Optical Polarization-Based Measurement Methods for Characterization of Self-Assembled Peptides’ and Amino Acids’ Micro- and Nanostructures
Abstract
:1. Introduction
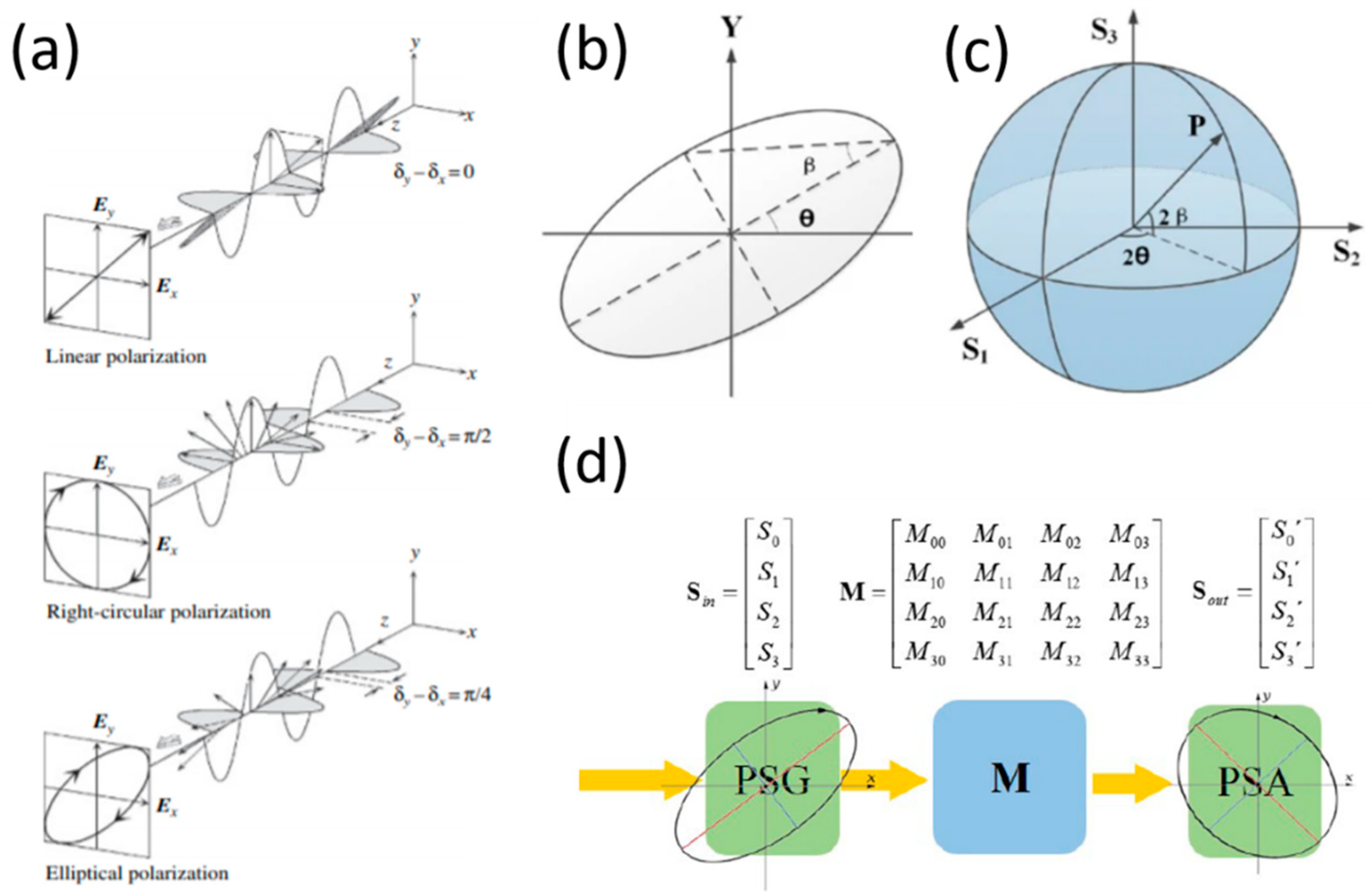
2. Key Concepts in Optical Polarization
3. Polarization-Based Measurement Methods for Characterization of Peptides and Amino Acids
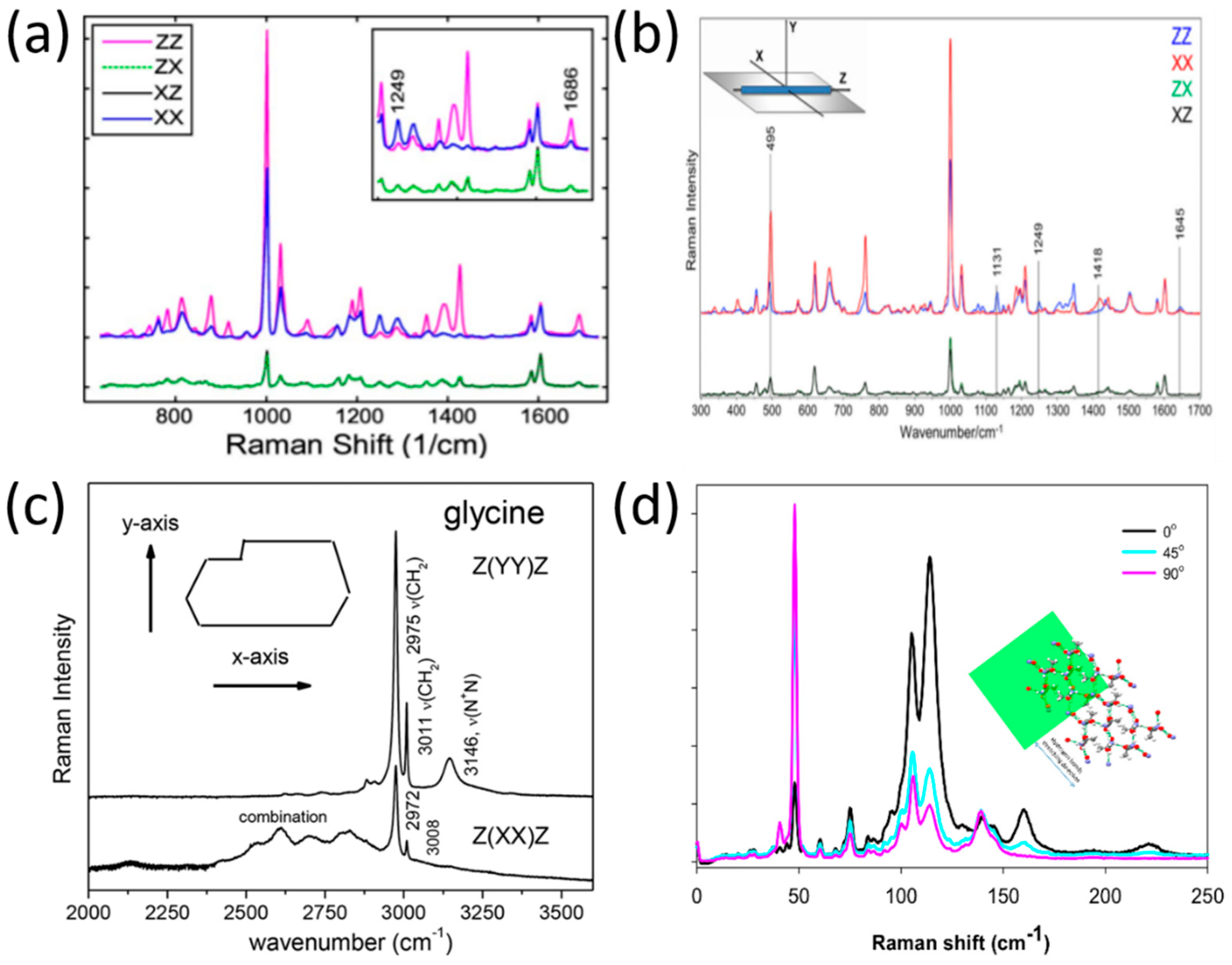
3.1. Polarized Raman Spectroscopy
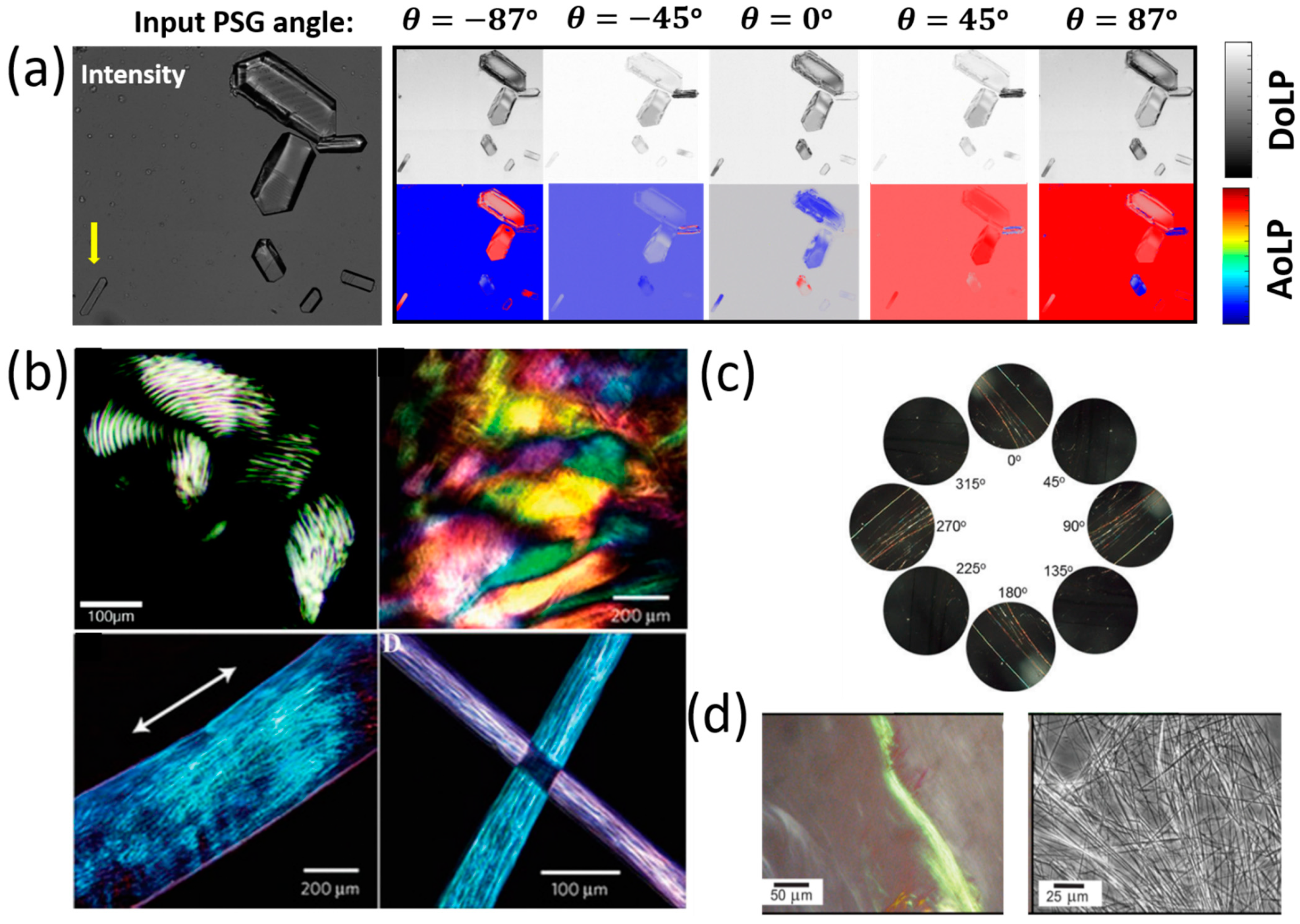
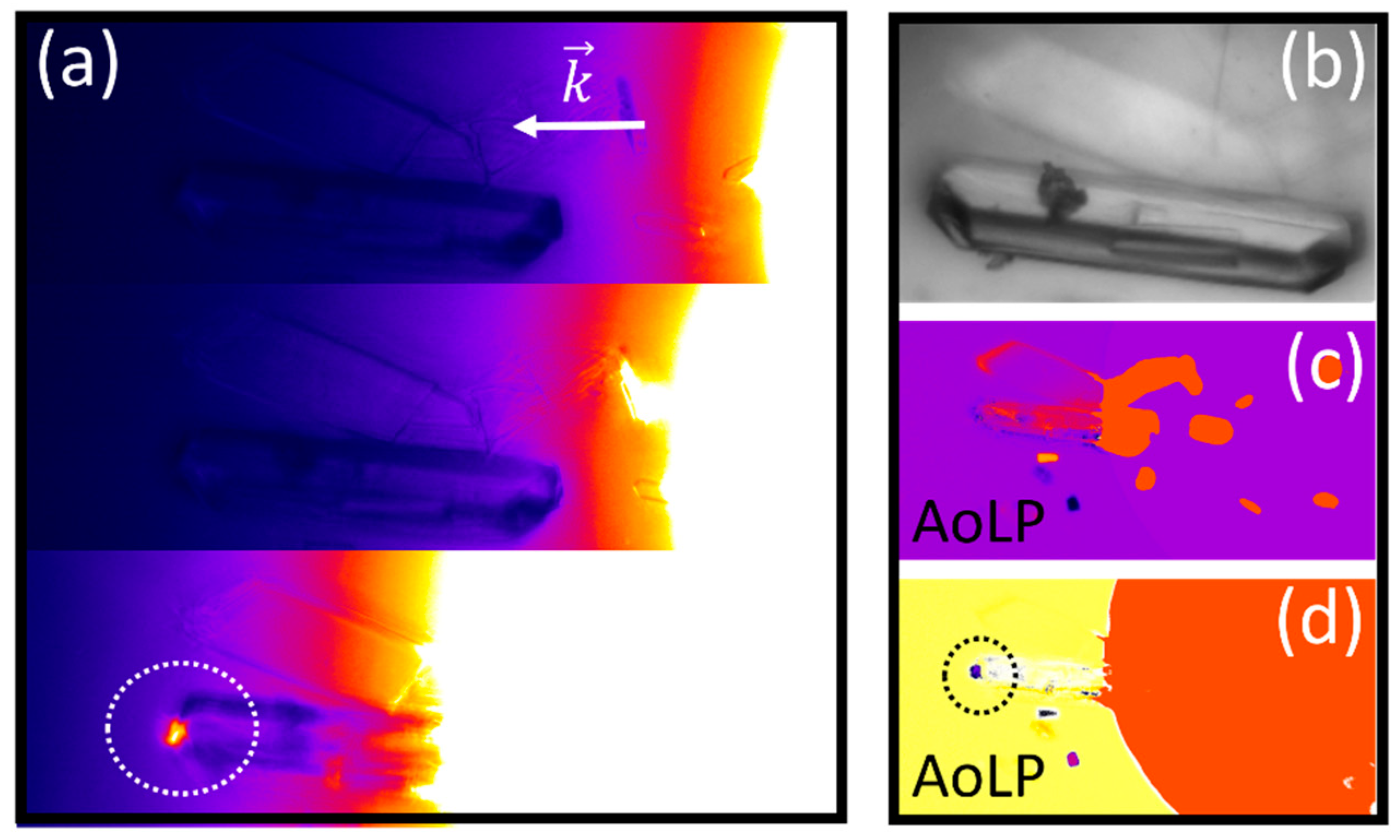
3.2. Polarized Imaging and Birefringence Monitoring
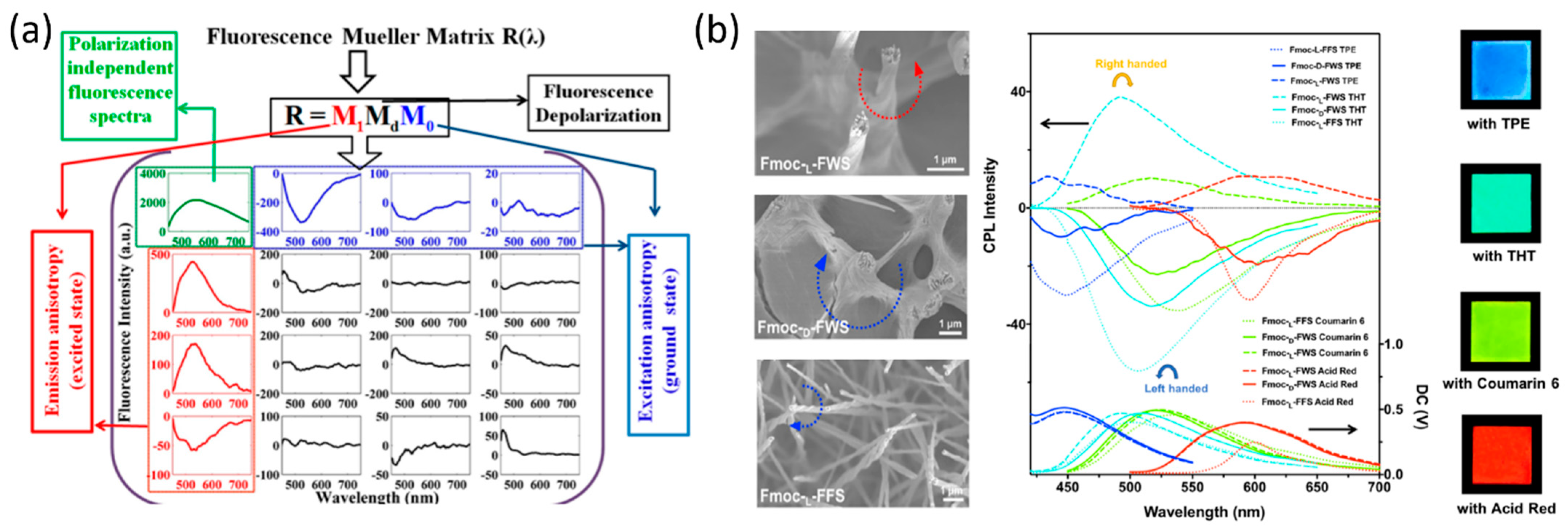
3.3. Polarization and Fluorescence
4. Polarized Waveguiding in Self-Assembled Amino Acid Microstructures
5. Final Remarks and Future Directions
Funding
Conflicts of Interest
References
- Xiong, R.; Luan, J.; Kang, S.; Ye, C.; Singamaneni, S.; Tsukruk, V.V. Vladimir, Biopolymeric photonic structures: Design, fabrication, and emerging applications. Chem. Soc. Rev. 2020, 49, 983–1031. [Google Scholar] [CrossRef] [PubMed]
- Clark, J.; Lanzani, G. Organic photonics for communications. Nat. Photonics 2010, 4, 438–446. [Google Scholar] [CrossRef]
- Chandrasekar, R. Organic photonics: Prospective nano/micro scale passive organic optical waveguides obtained from π-conjugated ligand molecules. Phys. Chem. Chem. Phys. 2014, 16, 7173–7183. [Google Scholar] [CrossRef] [PubMed]
- Mendes, A.C.L.; Baran, E.T.; Reis, R.L.; Azevedo, H.S. Self-assembly in nature: Using the principles of nature to create complex nanobiomaterials. WIREs Nanomed. Nanobiotechnol. 2013, 5, 582–612. [Google Scholar] [CrossRef]
- Stupp, S.I.; LeBonheur, V.; Walker, K.; Li, L.S.; Huggins, K.E.; Keser, M.; Amstutz, A. Supramolecular Materials: Self-Organized Nanostructures. Science 1997, 276, 384–389. [Google Scholar] [CrossRef]
- Lehn, J.M. Perspectives in supramolecular chemistry—From molecular recognition towards molecular information processing and self-organization. Angew. Chem. Int. Ed. Engl. 1990, 29, 1304–1319. [Google Scholar] [CrossRef]
- Gazit, E. Self-Assembled Peptide Nanostructures: The Design of Molecular Building Blocks and Their Technological Utilizatio. Chem. Soc. Rev. 2007, 36, 1263–1269. [Google Scholar] [CrossRef]
- Zhang, S. Fabrication of novel biomaterials through molecular self-assembly. Nat. Biotechnol. 2003, 21, 1171–1178. [Google Scholar] [CrossRef]
- Gao, X.; Matsui, H. Peptide-Based Nanotubes and Their Applications in Bionanotechnology. Adv. Mater. 2005, 17, 2037–2050. [Google Scholar] [CrossRef]
- d’Orlyé, F.; Trapiella-Alfonso, L.; Lescot, C.; Pinvidic, M.; Doan, B.-T.; Varenne, A. Synthesis, Characterization and Evaluation of Peptide Nanostructures for Biomedical Applications. Molecules 2021, 26, 4587. [Google Scholar] [CrossRef]
- Gatto, E.; Toniolo, C.; Venanzi, M. Peptide Self-Assembled Nanostructures: From Models to Therapeutic Peptides. Nanomaterials 2022, 12, 466. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.M.; Williams, R.J.; Tang, C.; Coppo, P.; Collins, R.F.; Turner, M.L.; Saiani, A.; Ulijn, R.V. Fmoc-Diphenylalanine Self Assembles to a Hydrogel via a Novel Architecture Based on π–π Interlocked β-Sheets. Adv. Mater. 2008, 20, 37–41. [Google Scholar] [CrossRef]
- Levin, A.; Mason, T.O.; Adler-Abramovich, L.; Buell, A.K.; Meisl, G.; Galvagnion, C.; Bram, Y.; Stratford, S.A.; Dobson, C.M.; Knowles, T.; et al. Ostwald’s rule of stages governs structural transitions and morphology of dipeptide supramolecular polymers. Nat. Commun. 2014, 5, 5219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Kwon, S.; Kim, S.H.; Lee, C.-K.; Lee, J.-H.; Cho, S.J.; Lee, H.-S.; Ihee, H. Microtubes with Rectangular Cross-Section by Self-Assembly of a Short β-Peptide Foldamer. J. Am. Chem. Soc. 2012, 134, 20573–20576. [Google Scholar] [CrossRef]
- Koley, P.; Pramanik, A. Multilayer Vesicles, Tubes, Various Porous Structures and Organo Gels through the Solvent-Assisted Self-Assembly of Two Modified Tripeptides and Their Different Applications. Soft Matter 2012, 8, 5364–5374. [Google Scholar] [CrossRef]
- Yuran, S.; Razvag, Y.; Reches, M. Coassembly of Aromatic Dipeptides into Biomolecular Necklaces. ACS Nano 2012, 6, 9559–9566. [Google Scholar] [CrossRef]
- Ghadiri, M.; Grania, J.; Milligan, R.; McRee, D.; Khazanovitch, N. Self-assembling Organic Nanotubes Based On a Cyclic Peptide Architecture. Nature 1993, 366, 324–327. [Google Scholar] [CrossRef]
- Qi, G.-B.; Gao, Y.-J.; Wang, L.; Wang, H. Self-Assembled Peptide-Based Nanomaterials for Biomedical Imaging and Therapy. Adv. Mater. 2018, 30, 1703444. [Google Scholar] [CrossRef]
- Habibi, N.; Kamaly, N.; Memic, A.; Shafiee, H. Self-assembled peptide-based nanostructures: Smart nanomaterials toward targeted drug delivery. Nano Today 2016, 11, 41–60. [Google Scholar] [CrossRef] [Green Version]
- Tao, K.; Makam, P.; Aizen, R.; Gazit, E. Self-assembling peptide semiconductors. Science 2017, 358, eaam9756. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Lu, X.-M.; Zhang, M.-R.; Hu, K.; Li, Z. Peptide-based nanomaterials: Self-assembly, properties and applications. Bioact. Mater. 2021, 11, 268–282. [Google Scholar] [CrossRef] [PubMed]
- Abramovich, L.A.; Gazit, E. The physical properties of supramolecular peptide assemblies: From building block association to technological applications. Chem. Soc. Rev. 2014, 43, 6881–6893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Handelman, A.; Kuritz, N.; Natan, A.; Rosenman, G. Reconstructive Phase Transition in Ultrashort Peptide Nanostructures and Induced Visible Photoluminescence. Langmuir 2015, 32, 2847–2862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamley, I.W. Peptide Nanotubes. Angew. Chem. Int. Ed. 2014, 53, 6866–6881. [Google Scholar] [CrossRef]
- Ikezoe, Y.; Washino, G.; Uemura, T.; Kitagawa, S.; Matsui, H. Autonomous motors of a metal–organic framework powered by reorganization of self-assembled peptides at interfaces. Nat. Mater. 2012, 11, 1081–1085. [Google Scholar] [CrossRef] [Green Version]
- Adler-Abramovich, L.; Aronov, D.; Beker, P.; Yevnin, M.; Stempler, S.; Buzhansky, L.; I Rosenman, G.; Gazit, E. Self-assembled arrays of peptide nanotubes by vapour deposition. Nat. Nanotechnol. 2009, 4, 849–854. [Google Scholar] [CrossRef] [PubMed]
- Berger, O.; Adler-Abramovich, L.; Levy-Sakin, M.; Grunwald, A.; Liebes-Peer, Y.; Bachar, M.; Buzhansky, L.; Mossou, E.; Forsyth, V.T.; Schwartz, T.; et al. Light-emitting self-assembled peptide nucleic acids exhibit both stacking interactions and Watson–Crick base pairing. Nat. Nanotechnol. 2015, 10, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.J.; Brat, G.A.; Medina, S.H.; Tong, D.; Huang, Y.; Grahammer, J.; Furtmüller, G.J.; Oh, B.C.; Nagy-Smith, K.J.; Walczak, P.; et al. A multiphase transitioning peptide hydrogel for suturing ultrasmall vessels. Nat. Nanotechnol. 2015, 11, 95–102. [Google Scholar] [CrossRef]
- Hu, K.; Xiong, W.; Li, H.; Zhang, P.-Y.; Yin, F.; Zhang, Q.; Geng, H.; Jiang, F.; Li, Z. Tuning peptide self-assembly by an in-tether chiral center. Sci. Adv. 2018, 4, eaar5907. [Google Scholar] [CrossRef] [Green Version]
- Hu, K.; Xiong, W.; Sun, C.; Li, J.; Yin, F.; Jiang, Y.; Zhang, M.-R.; Wang, X.; Li, Z. Self-Assembly of Constrained Cyclic Peptides Controlled by Ring Size. CCS Chem. 2020, 2, 42–51. [Google Scholar] [CrossRef]
- Hofmann, A.; Schmid, M.; Brütting, W. The Many Facets of Molecular Orientation in Organic Optoelectronics. Adv. Optical Mater. 2021, 9, 2101004. [Google Scholar] [CrossRef]
- Lafrance, C.-P.; Nabet, A.; Prud’homme, R.E.; Pézolet, M. On the relationship between the order parameter and the shape of orientation distributions. Can. J. Chem. 1995, 73, 1497–1505. [Google Scholar] [CrossRef]
- Tanaka, M.; Young, R.J. Review Polarised Raman spectroscopy for the study of molecular orientation distributions in polymers. J. Mater. Sci. 2006, 41, 963–991. [Google Scholar] [CrossRef]
- Mammadov, R.; Tekinay, A.B.; Dana, A.; Guler, M.O. Microscopic characterization of peptide nanostructures. Micron 2012, 43, 69–84. [Google Scholar] [CrossRef]
- Reches, M.; Gazit, E. Controlled patterning of aligned self-assembled peptide nanotubes. Nat. Nanotechnol. 2006, 1, 195–200. [Google Scholar] [CrossRef]
- Collett, E. Field Guide to Polarization; SPIE-Press: Bellingham, WA, USA, 2005. [Google Scholar]
- Fujiwara, H. Spectroscopic Ellipsometry: Principles and Applications; John Wiley & Sons: West Sussex, UK, 2007. [Google Scholar]
- Goldstein, D.H. Polarized Light, 3rd ed.; Taylor & Francis Group: Boca Raton, FL, USA, 2011. [Google Scholar]
- Sharma, K.K. Optics Principles and Applications; Elsevier Inc.: Burlington, MA, USA, 2006. [Google Scholar]
- Sun, X.; Geng, Y.; Zhu, Q.; Huang, W.; Zhang, Y.; Wang, W.; Liu, L. Unitary transformation for Poincaré beams on different parts of Poincaré sphere. Sci. Rep. 2020, 10, 14251. [Google Scholar] [CrossRef]
- Qi, J.; Elson, D.S. Mueller polarimetric imaging for surgical and diagnostic applications: A review. J. Biophotonics 2017, 10, 950–982. [Google Scholar] [CrossRef] [Green Version]
- Arteaga, O.; Kahr, B. Mueller matrix polarimetry of bianisotropic materials [Invited]. J. Opt. Soc. Am. B 2019, 36, F72–F83. [Google Scholar] [CrossRef]
- Jones, R.R.; Hooper, D.C.; Zhang, L.; Wolverson, D.; Valev, V.K. Raman Techniques: Fundamentals and Frontiers. Nanoscale Res. Lett. 2019, 14, 231. [Google Scholar] [CrossRef] [Green Version]
- Sourisseau, C. Polarization Measurements in Macro- and Micro-Raman Spectroscopies: Molecular Orientations in Thin Films and Azo-Dye Containing Polymer Systems. Chem. Rev. 2004, 104, 3851–3892. [Google Scholar] [CrossRef]
- Nielsen, A.S.; Pyrz, R. A novel approach to measure local strains in polymer matrix systems using polarised Raman microscopy. Comp. Sci. Technol. 2002, 62, 2219–2227. [Google Scholar] [CrossRef]
- Lekprasert, B.; Sedman, V.; Roberts, C.J.; Tedler, S.J.B.; Notingher, I. Nondestructive Raman and atomic force microscopy measurement of molecular structure for individual diphenylalanine nanotubes. Opt. Lett. 2010, 35, 4193–4195. [Google Scholar] [CrossRef] [PubMed]
- Sereda, V.; Ralbovsky, N.M.; Vasudev, M.C.; Naik, R.R.; Lednev, I.K. Polarized Raman Spectroscopy for Determining the Orientation of di-D-phenylalanine Molecules in a Nanotube. J. Raman Spectrosc. 2016, 47, 1056–1062. [Google Scholar] [CrossRef] [PubMed]
- Freire, P.T.; Barboza, F.M.; Lima, J.A.; Melo, F.E.; Filho, J.M. Raman Spectroscopy of Amino Acid Crystals. In Raman Spectroscopy and Applications [Internet]; Maaz, K., Ed.; IntechOpen: London, UK, 2017; Available online: https://www.intechopen.com/chapters/52839 (accessed on 8 February 2022).
- Nemtsov, I.; Aviv, H.; Mastai, Y.; Tischler, Y.R. Polarization Dependence of Low-Frequency Vibrations from Multiple Faces in an Organic Single Crystal. Crystals 2019, 9, 425. [Google Scholar] [CrossRef] [Green Version]
- Murphy, D.B.; Davidson, M.W. Fundamentals of Light Microscopy and Electronic Imaging, 2nd ed.; Wiley-Blackwel: Hoboken, NJ, USA, 2013. [Google Scholar]
- Tuchin, V.V. Polarized light interaction with tissues. J. Biomed. Opt. 2016, 21, 071114. [Google Scholar] [CrossRef] [Green Version]
- Le Gratiet, A.; Mohebi, A.; Callegari, F.; Bianchini, P.; Diaspro, A. Review on Complete Mueller Matrix Optical Scanning Microscopy Imaging. Appl. Sci. 2021, 11, 1632. [Google Scholar] [CrossRef]
- Gottlieb, D.; Arteaga, O. Mueller matrix imaging with a polarization camera: Application to microscopy. Opt. Express 2021, 29, 34723. [Google Scholar] [CrossRef]
- Cook, C.; Byrne, B.; d’Aubigny, C.D.; Viola, D.; Mikucki, J.; Ellis, W. Detection Limits for Chiral Amino Acids Using a Polarization Camera. Planet. Sci. J. 2020, 1, 46. [Google Scholar] [CrossRef]
- Wantha, L.; Flood, A.E. Growth and Dissolution Kinetics of A and B Polymorphs of L-Histidine. Chem. Eng. Technol. 2015, 38, 1022–1028. [Google Scholar] [CrossRef]
- Handelman, A.; Lapsker, I.; Jacob, A.; Laikhtman, A. Passive Polarized Light Guiding and Thermally Induced Visible Fluorescence in Histidine Microstructures with Optical Switching Function. Adv. Funct. Mater. 2021, 31, 2008183. [Google Scholar] [CrossRef]
- Zhang, S.; Greenfield, M.A.; Mata, A.; Palmer, L.; Bitton, R.; Mantei, J.R.; Aparicio, C.; de la Cruz, M.O.; Stupp, S.I. A self-assembly pathway to aligned monodomain gels. Nat. Mater. 2010, 9, 594–601. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Fei, J.; Wang, A.; Cui, W.; Zhu, P.; Li, L. Transformation of Dipeptide-Based Organogels into Chiral Crystals by Cryogenic Treatment. Angew. Chem. Int. Ed. 2017, 56, 2660. [Google Scholar] [CrossRef] [PubMed]
- Hamley, I.W. Lipopeptides: From self-assembly to bioactivity. Chem. Commun. 2015, 51, 8574–8583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, G.; Castelletto, V.; Jones, R.R.; Connon, C.J.; Hamley, I.W. Hydrogelation of self-assembling RGD-based peptides. Soft Matter 2010, 7, 1326–1333. [Google Scholar] [CrossRef]
- Apter, B.; Lapshina, N.; Lapsker, I.; Handelman, A.; Accardo, A.; Diaferia, C.; Morelli, G.; Rosenman, G. Fold-Sensitive Visible Fluorescence in β-Sheet Peptide Structures. Adv. Optical Mater. 2021, 9, 2002247. [Google Scholar] [CrossRef]
- Song, J.; Xing, R.; Jiao, T.; Peng, Q.; Yuan, C.; Möhwald, H.; Yan, X. Crystalline Dipeptide Nanobelts Based on Solid–Solid Phase Transformation Self-Assembly and Their Polarization Imaging of Cells. ACS Appl. Mater. Interfaces 2018, 10, 2368–2376. [Google Scholar] [CrossRef]
- Zhang, S.; Cortes, W.; Sasaki, T.; Asahina, S.; Natsuko, A.; Zhang, Q.; Zhang, Y. Mechanoresponsive Alignment of Molecular Self-Assembled Negatively Charged Nanofibrils. ACS Appl. Bio Mater. 2020, 3, 1698–1704. [Google Scholar] [CrossRef]
- Valeur, B. Molecular Fluorescence: Principles and Applications; Wiley-VCH Verlag GmbH: Weinheim, Germany, 2001. [Google Scholar]
- Chen, L.; Chen, X.; Yang, X.; He, C.; Wang, M.; Xi, P.; Gao, J. Advances of super-resolution fluorescence polarization microscopy and its applications in life sciences, Comp. Struct. Biotechnol. J. 2020, 18, 2209–2216. [Google Scholar]
- Mazumder, N.; Qiu, J.; Kao, F.-J.; Diaspro, A. Mueller matrix signature in advanced fluorescence microscopy imaging. J. Opt. 2017, 19, 025301. [Google Scholar] [CrossRef]
- Oura, M.; Yamamoto, J.; Ishikawa, H.; Mikuni, S.; Fukushima, R.; Kinjo, M. Polarization-dependent fluorescence correlation spectroscopy for studying structural properties of proteins in living cell. Sci. Rep. 2016, 6, 31091. [Google Scholar] [CrossRef] [Green Version]
- Lee, A.Y.S.; Bayly, A.; Lin, M.W. Evaluation of Polarized Light and Fluorescence Microscopy of Congo Red Stain in the Diagnosis of Renal Amyloidosis. Lab Med. 2021, 52, 574–577. [Google Scholar] [CrossRef] [PubMed]
- Duboisset, J.; Ferrand, P.; He, W.; Wang, X.; Rigneault, H.; Brasselet, S. Thioflavine-T and Congo Red Reveal the Polymorphism of Insulin Amyloid Fibrils When Probed by Polarization-Resolved Fluorescence Microscopy. J. Phys. Chem. B 2013, 117, 784–788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maji, K.; Saha, S.; Dey, R.; Ghosh, N.; Haldar, D. Mueller Matrix Fluorescence Spectroscopy for Probing Self-Assembled Peptide-Based Hybrid Supramolecular Structure and Orientation. J. Phys. Chem. C 2017, 121, 19519–19529. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, J.; Wang, Y.; Zhang, G.; Qi, W.; You, S.; Su, R.; He, Z. Self-Assembly of Peptide Hierarchical Helical Arrays with Sequence-Encoded Circularly Polarized Luminescence. Nano Lett. 2021, 21, 6406–6415. [Google Scholar] [CrossRef]
- Carr, R.; Evans, N.H.; Parker, D. Lanthanide complexes as chiral probes exploiting circularly polarized luminescence. Chem. Soc. Rev. 2012, 41, 7673–7686. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.-W.; Li, M.; Chen, C.-F. Recent advances in circularly polarized electroluminescence based on organic light-emitting diodes. Chem. Soc. Rev. 2020, 49, 1331–1343. [Google Scholar] [CrossRef]
- Ji, L.; Zhao, Y.; Tao, M.; Wang, H.-X.; Niu, D.; Ouyang, G.; Xia, A.; Liu, M. Dimension-Tunable Circularly Polarized Luminescent Nanoassemblies with Emerging Selective Chirality and Energy Transfer. ACS Nano 2020, 14, 2373–2384. [Google Scholar] [CrossRef]
- Fei, J.; Dai, L.; Gao, F.; Zhao, J.; Li, J. Assembled Vitamin B2 Nanocrystals with Optical Waveguiding and Photosensitizing Properties for Potential Biomedical Application. Angew. Chem. Int. Ed. 2019, 58, 7254–7258. [Google Scholar] [CrossRef]
- Wang, J.; Dong, J. Optical Waveguides and Integrated Optical Devices for Medical Diagnosis, Health Monitoring and Light Therapies. Sensors 2020, 20, 3981. [Google Scholar] [CrossRef]
- Qiao, X.; Qian, Z.; Li, J.; Sun, H.; Han, Y.; Xia, X.; Zhou, J.; Wang, C.; Wang, Y.; Wang, C. Synthetic Engineering of Spider Silk Fiber as Implantable Optical Waveguides for Low-Loss Light Guiding. ACS Appl. Mater. Interfaces 2017, 9, 14665–14676. [Google Scholar] [CrossRef]
- Humar, M.; Kwok, S.J.J.; Choi, M.; Yetisen, A.K.; Cho, S.; Yun, S.-H. Toward biomaterial-based implantable photonic devices. Nanophotonics 2017, 6, 414–434. [Google Scholar] [CrossRef]
- Lee, K.-L.; He, K.-Y. Effect of Micro-Structural Light Guide Plate on Source of Linearly Polarized Light. J. Light. Technol. 2011, 29, 3327–3330. [Google Scholar] [CrossRef]
- Yang, X.; Yan, Y.; Jin, G. Polarized light-guide plate for liquid crystal display. Opt. Express 2005, 13, 8349–8356. [Google Scholar] [CrossRef] [PubMed]
- Syu, S.-C.; Cheng, C.-H.; Wang, H.-Y.; Chi, Y.-C.; Wu, C.-I.; Lin, G.-R. Realizing multi-functional all-optical data processing on nanoscale SiC waveguides. Sci. Rep. 2018, 8, 14859. [Google Scholar] [CrossRef]
- Chen, S.; Zhuo, M.-P.; Wang, X.-D.; Wei, G.-Q.; Liao, L.-S. Optical waveguides based on one-dimensional organic crystals. PhotoniX 2021, 2, 1–24. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Handelman, A. Optical Polarization-Based Measurement Methods for Characterization of Self-Assembled Peptides’ and Amino Acids’ Micro- and Nanostructures. Molecules 2022, 27, 1802. https://doi.org/10.3390/molecules27061802
Handelman A. Optical Polarization-Based Measurement Methods for Characterization of Self-Assembled Peptides’ and Amino Acids’ Micro- and Nanostructures. Molecules. 2022; 27(6):1802. https://doi.org/10.3390/molecules27061802
Chicago/Turabian StyleHandelman, Amir. 2022. "Optical Polarization-Based Measurement Methods for Characterization of Self-Assembled Peptides’ and Amino Acids’ Micro- and Nanostructures" Molecules 27, no. 6: 1802. https://doi.org/10.3390/molecules27061802
APA StyleHandelman, A. (2022). Optical Polarization-Based Measurement Methods for Characterization of Self-Assembled Peptides’ and Amino Acids’ Micro- and Nanostructures. Molecules, 27(6), 1802. https://doi.org/10.3390/molecules27061802





