Abstract
The solubilization capacity of a series of sustainable phenylalanine-derived surface-active ionic liquids (SAILs) was evaluated towards polycyclic aromatic hydrocarbons—naphthalene, anthracene and pyrene. The key physico-chemical parameters of the studied systems (critical micelle concentration, spectral properties, solubilization parameters) were determined, analyzed and compared with conventional cationic surfactant, CTABr. For all studied PAH solubilization capacity increases with extension of alkyl chain length of PyPheOCn SAILs reaching the values comparable to CTABr for SAILs with n = 10–12. A remarkable advantage of the phenylalanine-derived SAILs PyPheOCn and PyPheNHCn is a possibility to cleave enzymatically ester and/or amide bonds under mild conditions, to separate polycyclic aromatic hydrocarbons in situ. A series of immobilized enzymes was tested to determine the most suitable candidates for tunable decomposition of SAILs. The decomposition pathway could be adjusted depending on the choice of the enzyme system, reaction conditions, and selection of SAILs type. The evaluated systems can provide selective cleavage of the ester and amide bond and help to choose the optimal decomposition method of SAILs for enzymatic recycling of SAILs transformation products or as a pretreatment towards biological mineralization. The concept of a possible practical application of studied systems for PAHs solubilization/separation was also discussed focusing on sustainability and a green chemistry approach.
1. Introduction
Ionic liquids (ILs) have been widely used in many industries [1,2,3] and are one of the core focuses of research over the past two decades [4,5]. ILs are proposed as more desirable than conventional volatile solvents in many physical and chemical processes, often referred as “green” solvents [6]. They can be of natural origin and be prepared by a “benign by design” approach [5,7]. Designing ILs that lead to a reduction in the losses of solvents as well as less damage to the environment is an important aspect in green chemistry [6]. Ionic liquids in general fulfil many of the 12 criteria as a green solvent related to the availability, price, recyclability, synthesis, toxicity, biodegradability, performance, stability, flammability, storage, and renewability [8]. Ionic liquids can offer a better alternative to volatile solvents, which has led to its massive use in industrial applications such as separation and purification, and as chemical catalysts, biorefinery concepts [3], extractions [1] and others [9,10,11,12]
Recently, research studies have revealed that some of these ILs demonstrate a significant toxicity level [13,14]. Though the toxicity evaluations of ILs have been extensively reported in the literature, biodegradation data are comparatively limited [15]. Biodegradation is considered as the cleanest ultimate fate for compounds in nature. Although ILs can be easily synthesized, often in only a few steps, it is important to ensure that they are fully mineralizable in case of their introduction into the environment. ILs’ persistence in nature, due to their high stability, would result in adverse environmental toxicity [16]. It can lead to problematic wastewater pollution upon release into the environment because ILs, which are highly water soluble and not consistently biodegradable in wastewater aeration tanks, can have varying degrees of biodegradability [17]. The toxicity of ILs is related to the sorption of a surfactant molecule to biological membranes, which is driven by the nonspecific hydrophobic interactions [18]. Such adsorption of surfactants disrupts the cellular membranes and results in acute or chronic effects in microbes. The hydrophobic/hydrophilic balance of the molecule and the cationic charge density ultimately results in antimicrobial activity [19].
The synthesis and investigation of ILs using natural structural elements is a prospective approach in designing new molecules [7,20,21,22,23,24]. The ILs based on the amino acids are considered as one of the green alternatives and have high demand in industrial applications, which lead to the further development of a benign by design approach for the elaboration of an L-phenylalanine ethyl ester platform for designing completely mineralizable ILs [23,25]. After all, biodegradation of a newly synthesized compound can be firstly screened by test methods recommended by OECD guidelines [26,27] and the development in analytical methods such as LC-MS and NMR make it easier to detect the degraded products with a limited sample volume.
In order to describe ILs as a greener solvent, one should show complete and rapid biotic/abiotic degradation in nature. It is worth noting that there is often very little correlation between (i) the rate of chemical and enzymatic hydrolysis and (ii) the rate of biodegradation [28,29]. The biodegradability of ILs depends on their molecular structure [30] and the length of the alkyl chain [7,31,32]. Some ILs are stable to a wide range of chemicals as well as to high temperature, which are not expected to be readily biodegradable [33]. Pyridinium, cholinium, and imidazolium cations are the most studied among IL cations [34,35,36]. Pyridinium ILs are, in general, biodegradable to a higher extent than imidazolium ILs [23,25]. Thus, in the work [25], it was reported that the combination of ionic head groups with readily biodegradable biomolecules did not necessarily lead to an increase in biodegradability or degradation of the compound. The imidazolium- and pyridinium-derived phenylalanine ethyl ester ILs were shown to be more biodegradable than the proline and choline derivatives [23]. Recent studies provided an insight into the biodegradability of the ethyl ester of cationic phenylanine-derived ILs [24]. The pyridinium derivatives are found to be the preferred greener IL based on synthesis, toxicity, and biodegradation considerations [7]. Among ILs, special interests are the compounds, which contain significant hydrophobic fragments in their structure and demonstrate remarkable adsorption onto surfaces and change their properties (so-called surface-active ionic liquids, SAILs) [32].
The extraction and separation of polycyclic aromatic hydrocarbons (PAHs) are challenging problems [4] having both technological and environmental impacts [37]. Applying a water/surfactant system instead of organic solvents can particularly solve the aforementioned problems and make systems “greener” [38]. Water solutions of low toxic and biodegradable SAILs are prospective alternatives to conventional surfactants in the creation of sustainable ecologically friendly mixed compositions [39].
In this study, we evaluated the efficacy of pyridinium SAILs in the solubilization of model representative PAHs (naphthalene, anthracene and pyrene) in water/SAIL systems (Figure 1) and proved a strategy for direct degradation/hydrolysis of these SAILs by commercial enzymes. This concept can be applied in the design of recyclable environmentally friendly systems for PAHs solubilization/separation.

Figure 1.
Structures of studied pyridinium SAILs and polycyclic aromatic hydrocarbons.
2. Results and Discussion
2.1. Study of Solubilization Capacity
The solubilization capacity of PyPheOCn surfactants (n = 4, 6, 8, 10, 12, 14, 16) was evaluated, related to model representative PAH (naphthalene, anthracene and pyrene) and compared with the solubilization capacity of the widely used conventional cationic surfactant cetyltrimethylammonium bromide (CTABr) (Table 1).

Table 1.
Critical micelle concentration (cmc), β parameter and solubilization capacity (S) for PyPheOCn surfactants, and cetyltrimethylammonium bromide (CTABr).
The observed regularities for dependencies “concentration of PyPheOCn–PAH absorbance” are typical for such types of systems [38,40,41,42]: below the cmc, no dramatical changes of PAH concentration (absorbance) in the solution appeared, but beyond the cmc, an increasing of PAH concentration (absorbance) in the solution was observed (Figure 2). The brake-point on dependencies “concentration of PyPheOCn–PAH absorbance” corresponds to the cmc of the studied SAILs. The cmc values for PyPheOCn SAILs and CTABr, obtained with different PAH, correspond well with each other and with the cmc of these compounds, determined using surface tension and conductivity measurements (Table 1; see also [32]).

Figure 2.
Dependence of the PAH absorbance vs. concentration of SAILs for the system PyPheOC10/pyrene: determination of cmc and β parameters.
For all studied PAH, the solubilization capacity increases with an extension of the alkyl chain length of PyPheOCn SAILs (Figure 3). The solubilization capacity, comparable with CTABr, demonstrates PyPheOCn with n = 10–12 (naphthalene, pyrene) and n = 12–14 (anthracene). It corresponds to aggregation properties of the studied compounds: the cmc of CTABr are similar to PyPheOCn with n = 10–12 (Table 1).
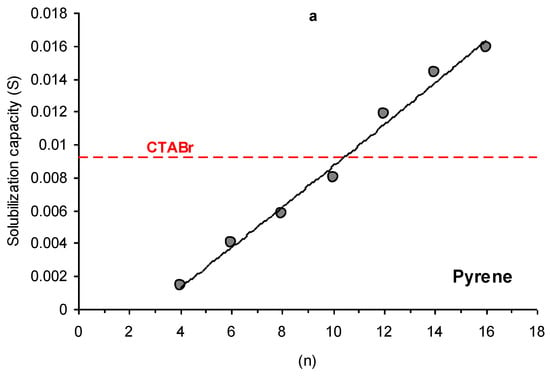

Figure 3.
Dependencies of solubilization capacity (S) on alkyl chain length (n) of PyPheOCn SAILs towards pyrene (a), naphthalene (b), and anthracene (c) compared to solubilization capacity of CTABr.
2.2. Study of Enzymatic Degradation/Hydrolysis of PyPheOCn SAILs
Selecting a suitable enzyme which has higher degradation potential over the chemical compounds is very important [43,44]. Amidase and protease are the most representative groups of enzymes belonging to the class hydrolases in the remediation of polluted environments. The breakdown of ester, amide and peptide bonds by esterases, amidases and proteases may lead to products with little or no toxicity [45]. A previous report by Neumann et al. [46] shows that primary biodegradation of the test samples cyanomethyl side chain and its transformation product was enabled by a microorganism. This suggests that hydrolysis happened via nitrile degrading enzymes such as nitrilases or nitrile hydratases together with amidase. These studies support the role of the amidase enzyme as a catalyst for the biodegradation. Similar studies also reported that enzymes such as nitrilases and amidase are commonly applied as catalysts in organic synthesis for the hydrolysis of nitrile groups in the pharmaceutical industry and for bioremediation purposes, amongst others [47,48]. Protease is one of the most important groups of industrial enzymes, having a unique catalytic mechanism, broad substrate specificity and high robustness, which has widened their application into bioremediation and waste management [49].
This study again reveals the use of direct enzymes to accelerate the process of degradation in the wastewater treatment plants before the release of ILs to the environment so that we can reduce the chances of adverse toxicity effects in the future. It will be a promising finding and provide insightful information for the chemical industries, where they use a specific class of ILs that do not have any alternative replacements and are more persistent in the environment. Once we have the information on the degradability data as described in the current study, it is more applicable to change the molecular structure of the ILs to synthesize 100% biodegradable compounds. This is because chemical modification of the IL side chains may compromise the ability of the enzymes to recognize the modified structure as a suitable substrate and effect the complete degradation of the compound [50].
The commercial amidase enzymes purchased were obtained from the source Escherichia coli and Achromobacter with a penicillin hydrolytic activity unit of NLT 850 U/g and NLT 250 U/g, respectively. Protease enzymes were obtained from bacteria, fungi and plant sources with different enzyme activity as given in Table 2. Amidase enzymes are a large group of hydrolytic enzymes that contain a conserved stretch of approximately 130 amino acids. They are widespread, being found in both prokaryotes and eukaryotes. Amidase enzymes catalyze the hydrolysis of amide bonds, although they have a wide range of substrate specificity and function. Nonetheless, these enzymes maintain a core alpha/beta/alpha structure, where the topologies of the N- and C-terminal halves are similar. These enzymes possess a unique, highly conserved Ser-Ser-Lys catalytic triad used for amide hydrolysis, although the catalytic mechanism for acyl-enzyme intermediate formation can differ between enzymes [51], whereas protease enzymes work by hydrolyzing the peptide bonds and have the highest market share among industrial enzymes [52]. Proteases are classified as acidic, alkaline and neutral proteases according to the pH, and they exhibit maximum efficacy within a specific pH range. Proteases (also known as proteinases or peptidases) hydrolyze the peptide bond between amino acid residues in a polypeptide chain. Proteases may be specific and limited to one or more sites within a protein, or they may be nonspecific, digesting proteins into individual amino acids. Proteases are found in all organisms and are involved in all areas of metabolism [53].

Table 2.
Sources of enzymes used for enzymatic hydrolysis of studied ionic liquids.
In the present study, we revealed the enzymatic hydrolysis of pyridinium-based ILs of PyPheOC4 (as a representative example of SAILs of PyPheOCn series), which have ester and amide bonds with amidase and protease enzymes. During the enzymatic hydrolysis, the enzymes enhanced the bond cleavage in molecules with water.
In theory, hydrolysis of an amide breaks the carbon–nitrogen bond and produces an acid and either ammonia or an amine. Though this reaction bears a resemblance to the hydrolysis of esters, there are, however, important differences. The hydrolysis of esters occurs relatively easily, whereas amides are much more resistant to hydrolysis. In the case of ester bonds, carboxylic esters readily hydrolyze to the parent carboxylic acid and an alcohol. Ester bond hydrolysis is the most prevalent type of biodegradation because they are a hydrolytically unstable functional group. Amides can be hydrolyzed only by heating for hours with a strong acid or strong base unless an enzyme is used. If amide hydrolysis occurs in a basic solution, the salt of the carboxylic acid forms, i.e., one mole of the base is required per mole of amide. If hydrolysis proceeds under acidic conditions, the ammonium salt of the amine is formed, and one mole of acid is required per mole of amide [54].
The possible way by which ester bond and amide bond hydrolysis proceeded in the PyPheOC4 compound is illustrated in Figure 4.

Figure 4.
Expected pathways of C-O and C-N bonds hydrolysis in PyPheOC4.
When 2% PyPheOC4 was treated with 0.2 g amidase enzymes (1 and 2) at the incubation temperature of 40 °C, pH 5.5, shaking speed of 170 rpm, and 3 days incubation time, showed 55 to 80% ester bond hydrolysis. With amidase enzyme 1, 80% of the ester bonds were hydrolyzed whereas, with amidase 2, only 55% ester bond hydrolysis was observed. In both cases, no significant (less than 5%) amide bond hydrolysis product was recorded. The high stability of amide bonds is subjected to its propensity to form a resonating structure, which brings a double bond character to the amide CO-N bond [54].
When the concentration of PyPheOC4 was reduced to 1% (w/v) while maintaining the same incubation conditions as mentioned before with both amidase (1 and 2), 95% of the ester bond was hydrolyzed in PyPheOC4 and no significant amounts of amide bond break occurred. These studies revealed that with PyPheOC4, it was easier to hydrolyze the ester bonds by amidase enzymes compared to amide bonds. Moreover, the reduced concentration further made it feasible for the amidase enzymes to open more active sites for ester bonds. The transformation products formed from the studies are given in Figure 4. Based on the results, more enzymes were applied in this study in order to find complete hydrolysis of the compounds. Experiments were further carried out with 0.1 g of 1–20 types of protease enzymes (Table 2) along with amidase 1 and 2.
Based on the results mentioned above, the concentration of PyPheOC4 was maintained as 1% followed by an incubation temperature of 50 °C, shaking speed of 100 rpm and incubation for 7 days.
The applied protease enzymes work better at slightly higher temperatures, therefore the temperature 50 °C was selected for these test trials. The shaking speed was reduced to 100 rpm due to the optimum shaking speed recommended for the enzymes with an extended incubation time to 7 days. Among the 20 different types of protease enzymes tested on PyPheOC4, 100% ester bond hydrolysis was observed for all the samples except enzymes P8, P9 (86%) and P18 (79%); P13 also gives ca. 8% amidolysis; and representative examples of the NMR spectra are given in Figure 5.

Figure 5.
1H NMR spectra of PyPheOC4 solutions after incubations with selected protease enzymes (concentration of PyPheOC4 1%). Spectrum 1 (bottom) corresponds to PyPheOC4 after incubation in buffer without enzymes; the spectra 2→11 represent incubation with enzymes P11→P20.
Based on these results, we further reduced the concentration of the tested IL PyPheOC4 to 0.5% in order to study the possibility of complete hydrolysis of ester and amide bonds (100%). The optimum activity of the protease enzyme was reported with a pH of 6–7 [55]. Therefore, the pH of the sodium acetate buffer was further increased to 6.5 to improve the degradation rate. Incubation temperature, shaking speed and time were maintained at 50 °C, 100 rpm and 7 days, respectively.
Only selected protease enzymes from 1–20 that have high enzyme activity (P1, P2, P3, P4, P6, P10, P11, P13, P14, P17) and amidase 1 and amidase 2 were selected based on the previous test results. When the selected protease enzymes were used (based on higher enzyme activity), all the samples showed 100% ester bond hydrolysis with PyPheOC4. Among those, only P13 enzymes showed about 10–20% amide bond hydrolysis (Figure 6). In the case of amidase 1 and 2, only ester bond hydrolysis (100%) was observed.

Figure 6.
1H NMR spectra of PyPheOC4 solutions after incubations with selected protease enzymes at pH 6.5 (concentration of PyPheOC4 0.5%). Spectrum 1 (bottom) corresponds to PyPheOC4 after incubation in buffer without enzymes; spectrum 2 recorded the model mixture of expected hydrolytic products; spectra 3–12 represent incubation with proteases P1, P2, P3, P4, P6, P10, P11, P13, P14, P17; spectra 13 and 14 represent incubation with amidase 1 and amidase 2, correspondingly.
PyPheOC4 in presence of the P13 enzyme showed a degradation pathway as presented in pathway A (90% of transformations) and pathway B (10% of transformations) (Figure 7). Transformations by pathway A are faster up to the first step (i.e., ester hydrolysis), whereas further hydrolysis of amide bonds in degradation product Py(+)-CH2-CO-NH-Phe-COOH is a very slow process. It can be connected to the reason that during the degradation process stabilizers, activators, or inhibitory products can form in the medium. which result from the material degradation or leaching out of enzyme additives, and those could affect the enzyme catalyzed reactions by influencing enzyme adsorption and activity, resulting from material degradation [56].

Figure 7.
Degradation pathways analysis for PyPheOC4 based on NMR data.
The intermediates and final products of enzymatic decomposition were not dependent on the degradation pathway (A or B) and were readily biodegradable compounds [24], which is critically important for the design of sustainable ecologically friendly systems.
2.3. Structural Modification of PyPheOC4 SAIL: Diamide Derivative PyPheNHC4
The change of ester bond in PyPheOC4 SAIL structure to amide bond can help to create compounds (Figure 8), which are more stable to alkaline hydrolysis [57,58] and can expand the possible application range of this type of compound, but also requires an evaluation of enzymatic decomposition.

Figure 8.
Structural modification of PyPheOC4 SAIL.
The rate of enzymatic hydrolysis reaction is influenced by the physicochemical properties of the substrate and also by the inherent characteristics of a specific enzyme, which can be enzyme activity and its stability, local concentration, amino acid composition, and 3D conformation. Moreover, it is again very important to consider the medium conditions such as pH and temperature, since they strongly influence the properties of the substrate and the enzyme.
2.4. Study of Enzymatic Degradation/Hydrolysis of PyPheNHC4 SAIL
It is expected that when compound PyPheNHC4 has been treated with enzymes, the hydrolysis may happen at either of these bonds, i.e., amide bond I or amide bond II, and ultimately lead to complete hydrolysis of the compound (Figure 9 and Figure 10).

Figure 9.
Expected ways of bonds hydrolysis in PyPheNHC4.

Figure 10.
1H NMR spectra of PyPheNHC4 solutions after incubations with selected protease enzymes (concentration of PyPheNHC4 1%). Spectrum 1 (bottom) corresponds to PyPheNHC4 after incubation in buffer without enzymes; the spectra 2→11 represent incubation with enzymes P11→P20.
The experimental trials and conditions explored and discussed before with PyPheOC4 have been tried with the new synthesized amide compounds. The first trial, i.e., 2% PyPheNHC4 with 0.2 g amidase enzymes (1 and 2) with an incubation temperature of 40 °C, pH of 5.5, shaking speed of 170 rpm, and 3 days incubation time, showed no evidence of amide bond hydrolysis with both the enzymes amidase 1 and 2. Thus, it is assumed that the stability of amide bonds is too strong to cleaved by amidase enzymes or is inaccessible to the active site. The high stability of amide bonds is subjected to its propensity to form a resonating structure, which brings a double bond character to the amide CO-N bond [54]. When the concentration of ionic liquids was reduced to 1% (w/v) by maintaining the same incubation conditions as mentioned before with both amidase enzymes (1 and 2) again, no traces of amide hydrolysis were observed. Experiments were further performed with 0.1 g of the 1–20 types of protease enzymes (Table 2) along with amidase 1 and 2. Among the 20 different types of protease enzymes tested with PyPheNHC4, the P1–P9 enzymes did not show any significant amide bond hydrolysis. Among the proteases P11–P20, the P13 enzyme showed 87% amide bond hydrolysis. The proteases P12, P15 and P16 demonstrated 10–20% amide bond hydrolysis (Figure 10). As shown in Table 2, all of the proteases were obtained from different sources. Among them, the P13 obtained from the source Aspergillus oryzae with an enzyme activity of 65 ELU/g worked well for hydrolysis of amide bonds in PyPheNHC4. It reveals that each enzyme has its optimum hydrolytic activity with the compounds under optimum conditions.
The pathway by which a protease enzyme breaks the amide bonds is illustrated in Figure 11.

Figure 11.
Degradation pathway analysis for PyPheNHC4 based on NMR data.
To enhance the complete degradation, the concentration of PyPheNHC4 was further reduced to 0.5% as investigated with PyPheOC4. The incubation temperature, shaking speed and time were maintained at 50 °C, 100 rpm and 7 days, respectively. Only selected protease enzymes from 1–20 that had high enzyme activity (P1, P2, P3, P4, P6, P10, P11, P13, P14, P17) and amidase 1 and 2 were selected based on the previous test results. With the reduced concentration (0.5%) of PyPheNHC4, the P13 enzyme showed 100% amide hydrolysis (Figure 12). PyPheNHC4 in the presence of the P13 enzyme showed 100% transformation by pathway B; i.e., hydrolysis of amide bond I. Transformations by pathway A by the hydrolysis of amide bond II is relatively slow in comparison with the rate of transformation by pathway B. The reaction showed a total cleavage of all amide bonds and observed Py(+)-CH2-COOH, phenylalanine and Bu-NH2 in the solution (Figure 11 and Figure 12).

Figure 12.
NMR spectra of PyPheNHC4 solutions after incubations with selected protease enzymes (concentration of PyPheNHC4 0.5%). Spectrum 1 (bottom) corresponds to PyPheNHC4 after incubation in buffer without enzymes; spectrum 2 recorded the model mixture of expected hydrolytic products; spectra 3–12 represent incubation with proteases P1, P2, P3, P4, P6, P10, P11, P13, P14, P17; spectra 13 and 14 represent incubation with amidase 1 and amidase 2, correspondingly.
2.5. Concept of PyPheOCn and PyPheNHCn SAILs Application to Solubilization of Polycyclic Aromatic Hydrocarbons
The application of surfactants and SAILs in the design of effective and ecologically friendly sustainable systems for the solubilization of polycyclic aromatic hydrocarbons is very often limited due to the negative influence of surfactants and SAILs on the environment [14] and technological problems connected with the separation of solubilized PAH from surfactant solutions [4]. Among the SAILs PyPheOCn and PyPheNHCn, which were the focus of our current study, there are several examples of compounds (with n = 4–8) that could be considered as low toxicity and readily biodegradable ILs [7]. An evaluation of the toxicity and biodegradability of potential hydrolytic decomposition products of PyPheOCn and PyPheNHCn, performed in our recent studies [10,24,32], also confirm a great potential of these SAILs as a platform for ecologically friendly sustainable systems. The solubilization capacity of PyPheOCn surfactants, evaluated related to model representative PAH (naphthalene, anthracene and pyrene), is comparable to the solubilization capacity of conventional cationic surfactant CTABr (see Table 1 and Figure 3; compare parameters for CTAB and SAILs with n = 8–12), but in the cases of PyPheOCn and PyPheNHCn, polycyclic aromatic hydrocarbons could be easily separated from the SAILs water solution using enzymatic decomposition of SAILs under mild conditions. The decomposition of SAILs could be adjusted via the choice of enzyme system, reaction conditions and/or choice of SAILs type. Despite the possibility of solubilized aromatic carbons to have an impact on the enzymatic degradation of phenylalanine-derived SAILs, we suggest that enzymatic cleavage of surfactant monomers occurs [49]. Since the monomer is in the equilibrium with dynamic micellar aggregates [40], the drop in the monomer concentration will affect the micelle concentration in the system and thus reduce the PAH solubilized by the surfactant aggregates. The PAH initially bound by the micelles will be released in the bulk aqueous solution and precipitate. After SAILs decomposition, PAH, immobilized enzymes, and the water solution of SAILs decomposition products could be separated from each other using filtration techniques. The water solution of SAILs decomposition products could be completely biodegraded by the microorganisms in the environment or used for enzymatic re-synthesis of SAILs from decomposition products with future usage of the obtained solution for the next cycle of PAH extraction/solubilization (Figure 13).

Figure 13.
Concept of PyPheOCn and PyPheNHCn SAILs application to solubilization of PAHs.
3. Experimental Section
3.1. Materials
Naphthalene (Nap), anthracene (Ant) and pyrene (Pyr), cetyltrimethylammonium bromide (CTABr), inorganic and organic salts for preparation of working buffer solutions, acids and organic solvents were purchased from Sigma-Aldrich/Merck KGaA or Acros Organics/Fisher Scientific. Deuterated solvents for NMR analysis were purchased from Deutero GmbH (Kastellaun, Germany). Deionized water was prepared using the Direct-Q UV 5 water purification system.
Synthesis of SAILs PyPheOCn is previously described [32]. Synthesis of PyPheNC4 was performed according to common synthetic procedures [10,24], confirmation of the structure and purity was performed using 1H, 13C, 1H-1H, 1H-13C, DETP 135 NMR techniques and by HRMS.
Amidase and protease enzymes (Table 2) were purchased from Fermenta Biotech Ltd. (Mumbai, India) and ChiralVision B.V. (Den Hoorn, The Netherlands), respectively. The enzymes were stored in a cold room until the start of the experiment.
3.2. Methods
3.2.1. Study of Solubilization Capacity
Solubilization capacity of micellar systems was evaluated by determining the maximal solubility of the substrates (naphthalene, anthracene and pyrene) in surfactant solutions [38,40]. To a series of corresponding PyPheOCn surfactant solutions (10–15 different concentrations in total, which cover concentration range before and after cmc; 3 mL of surfactant solution per each concentration) was added a fixed amount (5 mg per each concentration) of studied substrate, shaken intensively and leaved for equilibration for 48 h at 25 °C [38]. After equilibration, insoluble residue was filtered through Millipore filters (Durapore ® PVDF membrane, pore size 0.22 µm), the filtrate was placed in the quartz cell and the UV-vis spectrum was recorded using UV-Vis spectrophotometer JASCO V-730 in the wavelength range from 200 to 400 nm. For studied solutions obtained value of absorbance (A) was recalculated on a pathlength 10 mm at the 311 nm (ε = 320 M−1⋅cm−1) for naphthalene, 378 nm (ε = 15,100 M−1⋅cm−1) for anthracene and 336 nm (ε = 62,800 M−1⋅cm−1) for pyrene. Molar extinction coefficients (ε, M−1⋅cm−1) for substrates were determined in independent experiments in hexane. Solubilization capacity of the surfactant (S) was calculated from the ratio S = β/ε, where β is the slope of the linear part of the A vs. Csurf dependence [38,41].
3.2.2. Enzymatic Degradation/Hydrolysis Studies
In this experiment, two representative examples of pyridinium SAILs have been tested. Firstly, the experiments were started with compound PyPheOC4. Once the enzymatic degradability of this compound and its transformation products are obtained, the test trials were continued with the compound PyPheNHC4 (which is a result of structure improvement of PyPheOCn SAILs series). Degradation tests were performed with different reaction conditions, such as doses of enzymes, pH, temperature, shaking speed and incubation period to optimize the best parameters for the test. We started the test trials with ester-based SAIL, PyPheOC4, with a concentration of 2% was prepared in 0.1 M sodium acetate buffer (pH 5.5). Then, 0.2 g of enzymes such as amidase 1 and 2 were weighed in separate 15 mL tubes. To this, 1 mL of the samples (i.e., ionic liquid in sodium acetate buffer) was added. The tubes were then placed in an incubator with a shaking speed of 170 rpm and a temperature of 40 °C for 7 days. Respective controls were kept without the addition of enzymes. The supernatants were then collected and stored in fresh tubes and preserved for NMR analysis; before recording the 1H NMR spectra, up to 20% v/v of D2O was added to a sample for locking.
After obtaining the results, similar tests were performed with a further concentration of PyPheOC4 (1% and 0.5%), doses of amidase 1 and 2, and protease 1–20 enzymes (0.1 g), pH (5.5, 6.5), temperature (50 °C), and shaking speed (100 rpm) (see Table 3). Once the test results were analyzed, similar trials were performed with the compound PyPheNHC4.

Table 3.
Experiment set up and reaction conditions of ILs enzymatic degradation studies.
3.2.3. NMR Analysis of Enzymatic Degradation/Hydrolysis Products
The NMR spectra were recorded on a Bruker Avance III 400 MHz spectrometer. For analysis, 400 µL of clear supernatant was used, which was transferred to NMR tubes and mixed with 100 µL of deuterium oxide (99.9%). NMR spectra were recorded using the standard water suppression method. Identification of enzymatic hydrolysis products were performed by comparison of 1H NMR spectra of potential transformation products with 1H NMR spectra of reaction mixtures of corresponding SAILs after incubation with the immobilized enzymes.
4. Conclusions
The solubilization capacity of a series of sustainable phenylalanine-derived surface-active ionic liquids (SAILs) was evaluated towards polycyclic aromatic hydrocarbons—naphthalene, anthracene and pyrene. The key physico-chemical parameters of the studied systems (critical micelle concentration, spectral properties, solubilization parameters) were determined, analyzed and compared with a conventional cationic surfactant, CTABr. For all studied PAHs, the solubilization capacity increases with an extension of the alkyl chain length of PyPheOCn SAILs reaching the values comparable to CTABr for SAILs with n = 10–12. A remarkable advantage of the phenylalanine-derived SAILs PyPheOCn and PyPheNHCn consists in a possibility to cleave enzymatically ester and/or amide bonds under mild conditions, to separate polycyclic aromatic hydrocarbons in situ. A series of immobilized enzymes was tested to determine the most suitable candidates for tunable decomposition of SAILs. The decomposition pathway could be adjusted depending on the choice of enzyme system, reaction conditions and SAILs type. The evaluated systems can provide selective cleavage of the ester and/or amide bonds and help to choose optimal decomposition of SAILs for enzymatic recycling of SAILs transformation products or as a pretreatment towards biological mineralization. The concept of a possible practical application of the studied systems for PAHs solubilization/separation was also discussed, focusing on sustainability and green chemistry approaches.
Author Contributions
Conceptualization and idea of the research, Y.K., K.K.G., N.G. and V.K.G.; synthesis of compounds, methodology, I.V.K.; experimental techniques, data curation analysis of the results, conceptualization, methodology, T.Y., S.M.S. and I.V.K.; analysis and interpretation, S.M.S. and I.V.K.; formal analysis and interpretation, S.M.S.; writing—original draft preparation, I.V.K. and S.M.S.; writing—review and editing, Y.K., V.K.G., K.K.G. and N.G.; funding acquisition, Y.K. and N.G. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Estonian Research Council grants PUT1656 (for Y.V.K., N.G., Y.K.) and COVSG5 (Y.K.); ERDF Dora Plus program (for T.Y.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All the data gathered for this study are available in the article.
Acknowledgments
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compounds could be available from the authors upon request.
References
- Greer, A.J.; Jacquemin, J.; Hardacre, C. Industrial Applications of Ionic Liquids. Molecules 2020, 25, 5207. [Google Scholar] [CrossRef] [PubMed]
- Plechkova, N.V.; Seddon, K.R. Ionic Liquids: “Designer” Solvents for Green Chemistry. In Methods and Reagents for Green Chemistry; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2007; pp. 103–130. [Google Scholar] [CrossRef]
- Usmani, Z.; Sharma, M.; Gupta, P.; Karpichev, Y.; Gathergood, N.; Bhat, R.; Gupta, V.K. Ionic liquid based pretreatment of lignocellulosic biomass for enhanced bioconversion. Bioresour. Technol. 2020, 304, 123003. [Google Scholar] [CrossRef] [PubMed]
- Pillai, P.; Maiti, M.; Mandal, A. Mini-review on Recent Advances in the Application of Surface-Active Ionic Liquids: Petroleum Industry Perspective. Energy Fuels 2022, 36, 7925–7939. [Google Scholar] [CrossRef]
- Jordan, A.; Gathergood, N. Biodegradation of ionic liquids–a critical review. Chem. Soc. Rev. 2015, 44, 8200–8237. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, J.B.; Anastas, P.T.; Erythropel, H.C.; Leitner, W. Designing for a green chemistry future. Science 2020, 367, 397–400. [Google Scholar] [CrossRef]
- Suk, M.; Haiß, A.; Westphal, J.; Jordan, A.; Kellett, A.; Kapitanov, I.V.; Karpichev, Y.; Gathergood, N.; Kümmerer, K. Design rules for environmental biodegradability of phenylalanine alkyl ester linked ionic liquids. Green Chem. 2020, 22, 4498–4508. [Google Scholar] [CrossRef]
- Gu, Y.; Jérôme, F. Bio-based solvents: An emerging generation of fluids for the design of eco-efficient processes in catalysis and organic chemistry. Chem. Soc. Rev. 2013, 42, 9550–9570. [Google Scholar] [CrossRef]
- Kapitanov, I.V.; Mirgorodskaya, A.B.; Valeeva, F.G.; Gathergood, N.; Kuca, K.; Zakharova, L.Y.; Karpichev, Y. Physicochemical properties and esterolytic reactivity of oxime functionalized surfactants in pH-responsive mixed micellar system. Colloids Surf. A Physicochem. Eng. Asp. 2017, 524, 143–159. [Google Scholar] [CrossRef]
- Pandya, S.J.; Kapitanov, I.V.; Usmani, Z.; Sahu, R.; Sinha, D.; Gathergood, N.; Ghosh, K.K.; Karpichev, Y. An example of green surfactant systems based on inherently biodegradable IL-derived amphiphilic oximes. J. Mol. Liq. 2020, 305, 112857. [Google Scholar] [CrossRef]
- Banjare, M.K.; Behera, K.; Banjare, R.K.; Pandey, S.; Ghosh, K.K.; Karpichev, Y. Molecular interactions between novel synthesized biodegradable ionic liquids with antidepressant drug. Chem. Thermodyn. Therm. Anal. 2021, 3, 100012. [Google Scholar] [CrossRef]
- Pandya, S.J.; Kapitanov, I.V.; Banjare, M.K.; Behera, K.; Borovkov, V.; Ghosh, K.K.; Karpichev, Y. Mixed Oxime-Functionalized IL/16-s-16 Gemini Surfactants System: Physicochemical Study and Structural Transitions in the Presence of Promethazine as a Potential Chiral Pollutant. Chemosensors 2022, 10, 46. [Google Scholar] [CrossRef]
- Gonçalves, A.R.; Paredes, X.; Cristino, A.F.; Santos, F.J.; Queirós, C.S. Ionic Liquids—A Review of Their Toxicity to Living Organisms. Int. J. Mol. Sci. 2021, 22, 5612. [Google Scholar] [CrossRef] [PubMed]
- Kowalska, D.; Maculewicz, J.; Stepnowski, P.; Dołżonek, J. Ionic liquids as environmental hazards–Crucial data in view of future PBT and PMT assessment. J. Hazard. Mater. 2021, 403, 123896. [Google Scholar] [CrossRef]
- Amsel, A.-K.; Olsson, O.; Kümmerer, K. Inventory of biodegradation data of ionic liquids. Chemosphere 2022, 299, 134385. [Google Scholar] [CrossRef]
- Bubalo, M.C.; Radošević, K.; Redovniković, I.R.; Slivac, I.; Srček, V.G. Toxicity mechanisms of ionic liquids. Arch. Ind. Hyg. Toxicol. 2017, 68, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Docherty, K.M.; Aiello, S.W.; Buehler, B.K.; Jones, S.E.; Szymczyna, B.R.; Walker, K.A. Ionic liquid biodegradability depends on specific wastewater microbial consortia. Chemosphere 2015, 136, 160–166. [Google Scholar] [CrossRef]
- Kusumahastuti, D.K.; Sihtmäe, M.; Kapitanov, I.V.; Karpichev, Y.; Gathergood, N.; Kahru, A. Toxicity profiling of 24 l-phenylalanine derived ionic liquids based on pyridinium, imidazolium and cholinium cations and varying alkyl chains using rapid screening Vibrio fischeri bioassay. Ecotoxicol. Environ. Saf. 2019, 172, 556–565. [Google Scholar] [CrossRef]
- Pinazo, A.; Manresa, M.; Marques, A.; Bustelo, M.; Espuny, M.; Pérez, L. Amino acid–based surfactants: New antimicrobial agents. Adv. Colloid Interface Sci. 2016, 228, 17–39. [Google Scholar] [CrossRef]
- Mero, A.; Mezzetta, A.; Nowicki, J.; Łuczak, J.; Guazzelli, L. Betaine and l-carnitine ester bromides: Synthesis and comparative study of their thermal behaviour and surface activity. J. Mol. Liq. 2021, 334, 115988. [Google Scholar] [CrossRef]
- Mezzetta, A.; Łuczak, J.; Woch, J.; Chiappe, C.; Nowicki, J.; Guazzelli, L. Surface active fatty acid ILs: Influence of the hydrophobic tail and/or the imidazolium hydroxyl functionalization on aggregates formation. J. Mol. Liq. 2019, 289, 111155. [Google Scholar] [CrossRef]
- Rantamäki, A.H.; Ruokonen, S.-K.; Sklavounos, E.; Kyllönen, L.; King, A.W.T.; Wiedmer, S.K. Impact of Surface-Active Guanidinium-, Tetramethylguanidinium-, and Cholinium-Based Ionic Liquids on Vibrio Fischeri Cells and Dipalmitoylphosphatidylcholine Liposomes. Sci. Rep. 2017, 7, 46673. [Google Scholar] [CrossRef]
- Jordan, A.; Haiß, A.; Spulak, M.; Karpichev, Y.; Kümmerer, K.; Gathergood, N. Synthesis of a series of amino acid derived ionic liquids and tertiary amines: Green chemistry metrics including microbial toxicity and preliminary biodegradation data analysis. Green Chem. 2016, 18, 4374–4392. [Google Scholar] [CrossRef]
- Kapitanov, I.V.; Raba, G.; Špulák, M.; Vilu, R.; Karpichev, Y.; Gathergood, N. Design of sustainable ionic liquids based on l-phenylalanine and l-alanine dipeptides: Synthesis, toxicity and biodegradation studies. J. Mol. Liq. 2023, 374, 121285. [Google Scholar] [CrossRef]
- Haiß, A.; Jordan, A.; Westphal, J.; Logunova, E.; Gathergood, N.; Kümmerer, K. On the way to greener ionic liquids: Identification of a fully mineralizable phenylalanine-based ionic liquid. Green Chem. 2016, 18, 4361–4373. [Google Scholar] [CrossRef]
- OECD. OECD Test Guidelines for Chemicals; OECD: Paris, France, 1992. [Google Scholar]
- Friedrich, J.; Längin, A.; Kümmerer, K. Comparison of an Electrochemical and Luminescence-Based Oxygen Measuring System for Use in the Biodegradability Testing According to Closed Bottle Test (OECD 301D). Clean 2013, 41, 251–257. [Google Scholar] [CrossRef]
- Zubareva, T.M.; Anikeev, A.V.; Karpichev, E.A.; Red’ko, A.N.; Prokop’eva, T.M.; Popov, A.F. Cleavable dicationic surfactant micellar system for the decomposition of organophosphorus compounds. Theor. Exp. Chem. 2012, 47, 377–383. [Google Scholar] [CrossRef]
- Tehrani-Bagha, A.; Holmberg, K. Cleavable surfactants. Curr. Opin. Colloid Interface Sci. 2007, 12, 81–91. [Google Scholar] [CrossRef]
- Stolte, S.; Steudte, S.; Areitioaurtena, O.; Pagano, F.; Thöming, J.; Stepnowski, P.; Igartua, A. Ionic liquids as lubricants or lubrication additives: An ecotoxicity and biodegradability assessment. Chemosphere 2012, 89, 1135–1141. [Google Scholar] [CrossRef]
- Stolte, S.; Steudte, S.; Igartua, A.; Stepnowski, P. The Biodegradation of Ionic Liquids-the View from a Chemical Structure Perspective. Curr. Org. Chem. 2011, 15, 1946–1973. [Google Scholar] [CrossRef]
- Kapitanov, I.V.; Jordan, A.; Karpichev, Y.; Spulak, M.; Perez, L.; Kellett, A.; Kümmerer, K.; Gathergood, N. Synthesis, self-assembly, bacterial and fungal toxicity, and preliminary biodegradation studies of a series ofl-phenylalanine-derived surface-active ionic liquids. Green Chem. 2019, 21, 1777–1794. [Google Scholar] [CrossRef]
- Coleman, D.; Gathergood, N. Biodegradation studies of ionic liquids. Chem. Soc. Rev. 2010, 39, 600–637. [Google Scholar] [CrossRef]
- Gathergood, N.; Garcia, M.T.; Scammells, P.J. Biodegradable ionic liquids: Part I. Concept, preliminary targets and evaluation. Green Chem. 2004, 6, 166–175. [Google Scholar] [CrossRef]
- Garcia, M.T.; Gathergood, N.; Scammells, P.J. Biodegradable ionic liquids: Part II. Effect of the anion and toxicology. Green Chem. 2005, 7, 9–14. [Google Scholar] [CrossRef]
- Gathergood, N.; Scammells, P.J.; Garcia, M.T. Biodegradable ionic liquids: Part III. The first readily biodegradable ionic liquids. Green Chem. 2006, 8, 156–160. [Google Scholar] [CrossRef]
- Sharma, K.; Kumar, P.; Sharma, J.; Thapa, S.D.; Gupta, A.; Rajak, R.; Baruah, B.; Prakash, A.; Ranjan, R.K. Characterization of Polycyclic Aromatic Hydrocarbons (PAHs) associated with fine aerosols in ambient atmosphere of high-altitude urban environment in Sikkim Himalaya. Sci. Total Environ. 2023, 870, 161987. [Google Scholar] [CrossRef]
- Serdyuk, A.A.; Mirgorodskaya, A.B.; Kapitanov, I.V.; Gathergood, N.; Zakharova, L.Y.; Sinyashin, O.G.; Karpichev, Y. Effect of structure of polycyclic aromatic substrates on solubilization capacity and size of cationic monomeric and gemini 14-s-14 surfactant aggregates. Colloids Surf. A Physicochem. Eng. Asp. 2016, 509, 613–622. [Google Scholar] [CrossRef]
- Seitkalieva, M.M.; Kashin, A.S.; Egorova, K.S.; Ananikov, V.P. Ionic Liquids As Tunable Toxicity Storage Media for Sustainable Chemical Waste Management. ACS Sustain. Chem. Eng. 2018, 6, 719–726. [Google Scholar] [CrossRef]
- Holmberg, K. (Ed.) Handbook of Applied Surface and Colloid Chemistry; John Wiley & Sons: Chichester, UK, 2002. [Google Scholar]
- Zakharova, L.Y.; Serdyuk, A.A.; Mirgorodskaya, A.B.; Kapitanov, I.V.; Gainanova, G.A.; Karpichev, Y.; Gavrilova, E.L.; Sinyashin, O.G. Amino Acid-Functionalized Calix [4] Resorcinarene Solubilization by Mono- and Dicationic Surfactants. J. Surfactants Deterg. 2016, 19, 493–499. [Google Scholar] [CrossRef]
- Mirgorodskaya, A.B.; Karpichev, Y.; Zakharova, L.Y.; Yackevich, E.I.; Kapitanov, I.V.; Lukashenko, S.S.; Popov, A.F.; Konovalov, A.I. Aggregation behavior and interface properties of mixed surfactant systems gemini 14-s-14/CTABr. Colloids Surf. A Physicochem. Eng. Asp. 2014, 457, 425–432. [Google Scholar] [CrossRef]
- Konopka, A.; Zakharova, T.; Oliver, L.; Turco, R. Microbial biodegradation of organic wastes containing surfactants in a continuous-flow reactor. J. Ind. Microbiol. Biotechnol. 1997, 18, 235–240. [Google Scholar] [CrossRef]
- Sudheer, S.; Raba, G.; Kapitanov, I.; Karpichev, Y.; Gupta, V.K.; Vilu, R.; Gathergood, N. A Greener Approach to Hydrolyse Ionic Liquids. Basic Clin. Pharmacol. Toxicol. 2018, 124, 21. [Google Scholar]
- Ray, S.S. Environmentally friendly polymer matrices for composites. In Environmentally Friendly Polymer Nanocomposites; Elsevier: Amsterdam, The Netherlands, 2013; pp. 25–40. [Google Scholar] [CrossRef]
- Neumann, J.; Steudte, S.; Cho, C.-W.; Thöming, J.; Stolte, S. Biodegradability of 27 pyrrolidinium, morpholinium, piperidinium, imidazolium and pyridinium ionic liquid cations under aerobic conditions. Green Chem. 2014, 16, 2174–2184. [Google Scholar] [CrossRef]
- Kobayashi, M.; Shimizu, S. Metalloenzyme nitrile hydratase: Structure, regulation, and application to biotechnology. Nat. Biotechnol. 1998, 16, 733–736. [Google Scholar] [CrossRef] [PubMed]
- Mascharak, P.K. Structural and functional models of nitrile hydratase. Coord. Chem. Rev. 2002, 225, 201–214. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Salvesen, G. (Eds.) Handbook of Proteolytic Enzymes, 3rd ed.; Academic Press: Cambridge, MA, USA, 2013. [Google Scholar]
- Tomihata, K.; Ikada, Y. In vitro and in vivo degradation of films of chitin and its deacetylated derivatives. Biomaterials 1997, 18, 567–575. [Google Scholar] [CrossRef]
- Klebe, G. Inhibitors of Hydrolases with an Acyl–Enzyme Intermediate. In Drug Design; Springer: Berlin/Heidelberg, Germany, 2013; pp. 493–532. [Google Scholar] [CrossRef]
- Korhonen, H.; Pihlanto, A. Bioactive peptides: Production and functionality. Int. Dairy J. 2006, 16, 945–960. [Google Scholar] [CrossRef]
- López-Otín, C.; Bond, J.S. Proteases: Multifunctional Enzymes in Life and Disease. J. Biol. Chem. 2008, 283, 30433–30437. [Google Scholar] [CrossRef] [PubMed]
- Ouellette, R.J.; Rawn, J.D. Principles of Organic Chemistry; Elsevier: Amsterdam, The Netherlands, 2015. [Google Scholar] [CrossRef]
- Areekijseree, M.; Engkagul, A.; Kovitvadhi, U.; Thongpan, A.; Mingmuang, M.; Pakkong, P.; Rungruangsak-Torrissen, K. Temperature and pH characteristics of amylase and proteinase of adult freshwater pearl mussel, Hyriopsis (Hyriopsis) bialatus Simpson 1900. Aquaculture 2004, 234, 575–587. [Google Scholar] [CrossRef]
- Azevedo, H.S.; Reis, R.L. Understanding the Enzymatic Degradation of Biodegradable Polymers and Strategies to Control Their Degradation Rate. In Biodegradable Systems in Tissue Engineering and Regenerative Medicine; CRC Press: Boca Raton, FL, USA, 2004. [Google Scholar] [CrossRef]
- Bender, M.L.; Ginger, R.D.; Unik, J.P. Activation Energies of the Hydrolysis of Esters and Amides Involving Carbonyl Oxygen Exchange 1. J. Am. Chem. Soc. 1958, 80, 1044–1048. [Google Scholar] [CrossRef]
- Jencks, W.P.; Carriuolo, J. Reactivity of Nucleophilic Reagents toward Esters. J. Am. Chem. Soc. 1960, 82, 1778–1786. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).