Efflux Pump Inhibitors in Controlling Antibiotic Resistance: Outlook under a Heavy Metal Contamination Context
Abstract
1. Introduction
2. Co-Selection of Antibiotic and Metal Resistance Mechanism
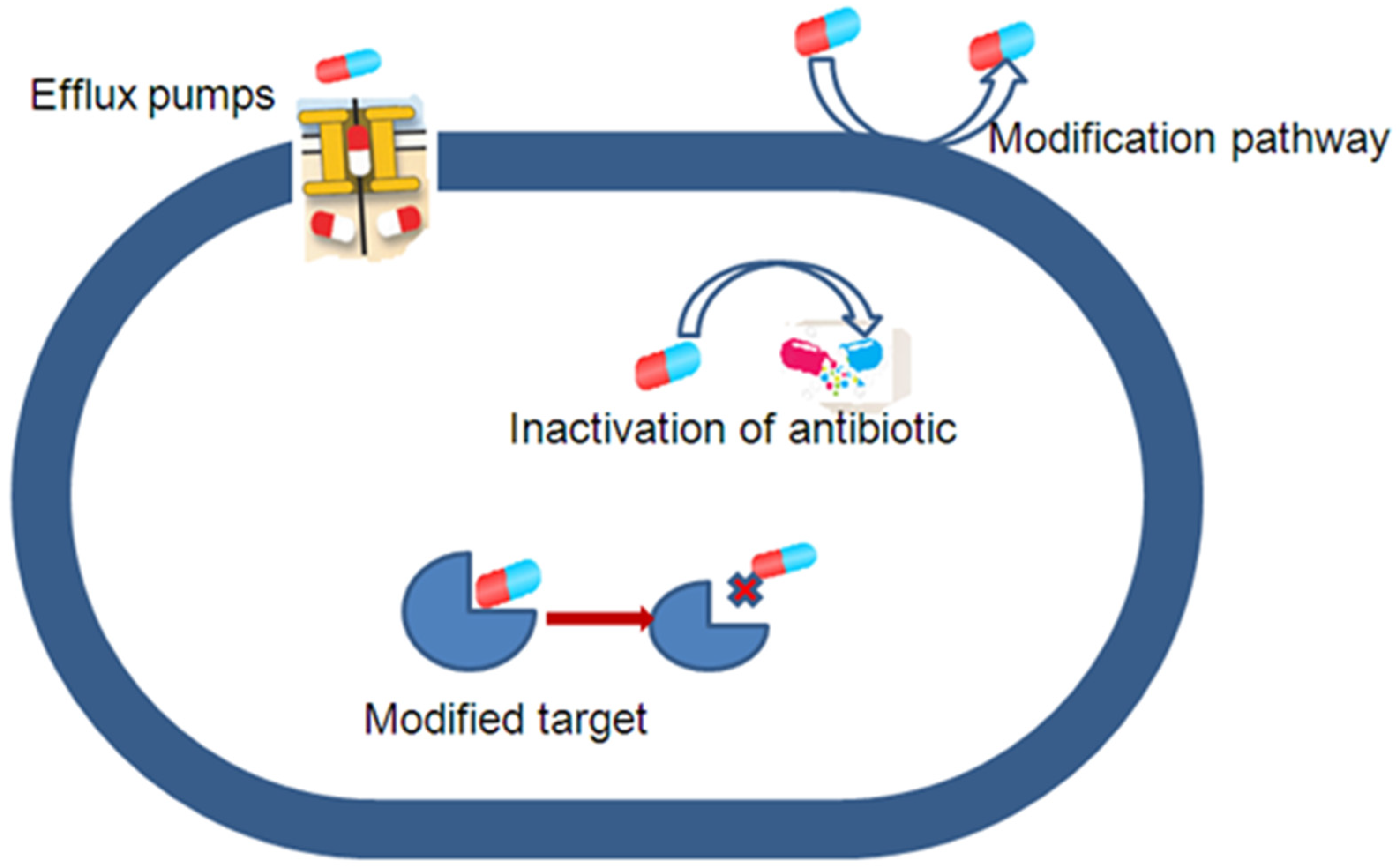
3. Antibiotic Resistance Caused by Efflux Pump Mechanisms
4. Secondary Metabolites Production of Plants as Potential EPIs
5. Challenges and Future
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, J.; Li, J.; Zhang, H.; Shi, W.; Liu, Y. Bacterial Heavy-Metal and Antibiotic Resistance Genes in a Copper Tailing Dam Area in Northern China. Front. Microbiol. 2019, 10, 1916. [Google Scholar] [CrossRef] [PubMed]
- Ndeddy Aka, R.J.; Babalola, O.O. Identification and characterization of Cr-, Cd-, and Ni-tolerant bacteria isolated from mine tailings. Bioremediation J. 2017, 21, 1–19. [Google Scholar] [CrossRef]
- van Hoek, A.; Mevius, D.; Guerra, B.; Mullany, P.; Roberts, A.; Aarts, H. Acquired Antibiotic Resistance Genes: An Overview. Front. Microbiol. 2011, 2, 203. [Google Scholar] [CrossRef]
- Baindara, P. Mechanism of Bacterial Co-resistance. In Bacterial Adaptation to Co-Resistance; Mandal, S.M., Paul, D., Eds.; Springer: Singapore, 2019; pp. 191–210. ISBN 978-981-13-8503-2. [Google Scholar]
- Reygaert, W.C. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 2018, 4, 482–501. [Google Scholar] [CrossRef] [PubMed]
- Blázquez, J.; Couce, A.; Rodríguez-Beltrán, J.; Rodríguez-Rojas, A. Antimicrobials as promoters of genetic variation. Curr. Opin. Microbiol. 2012, 15, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Imran, M.; Das, K.R.; Naik, M.M. Co-selection of multi-antibiotic resistance in bacterial pathogens in metal and microplastic contaminated environments: An emerging health threat. Chemosphere 2019, 215, 846–857. [Google Scholar] [CrossRef] [PubMed]
- Peterson, E.; Kaur, P. Antibiotic Resistance Mechanisms in Bacteria: Relationships Between Resistance Determinants of Antibiotic Producers, Environmental Bacteria, and Clinical Pathogens. Front. Microbiol. 2018, 9, 2928. [Google Scholar] [CrossRef]
- von Wintersdorff, C.J.H.; Penders, J.; van Niekerk, J.M.; Mills, N.D.; Majumder, S.; van Alphen, L.B.; Savelkoul, P.H.M.; Wolffs, P.F.G. Dissemination of Antimicrobial Resistance in Microbial Ecosystems through Horizontal Gene Transfer. Front. Microbiol. 2016, 7, 173. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Wright, M.S.; Stepanauskas, R.; McArthur, J.V. Co-selection of antibiotic and metal resistance. Trends Microbiol. 2006, 14, 176–182. [Google Scholar] [CrossRef]
- Alonso, A.; Sanchez, P.; Martínez, J.L. Stenotrophomonas maltophilia D457R contains a cluster of genes from gram-positive bacteria involved in antibiotic and heavy metal resistance. Antimicrob. Agents Chemother. 2000, 44, 1778–1782. [Google Scholar] [CrossRef]
- Allen, H.K.; Donato, J.; Wang, H.H.; Cloud-Hansen, K.A.; Davies, J.; Handelsman, J. Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010, 8, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Deredjian, A.; Alliot, N.; Blanchard, L.; Brothier, E.; Anane, M.; Cambier, P.; Jolivet, C.; Khelil, M.N.; Nazaret, S.; Saby, N.; et al. Occurrence of Stenotrophomonas maltophilia in agricultural soils and antibiotic resistance properties. Res. Microbiol. 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Seiler, C.; Berendonk, T.U. Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front. Microbiol. 2012, 3, 399. [Google Scholar] [CrossRef]
- Li, B.; Yang, Y.; Ma, L.; Ju, F.; Guo, F.; Tiedje, J.M.; Zhang, T. Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes. ISME J. 2015, 9, 2490–2502. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.C., IV; Oladeinde, A.; Kieran, T.J.; Finger, J.W., Jr.; Bayona-Vásquez, N.J.; Cartee, J.C.; Beasley, J.C.; Seaman, J.C.; McArthur, J.V.; Rhodes, O.E., Jr.; et al. Co-occurrence of antibiotic, biocide, and heavy metal resistance genes in bacteria from metal and radionuclide contaminated soils at the Savannah River Site. Microb. Biotechnol. 2020, 13, 1179–1200. [Google Scholar] [CrossRef] [PubMed]
- Li, L.-G.; Xia, Y.; Zhang, T. Co-occurrence of antibiotic and metal resistance genes revealed in complete genome collection. ISME J. 2017, 11, 651–662. [Google Scholar] [CrossRef]
- Yang, Q.E.; Agouri, S.R.; Tyrrell, J.M.; Walsh, T.R. Heavy Metal Resistance Genes Are Associated with blaNDM-1- and blaCTX-M-15-Carrying Enterobacteriaceae. Antimicrob. Agents Chemother. 2018, 62, e02642-17. [Google Scholar] [CrossRef]
- Yong, D.; Toleman, M.A.; Giske, C.G.; Cho, H.S.; Sundman, K.; Lee, K.; Walsh, T.R. Characterization of a New Metallo-β-Lactamase Gene, blaNDM-1, and a Novel Erythromycin Esterase Gene Carried on a Unique Genetic Structure in Klebsiella pneumoniae Sequence Type 14 from India. Antimicrob. Agents Chemother. 2009, 53, 5046–5054. [Google Scholar] [CrossRef]
- Zhang, L.; Lü, X.; Zong, Z. The emergence of blaCTX-M-15-carrying Escherichia coli of ST131 and new sequence types in Western China. Ann. Clin. Microbiol. Antimicrob. 2013, 12, 35. [Google Scholar] [CrossRef]
- Pham, H.-N. Metabolic Adaptation of Plants to Metal Stress: Consequences on Rhizospheric Bacterial Communities and the Selection of Antibiotic Resistant Populations. Ph.D. Thesis, University Paul Sabatier, Toulouse, French, 2017. [Google Scholar]
- Pham, H.N.; Michalet, S.; Bodillis, J.; Nguyen, T.D.; Nguyen, T.K.O.; Le, T.P.Q.; Haddad, M.; Nazaret, S.; Dijoux-Franca, M.-G. Impact of metal stress on the production of secondary metabolites in Pteris vittata L. and associated rhizosphere bacterial communities. Environ. Sci. Pollut. Res. Int. 2017, 24, 16735–16750. [Google Scholar] [CrossRef]
- Pham, H.N.; Pham, P.A.; Nguyen, T.T.H.; Meiffren, G.; Brothier, E.; Lamy, I.; Michalet, S.; Dijoux-Franca, M.-G.; Nazaret, S. Influence of metal contamination in soil on metabolic profiles of Miscanthus × giganteus belowground parts and associated bacterial communities. Appl. Soil Ecol. 2018, 125, 240–249. [Google Scholar] [CrossRef]
- Li, S.; Skov, R.L.; Han, X.; Larsen, A.R.; Larsen, J.; Sørum, M.; Wulf, M.; Voss, A.; Hiramatsu, K.; Ito, T. Novel Types of Staphylococcal Cassette Chromosome mec Elements Identified in Clonal Complex 398 Methicillin-Resistant Staphylococcus aureus Strains. Antimicrob. Agents Chemother. 2011, 55, 3046–3050. [Google Scholar] [CrossRef]
- Cantón, R.; Ruiz-Garbajosa, P. Co-resistance: An opportunity for the bacteria and resistance genes. Curr. Opin. Pharmacol. 2011, 11, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Chapman, J.S. Disinfectant resistance mechanisms, cross-resistance, and co-resistance. Int. Biodeterior. Biodegrad. 2003, 51, 271–276. [Google Scholar] [CrossRef]
- Pal, C.; Asiani, K.; Arya, S.; Rensing, C.; Stekel, D.J.; Larsson, D.G.J.; Hobman, J.L. Metal Resistance and Its Association with Antibiotic Resistance. Adv. Microb. Physiol. 2017, 70, 261–313. [Google Scholar] [CrossRef] [PubMed]
- Mata, M.T.; Baquero, F.; Pérez-Díaz, J.C. A multidrug efflux transporter in Listeria monocytogenes. FEMS Microbiol. Lett. 2000, 187, 185–188. [Google Scholar] [CrossRef] [PubMed]
- Perron, K.; Caille, O.; Rossier, C.; van Delden, C.; Dumas, J.-L.; Köhler, T. CzcR-CzcS, a Two-component System Involved in Heavy Metal and Carbapenem Resistance in Pseudomonas aeruginosa. J. Biol. Chem. 2004, 279, 8761–8768. [Google Scholar] [CrossRef]
- Alvarez-Ortega, C.; Olivares, J.; Martínez, J.L. RND multidrug efflux pumps: What are they good for? Front. Microbiol. 2013, 4, 7. [Google Scholar] [CrossRef]
- Forsberg, K.J.; Reyes, A.; Wang, B.; Selleck, E.M.; Sommer, M.O.A.; Dantas, G. The shared antibiotic resistome of soil bacteria and human pathogens. Science 2012, 337, 1107–1111. [Google Scholar] [CrossRef]
- Armalytė, J.; Skerniškytė, J.; Bakienė, E.; Krasauskas, R.; Šiugždinienė, R.; Kareivienė, V.; Kerzienė, S.; Klimienė, I.; Sužiedėlienė, E.; Ružauskas, M. Microbial Diversity and Antimicrobial Resistance Profile in Microbiota from Soils of Conventional and Organic Farming Systems. Front. Microbiol. 2019, 10, 892. [Google Scholar] [CrossRef]
- Berg, J.; Thorsen, M.K.; Holm, P.E.; Jensen, J.; Nybroe, O.; Brandt, K.K. Cu Exposure under Field Conditions Coselects for Antibiotic Resistance as Determined by a Novel Cultivation-Independent Bacterial Community Tolerance Assay. Environ. Sci. Technol. 2010, 44, 8724–8728. [Google Scholar] [CrossRef] [PubMed]
- Efflux Pump Inhibitors for Bacterial Pathogens: From Bench to Bedside. Semantic Scholar. Available online: https://www.semanticscholar.org/paper/Efflux-pump-inhibitors-for-bacterial-pathogens%3A-to-Sharma-Gupta/62202ea7844675b3d7aaa20099bdd5afead7b33c (accessed on 7 March 2023).
- Alibert, S.; N’gompaza Diarra, J.; Hernandez, J.; Stutzmann, A.; Fouad, M.; Boyer, G.; Pagès, J.-M. Multidrug efflux pumps and their role in antibiotic and antiseptic resistance: A pharmacodynamic perspective. Expert Opin. Drug Metab. Toxicol. 2017, 13, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Theuretzbacher, U. Accelerating resistance, inadequate antibacterial drug pipelines and international responses. Int. J. Antimicrob. Agents 2012, 39, 295–299. [Google Scholar] [CrossRef] [PubMed]
- Blair, J.M.; Richmond, G.E.; Piddock, L.J. Multidrug efflux pumps in Gram-negative bacteria and their role in antibiotic resistance. Future Microbiol. 2014, 9, 1165–1177. [Google Scholar] [CrossRef]
- Lomovskaya, O.; Bostian, K.A. Practical applications and feasibility of efflux pump inhibitors in the clinic—A vision for applied use. Biochem. Pharmacol. 2006, 71, 910–918. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Deng, Z.; Yan, A. Bacterial multidrug efflux pumps: Mechanisms, physiology and pharmacological exploitations. Biochem. Biophys. Res. Commun. 2014, 453, 254–267. [Google Scholar] [CrossRef]
- Maseda, H.; Hashida, Y.; Shirai, A.; Omasa, T.; Nakae, T. Mutation in the sdeS gene promotes expression of the sdeAB efflux pump genes and multidrug resistance in Serratia marcescens. Antimicrob. Agents Chemother. 2011, 55, 2922–2926. [Google Scholar] [CrossRef] [PubMed]
- Alcalde-Rico, M.; Hernando-Amado, S.; Blanco, P.; Martínez, J.L. Multidrug Efflux Pumps at the Crossroad between Antibiotic Resistance and Bacterial Virulence. Front. Microbiol. 2016, 7, 1483. [Google Scholar] [CrossRef]
- Youenou, B.; Favre-Bonté, S.; Bodilis, J.; Brothier, E.; Dubost, A.; Muller, D.; Nazaret, S. Comparative Genomics of Environmental and Clinical Stenotrophomonas maltophilia Strains with Different Antibiotic Resistance Profiles. Genome Biol. Evol. 2015, 7, 2484–2505. [Google Scholar] [CrossRef]
- Blanco, P.; Hernando-Amado, S.; Reales-Calderon, J.A.; Corona, F.; Lira, F.; Alcalde-Rico, M.; Bernardini, A.; Sanchez, M.B.; Martinez, J.L. Bacterial Multidrug Efflux Pumps: Much More Than Antibiotic Resistance Determinants. Microorganisms 2016, 4, 14. [Google Scholar] [CrossRef]
- Seukep, A.J.; Kuete, V.; Nahar, L.; Sarker, S.D.; Guo, M. Plant-derived secondary metabolites as the main source of efflux pump inhibitors and methods for identification. J. Pharm. Anal. 2020, 10, 277–290. [Google Scholar] [CrossRef]
- Lamut, A.; Peterlin Mašič, L.; Kikelj, D.; Tomašič, T. Efflux pump inhibitors of clinically relevant multidrug resistant bacteria. Med. Res. Rev. 2019, 39, 2460–2504. [Google Scholar] [CrossRef] [PubMed]
- Ashwath, P.; Sannejal, A.D. The Action of Efflux Pump Genes in Conferring Drug Resistance to Klebsiella Species and Their Inhibition. J. Health Allied Sci. NU 2022, 12, 24–31. [Google Scholar] [CrossRef]
- Deakin, G. How Efflux Pumps Affect the Sensitivity of Escherichia Coli Biofilms to Antibiotics. Master’s Thesis, Western Sydney University, Penrith, NSW, Australia, 2017. [Google Scholar]
- Henderson, P.J.F.; Maher, C.; Elbourne, L.D.H.; Eijkelkamp, B.A.; Paulsen, I.T.; Hassan, K.A. Physiological Functions of Bacterial “Multidrug” Efflux Pumps. Chem. Rev. 2021, 121, 5417–5478. [Google Scholar] [CrossRef] [PubMed]
- Impey, R.E.; Hawkins, D.A.; Sutton, J.M.; Soares da Costa, T.P. Overcoming Intrinsic and Acquired Resistance Mechanisms Associated with the Cell Wall of Gram-Negative Bacteria. Antibiotics 2020, 9, 623. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, P.; Alonso, A.; Martinez, J.L. Cloning and Characterization of SmeT, a Repressor of the Stenotrophomonas maltophilia Multidrug Efflux Pump SmeDEF. Antimicrob. Agents Chemother. 2002, 46, 3386–3393. [Google Scholar] [CrossRef]
- Michalet, S.; Rouifed, S.; Pellassa-Simon, T.; Fusade-Boyer, M.; Meiffren, G.; Nazaret, S.; Piola, F. Tolerance of Japanese knotweed s.l. to soil artificial polymetallic pollution: Early metabolic responses and performance during vegetative multiplication. Environ. Sci. Pollut. Res. 2017, 24, 20897. [Google Scholar] [CrossRef]
- Tseng, S.; Liang, C.; Chia, T.; Ton, S. Changes in the Composition of the Soil Bacterial Community in Heavy Metal-Contaminated Farmland. Int. J. Environ. Res. Public Health 2021, 18, 8661. [Google Scholar] [CrossRef]
- Teoh, E.S. Secondary Metabolites of Plants. In Medicinal Orchids of Asia; Teoh, E.S., Ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 59–73. ISBN 978-3-319-24274-3. [Google Scholar]
- Rao, M.; Padyana, S.; Dipin, K.M.; Kumar, S.; Nayak, B.B.; Varela, M.F. Antimicrobial Compounds of Plant Origin as Efflux Pump Inhibitors: New Avenues for Controlling Multidrug Resistant Pathogens. J. Antimicrob. Agents 2018, 4, 1000159. [Google Scholar] [CrossRef]
- Sruthi, D.; Jayabaskaran, C. Plant Secondary Metabolites—The Key Drivers of Plant’s Defence Mechanisms: A General Introduction. In Biotechnological Approaches to Enhance Plant Secondary Metabolites; CRC Press: Boca Raton, FL, USA, 2021; ISBN 978-1-00-303495-7. [Google Scholar]
- Vidal, C.; Larama, G.; Riveros, A.; Meneses, C.; Cornejo, P. Main Molecular Pathways Associated with Copper Tolerance Response in Imperata cylindrica by de novo Transcriptome Assembly. Plants 2021, 10, 357. [Google Scholar] [CrossRef]
- Anjitha, K.S.; Sameena, P.P.; Puthur, J.T. Functional aspects of plant secondary metabolites in metal stress tolerance and their importance in pharmacology. Plant Stress 2021, 2, 100038. [Google Scholar] [CrossRef]
- Thijs, S.; Sillen, W.; Rineau, F.; Weyens, N.; Vangronsveld, J. Towards an Enhanced Understanding of Plant–Microbiome Interactions to Improve Phytoremediation: Engineering the Metaorganism. Front. Microbiol. 2016, 7, 341. [Google Scholar] [CrossRef]
- Stermitz, F.R.; Lorenz, P.; Tawara, J.N.; Zenewicz, L.A.; Lewis, K. Synergy in a medicinal plant: Antimicrobial action of berberine potentiated by 5′-methoxyhydnocarpin, a multidrug pump inhibitor. Proc. Natl. Acad. Sci. USA 2000, 97, 1433–1437. [Google Scholar] [CrossRef] [PubMed]
- Stavri, M.; Piddock, L.J.V.; Gibbons, S. Bacterial efflux pump inhibitors from natural sources. J. Antimicrob. Chemother. 2007, 59, 1247–1260. [Google Scholar] [CrossRef]
- Nguyen, T.H.T.; Nguyen, N.A.T.; Nguyen, H.D.; Nguyen, T.T.H.; Le, M.H.; Pham, M.Q.; Do, H.N.; Hoang, K.C.; Michalet, S.; Dijoux-Franca, M.-G.; et al. Plant Secondary Metabolites on Efflux-Mediated Antibiotic Resistant Stenotrophomonas Maltophilia: Potential of Herbal-Derived Efflux Pump Inhibitors. Antibiotics 2023, 12, 421. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, A.K.; Mohanty, P. Bacterial Efflux Pumps Involved in Multidrug Resistance and their Inhibitors: Rejuvinating the Antimicrobial Chemotherapy. Recent Patents Anti-Infect. Drug Disc. 2012, 7, 73–89. [Google Scholar] [CrossRef]
- Charkhi, P.; Haghshenas, M.R.; Mirzaei, B.; Davoodi, L.; Norouzi Bazgir, Z.; Goli, H.R. Comparison of the Effect of Phenylalanine Arginine Beta Naphthylamide (PAβN) and Curcumin on Minimum Inhibitory Concentration of Aminoglycosides on Pseudomonas aeruginosa Clinical Isolates. J. Adv. Med. Biomed. Res. 2020, 28, 105–110. [Google Scholar] [CrossRef]
- Alugoju, P.; Tencomnao, T. Chapter 19—Production and role of plants secondary metabolites under various environmental pollution. In Plants and Their Interaction to Environmental Pollution; Husen, A., Ed.; Elsevier: Amsterdam, The Netherlands, 2023; pp. 379–410. ISBN 978-0-323-99978-6. [Google Scholar]
- Muniz, D.F.; Dos Santos Barbosa, C.R.; de Menezes, I.R.A.; de Sousa, E.O.; Pereira, R.L.S.; Júnior, J.T.C.; Pereira, P.S.; de Matos, Y.M.L.S.; da Costa, R.H.S.; de Morais Oliveira-Tintino, C.D.; et al. In vitro and in silico inhibitory effects of synthetic and natural eugenol derivatives against the NorA efflux pump in Staphylococcus aureus. Food Chem. 2021, 337, 127776. [Google Scholar] [CrossRef]
- Dhara, L.; Tripathi, A. The use of eugenol in combination with cefotaxime and ciprofloxacin to combat ESBL-producing quinolone-resistant pathogenic Enterobacteriaceae. J. Appl. Microbiol. 2020, 129, 1566–1576. [Google Scholar] [CrossRef]
- Zhang, Y.; Zheng, L.; Zheng, Y.; Xue, S.; Zhang, J.; Huang, P.; Zhao, Y.; Hao, X.; He, Z.; Hu, Z.; et al. Insight into the assembly of root-associated microbiome in the medicinal plant Polygonum cuspidatum. Ind. Crops Prod. 2020, 145, 112163. [Google Scholar] [CrossRef]
- Li, L.; Song, X.; Yin, Z.; Jia, R.; Li, Z.; Zhou, X.; Zou, Y.; Li, L.; Yin, L.; Yue, G.; et al. The antibacterial activity and action mechanism of emodin from Polygonum cuspidatum against Haemophilus parasuis in vitro. Microbiol. Res. 2016, 186–187, 139–145. [Google Scholar] [CrossRef]
- Huang, Y.-Q.; Huang, G.-R.; Wu, M.-H.; Tang, H.-Y.; Huang, Z.-S.; Zhou, X.-H.; Yu, W.-Q.; Su, J.-W.; Mo, X.-Q.; Chen, B.-P.; et al. Inhibitory effects of emodin, baicalin, schizandrin and berberine on hefA gene: Treatment of Helicobacter pylori-induced multidrug resistance. World J. Gastroenterol. WJG 2015, 21, 4225–4231. [Google Scholar] [CrossRef]
- Seukep, J.A.; Sandjo, L.P.; Ngadjui, B.T.; Kuete, V. Antibacterial and antibiotic-resistance modifying activity of the extracts and compounds from Nauclea pobeguinii against Gram-negative multi-drug resistant phenotypes. BMC Complement. Altern. Med. 2016, 16, 193. [Google Scholar] [CrossRef]
- Nguyen Thi, K.-O.; Nguyen, N.-L.; Pham, H.-N.; Sawada, Y.; Hirai, M.Y.; Dauwe, R.; Dijoux-Franca, M.-G. Development of a Pteris vittata L. compound database by widely targeted metabolomics profiling. Biomed. Chromatogr. 2021, 35, e5110. [Google Scholar] [CrossRef]
- Nguyen, N.-L.; Vu, C.-T.; To, H.-M.; Pham, H.-N.; Nguyen, H.-D.; Nguyen, T.-D.; Nguyen Thi, K.-O. The Interactions among the Heavy Metals in Soils and in Weeds and Their Antioxidant Capacity under the Mining Activities in Thai Nguyen Province, Vietnam. J. Chem. 2020, 2020, 8010376. [Google Scholar] [CrossRef]
- Paul, T.; Das, B.; Apte, K.G.; Banerjee, S.; Saxena, R.C. Hypoglycemic Activity of Pteris vittata L., a Fern on Alloxan Induced Diabetic Rats. Inven. Rapid Planta Act. 2012, 2, 88–91. [Google Scholar]
- Singh, M.; Govindarajan, R.; Rawat, A.K.S.; Khare, P.B. Antimicrobial Flavonoid Rutin from Pteris vittata L. against Pathogenic Gastrointestinal Microflora. Am. Fern J. 2008, 98, 98–103. [Google Scholar] [CrossRef]
- Nguyen, N.-L.; Bui, V.-H.; Pham, H.-N.; To, H.-M.; Dijoux-Franca, M.-G.; Vu, C.-T.; Nguyen, K.-O.T. Ionomics and metabolomics analysis reveal the molecular mechanism of metal tolerance of Pteris vittata L. dominating in a mining site in Thai Nguyen province, Vietnam. Environ. Sci. Pollut. Res. 2022, 29, 87268–87280. [Google Scholar] [CrossRef]
- Choi, J.-S.; Jo, B.-W.; Kim, Y.-C. Enhanced paclitaxel bioavailability after oral administration of paclitaxel or prodrug to rats pretreated with quercetin. Eur. J. Pharm. Biopharm. Off. J. Arb. Pharm. Verfahr. EV 2004, 57, 313–318. [Google Scholar] [CrossRef]
- Wang, Y.-H.; Chao, P.-D.L.; Hsiu, S.L.; Wen, K.-C.; Hou, Y.-C. Lethal quercetin-digoxin interaction in pigs. Life Sci. 2004, 74, 1191–1197. [Google Scholar] [CrossRef]
- Dupuy, J.; Larrieu, G.; Sutra, J.F.; Lespine, A.; Alvinerie, M. Enhancement of moxidectin bioavailability in lamb by a natural flavonoid: Quercetin. Vet. Parasitol. 2003, 112, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Dias, K.J.S.D.O.; Miranda, G.M.; Bessa, J.R.; Araújo, A.C.J.D.; Freitas, P.R.; Almeida, R.S.D.; Paulo, C.L.R.; Neto, J.B.D.A.; Coutinho, H.D.M.; Ribeiro-Filho, J. Terpenes as bacterial efflux pump inhibitors: A systematic review. Front. Pharmacol. 2022, 13, 953982. [Google Scholar] [CrossRef] [PubMed]
- Seukep, A.J.; Mbuntcha, H.G.; Kuete, V.; Chu, Y.; Fan, E.; Guo, M.-Q. What Approaches to Thwart Bacterial Efflux Pumps-Mediated Resistance? Antibiotics 2022, 11, 1287. [Google Scholar] [CrossRef] [PubMed]
- Waditzer, M.; Bucar, F. Flavonoids as Inhibitors of Bacterial Efflux Pumps. Molecules 2021, 26, 6904. [Google Scholar] [CrossRef]
- Poole, K. Efflux pumps as antimicrobial resistance mechanisms. Ann. Med. 2007, 39, 162–176. [Google Scholar] [CrossRef] [PubMed]
- AlMatar, M.; Albarri, O.; Makky, E.A.; Köksal, F. Efflux pump inhibitors: New updates. Pharmacol. Rep. 2021, 73, 1–16. [Google Scholar] [CrossRef]
- Shriram, V.; Khare, T.; Bhagwat, R.; Shukla, R.; Kumar, V. Inhibiting Bacterial Drug Efflux Pumps via Phyto-Therapeutics to Combat Threatening Antimicrobial Resistance. Front. Microbiol. 2018, 9, 2990. [Google Scholar] [CrossRef]

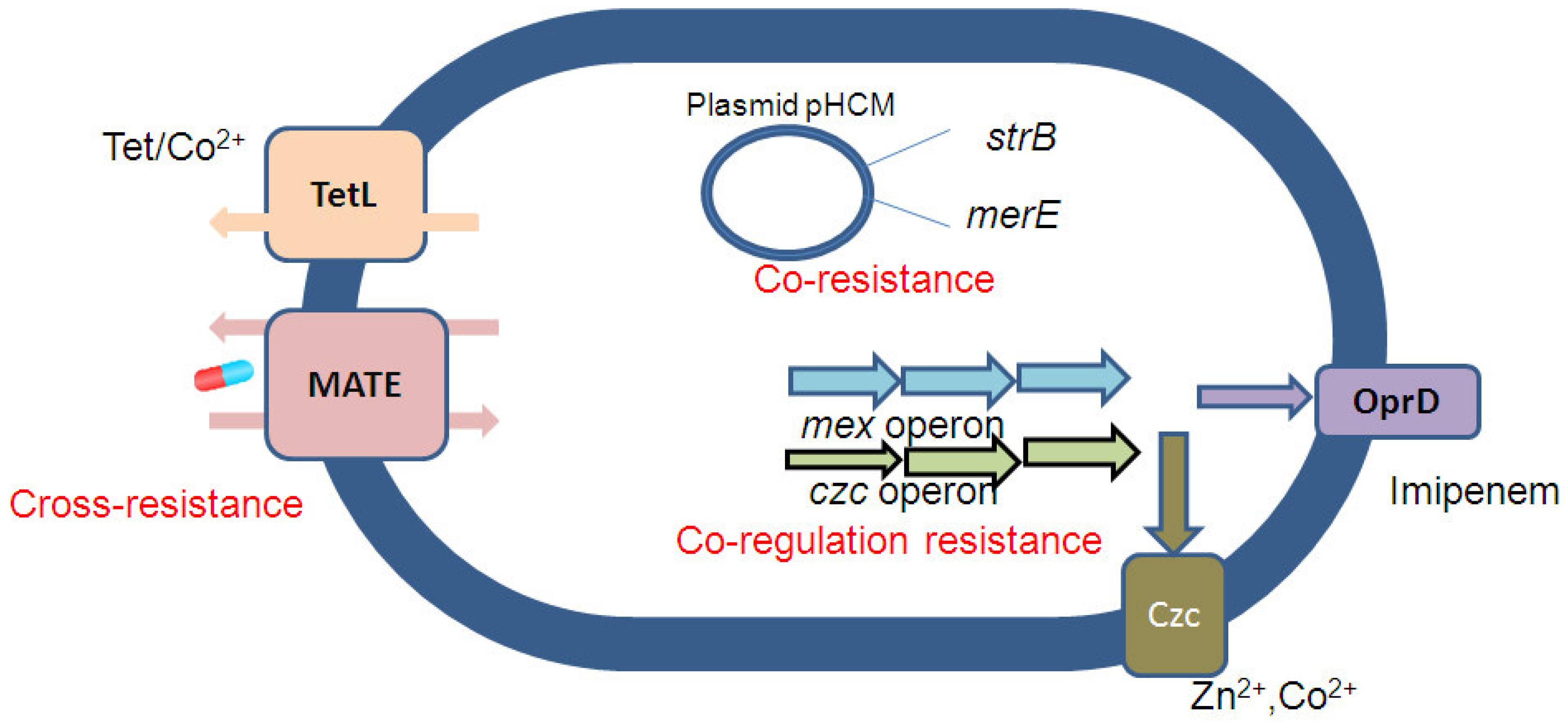

| Bacteria | Metal Ions | Antibiotics | Mechanism of Cross-Resistance |
|---|---|---|---|
| Burkholderia cepacia | Cd, Zn | Ofloxacin, erythromycin, kanamycin, novobiocin, β-lactams | Same efflux pumps systems |
| Salmonellaa Typhimurium | Cu, Zn, | β-lactams | Same efflux pumps systems |
| Listeria monocytogenes | Co, Zn, Cd | Erythromycin, clindamycin | Same efflux pumps systems |
| Bacteria | MFS | SMR | MATE | ABC | RND | Ref |
|---|---|---|---|---|---|---|
| Straphylococcus aureus | Nor A, Nor B, Nor C, MdeA, LmrS | QacA, QacB, QacD, Ebr | MepA | MrsA | FarE | [45] |
| Klebsiella pneumoniae | kmrA | kpnEF | kdeA | acrAB, kexD | [46] | |
| E. coli | EmrAB-TolC, Dep | EmrB Bcr EmrD EmrE | MacAB-TolC | AcrAD-TolC | [47] | |
| Pseudomonas aeuruginosa | EmrE | PmpM | MexAB-OprM MexCD-OprJ MexEF-OprN MeXY-OprM MeCD-OprJ MexAB-OprM | [45,48] |
| Plant | Active Compound | Bacterial Target | Efflux Pump Target |
|---|---|---|---|
| Caesalpinia spinosa | Tannic acids | Acinobacteria baumanii | Yo |
| Eucalyptus tereticornis | Ursolic acid and derivatives | E. coli | AcrAB, TolC, MacB, and YojI |
| Terminola chebula | Gallotannin 1,2,6-tri-O- galloyl-β-D | E. coli | ND |
| Thymus vulgaris | Baicalein | Salmonella enteridis | NorA |
| Dalea versicolor | Flavanoid, Phenolic | Staphylococcus aureus, Bacillus cereus | NorA |
| Berberis spp. | Berberin và Palmatin | Staphylococcus aureus, Pseudomonas aeruginosa | NorA, MexAB-OprM |
| Fallopia japonica | Resveratrol | Mycobacterium spp. | TetK |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, T.H.T.; Nguyen, H.D.; Le, M.H.; Nguyen, T.T.H.; Nguyen, T.D.; Nguyen, D.L.; Nguyen, Q.H.; Nguyen, T.K.O.; Michalet, S.; Dijoux-Franca, M.-G.; et al. Efflux Pump Inhibitors in Controlling Antibiotic Resistance: Outlook under a Heavy Metal Contamination Context. Molecules 2023, 28, 2912. https://doi.org/10.3390/molecules28072912
Nguyen THT, Nguyen HD, Le MH, Nguyen TTH, Nguyen TD, Nguyen DL, Nguyen QH, Nguyen TKO, Michalet S, Dijoux-Franca M-G, et al. Efflux Pump Inhibitors in Controlling Antibiotic Resistance: Outlook under a Heavy Metal Contamination Context. Molecules. 2023; 28(7):2912. https://doi.org/10.3390/molecules28072912
Chicago/Turabian StyleNguyen, Thi Huyen Thu, Hai Dang Nguyen, Mai Huong Le, Thi Thu Hien Nguyen, Thi Dua Nguyen, Duc Long Nguyen, Quang Huy Nguyen, Thi Kieu Oanh Nguyen, Serge Michalet, Marie-Geneviève Dijoux-Franca, and et al. 2023. "Efflux Pump Inhibitors in Controlling Antibiotic Resistance: Outlook under a Heavy Metal Contamination Context" Molecules 28, no. 7: 2912. https://doi.org/10.3390/molecules28072912
APA StyleNguyen, T. H. T., Nguyen, H. D., Le, M. H., Nguyen, T. T. H., Nguyen, T. D., Nguyen, D. L., Nguyen, Q. H., Nguyen, T. K. O., Michalet, S., Dijoux-Franca, M.-G., & Pham, H. N. (2023). Efflux Pump Inhibitors in Controlling Antibiotic Resistance: Outlook under a Heavy Metal Contamination Context. Molecules, 28(7), 2912. https://doi.org/10.3390/molecules28072912






