G-Quadruplexes Formation by the C9orf72 Nucleotide Repeat Expansion d(GGGGCC)n and Conformation Regulation by Fangchinoline
Abstract
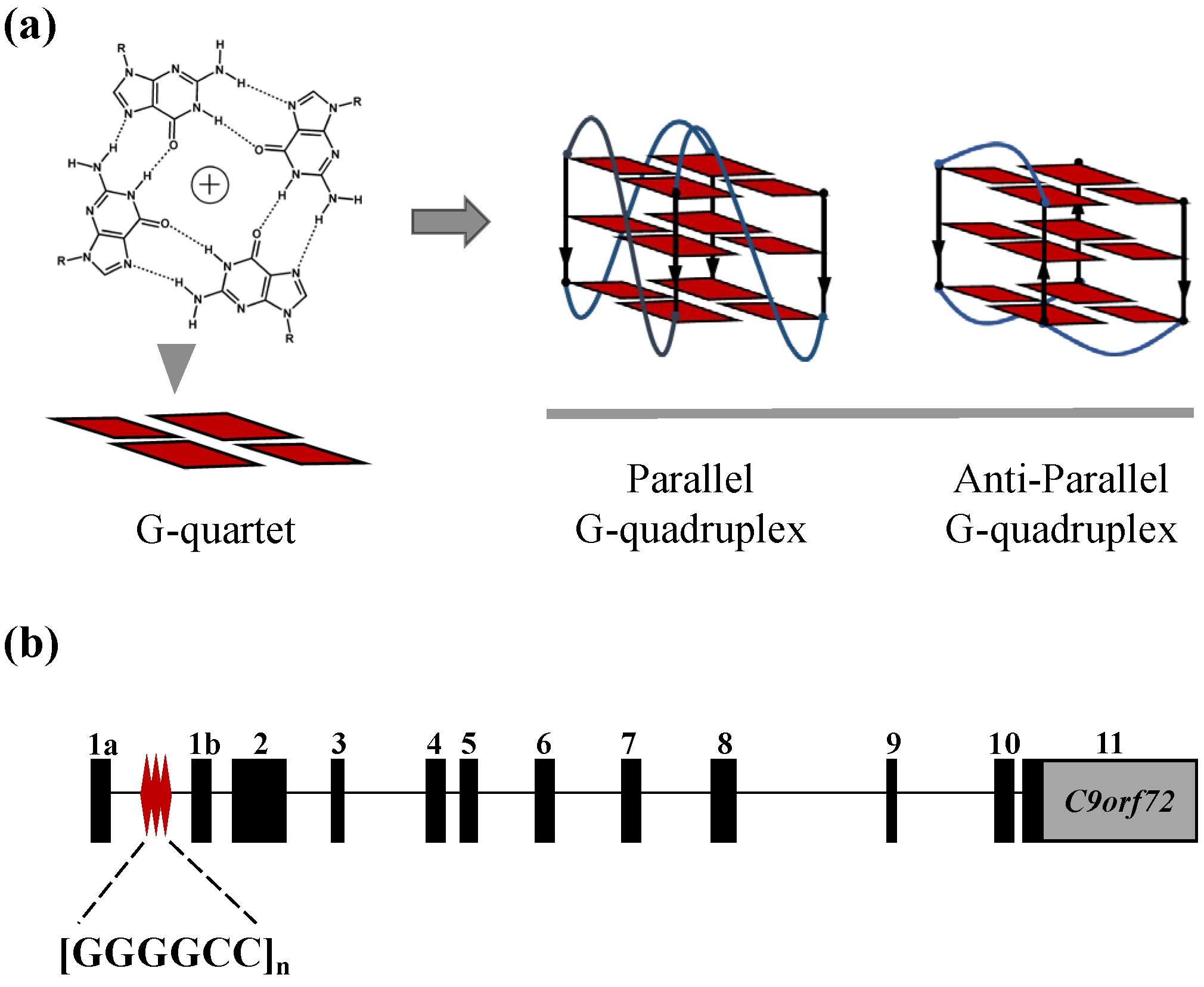
1. Introduction
2. Results
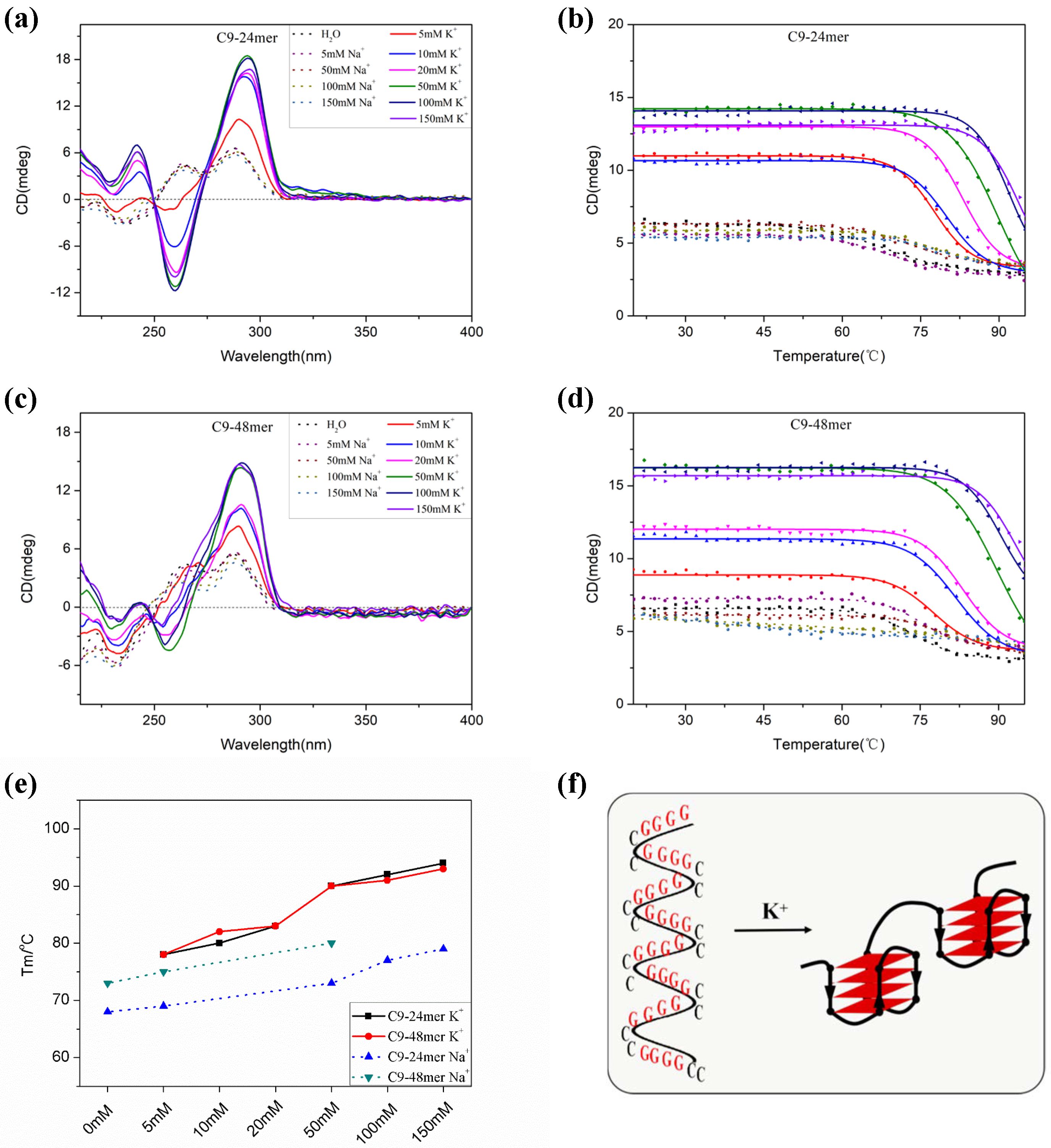
2.1. The C9-HRE Long Sequence Forms Unstacked Tandem AP-GQ Structure in a Bead-on-a-String Mode
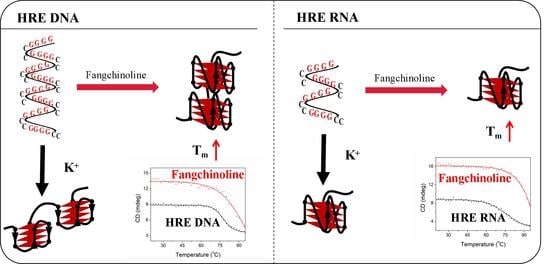
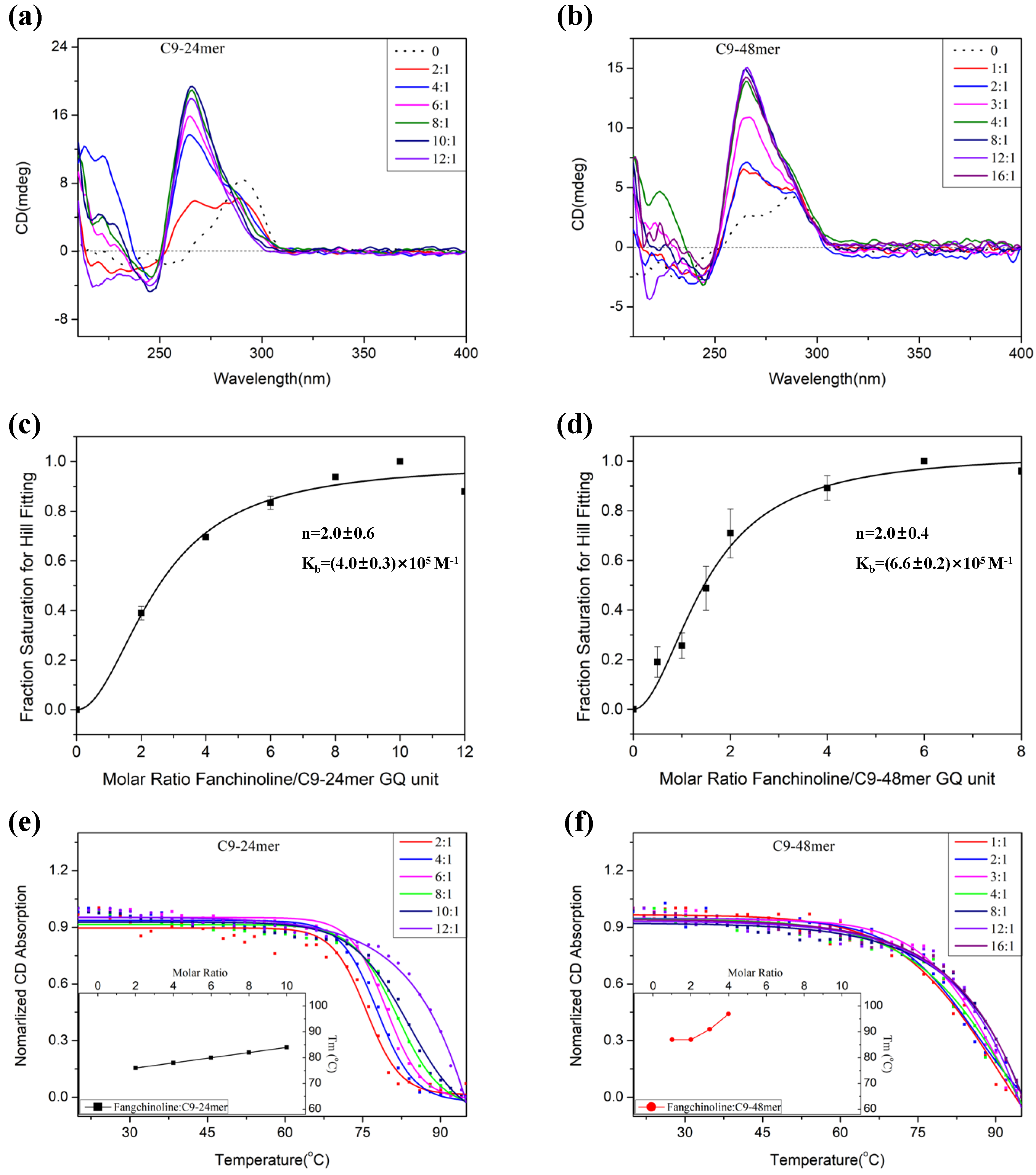
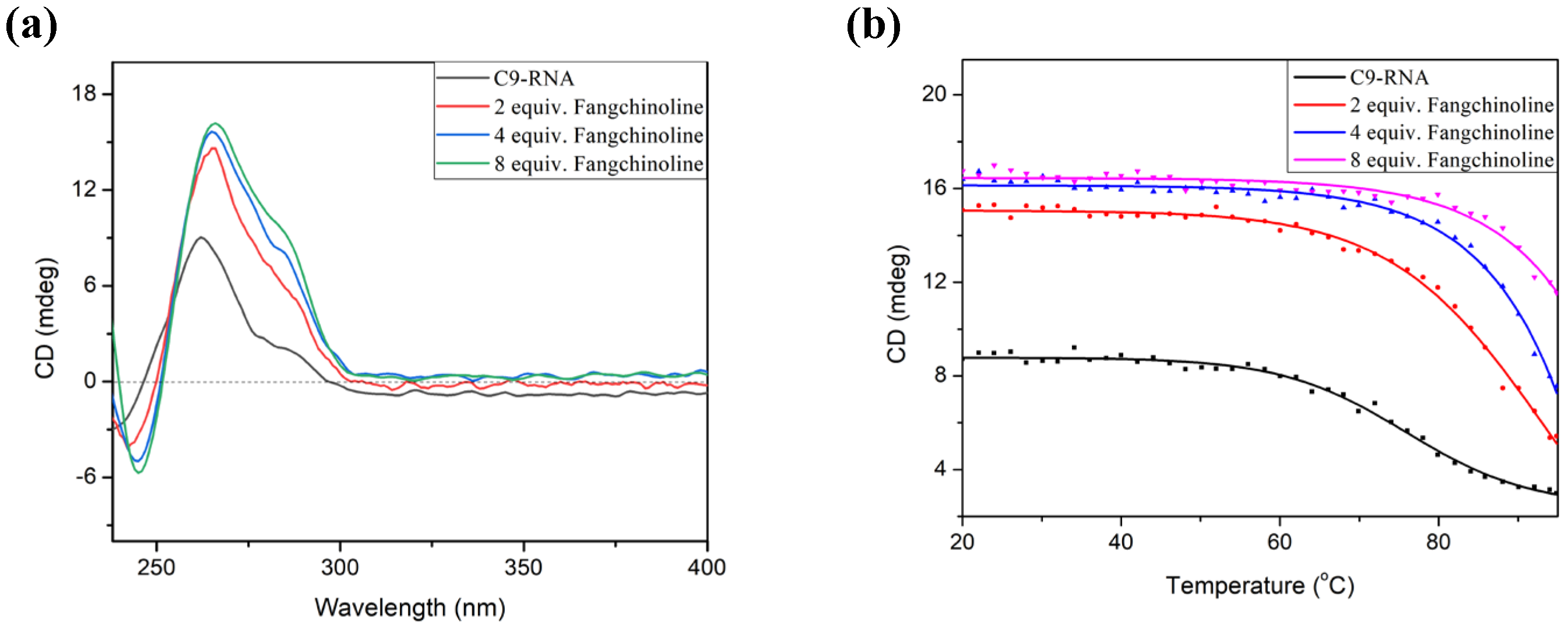
2.2. Fangchinoline Alters the C9-HRE Topology to Stable Parallel GQ
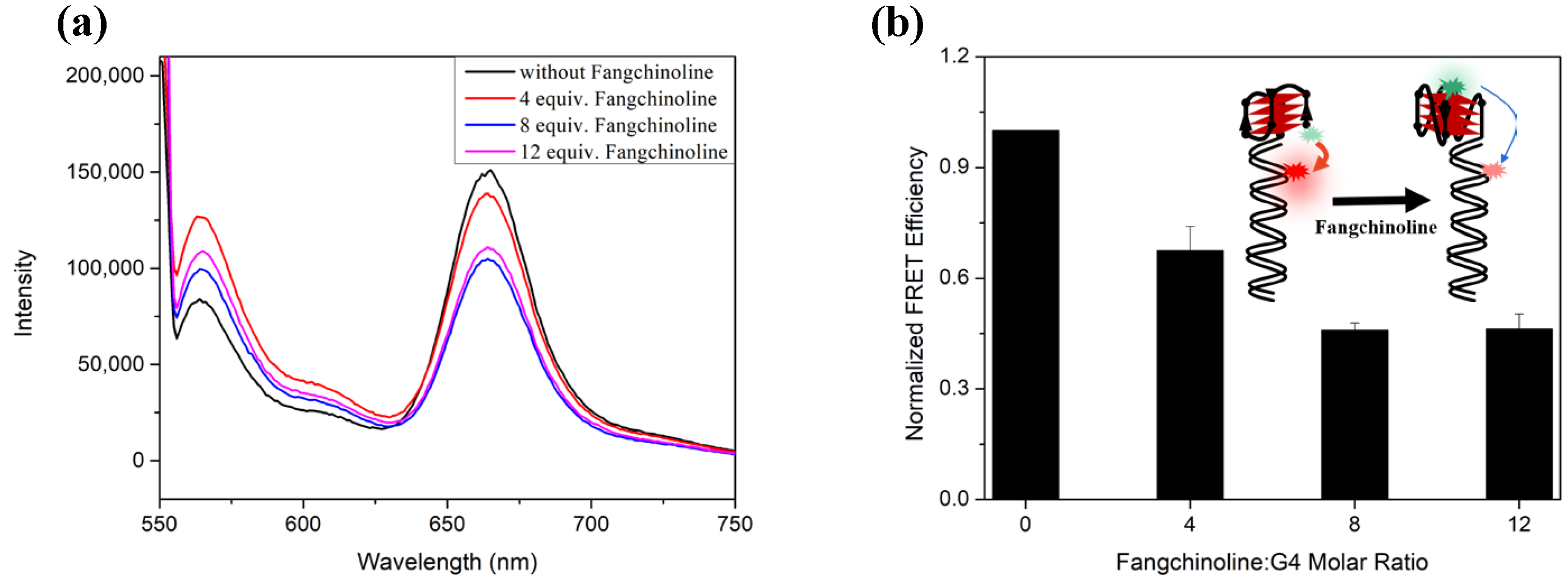
2.3. Fangchinoline Can Recognize and Stabilize C9-HRE RNA GQ
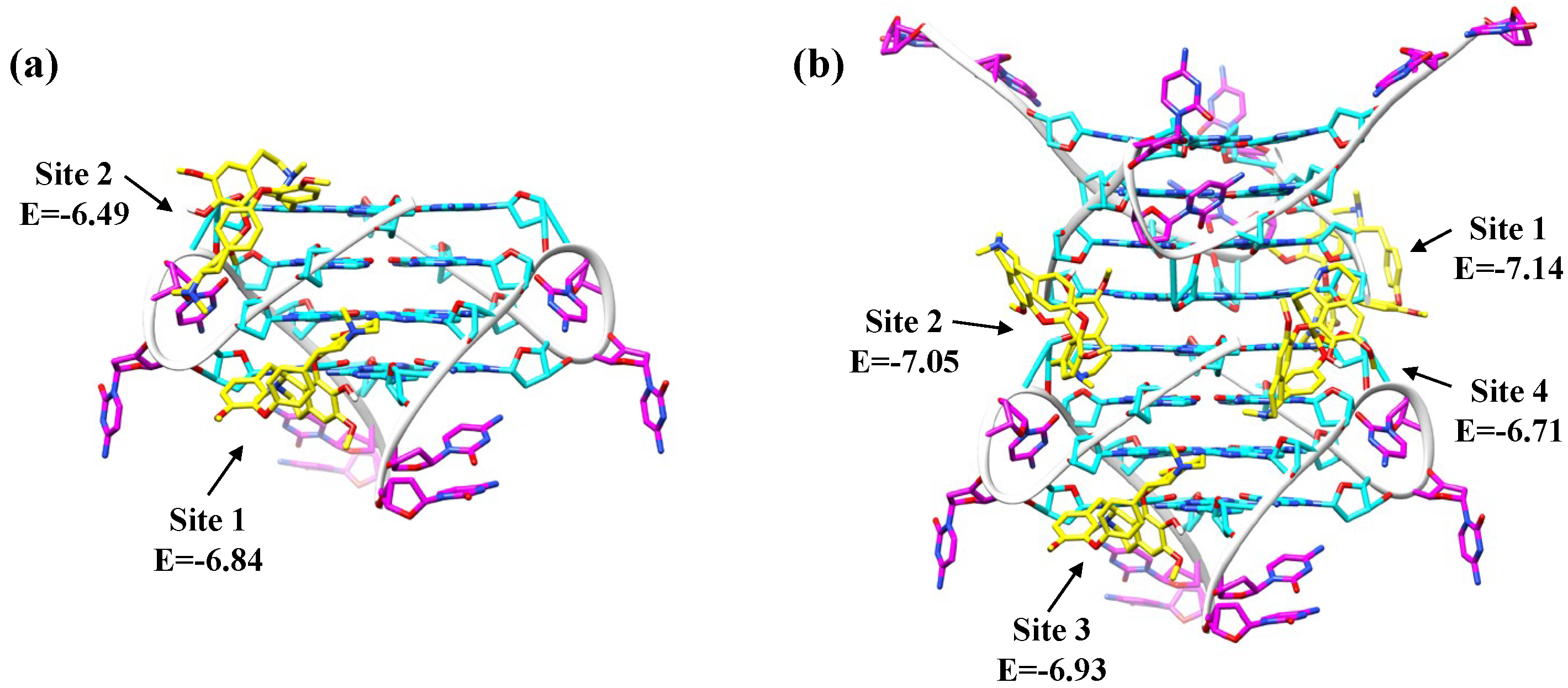
2.4. Fangchinoline Is a Groove-Binder to the Parallel C9-HRE GQ
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Circular Dichroism (CD) Spectroscopy
4.3. CD Melting Experiment
4.4. Polyacrylamide Gel Electrophoresis (PAGE) Experiment
4.5. Fluorescence Resonance Energy Transfer (FRET) Experiment
4.6. AutoDock Simulation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Bochman, M.L.; Paeschke, K.; Zakian, V.A. DNA secondary structures: Stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012, 13, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Bryan, T.M.; Baumann, P. G-quadruplexes: From guanine gels to chemotherapeutics. Methods Mol. Biol. 2010, 608, 1–16. [Google Scholar] [PubMed]
- Karsisiotis, A.I.; O’Kane, C.; da Silva, M.W. DNA quadruplex folding formalism—A tutorial on quadruplex topologies. Methods 2013, 64, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Chambers, V.S.; Marsico, G.; Boutell, J.M.; Di Antonio, M.; Smith, G.P.; Balasubramanian, S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015, 33, 877–881. [Google Scholar] [CrossRef]
- Hänsel-Hertsch, R.; Beraldi, D.; Lensing, S.V.; Marsico, G.; Zyner, K.; Parry, A.; Di Antonio, M.; Pike, J.; Kimura, H.; Narita, M.; et al. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016, 48, 1267–1272. [Google Scholar] [CrossRef]
- Rhodes, D.; Lipps, H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43, 8627–8637. [Google Scholar] [CrossRef]
- Fay, M.M.; Lyons, S.M.; Ivanov, P. RNA G-Quadruplexes in Biology: Principles and Molecular Mechanisms. J. Mol. Biol. 2017, 429, 2127–2147. [Google Scholar] [CrossRef]
- Neidle, S. Quadruplex Nucleic Acids as Novel Therapeutic Targets. J. Med. Chem. 2016, 59, 5987–6011. [Google Scholar] [CrossRef]
- Teng, F.-Y.; Jiang, Z.-Z.; Guo, M.; Tan, X.Z.; Chen, F.; Xi, X.-G.; Xu, Y. G-quadruplex DNA: A novel target for drug design. Cell. Mol. Life Sci. 2021, 78, 6557–6583. [Google Scholar] [CrossRef]
- Onel, B.; Lin, C.; Yang, D. DNA G-quadruplex and its potential as anticancer drug target. Sci. China Chem. 2014, 57, 1605–1614. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Hurley, L.H.; Neidle, S. Targeting G-quadruplexes in gene promoters: A novel anticancer strategy? Nat. Rev. Drug Discov. 2011, 10, 261–275. [Google Scholar] [CrossRef] [PubMed]
- Düchler, M. G-quadruplexes: Targets and tools in anticancer drug design. J. Drug Target. 2012, 20, 389–400. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Chen, H.; Zhang, Q.; Xu, M.; Yuan, G.; Zhou, J. Exploration of the Structure and Recognition of a G-quadruplex in the her2 Proto-oncogene Promoter and Its Transcriptional Regulation. Sci. Rep. 2019, 9, 3966. [Google Scholar] [CrossRef] [PubMed]
- Simone, R.; Fratta, P.; Neidle, S.; Parkinson, G.N.; Isaacs, A.M. G-quadruplexes: Emerging roles in neurodegenerative diseases and the non-coding transcriptome. FEBS Lett. 2015, 589, 1653–1668. [Google Scholar] [CrossRef]
- Cave, J.W.; Willis, D.E. G-quadruplex regulation of neural gene expression. FEBS J. 2022, 289, 3284–3303. [Google Scholar] [CrossRef]
- Asamitsu, S.; Takeuchi, M.; Ikenoshita, S.; Imai, Y.; Kashiwagi, H.; Shioda, N. Perspectives for Applying G-Quadruplex Structures in Neurobiology and Neuropharmacology. Int. J. Mol. Sci. 2019, 20, 2884. [Google Scholar] [CrossRef]
- Santoro, M.R.; Bray, S.M.; Warren, S.T. Molecular mechanisms of fragile X syndrome: A twenty-year perspective. Annu. Rev. Pathol. 2012, 7, 219–245. [Google Scholar] [CrossRef]
- Todd, P.K.; Oh, S.Y.; Krans, A.; He, F.; Sellier, C.; Frazer, M.; Renoux, A.J.; Chen, K.; Scaglione, K.M.; Basrur, V.; et al. CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron 2013, 78, 440–455. [Google Scholar] [CrossRef]
- Borel, C.; Migliavacca, E.; Letourneau, A.; Gagnebin, M.; Béna, F.; Sailani, M.R.; Dermitzakis, E.T.; Sharp, A.J.; Antonarakis, S.E. Tandem repeat sequence variation as causative cis-eQTLs for protein-coding gene expression variation: The case of CSTB. Hum. Mutat. 2012, 33, 1302–1309. [Google Scholar] [CrossRef]
- Li, X.; Liu, Y.; Yu, K.; Cui, X. Exploration of the Tandem G-quadruplexes Formed by the Dodecamer Repeat d[CGCGGGGCGGGG]n in the Promoter of the CSTB Gene. Chin. J. Biochem. Mol. Biol. 2020, 36, 1374–1380. [Google Scholar]
- Zarei, S.; Carr, K.; Reiley, L.; Diaz, K.; Guerra, O.; Altamirano, P.F.; Pagani, W.; Lodin, D.; Orozco, G.; Chinea, A. A comprehensive review of amyotrophic lateral sclerosis. Surg. Neurol. Int. 2015, 6, 171. [Google Scholar] [CrossRef] [PubMed]
- Cardarelli, R.; Kertesz, A.; Knebl, J.A. Frontotemporal dementia: A review for primary care physicians. Am. Fam. Physician 2010, 82, 1372–1377. [Google Scholar] [PubMed]
- Gao, F.-B.; Almeida, S.; Lopez-Gonzalez, R. Dysregulated molecular pathways in amyotrophic lateral sclerosis-frontotemporal dementia spectrum disorder. EMBO J. 2017, 36, 2931–2950. [Google Scholar] [CrossRef] [PubMed]
- McCauley, M.E.; Baloh, R.H. Inflammation in ALS/FTD pathogenesis. Acta Neuropathol. 2019, 137, 715–730. [Google Scholar] [CrossRef]
- Van Mossevelde, S.; van der Zee, J.; Cruts, M.; Van Broeckhoven, C. Relationship between C9orf72 repeat size and clinical phenotype. Curr. Opin. Genet. Dev. 2017, 44, 117–124. [Google Scholar] [CrossRef]
- Braems, E.; Swinnen, B.; Van Den Bosch, L. C9orf72 loss-of-function: A trivial, stand-alone or additive mechanism in C9 ALS/FTD? Acta Neuropathol. 2020, 140, 625–643. [Google Scholar] [CrossRef]
- Babić Leko, M.; Župunski, V.; Kirincich, J.; Smilović, D.; Hortobágyi, T.; Hof, P.R.; Šimić, G. Molecular Mechanisms of Neurodegeneration Related to C9orf72 Hexanucleotide Repeat Expansion. Behav. Neurol. 2019, 2019, 2909168. [Google Scholar] [CrossRef]
- Liu, Y.; Dodart, J.-C.; Tran, H.; Berkovitch, S.; Braun, M.; Byrne, M.; Durbin, A.F.; Hu, X.S.; Iwamoto, N.; Jang, H.G.; et al. Variant-selective stereopure oligonucleotides protect against pathologies associated with C9orf72-repeat expansion in preclinical models. Nat. Commun. 2021, 12, 847. [Google Scholar] [CrossRef]
- Liu, Y.; Andreucci, A.; Iwamoto, N.; Yin, Y.; Yang, H.; Liu, F.; Bulychev, A.; Hu, X.S.; Lin, X.; Lamore, S.; et al. Preclinical evaluation of WVE-004, aninvestigational stereopure oligonucleotide forthe treatment of C9orf72-associated ALS or FTD. Mol. Ther. Nucleic Acids 2022, 28, 558–570. [Google Scholar] [CrossRef]
- Zamiri, B.; Reddy, K.; Macgregor, R.B.; Pearson, C.E. TMPyP4 porphyrin distorts RNA G-quadruplex structures of the disease-associated r(GGGGCC)n repeat of the C9orf72 gene and blocks interaction of RNA-binding proteins. J. Biol. Chem. 2014, 289, 4653–4659. [Google Scholar] [CrossRef]
- Zhang, K.; Donnelly, C.J.; Haeusler, A.R.; Grima, J.C.; Machamer, J.B.; Steinwald, P.; Daley, E.L.; Miller, S.J.; Cunningham, K.M.; Vidensky, S.; et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 2015, 525, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Gotoh, S.; Yamashita, T.; Uozumi, R.; Kawabe, Y.; Tagami, S.; Kamp, F.; Nuscher, B.; Edbauer, D.; Haass, C.; et al. The porphyrin TMPyP4 inhibits elongation during the noncanonical translation of the FTLD/ALS-associated GGGGCC repeat in the C9orf72 gene. J. Biol. Chem. 2021, 297, 101120. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.; Liu, C.; Ye, W.; Huang, D.; She, W.; Liu, X.; Fung, C.P.; Xu, N.; Suen, M.C.; Ye, W.; et al. Selective C9orf72 G-Quadruplex-Binding Small Molecules Ameliorate Pathological Signatures of ALS/FTD Models. J. Med. Chem. 2022, 65, 12825–12837. [Google Scholar] [CrossRef]
- Simone, R.; Balendra, R.; Moens, T.G.; Preza, E.; Wilson, K.M.; Heslegrave, A.; Woodling, N.S.; Niccoli, T.; Gilbert-Jaramillo, J.; Abdelkarim, S.; et al. G-quadruplex-binding small molecules ameliorate C9orf72 FTD/ALS pathology in vitro and in vivo. EMBO Mol. Med. 2018, 10, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Zhang, Y.; Gendron, T.F.; Bauer, P.O.; Chew, J.; Yang, W.-Y.; Fostvedt, E.; Jansen-West, K.; Belzil, V.V.; Desaro, P.; et al. Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron 2014, 83, 1043–1050. [Google Scholar] [CrossRef] [PubMed]
- Ozcan, K.A.; Ghaffari, L.T.; Haeusler, A.R. The effects of molecular crowding and CpG hypermethylation on DNA G-quadruplexes formed by the C9orf72 nucleotide repeat expansion. Sci. Rep. 2021, 11, 23213. [Google Scholar] [CrossRef]
- Brčić, J.; Plavec, J. Solution structure of a DNA quadruplex containing ALS and FTD related GGGGCC repeat stabilized by 8-bromodeoxyguanosine substitution. Nucleic Acids Res. 2015, 43, 8590–8600. [Google Scholar] [CrossRef]
- Brcic, J.; Plavec, J. NMR structure of a G-quadruplex formed by four d(G4C2) repeats: Insights into structural polymorphism. Nucleic Acids Res. 2018, 46, 11605–11617. [Google Scholar]
- Brčić, J.; Plavec, J. ALS and FTD linked GGGGCC-repeat containing DNA oligonucleotide folds into two distinct G-quadruplexes. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1237–1245. [Google Scholar] [CrossRef]
- Zhou, B.; Liu, C.; Geng, Y.; Zhu, G. Topology of a G-quadruplex DNA formed by C9orf72 hexanucleotide repeats associated with ALS and FTD. Sci. Rep. 2015, 5, 16673. [Google Scholar] [CrossRef]
- Geng, Y.; Liu, C.; Cai, Q.; Luo, Z.; Miao, H.; Shi, X.; Xu, N.; Fung, C.P.; Choy, T.T.; Yan, B.; et al. Crystal structure of parallel G-quadruplex formed by the two-repeat ALS- and FTD-related GGGGCC sequence. Nucleic Acids Res. 2021, 49, 5881–5890. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.-Q.; Miyoshi, D.; Sugimoto, N. Characterization of structure and stability of long telomeric DNA G-quadruplexes. J. Am. Chem. Soc. 2006, 128, 15461–15468. [Google Scholar] [CrossRef] [PubMed]
- Zamiri, B.; Mirceta, M.; Bomsztyk, K.; Macgregor, R.B.; Pearson, C.E. Quadruplex formation by both G-rich and C-rich DNA strands of the C9orf72 (GGGGCC)8•(GGCCCC)8 repeat: Effect of CpG methylation. Nucleic Acids Res. 2015, 43, 10055–10064. [Google Scholar]
- Lim, K.W.; Alberti, P.; Guédin, A.; Lacroix, L.; Riou, J.-F.; Royle, N.J.; Mergny, J.-L.; Phan, A.T. Sequence variant (CTAGGG)n in the human telomere favors a G-quadruplex structure containing a G.C.G.C tetrad. Nucleic Acids Res. 2009, 37, 6239–6248. [Google Scholar] [CrossRef] [PubMed]
- Vorlíčková, M.; Kejnovská, I.; Bednářová, K.; Renčiuk, D.; Kypr, J. Circular dichroism spectroscopy of DNA: From duplexes to quadruplexes. Chirality 2012, 24, 691–698. [Google Scholar] [CrossRef]
- Tan, W.; Chen, H.; Zhou, J.; Xu, M.; Yuan, G. Induction of formation and conformational conversion of DNA G-quadruplexes by fangchinoline. Anal. Methods 2014, 6, 8476–8481. [Google Scholar] [CrossRef]
- Cui, X.; Lin, S.; Yuan, G. Spectroscopic probing of recognition of the G-quadruplex in c-kit promoter by small-molecule natural products. Int. J. Biol. Macromol. 2012, 50, 996–1001. [Google Scholar] [CrossRef]
- Ying, L.; Green, J.J.; Li, H.; Klenerman, D.; Balasubramanian, S. Studies on the structure and dynamics of the human telomeric G quadruplex by single-molecule fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. USA 2003, 100, 14629–14634. [Google Scholar] [CrossRef]
- Tippana, R.; Xiao, W.; Myong, S. G-quadruplex conformation and dynamics are determined by loop length and sequence. Nucleic Acids Res. 2014, 42, 8106–8114. [Google Scholar] [CrossRef]
- Sanner, M.F. Python: A programming language for software integration and development. J. Mol. Graph. Model. 1999, 17, 57–61. [Google Scholar]
- Ortiz de Luzuriaga, I.; Lopez, X.; Gil, A. Learning to Model G-Quadruplexes: Current Methods and Perspectives. Annu. Rev. Biophys. 2021, 50, 209–243. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-González, Á.; Bandeira, N.A.G.; Ortiz de Luzuriaga, I.; Martins, F.F.; Elleuchi, S.; Jarraya, K.; Lanuza, J.; Lopez, X.; Calhorda, M.J.; Gil, A. New Insights on the Interaction of Phenanthroline Based Ligands and Metal Complexes and Polyoxometalates with Duplex DNA and G-Quadruplexes. Molecules 2021, 26, 4737. [Google Scholar] [CrossRef] [PubMed]
- Ortiz de Luzuriaga, I.; Sánchez-González, Á.; Synoradzki, W.; Lopez, X.; Gil, A. Unravelling the binding affinity and selectivity of molybdenum(II) phenanthroline complexes with DNA G-quadruplexes by using linear-scaling DFT studies. The important role of ancillary ligands. Phys. Chem. Chem. Phys. 2022, 24, 25918–25929. [Google Scholar] [CrossRef]
- Bag, S.; Burman, M.D.; Bhowmik, S. Structural insights and shedding light on preferential interactions of dietary flavonoids with G-quadruplex DNA structures: A new horizon. Heliyon 2023, 9, e13959. [Google Scholar] [CrossRef] [PubMed]
- Belzil, V.V.; Bauer, P.O.; Prudencio, M.; Gendron, T.F.; Stetler, C.T.; Yan, I.K.; Pregent, L.; Daughrity, L.; Baker, M.C.; Rademakers, R.; et al. Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood. Acta Neuropathol. 2013, 126, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Viodé, A.; Fournier, C.; Camuzat, A.; Fenaille, F.; NeuroCEB Brain Bank; Latouche, M.; Elahi, F.; Le Ber, I.; Junot, C.; Lamari, F.; et al. New Antibody-Free Mass Spectrometry-Based Quantification Reveals That C9ORF72 Long Protein Isoform Is Reduced in the Frontal Cortex of Hexanucleotide-Repeat Expansion Carriers. Front. Neurosci. 2018, 12, 589. [Google Scholar] [CrossRef]
- Bauer, C.S.; Cohen, R.N.; Sironi, F.; Livesey, M.R.; Gillingwater, T.H.; Highley, J.R.; Fillingham, D.J.; Coldicott, I.; Smith, E.F.; Gibson, Y.B.; et al. An interaction between synapsin and C9orf72 regulates excitatory synapses and is impaired in ALS/FTD. Acta Neuropathol. 2022, 144, 437–464. [Google Scholar] [CrossRef]
- Mendoza, O.; Bourdoncle, A.; Boulé, J.-B.; Brosh, R.M.; Mergny, J.-L. G-quadruplexes and helicases. Nucleic Acids Res. 2016, 44, 1989–2006. [Google Scholar] [CrossRef]
- Smeyers, J.; Banchi, E.-G.; Latouche, M. C9ORF72: What It Is, What It Does, and Why It Matters. Front. Cell. Neurosci. 2021, 15, 661447. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Huang, J.; Yu, K.; Cui, X. G-Quadruplexes Formation by the C9orf72 Nucleotide Repeat Expansion d(GGGGCC)n and Conformation Regulation by Fangchinoline. Molecules 2023, 28, 4671. https://doi.org/10.3390/molecules28124671
Zhang Y, Huang J, Yu K, Cui X. G-Quadruplexes Formation by the C9orf72 Nucleotide Repeat Expansion d(GGGGCC)n and Conformation Regulation by Fangchinoline. Molecules. 2023; 28(12):4671. https://doi.org/10.3390/molecules28124671
Chicago/Turabian StyleZhang, Yun, Junliu Huang, Kainan Yu, and Xiaojie Cui. 2023. "G-Quadruplexes Formation by the C9orf72 Nucleotide Repeat Expansion d(GGGGCC)n and Conformation Regulation by Fangchinoline" Molecules 28, no. 12: 4671. https://doi.org/10.3390/molecules28124671
APA StyleZhang, Y., Huang, J., Yu, K., & Cui, X. (2023). G-Quadruplexes Formation by the C9orf72 Nucleotide Repeat Expansion d(GGGGCC)n and Conformation Regulation by Fangchinoline. Molecules, 28(12), 4671. https://doi.org/10.3390/molecules28124671