Oxoglaucine Suppresses Hepatic Fibrosis by Inhibiting TGFβ-Induced Smad2 Phosphorylation and ROS Generation
Abstract
1. Introduction
2. Results
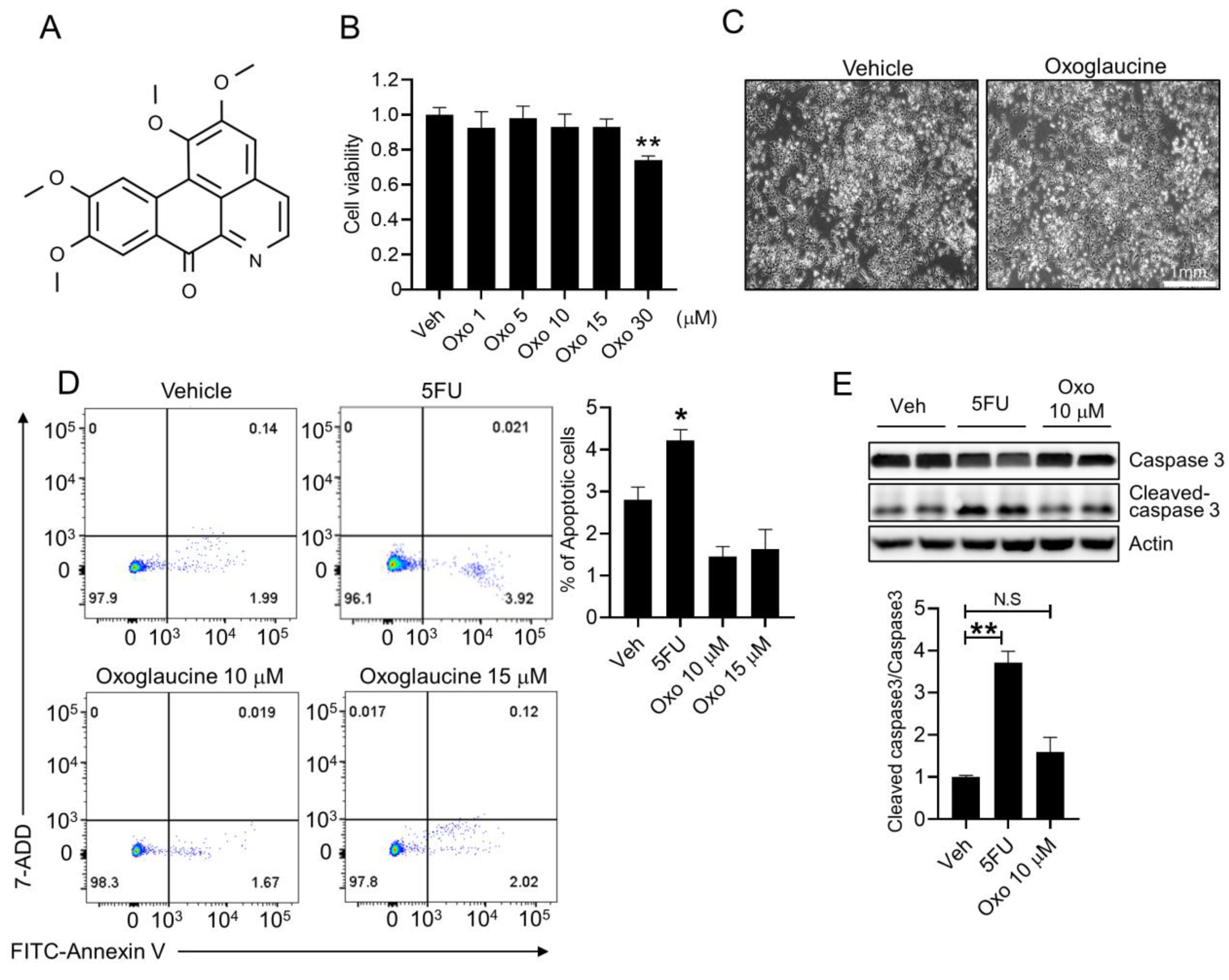
2.1. Effect of Oxoglaucine on Cellular Viability
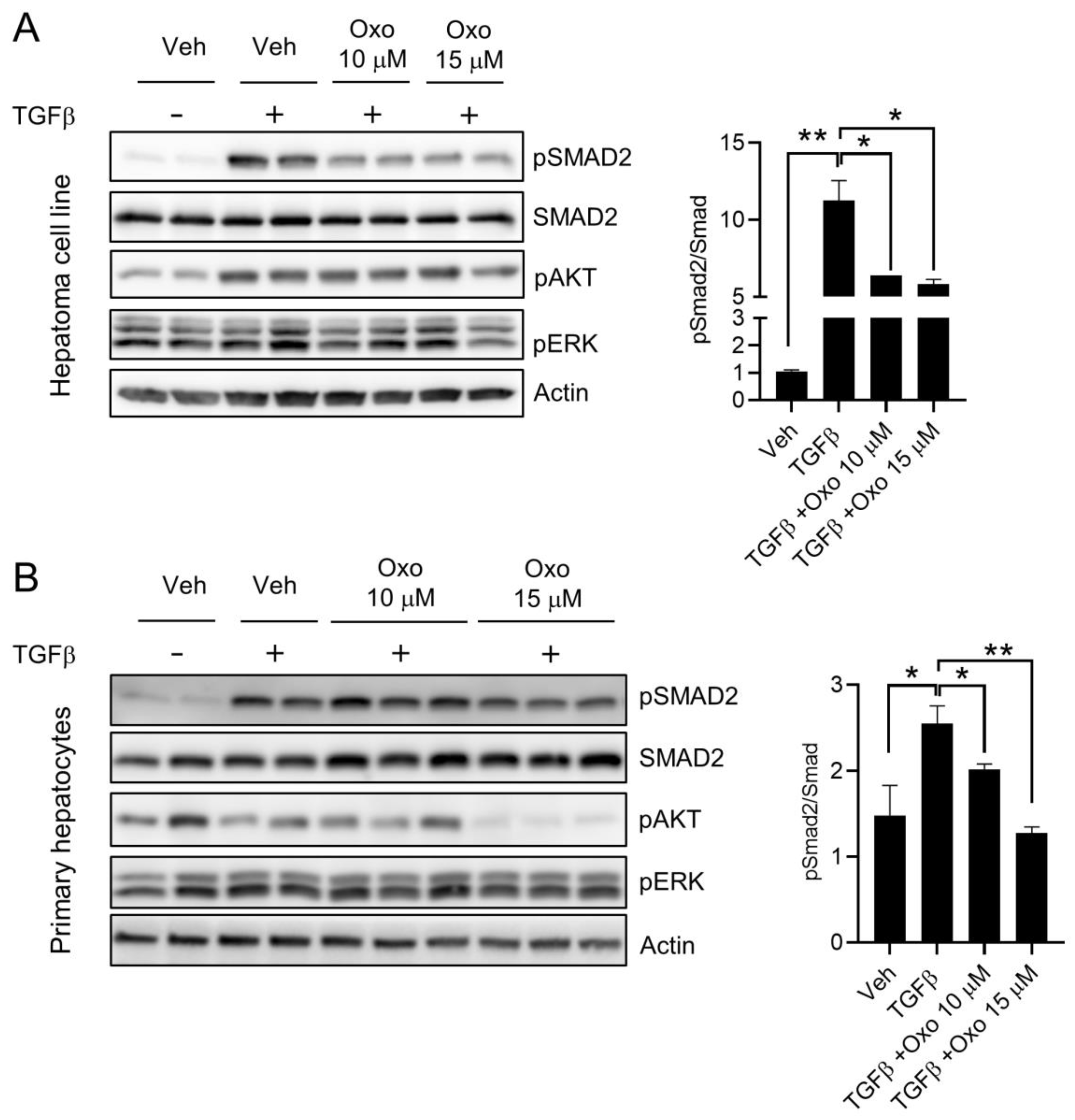
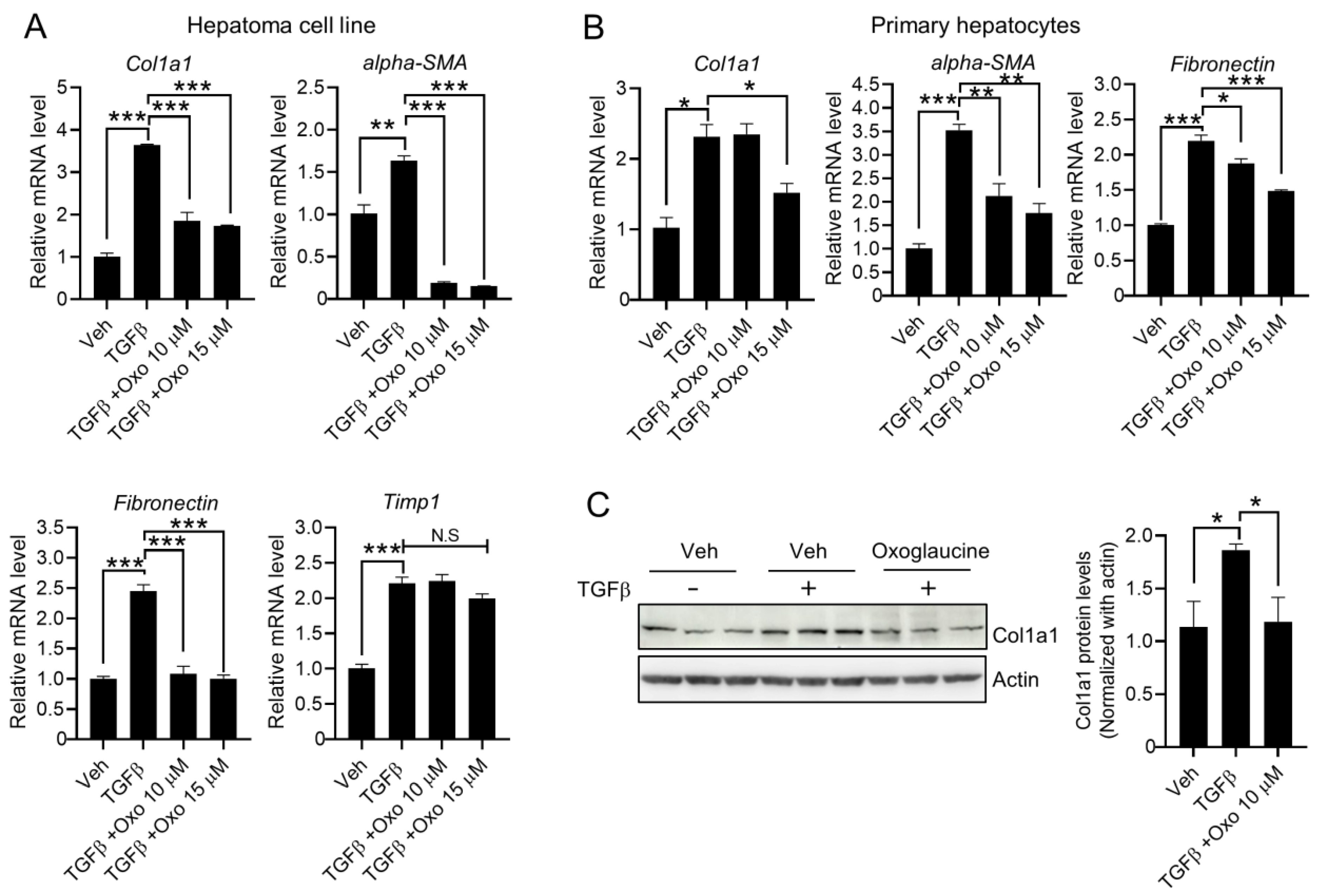
2.2. Oxoglaucine Attenuates TGFβ-Induced Phosphorylation of Smad2 and Fibrogenic Gene Expression
2.3. Oxoglaucine Suppresses the mRNA Levels of Pro-Inflammatory Cytokines
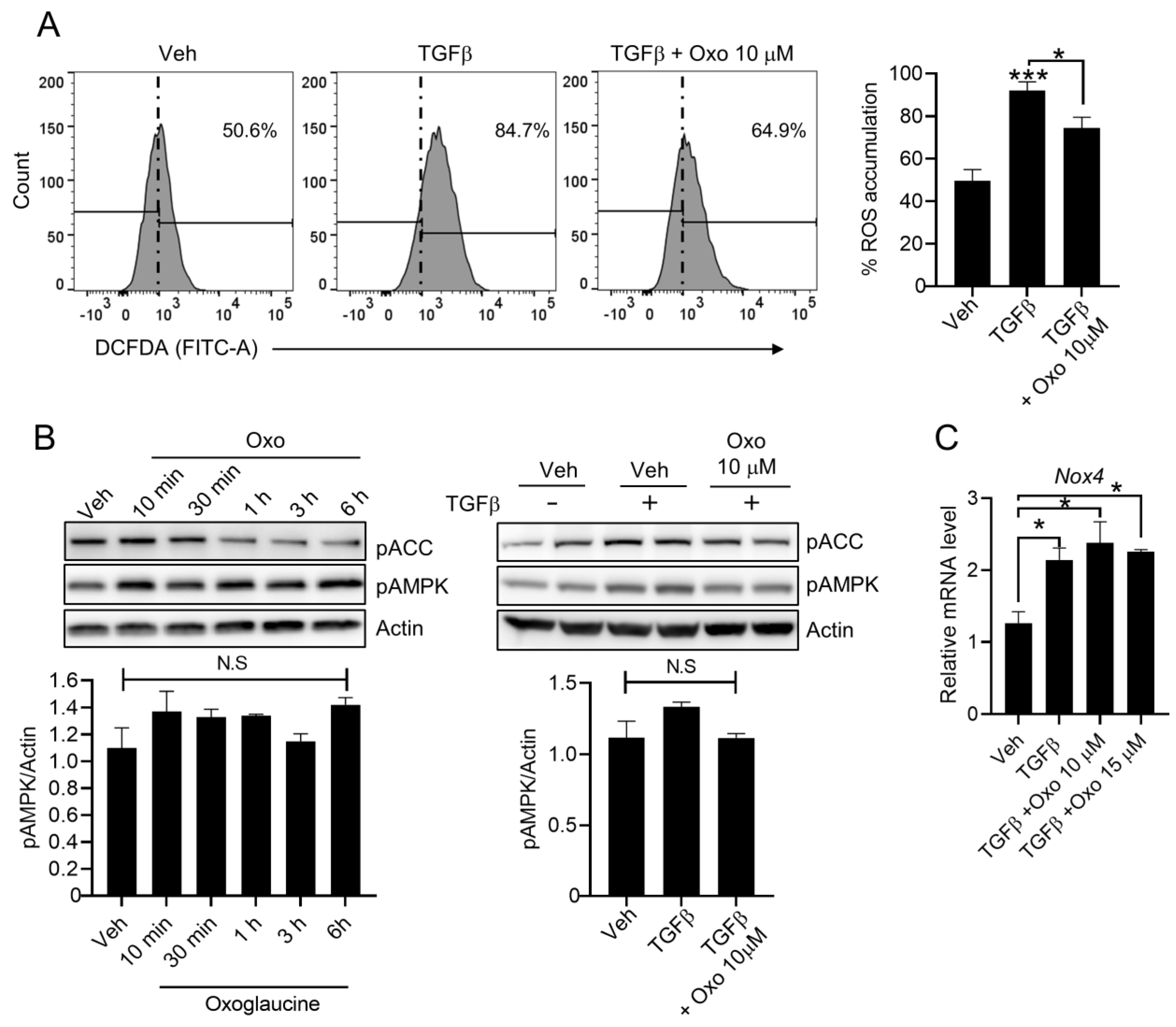
2.4. Effect of Oxoglaucine on ROS Levels in Hepatocytes
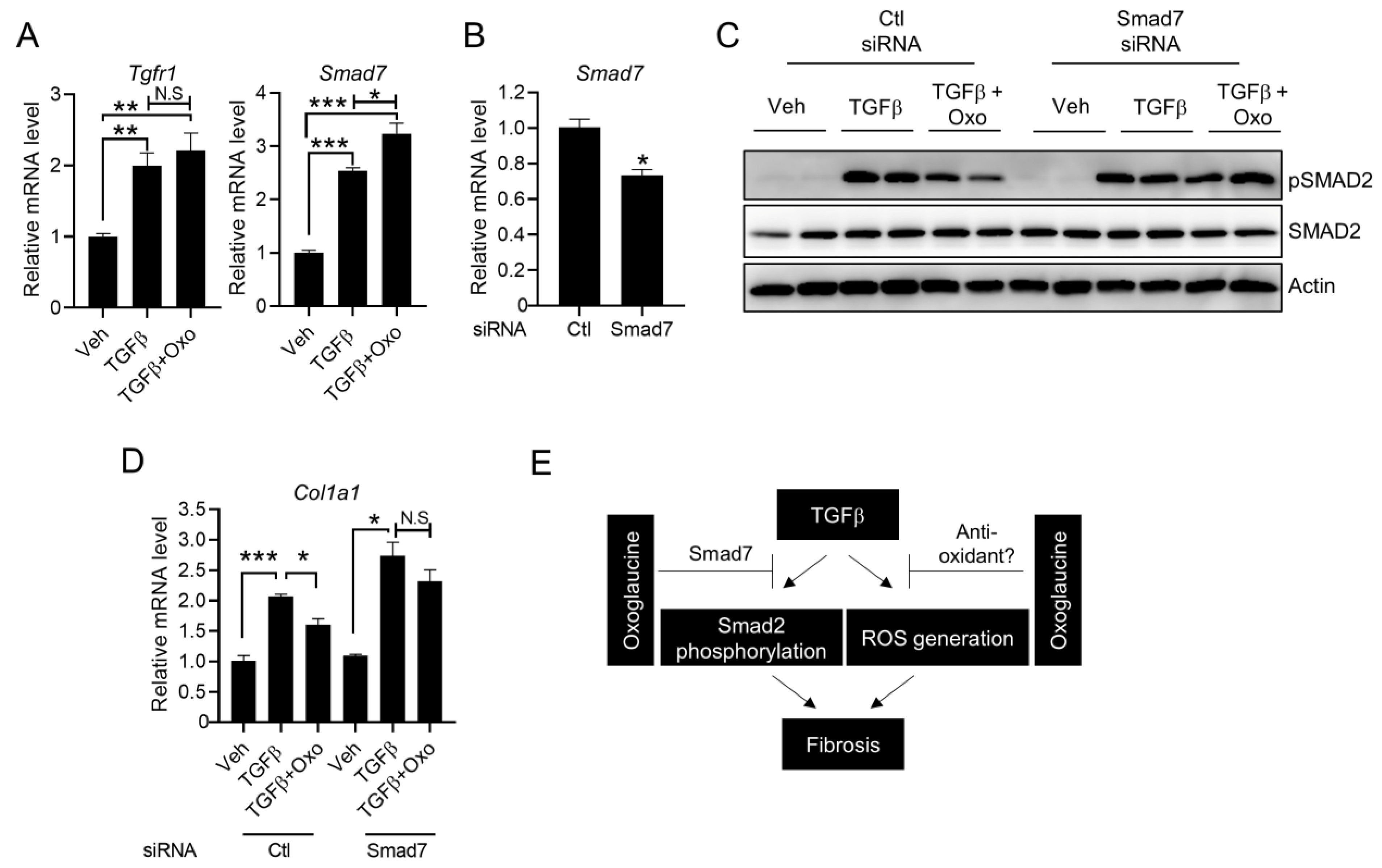
2.5. Suppression of Smad7 Blocks the Anti-Fibrogenic Effects of Oxoglaucine
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Material Treatment
4.2. Isolation of Primary Hepatocyte
4.3. WST1 Assay
4.4. Fluorescence-Activated Cell Sorting (FACS) Assay
4.5. RNAi Experiment
4.6. Immunoblotting
4.7. Quantitative RT-PCR
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bataller, R.; Brenner, D.A. Liver fibrosis. J. Clin. Investig. 2005, 115, 209–218. [Google Scholar] [CrossRef]
- Dongiovanni, P.; Romeo, S.; Valenti, L. Hepatocellular carcinoma in nonalcoholic fatty liver: Role of environmental and genetic factors. World J. Gastroenterol. 2014, 20, 12945–12955. [Google Scholar] [CrossRef]
- Friedman, S.L. Liver fibrosis—From bench to bedside. J. Hepatol. 2003, 38 (Suppl. 1), S38–S53. [Google Scholar] [CrossRef]
- Kisseleva, T.; Brenner, D.A. Mechanisms of fibrogenesis. Exp. Biol. Med. 2008, 233, 109–122. [Google Scholar] [CrossRef]
- Acharya, P.; Chouhan, K.; Weiskirchen, S.; Weiskirchen, R. Cellular Mechanisms of Liver Fibrosis. Front. Pharmacol. 2021, 12, 671640. [Google Scholar] [CrossRef] [PubMed]
- Mallat, A.; Lotersztajn, S. Cellular mechanisms of tissue fibrosis. 5. Novel insights into liver fibrosis. Am. J. Physiol. Cell. Physiol. 2013, 305, C789–C799. [Google Scholar] [CrossRef]
- Ying, H.Z.; Chen, Q.; Zhang, W.Y.; Zhang, H.H.; Ma, Y.; Zhang, S.Z.; Fang, J.; Yu, C. PDGF signaling pathway in hepatic fibrosis pathogenesis and therapeutics (Review). Mol. Med. Rep. 2017, 16, 7879–7889. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Yu, H.; Li, Q.-Y.; Wei, Y.-T.; Fu, J.; Dong, H.; Cao, D.; Guo, L.-N.; Chen, L.; Yang, Y.; et al. Hepatocyte-derived VEGFA accelerates the progression of non-alcoholic fatty liver disease to hepatocellular carcinoma via activating hepatic stellate cells. Acta Pharmacol. Sin. 2022, 43, 2917–2928. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.M.; Nikolic-Paterson, D.J.; Lan, H.Y. TGF-beta: The master regulator of fibrosis. Nat. Rev. Nephrol. 2016, 12, 325–338. [Google Scholar] [CrossRef]
- Stewart, A.G.; Thomas, B.; Koff, J. TGF-beta: Master regulator of inflammation and fibrosis. Respirology 2018, 23, 1096–1097. [Google Scholar] [CrossRef]
- Heldin, C.H.; Moustakas, A. Signaling Receptors for TGF-beta Family Members. Cold Spring Harb. Perspect Biol. 2016, 8, a022053. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Matsuzaki, K.; Mori, S.; Tahashi, Y.; Yamagata, H.; Furukawa, F.; Seki, T.; Nishizawa, M.; Fujisawa, J.; Okazaki, K. Transforming growth factor-beta and platelet-derived growth factor signal via c-Jun N-terminal kinase-dependent Smad2/3 phosphorylation in rat hepatic stellate cells after acute liver injury. Am. J. Pathol. 2005, 166, 1029–1039. [Google Scholar] [CrossRef]
- Massague, J.; Seoane, J.; Wotton, D. Smad transcription factors. Genes Dev. 2005, 19, 2783–2810. [Google Scholar] [CrossRef]
- Tsukada, S.; Westwick, J.K.; Ikejima, K.; Sato, N.; Rippe, R.A. SMAD and p38 MAPK signaling pathways independently regulate alpha1(I) collagen gene expression in unstimulated and transforming growth factor-beta-stimulated hepatic stellate cells. J. Biol. Chem. 2005, 280, 10055–10064. [Google Scholar] [CrossRef]
- Sanchez-Valle, V.; Chavez-Tapia, N.; Uribe, M.; Mendez-Sanchez, N. Role of oxidative stress and molecular changes in liver fibrosis: A review. Curr. Med. Chem. 2012, 19, 4850–4860. [Google Scholar] [CrossRef] [PubMed]
- Ebisawa, T.; Fukuchi, M.; Murakami, G.; Chiba, T.; Tanaka, K.; Imamura, T.; Miyazono, K. Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J. Biol. Chem. 2001, 276, 12477–12480. [Google Scholar] [CrossRef] [PubMed]
- Heuchel, R.; Itoh, S.; Kawabata, M.; Heldin, N.E.; Heldin, C.H.; ten Dijke, P. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature 1997, 389, 631–635. [Google Scholar]
- Zhang, S.; Fei, T.; Zhang, L.; Zhang, R.; Chen, F.; Ning, Y.; Han, Y.; Feng, X.; Meng, A.; Chen, Y. Smad7 antagonizes transforming growth factor beta signaling in the nucleus by interfering with functional Smad-DNA complex formation. Mol. Cell. Biol. 2007, 27, 4488–4499. [Google Scholar] [CrossRef] [PubMed]
- Cortijo, J.; Villagrasa, V.; Pons, R.; Berto, L.; Martí-Cabrera, M.; Martinez-Losa, M. Bronchodilator and anti-inflammatory activities of glaucine: In vitro studies in human airway smooth muscle and polymorphonuclear leukocytes. Br. J. Pharmacol. 1999, 127, 1641–1651. [Google Scholar] [CrossRef]
- Chang, F.R.; Wei, J.L.; Teng, C.M.; Wu, Y.C. Antiplatelet aggregation constituents from Annona purpurea. J. Nat. Prod. 1998, 61, 1457–1461. [Google Scholar] [CrossRef]
- Clark, A.M.; Watson, E.S.; Ashfaq, M.K.; Hufford, C.D. In vivo efficacy of antifungal oxoaporphine alkaloids in experimental disseminated candidiasis. Pharm. Res. 1987, 4, 495–498. [Google Scholar] [CrossRef]
- Wei, J.H.; Chen, Z.F.; Qin, J.L.; Liu, Y.C.; Li, Z.Q.; Khan, T.M.; Wang, M.; Jiang, Y.; Shen, W.; Liang, H. Water-soluble oxoglaucine-Y(III), Dy(III) complexes: In vitro and in vivo anticancer activities by triggering DNA damage, leading to S phase arrest and apoptosis. Dalton. Trans. 2015, 44, 11408–11419. [Google Scholar] [CrossRef]
- Gundogdu, G.; Gundogdu, K.; Nalci, K.A.; Demirkaya, A.K.; Tascı, S.Y.; Miloglu, F.D.; Senol, O.; Hacimuftuoglu, A. The Effect of Parietin Isolated from Rheum ribes L on In Vitro Wound Model Using Human Dermal Fibroblast Cells. Int. J. Low Extrem. Wounds 2019, 18, 56–64. [Google Scholar] [CrossRef]
- Tang, T.; Yin, L.; Yang, J.; Shan, G. Emodin, an anthraquinone derivative from Rheum officinale Baill, enhances cutaneous wound healing in rats. Eur. J. Pharmacol. 2007, 567, 177–185. [Google Scholar] [CrossRef]
- Xiao, D.; Zhang, Y.; Wang, R.; Fu, Y.; Zhou, T.; Diao, H.; Wang, Z.; Lin, Y.; Li, Z.; Wen, L.; et al. Emodin alleviates cardiac fibrosis by suppressing activation of cardiac fibroblasts via upregulating metastasis associated protein 3. Acta Pharm. Sin. B 2019, 9, 724–733. [Google Scholar] [CrossRef]
- Remichkova, M.; Dimitrova, P.; Philipov, S.; Ivanovska, N. Toll-like receptor-mediated anti-inflammatory action of glaucine and oxoglaucine. Fitoterapia 2009, 80, 411–414. [Google Scholar] [CrossRef] [PubMed]
- Zhong, G.; Long, H.; Chen, F.; Yu, Y. Oxoglaucine mediates Ca(2+) influx and activates autophagy to alleviate osteoarthritis through the TRPV5/calmodulin/CAMK-II pathway. Br. J. Pharmacol. 2021, 178, 2931–2947. [Google Scholar] [CrossRef] [PubMed]
- Ivanovska, N.; Philipov, S.; Georgieva, P. Immunopharmacological activity of aporphinoid alkaloid oxoglaucine. Pharmacol. Res. 1997, 35, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Koyama, Y.; Brenner, D.A. Liver inflammation and fibrosis. J. Clin. Investig. 2017, 127, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-C.; Chen, Z.-F.; Shi, Y.-F.; Huang, K.-B.; Geng, B.; Liang, H. Oxoglaucine-lanthanide complexes: Synthesis, crystal structure and cytotoxicity. Anticancer. Res. 2014, 34, 531–536. [Google Scholar]
- Bakin, A.V.; Tomlinson, A.K.; Bhowmick, N.A.; Moses, H.L.; Arteaga, C.L. Phosphatidylinositol 3-kinase function is required for transforming growth factor beta-mediated epithelial to mesenchymal transition and cell migration. J. Biol. Chem. 2000, 275, 36803–36810. [Google Scholar] [CrossRef]
- Frey, R.S.; Mulder, K.M. Involvement of extracellular signal-regulated kinase 2 and stress-activated protein kinase/Jun N-terminal kinase activation by transforming growth factor beta in the negative growth control of breast cancer cells. Cancer Res. 1997, 57, 628–633. [Google Scholar]
- Juhl, P.; Bondesen, S.; Hawkins, C.L.; Karsdal, M.A.; Bay-Jensen, A.C.; Davies, M.J.; Siebuhr, A.S. Dermal fibroblasts have different extracellular matrix profiles induced by TGF-beta, PDGF and IL-6 in a model for skin fibrosis. Sci. Rep. 2020, 10, 17300. [Google Scholar] [CrossRef] [PubMed]
- Seki, E.; Schwabe, R.F. Hepatic inflammation and fibrosis: Functional links and key pathways. Hepatology 2015, 61, 1066–1079. [Google Scholar] [CrossRef]
- Li, M.O.; Flavell, R.A. TGF-beta: A master of all T cell trades. Cell 2008, 134, 392–404. [Google Scholar] [CrossRef]
- Liu, R.M.; Desai, L.P. Reciprocal regulation of TGF-beta and reactive oxygen species: A perverse cycle for fibrosis. Redox. Biol. 2015, 6, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Mihaylova, M.M.; Shaw, R.J. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell. Biol. 2011, 13, 1016–1023. [Google Scholar] [CrossRef]
- Lin, H.; Li, N.; He, H.; Ying, Y.; Sunkara, S.; Luo, L.; Lv, N.; Huang, D.; Luo, Z. AMPK Inhibits the Stimulatory Effects of TGF-beta on Smad2/3 Activity, Cell Migration, and Epithelial-to-Mesenchymal Transition. Mol. Pharmacol. 2015, 88, 1062–1071. [Google Scholar] [CrossRef]
- Salminen, A.; Hyttinen, J.M.; Kaarniranta, K. AMP-activated protein kinase inhibits NF-kappaB signaling and inflammation: Impact on healthspan and lifespan. J. Mol. Med. 2011, 89, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Xue, X.; Fan, G.; Gu, Y.; Zhou, F.; Zheng, Q.; Liu, R.; Li, Y.; Ma, B.; Li, S. Ferulic Acid Ameliorates Hepatic Inflammation and Fibrotic Liver Injury by Inhibiting PTP1B Activity and Subsequent Promoting AMPK Phosphorylation. Front. Pharmacol. 2021, 12, 754976. [Google Scholar] [CrossRef]
- Siegert, A.; Ritz, E.; Orth, S.; Wagner, J. Differential regulation of transforming growth factor receptors by angiotensin II and transforming growth factor-beta1 in vascular smooth muscle. J. Mol. Med. 1999, 77, 437–445. [Google Scholar] [CrossRef]
- Yang, Y.; Sun, M.; Li, W.; Liu, C.; Jiang, Z.; Gu, P.; Li, J.; Wang, W.; You, R.; Ba, Q.; et al. Rebalancing TGF-beta/Smad7 signaling via Compound kushen injection in hepatic stellate cells protects against liver fibrosis and hepatocarcinogenesis. Clin. Transl. Med. 2021, 11, e410. [Google Scholar] [CrossRef]
- Yan, X.; Liao, H.; Cheng, M.; Shi, X.; Lin, X.; Feng, X.H.; Chen, Y.G. Smad7 Protein Interacts with Receptor-regulated Smads (R-Smads) to Inhibit Transforming Growth Factor-beta (TGF-beta)/Smad Signaling. J. Biol. Chem. 2016, 291, 382–392. [Google Scholar] [CrossRef]
- Suwanabol, P.A.; Seedial, S.M.; Zhang, F.; Shi, X.; Si, Y.; Liu, B.; Kent, K.C. TGF-beta and Smad3 modulate PI3K/Akt signaling pathway in vascular smooth muscle cells. Am. J. Physiol. Heart Circ. Physiol. 2012, 302, H2211-9. [Google Scholar] [CrossRef]
- Zhang, L.; Zhou, F.; ten Dijke, P. Signaling interplay between transforming growth factor-beta receptor and PI3K/AKT pathways in cancer. Trends Biochem. Sci. 2013, 38, 612–620. [Google Scholar] [CrossRef]
- Macáková, K.; Afonso, R.; Saso, L.; Mladěnka, P. The influence of alkaloids on oxidative stress and related signaling pathways. Free Radic. Biol. Med. 2019, 134, 429–444. [Google Scholar] [CrossRef]
- Mulcahy Levy, J.M.; Thorburn, A. Autophagy in cancer: Moving from understanding mechanism to improving therapy responses in patients. Cell. Death Differ. 2020, 27, 843–857. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Tan, L.; Hu, M. The role of autophagy in hepatic fibrosis. Am. J. Transl. Res. 2021, 13, 5747–5757. [Google Scholar] [PubMed]
- Lodder, J.; Denaës, T.; Chobert, M.-N.; Wan, J.; El-Benna, J.; Pawlotsky, J.-M.; Lotersztajn, S.; Teixeira-Clerc, F. Macrophage autophagy protects against liver fibrosis in mice. Autophagy 2015, 11, 1280–1292. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.W.; Tang, C.L.; Zheng, H.; Wu, J.X.; Wu, F.; Mo, Y.Y.; Liu, X.; Zhu, H.; Yin, C.; Cheng, B.; et al. Investigation of the hepatoprotective effect of Corydalis saxicola Bunting on carbon tetrachloride-induced liver fibrosis in rats by (1)H-NMR-based metabonomics and network pharmacology approaches. J. Pharm. Biomed. Anal. 2018, 159, 252–261. [Google Scholar] [CrossRef]
- Yu, Q.; Cheng, P.; Wu, J.; Guo, C. PPARgamma/NF-kappaB and TGF-beta1/Smad pathway are involved in the anti-fibrotic effects of levo-tetrahydropalmatine on liver fibrosis. J. Cell. Mol. Med. 2021, 25, 1645–1660. [Google Scholar] [CrossRef] [PubMed]
- Charni-Natan, M.; Goldstein, I. Protocol for Primary Mouse Hepatocyte Isolation. STAR Protoc. 2020, 1, 100086. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Azamov, B.; Lee, K.-M.; Hur, J.; Muradillaeva, S.; Shim, W.-S.; Lee, C.; Song, P. Oxoglaucine Suppresses Hepatic Fibrosis by Inhibiting TGFβ-Induced Smad2 Phosphorylation and ROS Generation. Molecules 2023, 28, 4971. https://doi.org/10.3390/molecules28134971
Azamov B, Lee K-M, Hur J, Muradillaeva S, Shim W-S, Lee C, Song P. Oxoglaucine Suppresses Hepatic Fibrosis by Inhibiting TGFβ-Induced Smad2 Phosphorylation and ROS Generation. Molecules. 2023; 28(13):4971. https://doi.org/10.3390/molecules28134971
Chicago/Turabian StyleAzamov, Bakhovuddin, Kwang-Min Lee, Jin Hur, Shakhnoza Muradillaeva, Wan-Seog Shim, Chanhee Lee, and Parkyong Song. 2023. "Oxoglaucine Suppresses Hepatic Fibrosis by Inhibiting TGFβ-Induced Smad2 Phosphorylation and ROS Generation" Molecules 28, no. 13: 4971. https://doi.org/10.3390/molecules28134971
APA StyleAzamov, B., Lee, K.-M., Hur, J., Muradillaeva, S., Shim, W.-S., Lee, C., & Song, P. (2023). Oxoglaucine Suppresses Hepatic Fibrosis by Inhibiting TGFβ-Induced Smad2 Phosphorylation and ROS Generation. Molecules, 28(13), 4971. https://doi.org/10.3390/molecules28134971






