, M., Miller, E. S., & Lindsey, J. S. (2023). Tolyporphins–Exotic Tetrapyrrole Pigments in a Cyanobacterium—A Review. Molecules, 28(16), 6132. https://doi.org/10.3390/molecules28166132







