Quinone Pool, a Key Target of Plant Flavonoids Inhibiting Gram-Positive Bacteria
Abstract
1. Introduction
2. Results
2.1. Calculated and Tested Minimum Inhibitory Concentrations (MICs)
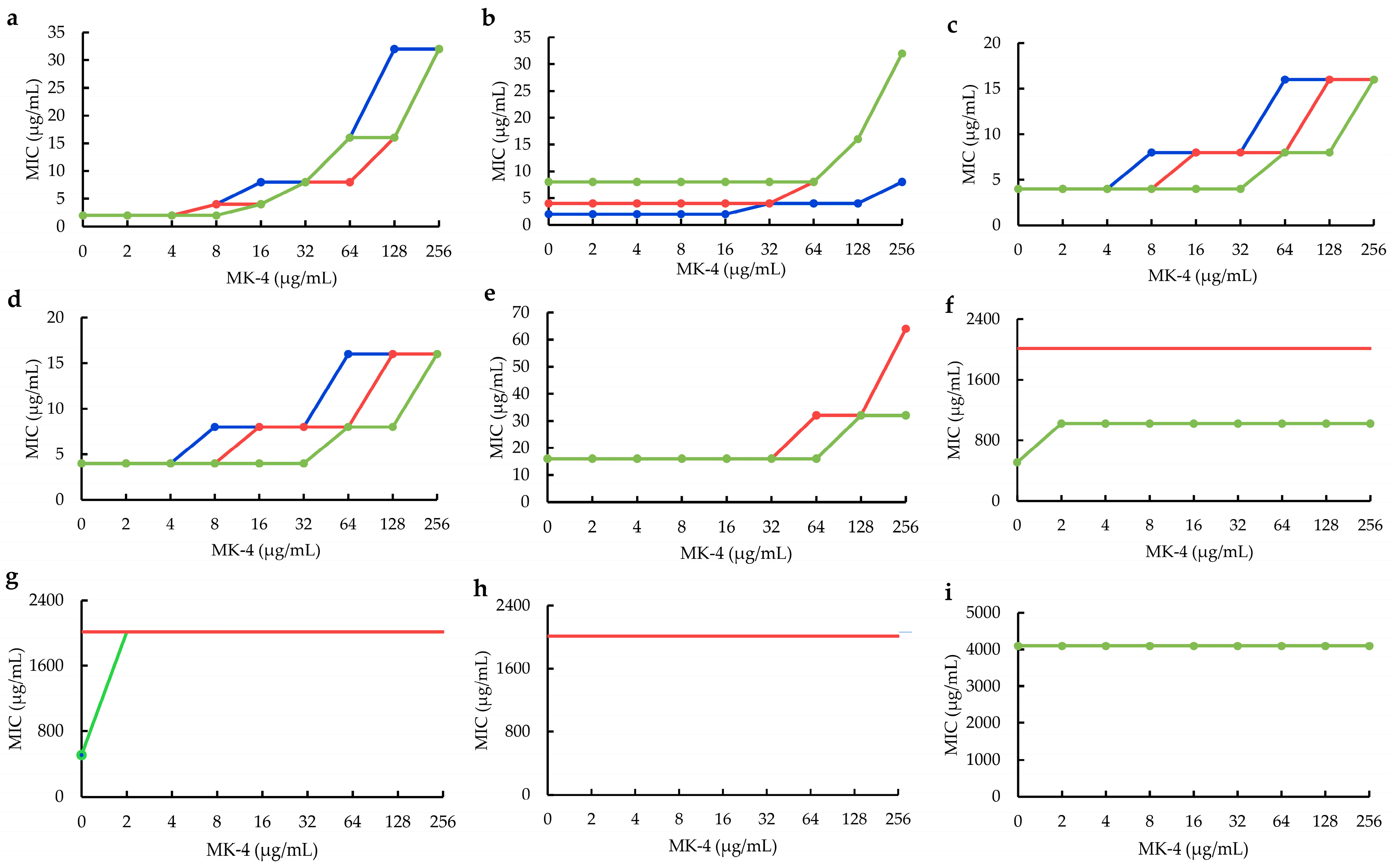
2.2. Influences of MK−4 on Plant Flavonoids against S. aureus
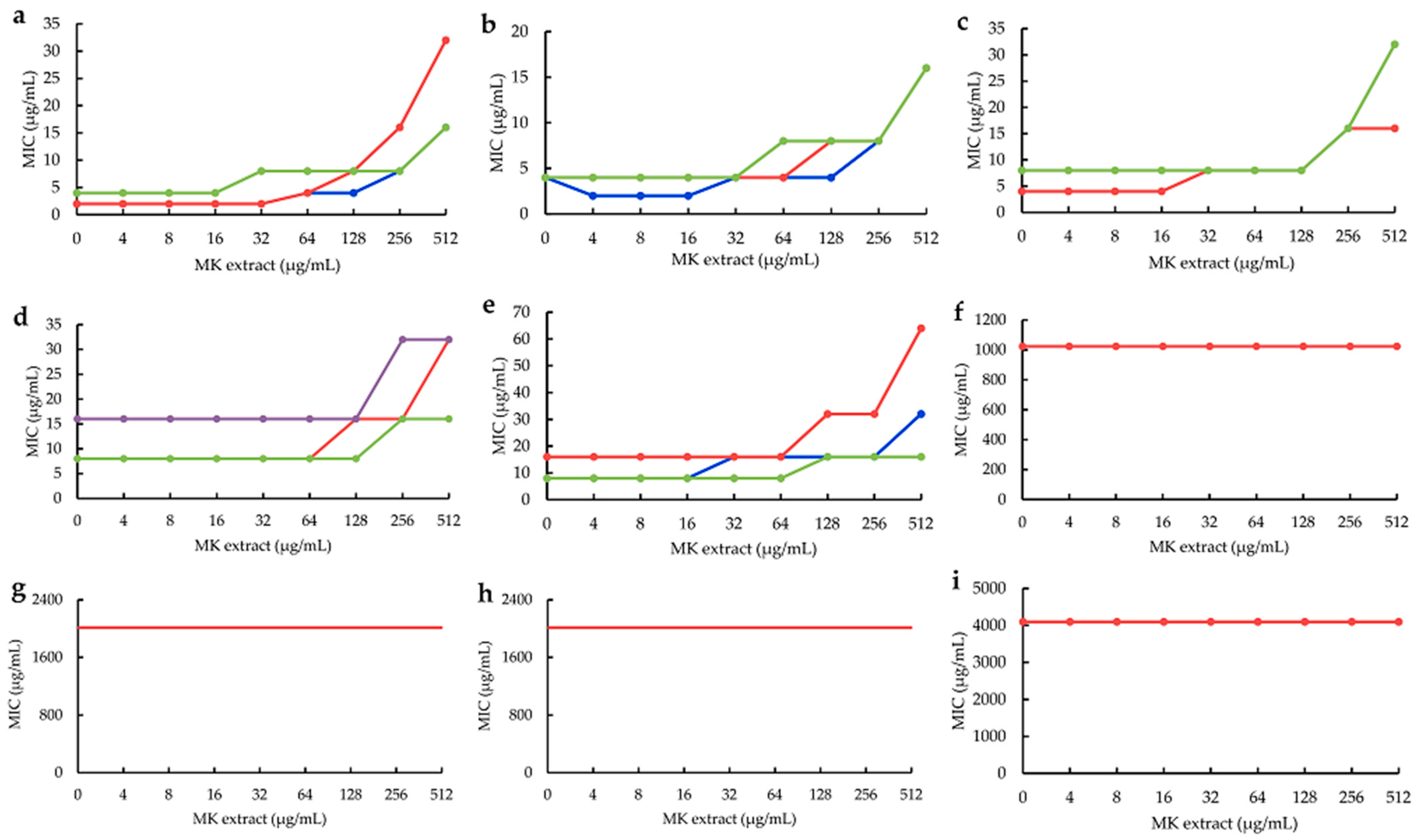
2.3. Influences of MK Extract on Plant Flavonoids against S. aureus
3. Discussion
4. Materials and Methods
4.1. Materials, Chemicals and Reagents
4.2. Bacterial Strains and Growth Condition
4.3. MIC Calculation
4.4. Antimicrobial Susceptibility Assay
4.5. Influences of MK−4 on Plant Flavonoids against S. aureus
4.6. Influences of MK Extract on Plant Flavonoids against S. aureus
4.6.1. MK Extract from S. aureus
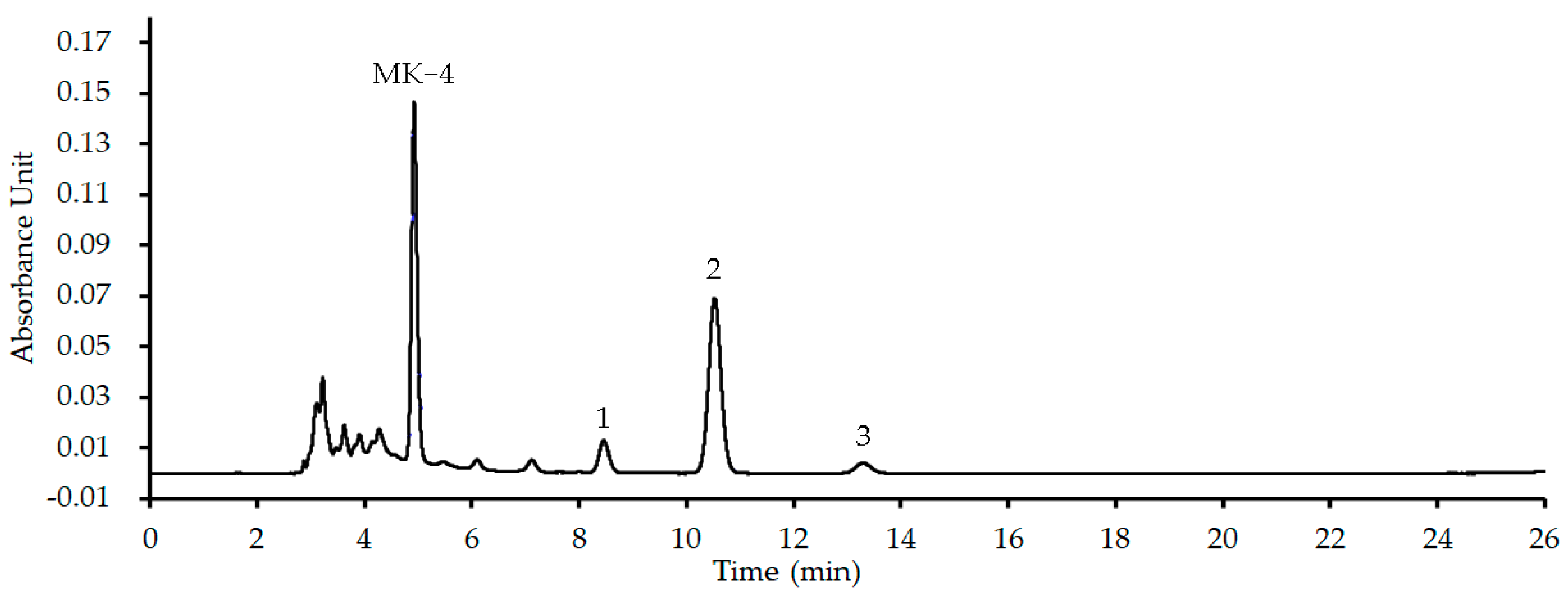
4.6.2. HPLC-UV Analyses for the MK Extract
4.6.3. MICs of Plant Flavonoids with the Interference of the MK Extract
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Laxminarayan, R.; Sridhar, D.; Blaser, M.; Wang, M.; Woolhouse, M. Achieving global targets for antimicrobial resistance. Science 2016, 353, 874–875. [Google Scholar] [CrossRef] [PubMed]
- Kurosu, M.; Siricilla, S.; Mitachi, K. Advances in MRSA drug discovery: Where are we and where do we need to be? Exp. Opin. Drug Discov. 2013, 8, 1095–1116. [Google Scholar] [CrossRef]
- Xu, X.; Xu, L.; Yuan, G.; Wang, Y.; Qu, Y.; Zhou, M. Synergistic combination of two antimicrobial agents closing each other’s mutant selection windows to prevent antimicrobial resistance. Sci. Rep. 2018, 8, 7237. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.; Ge, X.; Xua, H.; Ma, K.; Zhang, W.; Zan, Y.; Efferth, T.; Xue, Z.; Hua, X. Phytochemicals with activity against methicillin-resistant Staphylococcus aureus. Phytomedicine 2022, 100, 154073. [Google Scholar] [CrossRef] [PubMed]
- Panda, L.; Duarte-Sierra, A. Recent advancements in enhancing antimicrobial activity of plant-derived polyphenols by biochemical means. Horticulturae 2022, 8, 401. [Google Scholar] [CrossRef]
- Xie, Y.; Yang, W.; Tang, F.; Chen, X.; Ren, L. Antibacterial activities of flavonoids: Structure-activity relationship and mechanism. Curr. Med. Chem. 2015, 22, 132–149. [Google Scholar] [CrossRef]
- Tan, Z.; Deng, J.; Ye, Q.; Zhang, Z. The antibacterial activity of natural-derived flavonoids. Curr. Top. Med. Chem. 2022, 22, 1009–1019. [Google Scholar] [CrossRef]
- Song, L.; Hu, X.; Ren, X.; Liu, J.; Liu, X. Antibacterial modes of herbal flavonoids combat resistant bacteria. Front. Pharmacol. 2022, 13, 873374. [Google Scholar] [CrossRef]
- Wu, S.C.; Yang, Z.Q.; Liu, F.; Peng, W.J.; Qu, S.Q.; Li, Q.; Song, X.B.; Zhu, K.; Shen, J.Z. Antibacterial effect and mode of action of flavonoids from licorice against methicillin-resistant Staphylococcus aureus. Front. Microbiol. 2019, 10, 2489. [Google Scholar] [CrossRef]
- Zhou, K.; Yang, S.; Li, S.M. Naturally occurring prenylated chalcones from plants: Structural diversity, distribution, activities and biosynthesis. Nat. Prod. Rep. 2021, 38, 2236–2260. [Google Scholar] [CrossRef]
- Górniak, I.; Bartoszewski, R.; Króliczewski, J. Comprehensive review of antimicrobial activities of plant flavonoids. Phytochem. Rev. 2019, 18, 241–272. [Google Scholar] [CrossRef]
- Farhadi, F.; Khameneh, B.; Iranshahi, M.; Iranshahy, M. Antibacterial activity of flavonoids and their structure-activity relationship: An update review. Phytother. Res. 2019, 33, 13–40. [Google Scholar] [CrossRef] [PubMed]
- Yuan, G.; Guan, Y.; Yi, H.; Lai, S.; Sun, Y.; Cao, S. Antibacterial activity and mechanism of plant flavonoids to gram-positive bacteria predicted from their lipophilicities. Sci. Rep. 2021, 11, 10471. [Google Scholar] [CrossRef]
- Yuan, G.; Xia, X.; Guan, Y.; Yi, H.; Lai, S.; Sun, Y.; Cao, S. Antimicrobial quantitative relationship and mechanism of plant flavonoids to gram-positive bacteria. Pharmaceuticals 2022, 15, 1190. [Google Scholar] [CrossRef]
- Paudel, A.; Hamamoto, H.; Panthee, S.; Sekimizu, K. Menaquinone as a potential target of antibacterial agents. Drug Discov. Ther. 2016, 10, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Kurosu, M.; Begari, E. Vitamin K2 in electron transport system: Are enzymes involved in vitamin K2 biosynthesis promising drug targets? Molecules 2010, 15, 1531–1553. [Google Scholar] [CrossRef] [PubMed]
- Hamamoto, H.; Urai, M.; Ishii, K.; Yasukawa, J.; Paudel, A.; Murai, M.; Kaji, T.; Kuranaga, T.; Hamase, K.; Katsu, T.; et al. Lysocin E is a new antibiotic that targets menaquinone in the bacterial membrane. Nat. Chem. Biol. 2015, 11, 127–133. [Google Scholar] [CrossRef]
- Cao, S.; Du, X.; Li, P.; Yuan, G.; Chen, S.; Chen, W.; Song, X.; Kuang, B. A chemical screening method for menaquinone-producing strains based on HPLC-UV technology. J. Microbiol. Methods 2020, 172, 105907. [Google Scholar] [CrossRef]
- Shamsudin, N.F.; Ahmed, Q.U.; Mahmood, S.; Ali Shah, S.A.; Khatib, A.; Mukhtar, S.; Alsharif, M.A.; Parveen, H.; Zakaria, Z.A. Antibacterial effects of flavonoids and their structure-activity relationship study: A comparative interpretation. Molecules 2022, 27, 1149. [Google Scholar] [CrossRef]
- Jeong, K.; Lee, J.; Kang, D.; Lee, J.; Shin, S.Y.; Kim, Y. Screening of flavonoids as candidate antibiotics against Enterococcus faecalis. J. Nat. Prod. 2009, 72, 719–724. [Google Scholar] [CrossRef]
- Wu, D.; Kong, Y.; Han, C.; Chen, J.; Hu, L.; Jiang, H.; Shen, X. D-Alanine: D-alanine ligase as a new target for the flavonoids quercetin and apigenin. Int. J. Antimicrob. Agents 2008, 32, 421–426. [Google Scholar] [CrossRef]
- Elmasri, W.A.; Zhu, R.; Peng, W.; Al-Hariri, M.; Kobeissy, F.; Tran, P.; Hamood, A.N.; Hegazy, M.F.; Paré, P.W.; Mechref, Y. Multitargeted flavonoid inhibition of the pathogenic bacterium Staphylococcus aureus: A proteomic characterization. J. Proteome Res. 2017, 16, 2579–2586. [Google Scholar] [CrossRef]
- Ohemeng, K.A.; Schwender, C.F.; Fu, K.P.; Barrett, J.F. DNA gyrase inhibitory and antibacterial activity of some flavones (l). Bioorg. Med. Chem. Lett. 1993, 3, 225–230. [Google Scholar] [CrossRef]
- Iinuma, M.; Tosa, H.; Tanaka, T.; Asai, F.; Kobayashi, Y.; Shimano, R.; Miyauchi, K. Antibacterial activity of xanthones from guttiferaeous plants against methicillin-resistant Staphylococcus aureus. J. Pharm. Pharmacol. 1996, 48, 861–865. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.H.; Zhang, K.J.; Gu, Q.L.; Bi, X.L.; Wang, J.X. Pharmacology of mangostins and their derivatives: A comprehensive review. Chin. J. Nat. Med. 2017, 15, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Liu, Y.; Li, T.; Liu, X.; Hao, Z.; Ding, S.; Panichayupakaranant, P.; Zhu, K.; Shen, J. Plant natural flavonoids against multidrug resistant pathogens. Adv. Sci. 2021, 8, e2100749. [Google Scholar] [CrossRef]
- Lin, S.; Zhu, C.; Li, H.; Chen, Y.; Liu, S. Potent in vitro and in vivo antimicrobial activity of semisynthetic amphiphilic γ-mangostin derivative LS02 against Gram-positive bacteria with destructive effect on bacterial membrane. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183353. [Google Scholar] [CrossRef] [PubMed]
- Koh, J.J.; Qiu, S.; Zou, H.; Lakshminarayanan, R.; Li, J.; Zhou, X.; Tang, C.; Saraswathi, P.; Verma, C.; Tan, D.T.; et al. Rapid bactericidal action of α-mangostin against MRSA as an outcome of membrane targeting. Biochim. Biophys. Acta 2013, 1828, 834–844. [Google Scholar] [CrossRef] [PubMed]
- Haraguchi, H.; Tanimoto, K.; Tamura, Y.; Mizutani, K.; Kinoshita, T. Mode of antibacterial action of retrochalcones from Glycyrrhiza inflata. Phytochemistry 1998, 48, 125–129. [Google Scholar] [CrossRef]
- Clinical and Laboratory and Standards Institute (CLSI). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 10th ed.; Approved Standards, CLSI Document M07-A10; Clinical and Laboratory and Standards Institute: Wayne, PA, USA, 2015. [Google Scholar]
- Wu, X.; Xu, L.; Li, P.; Wang, Y.; Yuan, G. The anti-methicillin-resistant Staphylococcus aureus activities of azalomycin F derivatives and those of them combined with vitamin K3. Chin. J. Antibiot. 2016, 41, 584–589. [Google Scholar]
- Schurig-Briccio, L.A.; Yano, T.; Rubin, H.; Gennis, R.B. Characterization of the type 2 NADH:menaquinone oxidoreductases from Staphylococcus aureus and the bactericidal action of phenothiazines. Biochim. Biophys. Acta. 2014, 1837, 954–963. [Google Scholar] [CrossRef] [PubMed]
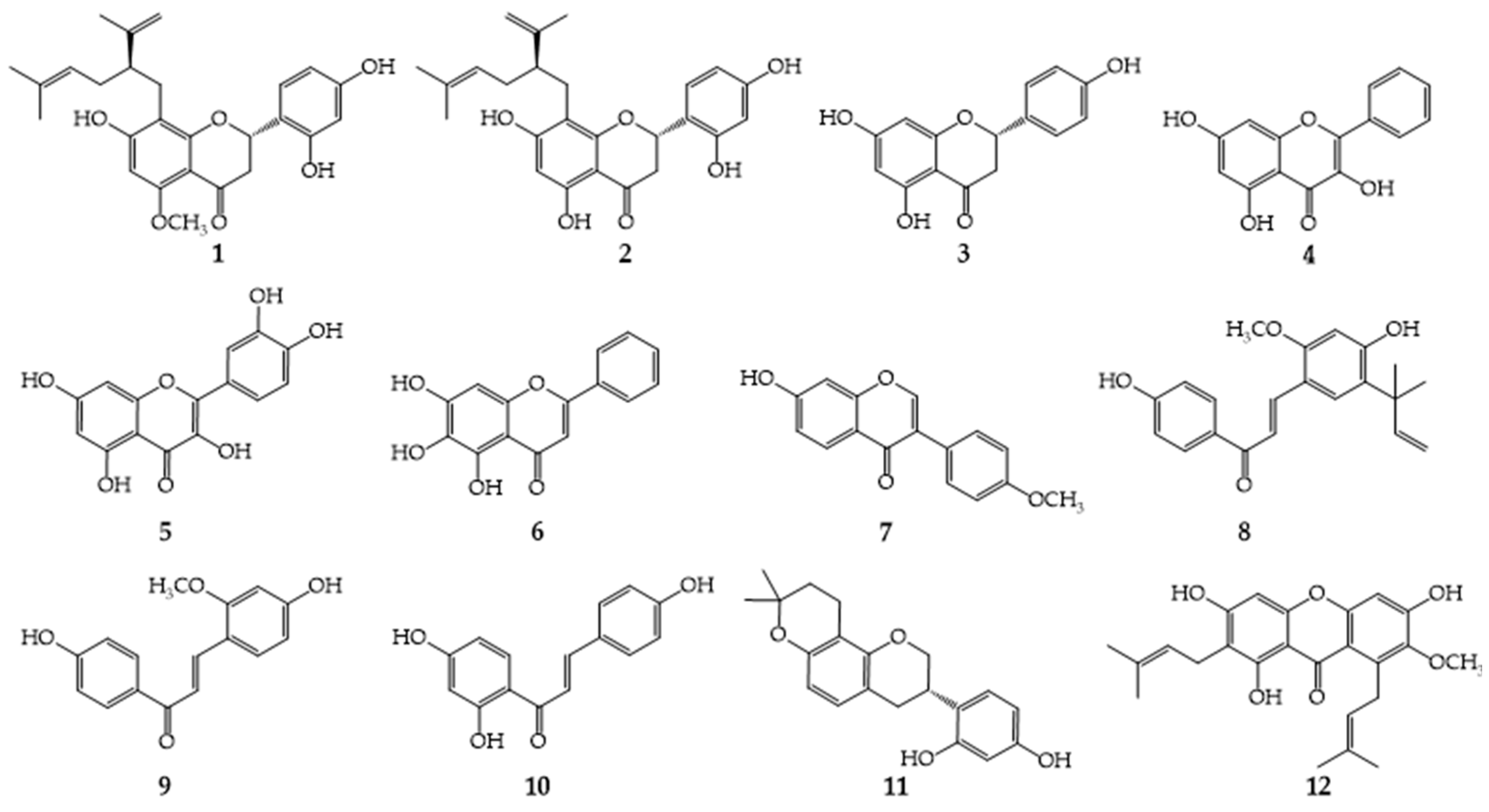
| Compounds | LogP | Calculated MICs a | Tested MICs (μg/mL) b | Compounds | LogP | Calculated MICs | Tested MICs (μg/mL) | ||
|---|---|---|---|---|---|---|---|---|---|
| μmol/L | μg/mL | μmol/L | μg/mL | ||||||
| Kurarinone | 6.30 | 28.92 | 12.67 | 8 | Formononetin | 3.15 | 549.61 | 147.35 | ˃1024 |
| Sophoraflavanone G | 6.52 | 16.37 | 6.95 | 2~4 | Licochalcone A | 4.95 | 74.44 | 25.17 | 4 |
| Naringenin | 3.19 | 509.64 | 138.67 | 512 | Echinatin | 3.23 | 472.20 | 127.54 | ˃1024 |
| Galangin | 2.83 | 974.47 | 263.15 | >1024 | Isoliquiritigenin | 3.40 | 338.78 | 86.76 | 512~1024 |
| Quercetin | 2.07 | 3063.61 | 925.21 | 4096 | Glabridin | 4.39 | 74.74 | 24.38 | 8~16 |
| Baicalein | 3.31 | 404.47 | 109.23 | 512~˃1024 | α-Mangostin | 6.70 | 8.17 | 3.35 | 2 |
| Compounds | MICAlone (μg/mL) | MIC Change (Times) b | Compounds | MICAlone (μg/mL) | MIC Change (Times) | ||
|---|---|---|---|---|---|---|---|
| MK−4 | MK Extract | MK−4 | MK Extract | ||||
| α-Mangostin | 2 | 16 | 8~16 | Isoliquiritigenin | 512~˃1024 | 2/- c | 1 |
| Sophoraflavanone G | 2~4 | 4~8 | 4~8 | Baicalein | 512~˃1024 | - | - |
| Licochalcone A | 4 | 4 | 4~8 | Echinatin | ˃1024 | - | - |
| Kurarinone | 8 | 2~4 | 2~4 | Quercetin | 4096 | 1 | 1 |
| Glabridin | 8~16 | 2~4 | 2~4 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Yan, Y.; Zhu, J.; Xia, X.; Yuan, G.; Li, S.; Deng, B.; Luo, X. Quinone Pool, a Key Target of Plant Flavonoids Inhibiting Gram-Positive Bacteria. Molecules 2023, 28, 4972. https://doi.org/10.3390/molecules28134972
Zhang L, Yan Y, Zhu J, Xia X, Yuan G, Li S, Deng B, Luo X. Quinone Pool, a Key Target of Plant Flavonoids Inhibiting Gram-Positive Bacteria. Molecules. 2023; 28(13):4972. https://doi.org/10.3390/molecules28134972
Chicago/Turabian StyleZhang, Li, Yu Yan, Jianping Zhu, Xuexue Xia, Ganjun Yuan, Shimin Li, Beibei Deng, and Xinrong Luo. 2023. "Quinone Pool, a Key Target of Plant Flavonoids Inhibiting Gram-Positive Bacteria" Molecules 28, no. 13: 4972. https://doi.org/10.3390/molecules28134972
APA StyleZhang, L., Yan, Y., Zhu, J., Xia, X., Yuan, G., Li, S., Deng, B., & Luo, X. (2023). Quinone Pool, a Key Target of Plant Flavonoids Inhibiting Gram-Positive Bacteria. Molecules, 28(13), 4972. https://doi.org/10.3390/molecules28134972






