Chemical Profile, Antibacterial, Antibiofilm, and Antiviral Activities of Pulicaria crispa Most Potent Fraction: An In Vitro and In Silico Study
Abstract
1. Introduction
2. Results
2.1. Antibacterial Activity
2.1.1. Preliminary Antibacterial Screening of P. crispa Crude Extract and Fractions
2.1.2. Evaluation of Antibacterial Activity of P. crispa Most Potent Fraction (HF)
2.1.3. Gene Expression Analysis
2.2. Antibiofilm Activity
2.2.1. Biofilm Forming Ability
2.2.2. Biofilm Inhibition Activity of P. crispa HF Fraction
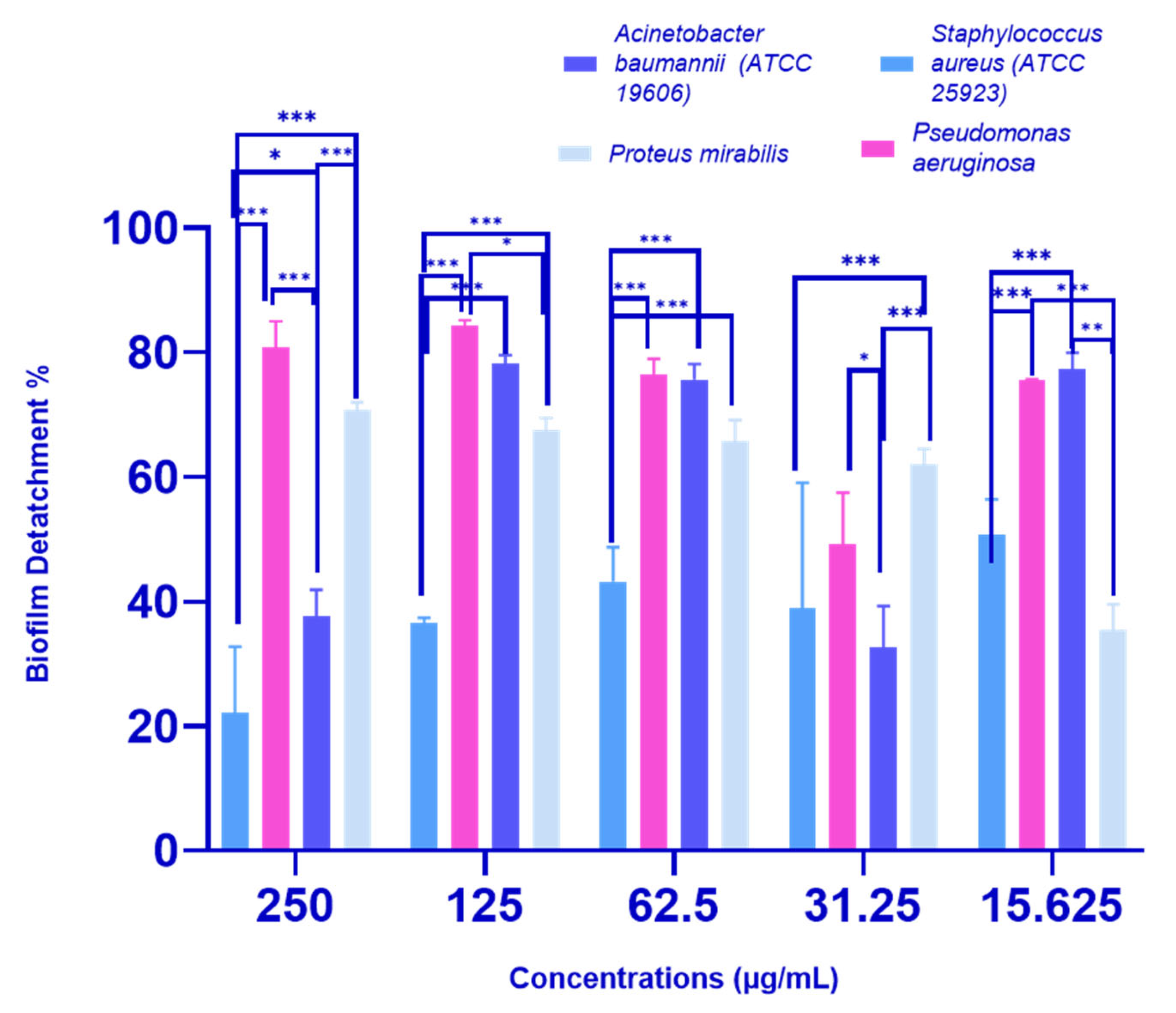
2.2.3. Biofilm Detachment Activity of P. crispa HF Fraction
2.3. Antiviral Activity
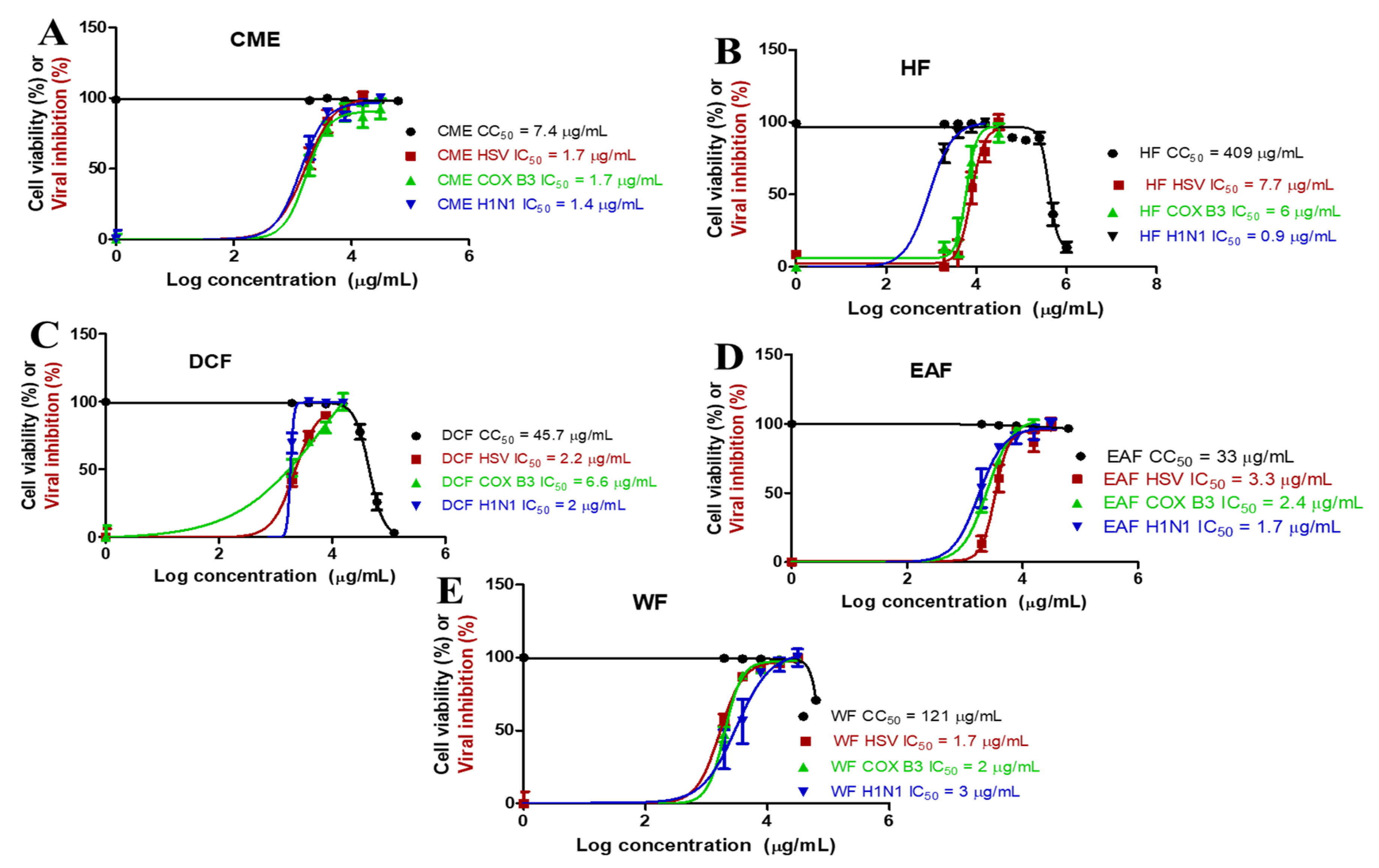
2.3.1. Cytotoxicity Activity
2.3.2. Antiviral Screening (Determination of 50% Inhibitory Concentration, IC50)
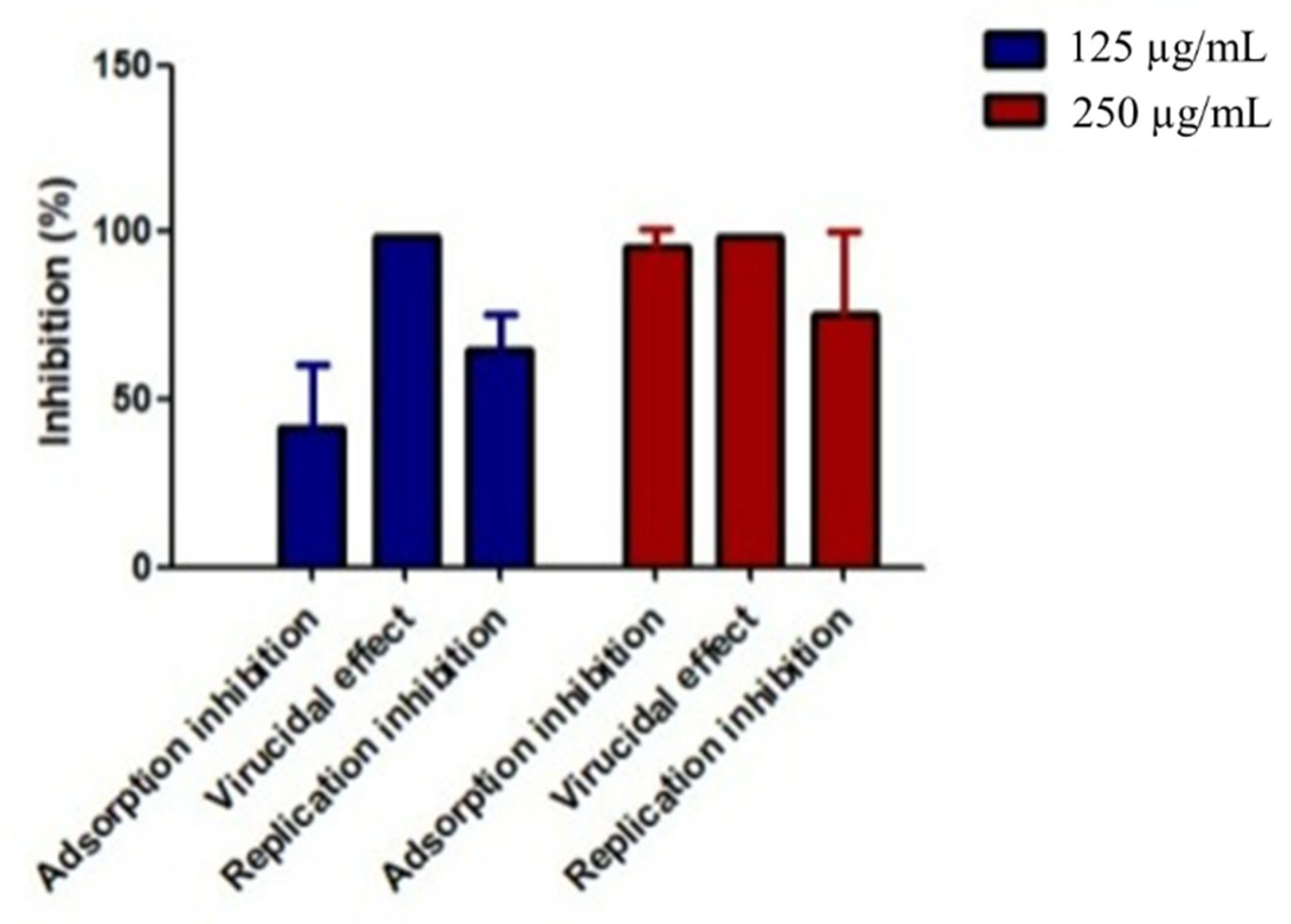
2.3.3. Time of Addition (TOA) Effect of P. crispa HF Fraction on Influenza A Virus
2.4. Chemical Profiling of P. crispa HF Fraction Using GC/MS Analysis
2.5. In Silico Study
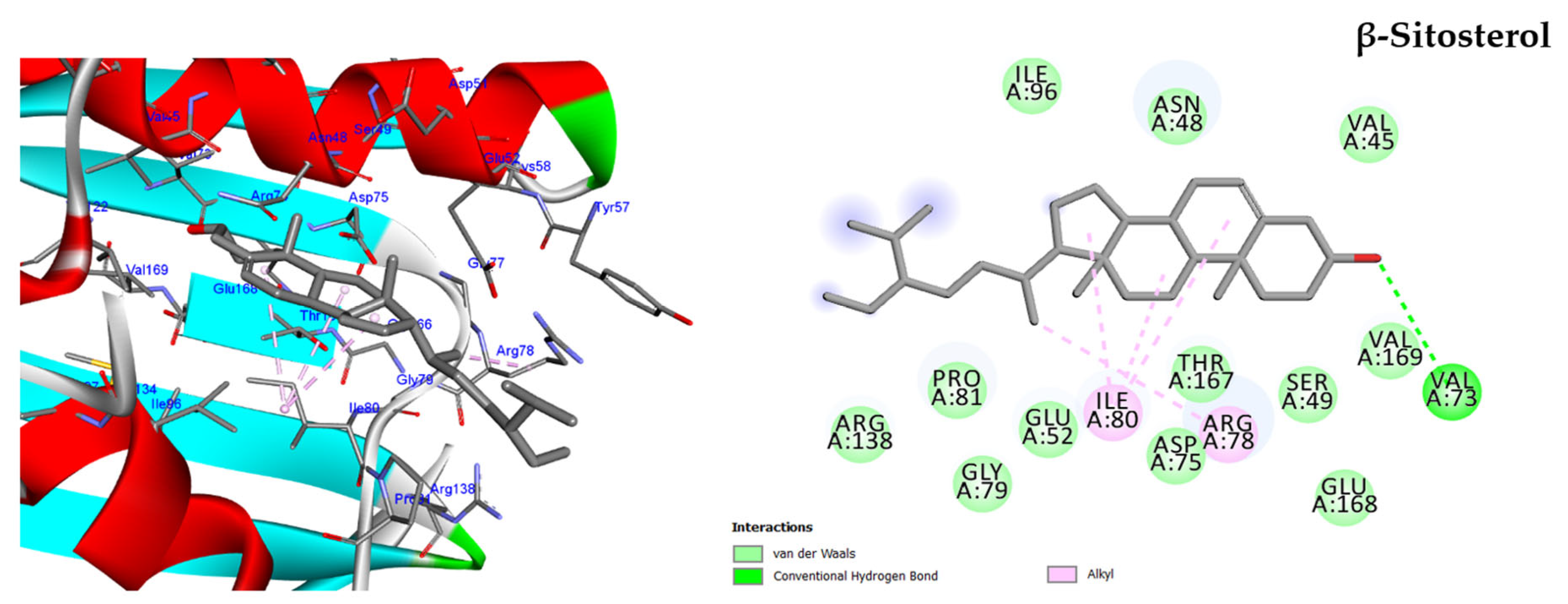
2.5.1. Molecular Docking against DNA Gyrase B
2.5.2. Molecular Docking against Penicillin-Binding Protein (PBP2A)
2.5.3. Molecular Docking against Influenza A Virus Nucleoprotein (NP)
2.5.4. Molecular Docking against Influenza A Virus Polymerase
3. Discussion
4. Material and Methods
4.1. Plant Material
4.2. Extraction and Fractionation
4.3. Antibacterial Activity
4.3.1. Determination of Minimum Inhibitory Concentration (MIC)
4.3.2. Determination of Minimum Bactericidal Concentrations (MBC)
4.3.3. Real Time-Polymerase Chain Reaction (RT-qPCR) Analysis
4.4. Antibiofilm Activity
4.4.1. Biofilm Inhibition Assay
4.4.2. Biofilm Detachment Assay
4.5. Antiviral Activity
4.5.1. Cell Lines
4.5.2. Viruses
4.5.3. Cytotoxicity Assay
4.5.4. Antiviral Screening Assay (Determination of 50% Inhibitory Concentration, IC50)
4.5.5. Time of Addition (TOA) Assay
- For the pretreatment protocol, we incubated MDCK cells with HF fraction at 125 and 250 µg/mL for 2 h at 37 °C before infecting them with the influenza A virus. After removing the supernatant, we added the virus to the cells and allowed it to incubate for 1.5 h. Then, any unabsorbed virus was removed with PBS and replaced with a fresh medium.
- In the postinfection protocol, we infected MDCK cells with the influenza A virus and added P. crispa HF fraction at 125 and 250 µg/mL 1.5 h later.
- In the competition protocol, we added HF fraction at 125 and 250 µg/mL to the virus during the adsorption period for 1.5 h. Then, any unbound viruses and HF were removed and cultured in the infected MDCK cells in a fresh medium at 37 °C.
4.6. Gas Chromatography-Mass Spectrometry (GC-MS) Analysis of P. crispa HF Fraction
4.7. In Silico Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Ambavade, S.D.; Misar, A.V.; Ambavade, P.D. Pharmacological, nutritional, and analytical aspects of β-sitosterol: A review. Orient. Pharm. Exp. Med. 2014, 14, 193–211. [Google Scholar] [CrossRef]
- Lee, W.; Woo, E.-R.; Lee, D.G. Phytol has antibacterial property by inducing oxidative stress response in Pseudomonas aeruginosa. Free Radic. Res. 2016, 50, 1309–1318. [Google Scholar] [CrossRef] [PubMed]
- Pejin, B.; Kojic, V.; Bogdanovic, G. An insight into the cytotoxic activity of phytol at in vitro conditions. Nat. Prod. Res. 2014, 28, 2053–2056. [Google Scholar] [CrossRef]
- Santoyo, S.; Plaza, M.; Jaime, L.; Ibañez, E.; Reglero, G.; Señorans, F.J. Pressurized Liquid Extraction as an Alternative Process To Obtain Antiviral Agents from the Edible Microalga Chlorella vulgaris. J. Agric. Food Chem. 2010, 58, 8522–8527. [Google Scholar] [CrossRef] [PubMed]
- Pejin, B.; Savic, A.; Sokovic, M.; Glamoclija, J.; Ciric, A.; Nikolic, M.; Radotic, K.; Mojovic, M. Further in vitro evaluation of antiradical and antimicrobial activities of phytol. Nat. Prod. Res. 2014, 28, 372–376. [Google Scholar] [CrossRef]
- Mailafiya Maria, M.; Yusuf Amina, J.; Abdullahi Musa, I.; Aleku Godwin, A.; Ibrahim Ilyasu, A.A.; Yahaya, M.; Abubakar, H.; Sanusi, A.; Adamu, H.W.; Alebiosu, C.O. Antimicrobial activity of stigmasterol from the stem bark of Neocarya macrophylla. J. Med. Plants Econ. Dev. 2018, 2, 1–5. [Google Scholar]
- Morgan, L.V.; Petry, F.; Scatolin, M.; de Oliveira, P.V.; Alves, B.O.; Zilli, G.A.L.; Volfe, C.R.B.; Oltramari, A.R.; de Oliveira, D.; Scapinello, J.; et al. Investigation of the anti-inflammatory effects of stigmasterol in mice: Insight into its mechanism of action. Behav. Pharmacol. 2021, 32, 640–651. [Google Scholar] [CrossRef] [PubMed]
- Abid Ali Khan, M.M.; Jain, D.C.; Bhakuni, R.S.; Zaim, M.; Thakur, R.S. Occurrence of some antiviral sterols in Artemisia annua. Plant Sci. 1991, 75, 161–165. [Google Scholar] [CrossRef]
- Kangsamaksin, T.; Chaithongyot, S.; Wootthichairangsan, C.; Hanchaina, R.; Tangshewinsirikul, C.; Svasti, J. Lupeol and stigmasterol suppress tumor angiogenesis and inhibit cholangiocarcinoma growth in mice via downregulation of tumor necrosis factor-α. PLoS ONE 2017, 12, e0189628. [Google Scholar] [CrossRef]
- Gallo, M.B.; Sarachine, M.J. Biological activities of lupeol. Int. J. Biomed. Pharm. Sci. 2009, 3, 46–66. [Google Scholar]
- Hu, Y.; Shi, H.; Zhou, M.; Ren, Q.; Zhu, W.; Zhang, W.; Zhang, Z.; Zhou, C.; Liu, Y.; Ding, X.; et al. Discovery of pyrido [2, 3-b] indole derivatives with Gram-negative activity targeting both DNA gyrase and topoisomerase IV. J. Med. Chem. 2020, 63, 9623–9649. [Google Scholar] [CrossRef]
- Lovering, A.L.; Gretes, M.C.; Safadi, S.S.; Danel, F.; de Castro, L.; Page, M.G.P.; Strynadka, N.C.J. Structural insights into the anti-methicillin-resistant Staphylococcus aureus (MRSA) activity of ceftobiprole. J. Biol. Chem. 2012, 287, 32096–32102. [Google Scholar] [CrossRef] [PubMed]
- Urvashi, J.B.S.K.; Das, P.; Tandon, V. Development of Azaindole-Based Frameworks as Potential Antiviral Agents and Their Future Perspectives. J. Med. Chem. 2022, 65, 6454–6495. [Google Scholar] [CrossRef] [PubMed]
- Clark, M.P.; Ledeboer, M.W.; Davies, I.; Byrn, R.A.; Jones, S.M.; Perola, E.; Tsai, A.; Jacobs, M.; Nti-Addae, K.; Bandarage, U.K.; et al. Discovery of a novel, first-in-class, orally bioavailable azaindole inhibitor (VX-787) of influenza PB2. J. Med. Chem. 2014, 57, 6668–6678. [Google Scholar] [CrossRef] [PubMed]
- Mobarki, N.; Almerabi, B.; Hattan, A. Antibiotic resistance crisis. Int. J. Med. Dev. Ctries 2019, 40, 561–564. [Google Scholar] [CrossRef]
- Elshiekh, Y.H.; Mona, A.M. Gas chromatography–mass spectrometry analysis of Pulicaria crispa (whole plant) petroleum ether extracts. Am. J. Res. Commun. 2015, 3, 58–67. [Google Scholar]
- Mohamed, E.A.A.; Muddathir, A.M.; Osman, M.A. Antimicrobial activity, phytochemical screening of crude extracts, and essential oils constituents of two Pulicaria spp. growing in Sudan. Sci. Rep. 2020, 10, 17148. [Google Scholar] [CrossRef]
- Konaté, K.; Mavoungou, J.F.; Lepengué, A.N.; Aworet-Samseny, R.R.R.; Hilou, A.; Souza, A.; Dicko, M.H.; M’Batchi, B. Antibacterial activity against β- lactamase producing Methicillin and Ampicillin-resistants Staphylococcus aureus: Fractional Inhibitory Concentration Index (FICI) determination. Ann. Clin. Microbiol. Antimicrob. 2012, 11, 18. [Google Scholar] [CrossRef]
- Shalaby, M.-A.W.; Dokla, E.M.E.; Serya, R.A.T.; Abouzid, K.A.M. Penicillin binding protein 2a: An overview and a medicinal chemistry perspective. Eur. J. Med. Chem. 2020, 199, 112312. [Google Scholar] [CrossRef]
- Khan, T.; Sankhe, K.; Suvarna, V.; Sherje, A.; Patel, K.; Dravyakar, B. DNA gyrase inhibitors: Progress and synthesis of potent compounds as antibacterial agents. Biomed. Pharmacother. 2018, 103, 923–938. [Google Scholar] [CrossRef]
- Morsy, B.M.; Hamed, M.A.; Abd-Alla, H.I.; Aziz, W.M.; Kamel, S.N. Downregulation of fibrosis and inflammatory signalling pathways in rats liver via Pulicaria crispa aerial parts ethanol extract. Biomarkers 2021, 26, 665–673. [Google Scholar] [CrossRef]
- Kamaraj, C.; Ragavendran, C.; Kumar, R.C.S.; Ali, A.; Khan, S.U.; Mashwani, Z.-R.; Luna-Arias, J.P.; Pedroza, J.P.R. Antiparasitic potential of asteraceae plants: A comprehensive review on therapeutic and mechanistic aspects for biocompatible drug discovery. Phytomed. Plus 2022, 2, 100377. [Google Scholar] [CrossRef]
- Bowler, P.; Murphy, C.; Wolcott, R. Biofilm exacerbates antibiotic resistance: Is this a current oversight in antimicrobial stewardship? Antimicrob. Resist. Infect. Control 2020, 9, 162. [Google Scholar] [CrossRef]
- Adeyemo, R.O.; Famuyide, I.M.; Dzoyem, J.P.; Lyndy Joy, M. Anti-Biofilm, Antibacterial, and Anti-Quorum Sensing Activities of Selected South African Plants Traditionally Used to Treat Diarrhoea. Evid. Based Complement. Altern. Med. 2022, 2022, 1307801. [Google Scholar] [CrossRef] [PubMed]
- Marklund Jesper, K.; Ye, Q.; Dong, J.; Tao Yizhi, J.; Krug Robert, M. Sequence in the Influenza A Virus Nucleoprotein Required for Viral Polymerase Binding and RNA Synthesis. J. Virol. 2012, 86, 7292–7297. [Google Scholar] [CrossRef]
- Karimi, A.; Moradi, M.-T.; Rabiei, M.; Alidadi, S. In vitro anti-adenoviral activities of ethanol extract, fractions, and main phenolic compounds of pomegranate (Punica granatum L.) peel. Antivir. Chem. Chemother. 2020, 28, 2040206620916571. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: 20th Informational Supplement; CLSI Document M100-S20; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2010. [Google Scholar]
- Lanoix, D.; Ouellette, R.; Vaillancourt, C. Expression of melatoninergic receptors in human placental choriocarcinoma cell lines. Hum. Reprod. 2006, 21, 1981–1989. [Google Scholar] [CrossRef]
- Wang, H.; Miao, F.; Ning, D.; Shan, C. Ellagic acid Alleviates hepatic ischemia–reperfusion injury in C57 mice via the Caspase-1-GSDMD pathway. BMC Vet. Res. 2022, 18, 229. [Google Scholar] [CrossRef]
- Sanbongi, Y.; Ida, T.; Ishikawa, M.; Osaki, Y.; Kataoka, H.; Suzuki, T.; Kondo, K.; Ohsawa, F.; Yonezawa, M. Complete sequences of six penicillin-binding protein genes from 40 Streptococcus pneumoniae clinical isolates collected in Japan. Antimicrob. Agents Chemother. 2004, 48, 2244–2250. [Google Scholar] [CrossRef]
- Wen, S.; Chen, X.; Xu, F.; Sun, H. Validation of reference genes for real-time quantitative PCR (qPCR) analysis of Avibacterium paragallinarum. PLoS ONE 2016, 11, e0167736. [Google Scholar] [CrossRef]
- Tawfick, M.M.; El Menofy, N.G.; Omran, M.E.; Alsharony, O.A.; Abo-Shady, M.A. Phenotypic and molecular characterization of plasmidmediated virulence and antimicrobial resistance traits among multidrug resistant Enterococcus spp. Egypt, J. Pure Appl. Microbiol. 2020, 14, 1649–1661. [Google Scholar] [CrossRef]
- Nassima, B.; Nassima, B.; Riadh, K. Antimicrobial and antibiofilm activities of phenolic compounds extracted from Populus nigra and Populus alba buds (Algeria). Braz. J. Pharm. Sci. 2019, 55. [Google Scholar] [CrossRef]
- Schmidtke, M.; Schnittler, U.; Jahn, B.; Dahse, H.M.; Stelzner, A. A rapid assay for evaluation of antiviral activity against coxsackie virus B3, influenza virus A, and herpes simplex virus type 1. J. Virol. Methods 2001, 95, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Chen, Y.; Wang, D.; Ma, L.; Guo, M.; Zhou, C.; Dou, J. The inhibitory effect of sodium baicalin on oseltamivir-resistant influenza A virus via reduction of neuraminidase activity. Arch. Pharmacal Res. 2018, 41, 664–676. [Google Scholar] [CrossRef] [PubMed]



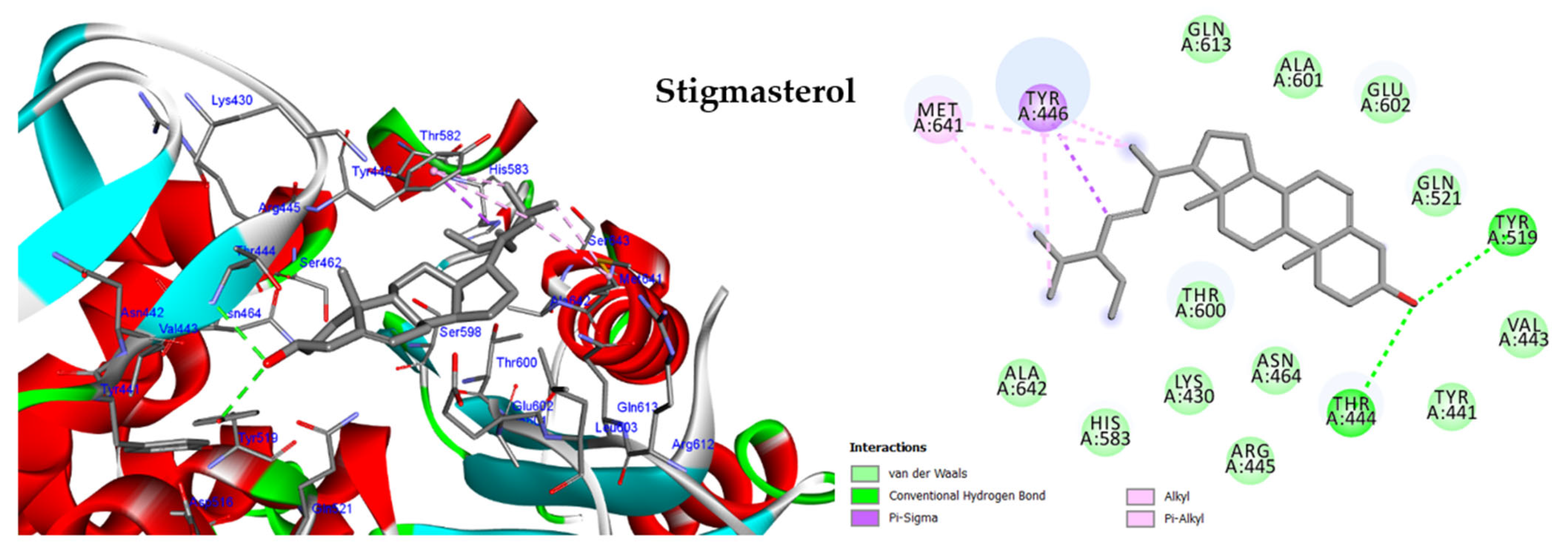
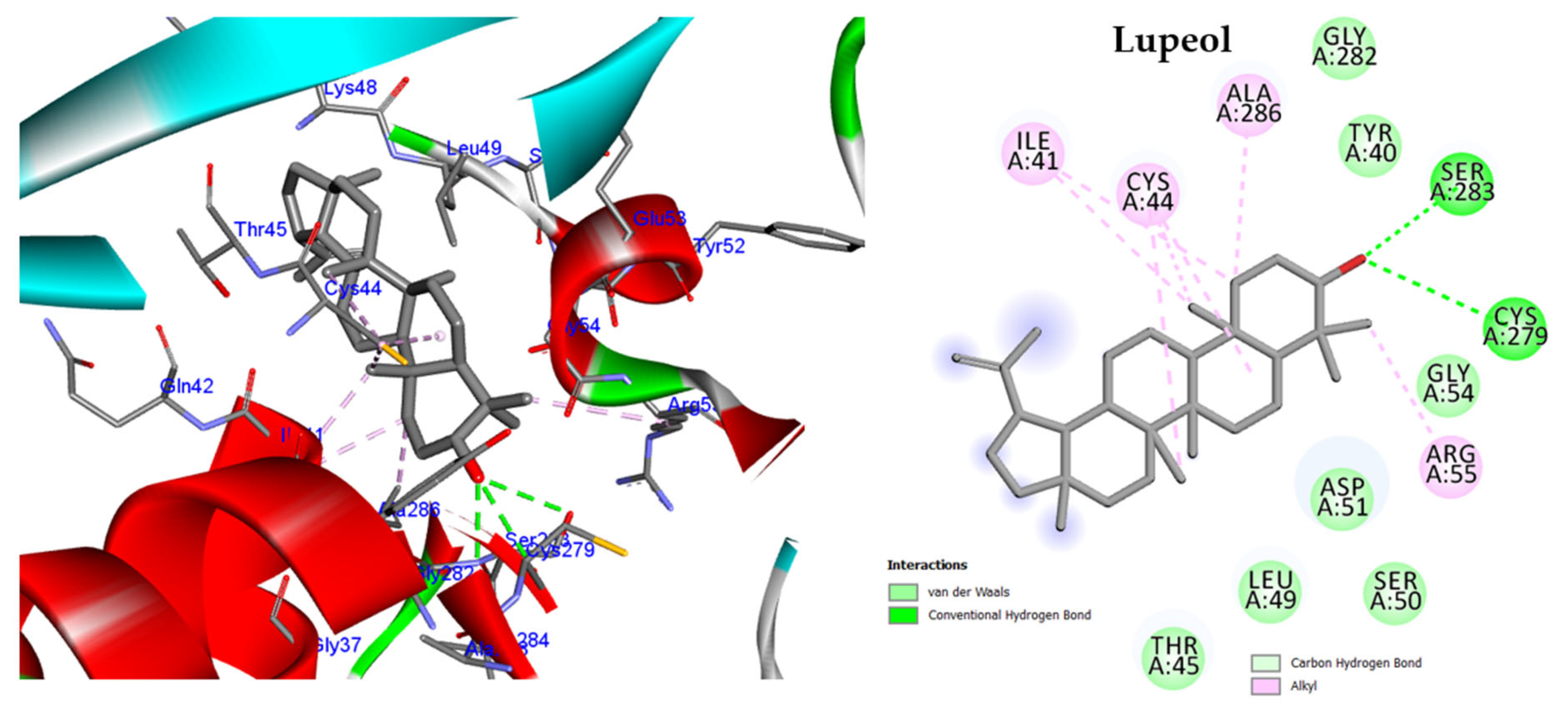
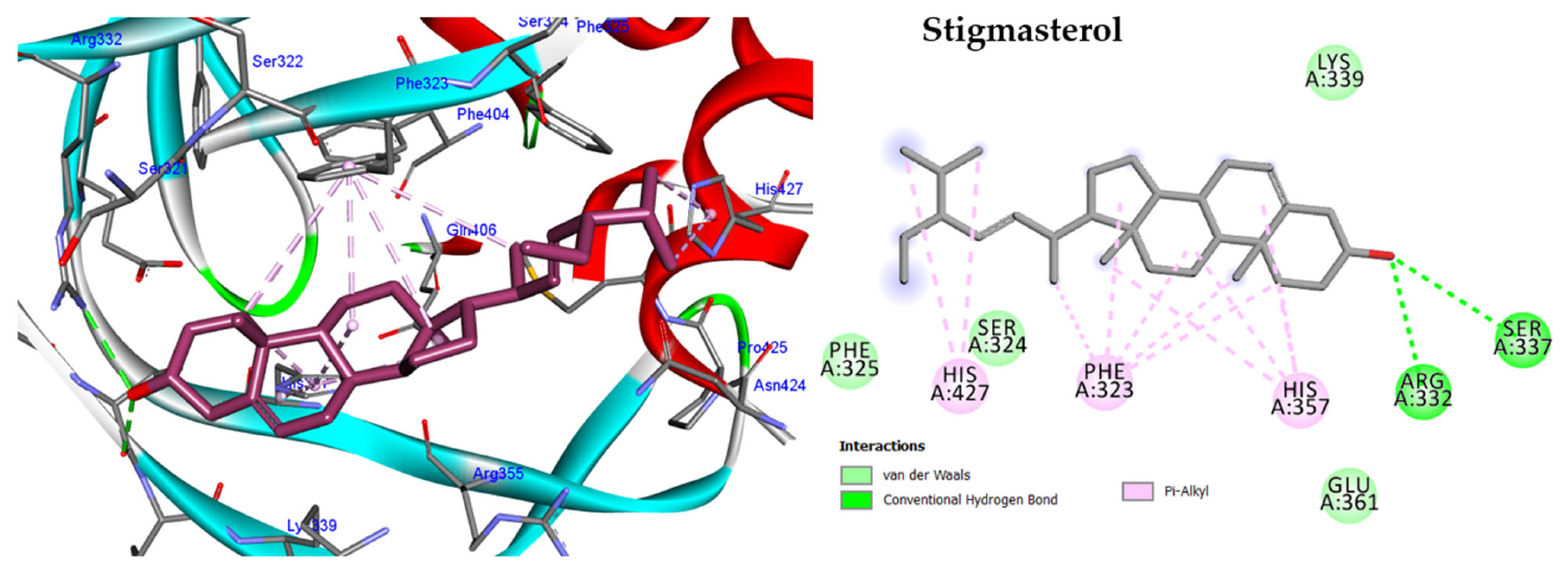
| Bacterial Strain | MIC (µg/mL) | |||||
|---|---|---|---|---|---|---|
| CME | HF | DCF | EAF | WF | Amoxicillin | |
| Staphylococcus aureus (Clinical isolate) | 125 | 62.5 | 62.5 | 125 | 125 | >500 |
| Klebsiella pneumonia (ATCC 700603) | 125 | 125 | 125 | 125 | 125 | >500 |
| Pseudomonas aeruginosa (ATCC 29853) | 125 | 62.5 | 125 | 125 | 125 | 125 |
| Escherichia coli (Clinical isolate) | 125 | 125 | 125 | 125 | 125 | 62.5 |
| Bacterial Strain | HF | Amoxicillin | ||
|---|---|---|---|---|
| MIC (µg/mL) | MBC (µg/mL) | MBC/MIC | MIC (µg/mL) | |
| Gram-negative bacterial isolates | ||||
| Escherichia coli (ATCC 25922) | 125 | 250 | 2 | 62.5 |
| Acinetobacter baumannii (ATCC 19606) | 62.5 | 125 | 2 | >500 |
| Pseudomonas aeruginosa (Clinical isolate) | 15.6 | 31.25 | 2 | 62.5 |
| Pseudomonas aeruginosa (Biofilm producer) | 125 | 250 | 2 | 62.5 |
| Proteus mirabilias (Biofilm producer). | 125 | 250 | 2 | >500 |
| Gram-positive bacterial isolates | ||||
| Staphylococcus aureus (ATCC 25923) | 62.5 | 125 | 2 | ≤7.8125 |
| MRSA (ATCC 43300) | 125 | 250 | 2 | >500 |
| Sample | CC50 (µg/mL) | IC50 (µg/mL) | SI |
|---|---|---|---|
| CME | 7.4 | 1.4 | 5.3 |
| HF | 409 | 0.9 | 454.4 |
| DCF | 45.7 | 2 | 22.85 |
| EAF | 33 | 1.7 | 19.4 |
| WF | 121 | 3 | 40.3 |
| Peak No. | Rt (min.) | Molecular Formula | Relative Area (%) | Compound |
|---|---|---|---|---|
| 1 | 18.328 | C15H26O | 0.1 | Farnesol |
| 2 | 24.027 | C16H34O | 0.19 | 1-Hexadecanol |
| 3 | 25.530 | C20H42O | 0.86 | Dihydrophytol |
| 4 | 25.848 | C18H38O | 0.29 | 1-Octadecanol |
| 5 | 26.076 | C20H40O | 15.65 | Phytol |
| 6 | 28.846 | C13H18O6 | 0.48 | 1-Monoferuloylglycerol |
| 7 | 29.036 | C22H46O | 0.35 | Docosanol |
| 8 | 29.430 | C19H38O4 | 0.11 | 1-Monopalmitin |
| 9 | 30.113 | C24H50 | 2.25 | Tetracosane |
| 10 | 30.500 | C24H50O | 3.81 | 1-Tetracosanol |
| 11 | 30.811 | C21H42O4 | 0.71 | Glycerol monostearate |
| 12 | 31.555 | C30H50 | 12.05 | Squalene |
| 13 | 31.836 | C30H62 | 0.84 | Triacontane |
| 14 | 32.162 | C32H66 | 0.83 | Dotriacontane |
| 15 | 32.838 | C35H72 | 5.97 | Pentatriacontane |
| 16 | 33.111 | C29H50O2 | 0.69 | α-Tocopherol |
| 17 | 33.786 | C28H48O | 2.7 | Campesterol |
| 18 | 34.021 | C29H48O | 13.13 | Stigmasterol |
| 19 | 34.272 | C30H50O | 1.00 | β-Amyrin |
| 20 | 34.386 | C29H50O | 17.89 | β-Sitosterol |
| 21 | 34.454 | C29H48O | 1.41 | Isofucosterol |
| 22 | 34.568 | C30H50O | 4.38 | α-Amyrin |
| 23 | 34.773 | C30H50O | 0.29 | Cycloartenol |
| 24 | 35.023 | C37H76O | 0.25 | 1-Heptatriacotanol |
| 25 | 35.160 | C30H50O | 12.89 | Lupeol |
| 26 | 35.266 | C21H38O4 | 0.65 | 1-Monolinolein |
| 27 | 35.334 | C26H44O5 | 0.23 | Ethyl iso-allocholate |
| Compound | Biological Activity | Reference |
|---|---|---|
β-Sitosterol | Antibacterial, antifungal, anti-inflammatory, anticancer, antihyperlipidemic, anti-atherosclerosis, and antidiabetic activities. | [1] |
Phytol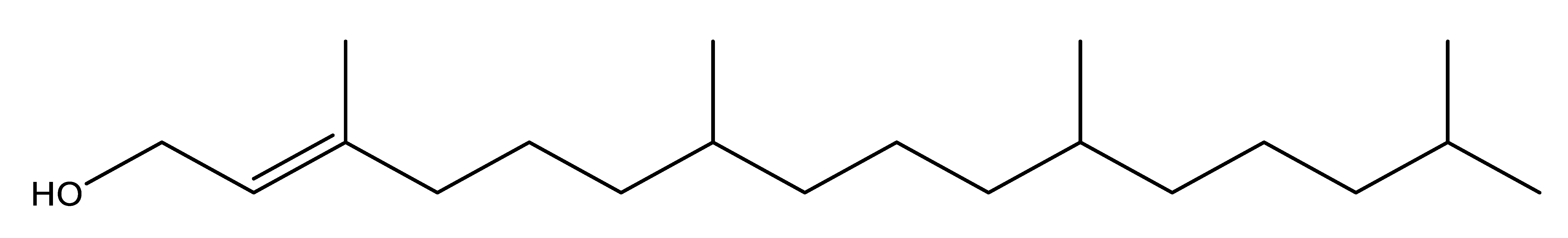 | Antibacterial activity via inducing bacterial oxidative cell death. It elevates the level of bacterial intracellular reactive oxygen species (ROS) and transient NADH depletion. | [2] |
| Cytotoxic activity on different cancer cell lines in a concentration dependent manner, particularly breast cancer. | [3] | |
| Antiviral activity against HSV (Herpes Simplex virus). | [4] | |
| Antioxidant and antifungal activity. | [5] | |
Stigmasterol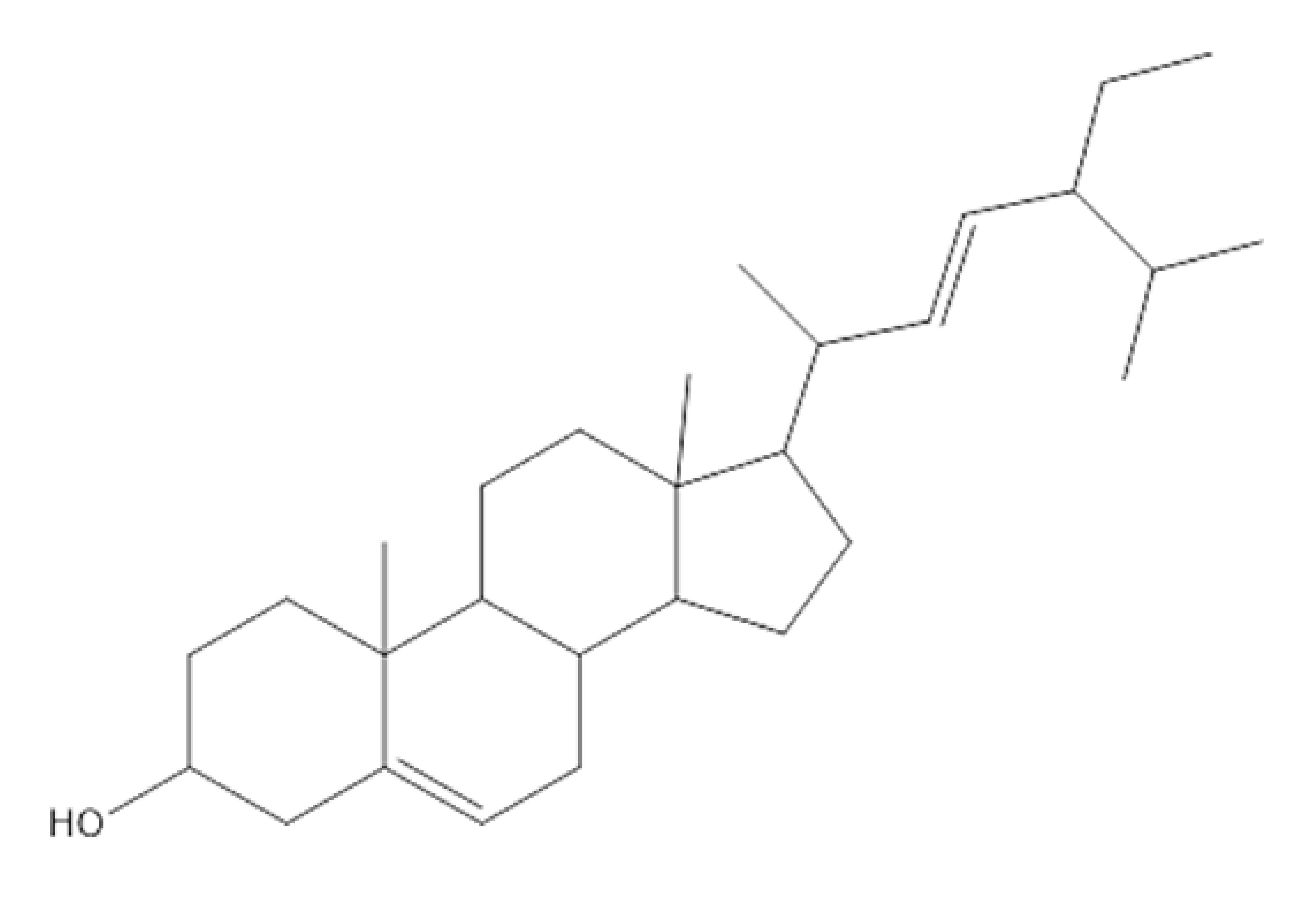 | Antibacterial and antifungal activity. | [6] |
| Anti-inflammatory activity. | [7] | |
| Antiviral activity. | [8] | |
| Antitumor activity. | [9] | |
Lupeol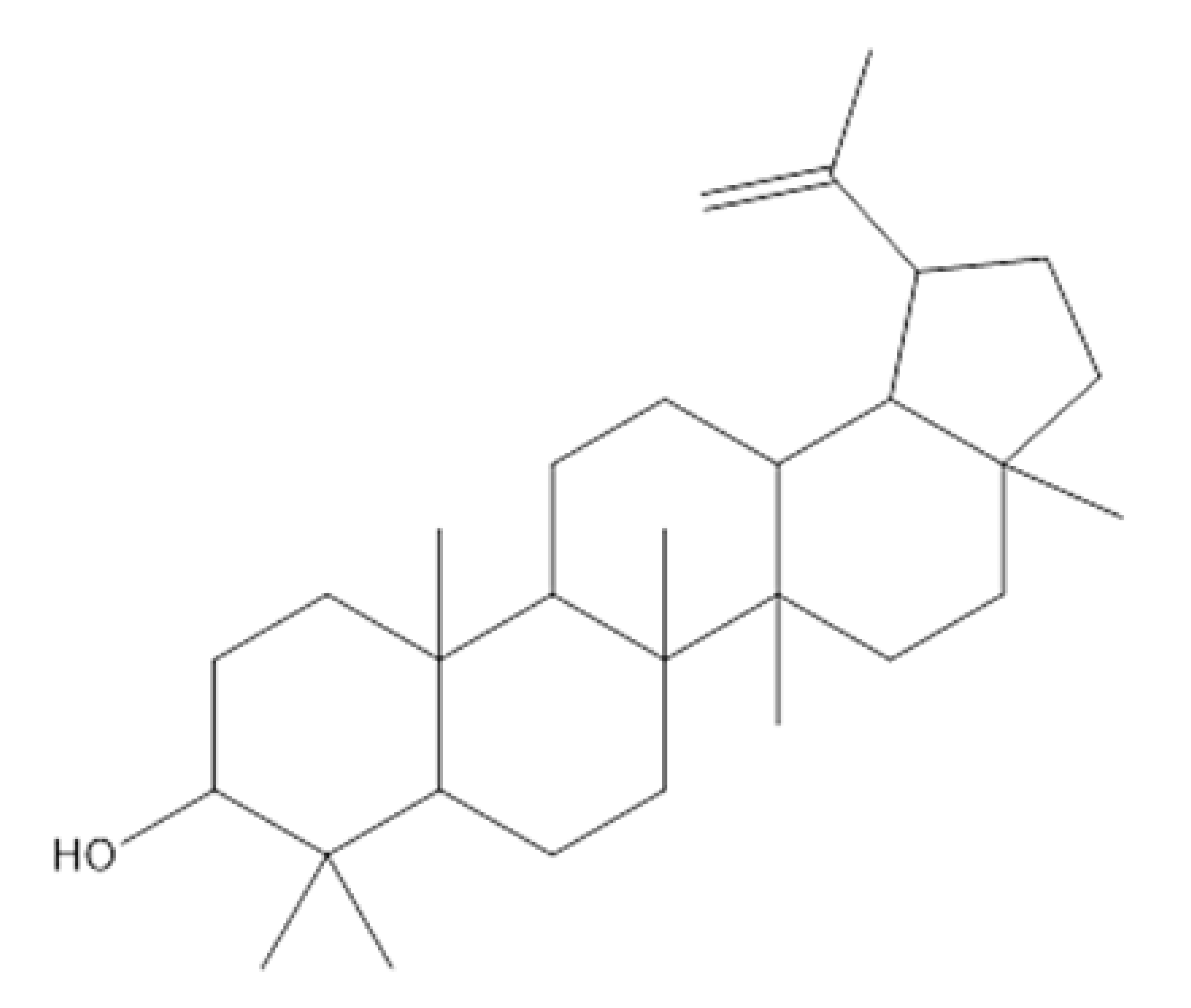 | Antimicrobial, antiprotozoal, anti-inflammatory, antitumor, cardioprotective, and hepatoprotective activities. | [10] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abo-Elghiet, F.; Rushdi, A.; Ibrahim, M.H.; Mahmoud, S.H.; Rabeh, M.A.; Alshehri, S.A.; El Menofy, N.G. Chemical Profile, Antibacterial, Antibiofilm, and Antiviral Activities of Pulicaria crispa Most Potent Fraction: An In Vitro and In Silico Study. Molecules 2023, 28, 4184. https://doi.org/10.3390/molecules28104184
Abo-Elghiet F, Rushdi A, Ibrahim MH, Mahmoud SH, Rabeh MA, Alshehri SA, El Menofy NG. Chemical Profile, Antibacterial, Antibiofilm, and Antiviral Activities of Pulicaria crispa Most Potent Fraction: An In Vitro and In Silico Study. Molecules. 2023; 28(10):4184. https://doi.org/10.3390/molecules28104184
Chicago/Turabian StyleAbo-Elghiet, Fatma, Areej Rushdi, Mona H. Ibrahim, Sara H. Mahmoud, Mohamed A. Rabeh, Saad Ali Alshehri, and Nagwan Galal El Menofy. 2023. "Chemical Profile, Antibacterial, Antibiofilm, and Antiviral Activities of Pulicaria crispa Most Potent Fraction: An In Vitro and In Silico Study" Molecules 28, no. 10: 4184. https://doi.org/10.3390/molecules28104184
APA StyleAbo-Elghiet, F., Rushdi, A., Ibrahim, M. H., Mahmoud, S. H., Rabeh, M. A., Alshehri, S. A., & El Menofy, N. G. (2023). Chemical Profile, Antibacterial, Antibiofilm, and Antiviral Activities of Pulicaria crispa Most Potent Fraction: An In Vitro and In Silico Study. Molecules, 28(10), 4184. https://doi.org/10.3390/molecules28104184







