Self-Assembling Peptides and Carbon Nanomaterials Join Forces for Innovative Biomedical Applications
Abstract
:1. Introduction
1.1. Self-Assembling Peptides
1.2. Carbon Nanomaterials
1.3. Combination of Self-Assembling Peptides and Carbon Nanomaterials
2. Research on the Interaction between Self-Assembling Peptides and Nanocarbons
2.1. Fullerenes
2.2. Carbon Dots
2.3. Carbon Nanotubes (CNTs)
2.4. Graphene-Based Materials
2.4.1. Sensing Applications
2.4.2. Energy-Harvesting Systems for Bioelectronics
2.4.3. Biomaterials for Wound Healing and Tissue Regeneration
2.4.4. Drug Release
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Levin, A.; Hakala, T.A.; Schnaider, L.; Bernardes, G.J.L.; Gazit, E.; Knowles, T.P.J. Biomimetic peptide self-assembly for functional materials. Nat. Rev. Chem. 2020, 4, 615–634. [Google Scholar] [CrossRef]
- Lampel, A. Biology-Inspired Supramolecular Peptide Systems. Chem 2020, 6, 1222–1236. [Google Scholar] [CrossRef]
- Ding, Y.; Ting, J.P.; Liu, J.; Al-Azzam, S.; Pandya, P.; Afshar, S. Impact of non-proteinogenic amino acids in the discovery and development of peptide therapeutics. Amino Acids 2020, 52, 1207–1226. [Google Scholar] [CrossRef]
- Brittain, W.D.; Lloyd, C.M.; Cobb, S.L. Synthesis of complex unnatural fluorine-containing amino acids. J. Fluor. Chem. 2020, 239, 109630. [Google Scholar] [CrossRef]
- Jaradat, D.M.M. Thirteen decades of peptide synthesis: Key developments in solid phase peptide synthesis and amide bond formation utilized in peptide ligation. Amino Acids 2018, 50, 39–68. [Google Scholar] [CrossRef] [PubMed]
- Hamley, I.W. Small Bioactive Peptides for Biomaterials Design and Therapeutics. Chem. Rev. 2017, 117, 14015–14041. [Google Scholar] [CrossRef] [Green Version]
- Ahn, W.; Lee, J.-H.; Kim, S.R.; Lee, J.; Lee, E.J. Designed protein- and peptide-based hydrogels for biomedical sciences. J. Mater. Chem. B 2021, 9, 1919–1940. [Google Scholar] [CrossRef] [PubMed]
- Osorno, L.L.; Brandley, A.N.; Maldonado, D.E.; Yiantsos, A.; Mosley, R.J.; Byrne, M.E. Review of Contemporary Self-Assembled Systems for the Controlled Delivery of Therapeutics in Medicine. Nanomaterials 2021, 11, 278. [Google Scholar] [CrossRef] [PubMed]
- Kurbasic, M.; Parisi, E.; Garcia, A.M.; Marchesan, S. Self-Assembling, Ultrashort Peptide Gels as Antimicrobial Biomaterials. Curr. Top. Med. Chem. 2020, 20, 1300–1309. [Google Scholar] [CrossRef]
- Gao, J.; Zhan, J.; Yang, Z. Enzyme-Instructed Self-Assembly (EISA) and Hydrogelation of Peptides. Adv. Mater. 2020, 32, 1805798. [Google Scholar] [CrossRef]
- Lyu, Y.; Azevedo, H.S. Supramolecular Hydrogels for Protein Delivery in Tissue Engineering. Molecules 2021, 26, 873. [Google Scholar] [CrossRef]
- Nguyen, Q.D.; Kikuchi, K.; Maity, B.; Ueno, T. The Versatile Manipulations of Self-Assembled Proteins in Vaccine Design. Int. J. Mol. Sci. 2021, 22, 1934. [Google Scholar] [CrossRef]
- Wang, Y.; Xia, K.; Wang, L.; Wu, M.; Sang, X.; Wan, K.; Zhang, X.; Liu, X.; Wei, G. Peptide-Engineered Fluorescent Nanomaterials: Structure Design, Function Tailoring, and Biomedical Applications. Small 2021, 17, 2005578. [Google Scholar] [CrossRef] [PubMed]
- Apter, B.; Lapshina, N.; Lapsker, I.; Handelman, A.; Accardo, A.; Diaferia, C.; Morelli, G.; Rosenman, G. Fold-Sensitive Visible Fluorescence in β-Sheet Peptide Structures. Adv. Opt. Mater. 2021, 2002247. [Google Scholar] [CrossRef]
- Balasco, N.; Diaferia, C.; Morelli, G.; Vitagliano, L.; Accardo, A. Amyloid-Like Aggregation in Diseases and Biomaterials: Osmosis of Structural Information. Front. Bioeng. Biotechnol. 2021, 9, 641372. [Google Scholar] [CrossRef] [PubMed]
- Chu, B.; He, J.-M.; Liu, L.-L.; Wu, C.-X.; You, L.-L.; Li, X.-L.; Wang, S.; Chen, C.-S.; Tu, M. Proangiogenic Peptide Nanofiber Hydrogels for Wound Healing. ACS Biomater. Sci. Eng. 2021, 7, 1100–1110. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ma, B.; Wang, W. Peptide-Based Nanomaterials for Tumor Immunotherapy. Molecules 2021, 26, 132. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Zhan, W.; Liang, G. Intracellular Self-Assembly of Peptide Conjugates for Tumor Imaging and Therapy. Adv. Healthc. Mater. 2021, 10, 2001211. [Google Scholar] [CrossRef]
- Ke, P.C.; Zhou, R.; Serpell, L.C.; Riek, R.; Knowles, T.P.J.; Lashuel, H.A.; Gazit, E.; Hamley, I.W.; Davis, T.P.; Fändrich, M.; et al. Half a century of amyloids: Past, present and future. Chem. Soc. Rev. 2020, 49, 5473–5509. [Google Scholar] [CrossRef]
- Chatani, E.; Yuzu, K.; Ohhashi, Y.; Goto, Y. Current Understanding of the Structure, Stability and Dynamic Properties of Amyloid Fibrils. Int. J. Mol. Sci. 2021, 22, 4349. [Google Scholar] [CrossRef]
- Mayans, E.; Alemán, C. Revisiting the Self-Assembly of Highly Aromatic Phenylalanine Homopeptides. Molecules 2020, 25, 6037. [Google Scholar] [CrossRef] [PubMed]
- Reches, M.; Gazit, E. Casting metal nanowires within discrete self-assembled peptide nanotubes. Science 2003, 300, 625–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, X.; Zhou, J.; Shi, J.; Xu, B. Supramolecular Hydrogelators and Hydrogels: From Soft Matter to Molecular Biomaterials. Chem. Rev. 2015, 115, 13165–13307. [Google Scholar] [CrossRef]
- Das, A.K.; Gavel, P.K. Low molecular weight self-assembling peptide-based materials for cell culture, antimicrobial, anti-inflammatory, wound healing, anticancer, drug delivery, bioimaging and 3D bioprinting applications. Soft Matter 2020, 16, 10065–10095. [Google Scholar] [CrossRef]
- Mougin, J.; Bourgaux, C.; Couvreur, P. Elongated self-assembled nanocarriers: From molecular organization to therapeutic applications. Adv. Drug Deliv. Rev. 2021, 172, 127–147. [Google Scholar] [CrossRef]
- Zhang, Z.; Ai, S.; Yang, Z.; Li, X. Peptide-based supramolecular hydrogels for local drug delivery. Adv. Drug Deliv. Rev. 2021, 174, 482–503. [Google Scholar] [CrossRef]
- Diaferia, C.; Rosa, E.; Accardo, A.; Morelli, G. Peptide-based hydrogels as delivery systems for doxorubicin. J. Pept. Sci. 2021, e3301. [Google Scholar] [CrossRef]
- Karavasili, C.; Fatouros, D.G. Self-assembling peptides as vectors for local drug delivery and tissue engineering applications. Adv. Drug Deliv. Rev. 2021, 174, 387–405. [Google Scholar] [CrossRef]
- Da Silva, K.; Kumar, P.; Choonara, Y.E.; du Toit, L.C.; Pillay, V. Three-dimensional printing of extracellular matrix (ECM)-mimicking scaffolds: A critical review of the current ECM materials. J. Biomed. Mater. Res. A 2020, 108, 2324–2350. [Google Scholar] [CrossRef]
- Alzanbaki, H.; Moretti, M.; Hauser, C.A.E. Engineered Microgels-Their Manufacturing and Biomedical Applications. Micromachines 2021, 12, 45. [Google Scholar] [CrossRef] [PubMed]
- Carratalá, J.V.; Serna, N.; Villaverde, A.; Vázquez, E.; Ferrer-Miralles, N. Nanostructured antimicrobial peptides: The last push towards clinics. Biotechnol. Adv. 2020, 44, 107603. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Zhang, Z.; Sun, J.; Li, K.; Li, Y.; Ren, C.; Meng, Q.; Yang, J. Self-Assembling Peptide-Based Hydrogels in Angiogenesis. Int. J. Nanomed. 2020, 15, 10257–10269. [Google Scholar] [CrossRef] [PubMed]
- Tarvirdipour, S.; Huang, X.; Mihali, V.; Schoenenberger, C.A.; Palivan, C.G. Peptide-Based Nanoassemblies in Gene Therapy and Diagnosis: Paving the Way for Clinical Application. Molecules 2020, 25, 3482. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Díaz, L.A.; Ruiz-Pacheco, J.A.; Elsawy, M.A.; Reyes-Martínez, J.E.; Enríquez-Rodríguez, A.I. Self-Assembling Peptides as an Emerging Platform for the Treatment of Metabolic Syndrome. Int. J. Nanomed. 2020, 15, 10349–10370. [Google Scholar] [CrossRef]
- Petit, N.; Dyer, J.M.; Clerens, S.; Gerrard, J.A.; Domigan, L.J. Oral delivery of self-assembling bioactive peptides to target gastrointestinal tract disease. Food Funct. 2020, 11, 9468–9488. [Google Scholar] [CrossRef]
- Chen, C.H.; Hsu, E.L.; Stupp, S.I. Supramolecular self-assembling peptides to deliver bone morphogenetic proteins for skeletal regeneration. Bone 2020, 141, 115565. [Google Scholar] [CrossRef]
- Garifullin, R.; Guler, M.O. Electroactive peptide-based supramolecular polymers. Mater. Today Bio 2021, 10, 100099. [Google Scholar] [CrossRef]
- Sharma, P.; Pal, V.K.; Roy, S. An overview of latest advances in exploring bioactive peptide hydrogels for neural tissue engineering. Biomater. Sci. 2021, 9, 3911–3938. [Google Scholar] [CrossRef]
- Zhang, L.; Lu, J.R.; Waigh, T.A. Electronics of peptide- and protein-based biomaterials. Adv. Colloid Interface Sci. 2021, 287, 102319. [Google Scholar] [CrossRef]
- O’Neill, C.L.; Shrimali, P.C.; Clapacs, Z.P.; Files, M.A.; Rudra, J.S. Peptide-Based Supramolecular Vaccine Systems. Acta Biomater. 2021. [Google Scholar] [CrossRef]
- Görbitz, C.H. Nanotube formation by hydrophobic dipeptides. Chem. Eur. J. 2001, 7, 5153–5159. [Google Scholar] [CrossRef]
- Kralj, S.; Bellotto, O.; Parisi, E.; Garcia, A.M.; Iglesias, D.; Semeraro, S.; Deganutti, C.; D’Andrea, P.; Vargiu, A.V.; Geremia, S.; et al. Heterochirality and Halogenation Control Phe-Phe Hierarchical Assembly. ACS Nano 2020, 14, 16951–16961. [Google Scholar] [CrossRef] [PubMed]
- Aslam Khan, M.U.; Abd Razak, S.I.; Al Arjan, W.S.; Nazir, S.; Sahaya Anand, T.J.; Mehboob, H.; Amin, R. Recent Advances in Biopolymeric Composite Materials for Tissue Engineering and Regenerative Medicines: A Review. Molecules 2021, 26, 619. [Google Scholar] [CrossRef] [PubMed]
- Pushpavanam, K.; Ma, J.; Cai, Y.; Naser, N.Y.; Baneyx, F. Solid-Binding Proteins: Bridging Synthesis, Assembly, and Function in Hybrid and Hierarchical Materials Fabrication. Annu. Rev. Chem. Biomol. Eng. 2021, 12, 333–357. [Google Scholar] [CrossRef] [PubMed]
- Mondal, S.; Das, S.; Nandi, A.K. A review on recent advances in polymer and peptide hydrogels. Soft Matter 2020, 16, 1404–1454. [Google Scholar] [CrossRef] [PubMed]
- Song, H.-Q.; Fan, Y.; Hu, Y.; Cheng, G.; Xu, F.-J. Polysaccharide–Peptide Conjugates: A Versatile Material Platform for Biomedical Applications. Adv. Funct. Mater. 2021, 31, 2005978. [Google Scholar] [CrossRef]
- Higashi, S.L.; Rozi, N.; Hanifah, S.A.; Ikeda, M. Supramolecular Architectures of Nucleic Acid/Peptide Hybrids. Int. J. Mol. Sci. 2020, 21, 9458. [Google Scholar] [CrossRef] [PubMed]
- Bellotto, O.; Cringoli, M.C.; Perathoner, S.; Fornasiero, P.; Marchesan, S. Peptide Gelators to Template Inorganic Nanoparticle Formation. Gels 2021, 7, 14. [Google Scholar] [CrossRef]
- Pigliacelli, C.; Sánchez-Fernández, R.; García, M.D.; Peinador, C.; Pazos, E. Self-assembled peptide-inorganic nanoparticle superstructures: From component design to applications. Chem. Commun. 2020, 56, 8000–8014. [Google Scholar] [CrossRef]
- Gao, P.; Wu, Y.; Wu, L. Co-assembly of polyoxometalates and peptides towards biological applications. Soft Matter 2016, 12, 8464–8479. [Google Scholar] [CrossRef]
- Kieffer, M.; Garcia, A.M.; Haynes, C.J.E.; Kralj, S.; Iglesias, D.; Nitschke, J.R.; Marchesan, S. Embedding and Positioning of Two FeII4L4 Cages in Supramolecular Tripeptide Gels for Selective Chemical Segregation. Angew. Chem. Int. Ed. 2019, 58, 7982–7986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Georgakilas, V.; Perman, J.A.; Tucek, J.; Zboril, R. Broad Family of Carbon Nanoallotropes: Classification, Chemistry, and Applications of Fullerenes, Carbon Dots, Nanotubes, Graphene, Nanodiamonds, and Combined Superstructures. Chem. Rev. 2015, 115, 4744–4822. [Google Scholar] [CrossRef] [PubMed]
- McHedlov-Petrossyan, N.O. Fullerenes in Liquid Media: An Unsettling Intrusion into the Solution Chemistry. Chem. Rev. 2013, 113, 5149–5193. [Google Scholar] [CrossRef] [PubMed]
- Silvia, G.; Adalberto, C.; Viviana, M. Carbon Nano-onions: A Valuable Class of Carbon Nanomaterials in Biomedicine. Curr. Med. Chem. 2019, 26, 6915–6929. [Google Scholar] [CrossRef]
- Liu, J.; Li, R.; Yang, B. Carbon Dots: A New Type of Carbon-Based Nanomaterial with Wide Applications. ACS Centr. Sci. 2020, 6, 2179–2195. [Google Scholar] [CrossRef]
- Yang, F.; Wang, M.; Zhang, D.; Yang, J.; Zheng, M.; Li, Y. Chirality pure carbon nanotubes: Growth, sorting, and characterization. Chem. Rev. 2020, 120, 2693–2758. [Google Scholar] [CrossRef]
- Bottari, G.; Herranz, M.; Wibmer, L.; Volland, M.; Rodríguez-Pérez, L.; Guldi, D.M.; Hirsch, A.; Martín, N.; D’Souza, F.; Torres, T. Chemical functionalization and characterization of graphene-based materials. Chem. Soc. Rev. 2017, 46, 4464–4500. [Google Scholar] [CrossRef] [Green Version]
- Karousis, N.; Suarez-Martinez, I.; Ewels, C.P.; Tagmatarchis, N. Structure, Properties, Functionalization, and Applications of Carbon Nanohorns. Chem. Rev. 2016, 116, 4850–4883. [Google Scholar] [CrossRef]
- Basso, L.; Cazzanelli, M.; Orlandi, M.; Miotello, A. Nanodiamonds: Synthesis and application in sensing, catalysis, and the possible connection with some processes occurring in space. Appl. Sci. 2020, 10, 4094. [Google Scholar] [CrossRef]
- Bondon, N.; Raehm, L.; Charnay, C.; Boukherroub, R.; Durand, J.O. Nanodiamonds for bioapplications, recent developments. J. Mater. Chem. B 2020, 8, 10878–10896. [Google Scholar] [CrossRef]
- Adorinni, S.; Cringoli, M.C.; Perathoner, S.; Fornasiero, P.; Marchesan, S. Green Approaches to Carbon Nanostructure-Based Biomaterials. Appl. Sci. 2021, 11, 2490. [Google Scholar] [CrossRef]
- Ugarte, D. Onion-like graphitic particles. Carbon 1995, 33, 989–993. [Google Scholar] [CrossRef]
- Piovesana, S.; Iglesias, D.; Melle-Franco, M.; Kralj, S.; Cavaliere, C.; Melchionna, M.; Laganà, A.; Capriotti, A.L.; Marchesan, S. Carbon nanostructure morphology templates nanocomposites for phosphoproteomics. Nano Res. 2020, 13, 380–388. [Google Scholar] [CrossRef]
- Tonellato, M.; Piccione, M.; Gasparotto, M.; Bellet, P.; Tibaudo, L.; Vicentini, N.; Bergantino, E.; Menna, E.; Vitiello, L.; Di Liddo, R.; et al. Commitment of autologous human multipotent stem cells on biomimetic poly-l-lactic acid-based scaffolds is strongly influenced by structure and concentration of carbon nanomaterial. Nanomaterials 2020, 10, 415. [Google Scholar] [CrossRef] [Green Version]
- Vicentini, N.; Gatti, T.; Salerno, M.; Hernandez Gomez, Y.S.; Bellon, M.; Gallio, S.; Marega, C.; Filippini, F.; Menna, E. Effect of different functionalized carbon nanostructures as fillers on the physical properties of biocompatible poly(L-lactic acid) composites. Mater. Chem. Phys. 2018, 214, 265–276. [Google Scholar] [CrossRef]
- Iglesias, D.; Melle-Franco, M.; Kurbasic, M.; Melchionna, M.; Abrami, M.; Grassi, M.; Prato, M.; Marchesan, S. Oxidized Nanocarbons-Tripeptide Supramolecular Hydrogels: Shape Matters! ACS Nano 2018, 12, 5530–5538. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhu, Z.; Bortolini, C.; Hoffmann, S.V.; Amari, A.; Zhang, H.X.; Liu, L.; Dong, M.D. Dimensionality of carbon nanomaterial impacting on the modulation of amyloid peptide assembly. Nanotechnology 2016, 27, 304001. [Google Scholar] [CrossRef]
- Pinals, R.L.; Yang, D.; Lui, A.; Cao, W.; Landry, M.P. Corona exchange dynamics on carbon nanotubes by multiplexed fluorescence monitoring. J. Am. Chem. Soc. 2019, 142, 1254–1264. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, V.; Perini, G.; De Spirito, M.; Papi, M. Graphene oxide touches blood: In Vivo interactions of bio-coronated 2D materials. Nanoscale Horiz. 2019, 4, 273–290. [Google Scholar] [CrossRef]
- Duke, K.S.; Bonner, J.C. Mechanisms of carbon nanotube-induced pulmonary fibrosis: A physicochemical characteristic perspective. Wiley Int. Rev. Nanomed. Nanobiotechnol. 2018, 10, e1498. [Google Scholar] [CrossRef]
- Chen, M.; Qin, X.; Zeng, G. Biodegradation of carbon nanotubes, graphene, and their derivatives. Trends Biotechnol. 2017, 35, 836–846. [Google Scholar] [CrossRef]
- Keshavan, S.; Calligari, P.; Stella, L.; Fusco, L.; Delogu, L.G.; Fadeel, B. Nano-bio interactions: A neutrophil-centric view. Cell Death Dis. 2019, 10, 569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, K.; Wang, A.Z.; Yin, L.; Cheng, J. Bio-nano interface: The impact of biological environment on nanomaterials and their delivery properties. J. Control. Release 2017, 263, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Panwar, N.; Soehartono, A.M.; Chan, K.K.; Zeng, S.; Xu, G.; Qu, J.; Coquet, P.; Yong, K.-T.; Chen, X. Nanocarbons for Biology and Medicine: Sensing, Imaging, and Drug Delivery. Chem. Rev. 2019, 119, 9559–9656. [Google Scholar] [CrossRef]
- Zhuang, W.R.; Wang, Y.; Cui, P.F.; Xing, L.; Lee, J.; Kim, D.; Jiang, H.L.; Oh, Y.K. Applications of π-π stacking interactions in the design of drug-delivery systems. J. Control. Release 2019, 294, 311–326. [Google Scholar] [CrossRef] [PubMed]
- Marchesan, S.; Melchionna, M.; Prato, M. Carbon Nanostructures for Nanomedicine: Opportunities and Challenges. Fuller. Nanotub. Carbon Nanostruct. 2014, 22, 190–195. [Google Scholar] [CrossRef]
- Mehra, N.K.; Jain, A.K.; Nahar, M. Carbon nanomaterials in oncology: An expanding horizon. Drug Discov. Today 2018, 23, 1016–1025. [Google Scholar] [CrossRef] [PubMed]
- Xin, Q.; Shah, H.; Nawaz, A.; Xie, W.; Akram, M.Z.; Batool, A.; Tian, L.; Jan, S.U.; Boddula, R.; Guo, B. Antibacterial carbon-based nanomaterials. Adv. Mater. 2019, 31, 1804838. [Google Scholar] [CrossRef] [PubMed]
- Ku, S.H.; Lee, M.; Park, C.B. Carbon-based nanomaterials for tissue engineering. Adv. Healthc. Mater. 2013, 2, 244–260. [Google Scholar] [CrossRef] [PubMed]
- Marchesan, S.; Ballerini, L.; Prato, M. Nanomaterials for stimulating nerve growth. Science 2017, 356, 1010–1011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aydin, T.; Gurcan, C.; Taheri, H.; Yilmazer, A. Graphene Based Materials in Neural Tissue Regeneration. Adv. Exp. Med. Biol. 2018, 1107, 129–142. [Google Scholar] [CrossRef] [PubMed]
- Marchesan, S.; Bosi, S.; Alshatwi, A.; Prato, M. Carbon nanotubes for organ regeneration: An electrifying performance. Nano Today 2016, 11, 398–401. [Google Scholar] [CrossRef]
- Raja, A.; Rajasekaran, P.; Selvakumar, K.; Arivanandhan, M.; Asath Bahadur, S.; Swaminathan, M. Efficient Photoreduction of Hexavalent Chromium Using the Reduced Graphene Oxide-Sm(2)MoO(6)-TiO(2) Catalyst under Visible Light Illumination. ACS Omega 2020, 5, 6414–6422. [Google Scholar] [CrossRef] [Green Version]
- Peng, Z.; Zhao, T.; Zhou, Y.; Li, S.; Li, J.; Leblanc, R.M. Bone Tissue Engineering via Carbon-Based Nanomaterials. Adv. Healthc. Mater. 2020, 9, e1901495. [Google Scholar] [CrossRef]
- Sainio, S.; Leppänen, E.; Mynttinen, E.; Palomäki, T.; Wester, N.; Etula, J.; Isoaho, N.; Peltola, E.; Koehne, J.; Meyyappan, M. Integrating Carbon Nanomaterials with Metals for Bio-sensing Applications. Mol. Neurobiol. 2020, 57, 179–190. [Google Scholar] [CrossRef] [Green Version]
- Loh, K.P.; Ho, D.; Chiu, G.N.C.; Leong, D.T.; Pastorin, G.; Chow, E.K.-H. Clinical Applications of Carbon Nanomaterials in Diagnostics and Therapy. Adv. Mater. 2018, 30, 1802368. [Google Scholar] [CrossRef]
- Madannejad, R.; Shoaie, N.; Jahanpeyma, F.; Darvishi, M.H.; Azimzadeh, M.; Javadi, H. Toxicity of carbon-based nanomaterials: Reviewing recent reports in medical and biological systems. Chem. Biol. Interact. 2019, 307, 206–222. [Google Scholar] [CrossRef]
- Gupta, N.; Rai, D.B.; Jangid, A.K.; Kulhari, H. A review of theranostics applications and toxicities of carbon nanomaterials. Curr. Drug Metab. 2019, 20, 506–532. [Google Scholar] [CrossRef]
- Fadeel, B.; Kostarelos, K. Grouping all carbon nanotubes into a single substance category is scientifically unjustified. Nat. Nanotechnol. 2020, 15, 164. [Google Scholar] [CrossRef] [Green Version]
- Graphene Standards. Available online: https://www.thegraphenecouncil.org/page/GrapheneStandards (accessed on 30 June 2021).
- Xiarchos, I.; Morozinis, A.K.; Kavouras, P.; Charitidis, C.A. Nanocharacterization, Materials Modeling, and Research Integrity as Enablers of Sound Risk Assessment: Designing Responsible Nanotechnology. Small 2020, 16, 2001590. [Google Scholar] [CrossRef]
- Romanos, N.; Kalogerini, M.; Koumoulos, E.P.; Morozinis, A.K.; Sebastiani, M.; Charitidis, C. Innovative Data Management in advanced characterization: Implications for materials design. Mater. Today Commun. 2019, 20, 100541. [Google Scholar] [CrossRef]
- Kotzabasaki, M.; Sotiropoulos, I.; Charitidis, C.; Sarimveis, H. Machine learning methods for multi-walled carbon nanotubes (MWCNT) genotoxicity prediction. Nanoscale Adv. 2021, 3, 3167–3176. [Google Scholar] [CrossRef]
- Tong, T.; Wang, L.; You, X.; Wu, J. Nano and microscale delivery platforms for enhanced oral peptide/protein bioavailability. Biomater. Sci. 2020, 8, 5804–5823. [Google Scholar] [CrossRef] [PubMed]
- Katyal, P.; Mahmoudinobar, F.; Montclare, J.K. Recent trends in peptide and protein-based hydrogels. Curr. Opin. Struct. Biol. 2020, 63, 97–105. [Google Scholar] [CrossRef]
- Iglesias, D.; Bosi, S.; Melchionna, M.; Da Ros, T.; Marchesan, S. The Glitter of Carbon Nanostructures in Hybrid/Composite Hydrogels for Medicinal Use. Curr. Top. Med. Chem. 2016, 16, 1976–1989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Speranza, G. Carbon Nanomaterials: Synthesis, Functionalization and Sensing Applications. Nanomaterials 2021, 11, 967. [Google Scholar] [CrossRef]
- Sheikholeslam, M.; Wheeler, S.D.; Duke, K.G.; Marsden, M.; Pritzker, M.; Chen, P. Peptide and peptide-carbon nanotube hydrogels as scaffolds for tissue & 3D tumor engineering. Acta Biomater. 2018, 69, 107–119. [Google Scholar] [CrossRef] [Green Version]
- El-Mahdy, A.F.M.; Kuo, S.-W. Diphenylpyrenylamine-functionalized polypeptides: Secondary structures, aggregation-induced emission, and carbon nanotube dispersibility. RSC Adv. 2018, 8, 15266–15281. [Google Scholar] [CrossRef] [Green Version]
- Garriga, R.; Jurewicz, I.; Seyedin, S.; Bardi, N.; Totti, S.; Matta-Domjan, B.; Velliou, E.G.; Alkhorayef, M.A.; Cebolla, V.L.; Razal, J.M.; et al. Multifunctional, biocompatible and pH-responsive carbon nanotube- and graphene oxide/tectomer hybrid composites and coatings. Nanoscale 2017, 9, 7791–7804. [Google Scholar] [CrossRef] [Green Version]
- Lian, Z.; Ji, T. Functional peptide-based drug delivery systems. J. Mater. Chem. B 2020, 8, 6517–6529. [Google Scholar] [CrossRef]
- Khan, M.M.; Filipczak, N.; Torchilin, V.P. Cell penetrating peptides: A versatile vector for co-delivery of drug and genes in cancer. J. Control. Release 2021, 330, 1220–1228. [Google Scholar] [CrossRef]
- Jin, J.; Wu, Y.; Chen, J.; Shen, Y.; Zhang, L.; Zhang, H.; Chen, L.; Yuan, H.; Chen, H.; Zhang, W.; et al. The peptide PROTAC modality: A novel strategy for targeted protein ubiquitination. Theranostics 2020, 10, 10141–10153. [Google Scholar] [CrossRef] [PubMed]
- Maity, D. Selected peptide-based fluorescent probes for biological applications. Beilstein J. Org. Chem. 2020, 16, 2971–2982. [Google Scholar] [CrossRef]
- Calvaresi, M.; Zerbetto, F. The Devil and Holy Water: Protein and Carbon Nanotube Hybrids. Acc. Chem. Res. 2013, 46, 2454–2463. [Google Scholar] [CrossRef]
- Henna, T.K.; Raphey, V.R.; Sankar, R.; Ameena Shirin, V.K.; Gangadharappa, H.V.; Pramod, K. Carbon nanostructures: The drug and the delivery system for brain disorders. Int. J. Pharm. 2020, 587, 119701. [Google Scholar] [CrossRef]
- Zhang, X.; Gong, C.; Akakuru, O.U.; Su, Z.; Wu, A.; Wei, G. The design and biomedical applications of self-assembled two-dimensional organic biomaterials. Chem. Soc. Rev. 2019, 48, 5564–5595. [Google Scholar] [CrossRef]
- Gholami, A.; Hashemi, S.A.; Yousefi, K.; Mousavi, S.M.; Chiang, W.-H.; Ramakrishna, S.; Mazraedoost, S.; Alizadeh, A.; Omidifar, N.; Behbudi, G.; et al. 3D Nanostructures for Tissue Engineering, Cancer Therapy, and Gene Delivery. J. Nanomater. 2020, 2020, 1852946. [Google Scholar] [CrossRef]
- Mazzier, D.; Carraro, F.; Crisma, M.; Rancan, M.; Toniolo, C.; Moretto, A. A terminally protected dipeptide: From crystal structure and self-assembly, through co-assembly with carbon-based materials, to a ternary catalyst for reduction chemistry in water. Soft Matter 2016, 12, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Garriga, R.; Jurewicz, I.; Seyedin, S.; Tripathi, M.; Pearson, J.R.; Cebolla, V.L.; Dalton, A.B.; Razal, J.M.; Muñoz, E. Two-dimensional oligoglycine tectomer adhesives for graphene oxide fiber functionalization. Carbon 2019, 147, 460–475. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, W.; Zhang, L.; Li, J.; Su, Z.; Wei, G. Sequence-Designed Peptide Nanofibers Bridged Conjugation of Graphene Quantum Dots with Graphene Oxide for High Performance Electrochemical Hydrogen Peroxide Biosensor. Adv. Mater. Interfaces 2017, 4, 1600895. [Google Scholar] [CrossRef]
- Liu, Z.; Zou, Y.; Zhang, Q.; Chen, P.; Liu, Y.; Qian, Z. Distinct Binding Dynamics, Sites and Interactions of Fullerene and Fullerenols with Amyloid-β Peptides Revealed by Molecular Dynamics Simulations. Int. J. Mol. Sci. 2019, 20, 2048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Qian, Z.; Wei, G. The inhibitory mechanism of a fullerene derivative against amyloid-β peptide aggregation: An atomistic simulation study. Phys. Chem. Chem. Phys. 2016, 18, 12582–12591. [Google Scholar] [CrossRef] [PubMed]
- Lamas, A.; Guerra, A.; Amorín, M.; Granja, J.R. New self-assembling peptide nanotubes of large diameter using δ-amino acids. Chem. Sci. 2018, 9, 8228–8233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Zhang, H.; Zou, Q.; Xing, R.; Jiao, T.; Yan, X. An injectable dipeptide–fullerene supramolecular hydrogel for photodynamic antibacterial therapy. J. Mater. Chem. B 2018, 6, 7335–7342. [Google Scholar] [CrossRef]
- Cringoli, M.C.; Kralj, S.; Kurbasic, M.; Urban, M.; Marchesan, S. Luminescent supramolecular hydrogels from a tripeptide and nitrogen-doped carbon nanodots. Beilstein J. Nanotechnol. 2017, 8, 1553–1562. [Google Scholar] [CrossRef] [Green Version]
- Chung, Y.J.; Lee, B.I.; Park, C.B. Multifunctional carbon dots as a therapeutic nanoagent for modulating Cu(II)-mediated β-amyloid aggregation. Nanoscale 2019, 11, 6297–6306. [Google Scholar] [CrossRef]
- Chung, Y.J.; Lee, C.H.; Lim, J.; Jang, J.; Kang, H.; Park, C.B. Photomodulating Carbon Dots for Spatiotemporal Suppression of Alzheimer’s β-Amyloid Aggregation. ACS Nano 2020, 14, 16973–16983. [Google Scholar] [CrossRef]
- Huang, H.; Li, P.; Zhang, M.; Yu, Y.; Huang, Y.; Gu, H.; Wang, C.; Yang, Y. Graphene quantum dots for detecting monomeric amyloid peptides. Nanoscale 2017, 9, 5044–5048. [Google Scholar] [CrossRef]
- Wang, Y.; Kadiyala, U.; Qu, Z.; Elvati, P.; Altheim, C.; Kotov, N.A.; Violi, A.; VanEpps, J.S. Anti-Biofilm Activity of Graphene Quantum Dots via Self-Assembly with Bacterial Amyloid Proteins. ACS Nano 2019, 13, 4278–4289. [Google Scholar] [CrossRef]
- Su, Z.; Shen, H.; Wang, H.; Wang, J.; Li, J.; Nienhaus, G.U.; Shang, L.; Wei, G. Motif-Designed Peptide Nanofibers Decorated with Graphene Quantum Dots for Simultaneous Targeting and Imaging of Tumor Cells. Adv. Funct. Mater. 2015, 25, 5472–5478. [Google Scholar] [CrossRef]
- He, L.; Xiao, Q.; Zhao, Y.; Li, J.; Reddy, S.; Shi, X.; Su, X.; Chiu, K.; Ramakrishna, S. Engineering an Injectable Electroactive Nanohybrid Hydrogel for Boosting Peripheral Nerve Growth and Myelination in Combination with Electrical Stimulation. ACS Appl. Mater. Interfaces 2020, 12, 53150–53163. [Google Scholar] [CrossRef]
- Dinesh, B.; Squillaci, M.A.; Ménard-Moyon, C.; Samorì, P.; Bianco, A. Self-assembly of diphenylalanine backbone homologues and their combination with functionalized carbon nanotubes. Nanoscale 2015, 7, 15873–15879. [Google Scholar] [CrossRef] [Green Version]
- Rehak, P.; Král, P. Hybridization of Biomolecular Crystals and Low-Dimensional Materials. ACS Nano 2021, 15, 6678–6683. [Google Scholar] [CrossRef]
- Guilbaud-Chéreau, C.; Dinesh, B.; Schurhammer, R.; Collin, D.; Bianco, A.; Ménard-Moyon, C. Protected Amino Acid–Based Hydrogels Incorporating Carbon Nanomaterials for Near-Infrared Irradiation-Triggered Drug Release. ACS Appl. Mater. Interfaces 2019, 11, 13147–13157. [Google Scholar] [CrossRef]
- Rissanou, A.N.; Keliri, A.; Arnittali, M.; Harmandaris, V. Self-assembly of diphenylalanine peptides on graphene via detailed atomistic simulations. Phys. Chem. Chem. Phys. 2020, 22, 27645–27657. [Google Scholar] [CrossRef] [PubMed]
- Khatayevich, D.; Page, T.; Gresswell, C.; Hayamizu, Y.; Grady, W.; Sarikaya, M. Selective Detection of Target Proteins by Peptide-Enabled Graphene Biosensor. Small 2014, 10, 1505–1513. [Google Scholar] [CrossRef] [PubMed]
- Thodkar, K.; Cazade, P.-A.; Bergmann, F.; Lopez-Calle, E.; Thompson, D.; Heindl, D. Self-Assembled Pyrene Stacks and Peptide Monolayers Tune the Electronic Properties of Functionalized Electrolyte-Gated Graphene Field-Effect Transistors. ACS Appl. Mater. Interfaces 2021, 13, 9134–9142. [Google Scholar] [CrossRef]
- Li, P.; Sakuma, K.; Tsuchiya, S.; Sun, L.; Hayamizu, Y. Fibroin-like Peptides Self-Assembling on Two-Dimensional Materials as a Molecular Scaffold for Potential Biosensing. ACS Appl. Mater. Interfaces 2019, 11, 20670–20677. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Zhang, Z.; Li, D.; Zhang, W.; Yu, X.; Liu, W.; Gong, C.; Wei, G.; Su, Z. Biomimetic ultralight, highly porous, shape-adjustable, and biocompatible 3D graphene minerals via incorporation of self-assembled peptide nanosheets. Adv. Funct. Mater. 2018, 28, 1056. [Google Scholar] [CrossRef]
- Almohammed, S.; Zhang, F.; Rodriguez, B.J.; Rice, J.H. Electric Field-Induced Chemical Surface-Enhanced Raman Spectroscopy Enhancement from Aligned Peptide Nanotube-Graphene Oxide Templates for Universal Trace Detection of Biomolecules. J. Phys. Chem. Lett. 2019, 10, 1878–1887. [Google Scholar] [CrossRef] [Green Version]
- Trapani, G.; Caruso, V.C.L.; Cucci, L.M.; Attanasio, F.; Tabbì, G.; Forte, G.; La Mendola, D.; Satriano, C. Graphene Oxide Nanosheets Tailored With Aromatic Dipeptide Nanoassemblies for a Tuneable Interaction With Cell Membranes. Front. Bioeng. Biotechnol. 2020, 8. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, D.; Xu, G.; Xu, Y.; Lu, Y.; Li, S.; Liu, Q. Spectroscopic detection of thrombin with peptides self-assembled on gold nanoparticles hybridized graphene oxide. Sens. Actuators B Chem. 2017, 242, 443–449. [Google Scholar] [CrossRef]
- Castelletto, V.; Kaur, A.; Hamley, I.; Barnes, R.; Karatzas, K.; Hermida-Merino, D.; Swioklo, S.; Connon, C.; Stasiak, J.; Reza, M.; et al. Hybrid membrane biomaterials from self-assembly in polysaccharide and peptide amphiphile mixtures: Controllable structural and mechanical properties and antimicrobial activity. RSC Adv. 2017, 7, 8366–8375. [Google Scholar] [CrossRef] [Green Version]
- Ligorio, C.; Zhou, M.; Wychowaniec, J.K.; Zhu, X.; Bartlam, C.; Miller, A.F.; Vijayaraghavan, A.; Hoyland, J.A.; Saiani, A. Graphene oxide containing self-assembling peptide hybrid hydrogels as a potential 3D injectable cell delivery platform for intervertebral disc repair applications. Acta Biomater. 2019, 92, 92–103. [Google Scholar] [CrossRef]
- Wang, J.; Ouyang, Z.; Ren, Z.; Li, J.; Zhang, P.; Wei, G.; Su, Z. Self-assembled peptide nanofibers on graphene oxide as a novel nanohybrid for biomimetic mineralization of hydroxyapatite. Carbon 2015, 89, 20–30. [Google Scholar] [CrossRef]
- Yang, J.-K.; Kwak, S.-Y.; Jeon, S.-J.; Lee, E.; Ju, J.-M.; Kim, H.-I.; Lee, Y.-S.; Kim, J.-H. Proteolytic disassembly of peptide-mediated graphene oxide assemblies for turn-on fluorescence sensing of proteases. Nanoscale 2016, 8, 12272–12281. [Google Scholar] [CrossRef] [PubMed]
- Lei, W.-W.; Shi, L.-Y.; Li, H.; Li, C.-X.; Diao, Y.-F.; Zhang, Y.-L.; Ran, R. A novel self-assembled hybrid organogel of polypeptide-based block copolymers with inclusion of polypeptide-functionalized graphene. RSC Adv. 2017, 7, 1471–1479. [Google Scholar] [CrossRef] [Green Version]
- Wychowaniec, J.K.; Iliut, M.; Zhou, M.; Moffat, J.; Elsawy, M.A.; Pinheiro, W.A.; Hoyland, J.A.; Miller, A.F.; Vijayaraghavan, A.; Saiani, A. Designing Peptide/Graphene Hybrid Hydrogels through Fine-Tuning of Molecular Interactions. Biomacromolecules 2018, 19, 2731–2741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chetia, M.; Debnath, S.; Chowdhury, S.; Chatterjee, S. Self-assembly and multifunctionality of peptide organogels: Oil spill recovery, dye absorption and synthesis of conducting biomaterials. RSC Adv. 2020, 10, 5220–5233. [Google Scholar] [CrossRef] [Green Version]
- Eckhart, K.E.; Holt, B.D.; Laurencin, M.G.; Sydlik, S.A. Covalent conjugation of bioactive peptides to graphene oxide for biomedical applications. Biomater. Sci. 2019, 7, 3876–3885. [Google Scholar] [CrossRef]
- Yao, S.; Yuan, X.; Jiang, L.; Xiong, T.; Zhang, J. Recent Progress on Fullerene-Based Materials: Synthesis, Properties, Modifications, and Photocatalytic Applications. Materials 2020, 13, 2924. [Google Scholar] [CrossRef]
- Antoku, D.; Sugikawa, K.; Ikeda, A. Photodynamic Activity of Fullerene Derivatives Solubilized in Water by Natural-Product-Based Solubilizing Agents. Chem. Eur. J. 2019, 25, 1854–1865. [Google Scholar] [CrossRef] [PubMed]
- Galvan, Y.P.; Alperovich, I.; Zolotukhin, P.; Prazdnova, E.; Mazanko, M.; Belanova, A.; Chistyakov, V. Fullerenes as Anti-Aging Antioxidants. Curr. Aging Sci. 2017, 10, 56–67. [Google Scholar] [CrossRef]
- Akhtar, M.J.; Ahamed, M.; Alhadlaq, H.A.; Alshamsan, A. Mechanism of ROS scavenging and antioxidant signalling by redox metallic and fullerene nanomaterials: Potential implications in ROS associated degenerative disorders. Biochim. Biophys. Acta 2017, 1861, 802–813. [Google Scholar] [CrossRef] [PubMed]
- Hamblin, M.R. Antimicrobial photodynamic inactivation: A bright new technique to kill resistant microbes. Curr. Opin. Microbiol. 2016, 33, 67–73. [Google Scholar] [CrossRef] [Green Version]
- Kazemzadeh, H.; Mozafari, M. Fullerene-based delivery systems. Drug Discov. Today 2019, 24, 898–905. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Raza, K. C60-fullerenes as Drug Delivery Carriers for Anticancer Agents: Promises and Hurdles. Pharm. Nanotechnol. 2017, 5, 169–179. [Google Scholar] [CrossRef]
- Zhao, Y.; Shen, X.; Ma, R.; Hou, Y.; Qian, Y.; Fan, C. Biological and biocompatible characteristics of fullerenols nanomaterials for tissue engineering. Histol. Histopathol. 2021. [Google Scholar] [CrossRef]
- Szymański, M.P.; Czajka, J.S.; Cmoch, P.; Iwanek, W.; Szumna, A. Interlaced capsules by self-assembly of cavitands substituted with tripeptides and tetrapeptides. Supramol. Chem. 2018, 30, 430–437. [Google Scholar] [CrossRef]
- Bjelaković, M.; Kop, T.; Maslak, V.; Milić, D. Synthesis and characterization of highly ordered self-assembled bioactive fulleropeptides. J. Mater. Sci. 2016, 51, 739–747. [Google Scholar] [CrossRef]
- Bjelaković, M.S.; Kop, T.J.; Đorđević, J.; Milić, D.R. Fulleropeptide esters as potential self-assembled antioxidants. Beilstein J. Nanotechnol. 2015, 6, 1065–1071. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Mintz, K.J.; Sharma, S.K.; Leblanc, R.M. Carbon Dots: Diverse Preparation, Application, and Perspective in Surface Chemistry. Langmuir 2019, 35, 9115–9132. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.J.; Kim, J.; Park, C.B. Photonic Carbon Dots as an Emerging Nanoagent for Biomedical and Healthcare Applications. ACS Nano 2020, 14, 6470–6497. [Google Scholar] [CrossRef] [PubMed]
- Anwar, S.; Ding, H.; Xu, M.; Hu, X.; Li, Z.; Wang, J.; Liu, L.; Jiang, L.; Wang, D.; Dong, C.; et al. Recent Advances in Synthesis, Optical Properties, and Biomedical Applications of Carbon Dots. ACS Appl. Bio Mater. 2019, 2, 2317–2338. [Google Scholar] [CrossRef]
- Ji, C.; Zhou, Y.; Leblanc, R.M.; Peng, Z. Recent Developments of Carbon Dots in Biosensing: A Review. ACS Sens. 2020, 5, 2724–2741. [Google Scholar] [CrossRef] [PubMed]
- Apter, B.; Lapshina, N.; Barhom, H.; Fainberg, B.; Handelman, A.; Accardo, A.; Diaferia, C.; Ginzburg, P.; Morelli, G.; Rosenman, G. Fluorescence Phenomena in Amyloid and Amyloidogenic Bionanostructures. Crystals 2020, 10, 668. [Google Scholar] [CrossRef]
- Kosolapova, A.O.; Antonets, K.S.; Belousov, M.V.; Nizhnikov, A.A. Biological Functions of Prokaryotic Amyloids in Interspecies Interactions: Facts and Assumptions. Int. J. Mol. Sci. 2020, 21, 7240. [Google Scholar] [CrossRef] [PubMed]
- Tasis, D.; Tagmatarchis, N.; Bianco, A.; Prato, M. Chemistry of Carbon Nanotubes. Chem. Rev. 2006, 106, 1105–1136. [Google Scholar] [CrossRef]
- Yomogida, Y.; Tanaka, T.; Tsuzuki, M.; Wei, X.; Kataura, H. Chirality Pure Carbon Nanotubes: Growth, Sorting, and Characterization. ACS Appl. Nano Mater. 2020, 3, 11289–11297. [Google Scholar] [CrossRef]
- Bati, A.S.R.; Yu, L.; Batmunkh, M.; Shapter, J.G. Recent Advances in Applications of Sorted Single-Walled Carbon Nanotubes. Adv. Funct. Mater. 2019, 29, 1902273. [Google Scholar] [CrossRef]
- He, M.; Zhang, S.; Zhang, J. Horizontal single-walled carbon nanotube arrays: Controlled synthesis, characterizations, and applications. Chem. Rev. 2020, 120, 12592–12684. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Chen, C.; Sun, X.; Peng, H. Implantable Fiber Biosensors Based on Carbon Nanotubes. Acc. Mater. Res. 2021, 2, 138–146. [Google Scholar] [CrossRef]
- Mikhalchan, A.; Vilatela, J.J. A perspective on high-performance CNT fibres for structural composites. Carbon 2019, 150, 191–215. [Google Scholar] [CrossRef]
- Zhang, X.; Lu, W.; Zhou, G.; Li, Q. Understanding the Mechanical and Conductive Properties of Carbon Nanotube Fibers for Smart Electronics. Adv. Mater. 2020, 32, 1902028. [Google Scholar] [CrossRef]
- Iglesias, D.; Senokos, E.; Alemán, B.; Cabana, L.; Navío, C.; Marcilla, R.; Prato, M.; Vilatela, J.J.; Marchesan, S. Gas-Phase Functionalization of Macroscopic Carbon Nanotube Fiber Assemblies: Reaction Control, Electrochemical Properties, and Use for Flexible Supercapacitors. ACS Appl. Mater. Interfaces 2018, 10, 5760–5770. [Google Scholar] [CrossRef]
- Anaya-Plaza, E.; Shaukat, A.; Lehtonen, I.; Kostiainen, M.A. Biomolecule-Directed Carbon Nanotube Self-Assembly. Adv. Healthc. Mater. 2021, 10, 2001162. [Google Scholar] [CrossRef]
- Kang, N.; Hua, J.; Gao, L.; Zhang, B.; Pang, J. The Interplay between Whey Protein Fibrils with Carbon Nanotubes or Carbon Nano-Onions. Materials 2021, 14, 608. [Google Scholar] [CrossRef]
- Zhang, N.; Yeo, J.; Lim, Y.; Guan, P.; Zeng, K.; Hu, X.; Cheng, Y. Tuning the structure of monomeric amyloid beta peptide by the curvature of carbon nanotubes. Carbon 2019, 153, 717–724. [Google Scholar] [CrossRef]
- Antonucci, A.; Kupis-Rozmysłowicz, J.; Boghossian, A.A. Noncovalent Protein and Peptide Functionalization of Single-Walled Carbon Nanotubes for Biodelivery and Optical Sensing Applications. ACS Appl. Mater. Interfaces 2017, 9, 11321–11331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de la Zerda, A.; Bodapati, S.; Teed, R.; May, S.Y.; Tabakman, S.M.; Liu, Z.; Khuri-Yakub, B.T.; Chen, X.; Dai, H.; Gambhir, S.S. Family of Enhanced Photoacoustic Imaging Agents for High-Sensitivity and Multiplexing Studies in Living Mice. ACS Nano 2012, 6, 4694–4701. [Google Scholar] [CrossRef] [Green Version]
- Wick, P.; Louw-Gaume, A.E.; Kucki, M.; Krug, H.F.; Kostarelos, K.; Fadeel, B.; Dawson, K.A.; Salvati, A.; Vázquez, E.; Ballerini, L. Classification framework for graphene-based materials. Angew. Chem. Int. Ed. 2014, 53, 7714–7718. [Google Scholar] [CrossRef] [Green Version]
- Catania, F.; Marras, E.; Giorcelli, M.; Jagdale, P.; Lavagna, L.; Tagliaferro, A.; Bartoli, M. A Review on Recent Advancements of Graphene and Graphene-Related Materials in Biological Applications. Appl. Sci. 2021, 11, 614. [Google Scholar] [CrossRef]
- Bellet, P.; Gasparotto, M.; Pressi, S.; Fortunato, A.; Scapin, G.; Mba, M.; Menna, E.; Filippini, F. Graphene-Based Scaffolds for Regenerative Medicine. Nanomaterials 2021, 11, 404. [Google Scholar] [CrossRef] [PubMed]
- Dasetty, S.; Sarupria, S. Advancing Rational Control of Peptide-Surface Complexes. J. Phys. Chem. B 2021, 125, 2644–2657. [Google Scholar] [CrossRef]
- Bolat, G.; Akbal Vural, O.; Tugce Yaman, Y.; Abaci, S. Label-free impedimetric miRNA-192 genosensor platform using graphene oxide decorated peptide nanotubes composite. Microchem. J. 2021, 166, 106218. [Google Scholar] [CrossRef]
- Shang, G.; Mi, Y.; Mei, Y.; Wang, G.; Wang, Y.; Li, X.; Wang, Y.; Li, Y.; Zhao, G. MicroRNA-192 inhibits the proliferation, migration and invasion of osteosarcoma cells and promotes apoptosis by targeting matrix metalloproteinase-11. Oncol. Lett. 2018, 15, 7265–7272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, V.; Zhu, R.; Jenkins, K.; Yang, R. Self-assembly of diphenylalanine peptide with controlled polarization for power generation. Nat. Commun. 2016, 7, 13566. [Google Scholar] [CrossRef] [Green Version]
- Almohammed, S.; Thampi, A.; Bazaid, A.; Zhang, F.; Moreno, S.; Keogh, K.; Minary-Jolandan, M.; Rice, J.H.; Rodriguez, B.J. Energy harvesting with peptide nanotube-graphene oxide flexible substrates prepared with electric field and wettability assisted self-assembly. J. Appl. Phys. 2020, 128, 115101. [Google Scholar] [CrossRef]
- Joshi, S.; Siddiqui, R.; Sharma, P.; Kumar, R.; Verma, G.; Saini, A. Green synthesis of peptide functionalized reduced graphene oxide (rGO) nano bioconjugate with enhanced antibacterial activity. Sci. Rep. 2020, 10, 9441. [Google Scholar] [CrossRef]
- Eckhart, K.E.; Starvaggi, F.A.; Sydlik, S.A. One-Shot Synthesis of Peptide Amphiphiles with Applications in Directed Graphenic Assembly. Biomacromolecules 2020, 21, 3878–3886. [Google Scholar] [CrossRef]
- Schnaider, L.; Brahmachari, S.; Schmidt, N.W.; Mensa, B.; Shaham-Niv, S.; Bychenko, D.; Adler-Abramovich, L.; Shimon, L.J.W.; Kolusheva, S.; DeGrado, W.F.; et al. Self-assembling dipeptide antibacterial nanostructures with membrane disrupting activity. Nat. Commun. 2017, 8, 1365. [Google Scholar] [CrossRef]
- Balu, R.; Dorishetty, P.; Mata, J.P.; Hill, A.J.; Dutta, N.K.; Choudhury, N.R. Tuning the Hierarchical Structure and Resilience of Resilin-like Polypeptide Hydrogels Using Graphene Oxide. ACS Appl. Bio Mater. 2020, 3, 8688–8697. [Google Scholar] [CrossRef]
- Giuri, D.; Barbalinardo, M.; Zanna, N.; Paci, P.; Montalti, M.; Cavallini, M.; Valle, F.; Calvaresi, M.; Tomasini, C. Tuning Mechanical Properties of Pseudopeptide Supramolecular Hydrogels by Graphene Doping. Molecules 2019, 24, 4345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Deng, Y.; Zhang, B.; Hua, W.; Wang, X.; Qi, Q.; Lin, Q.; Lv, W. An interlayer composed of a porous carbon sheet embedded with TiO2 nanoparticles for stable and high rate lithium–sulfur batteries. Nanoscale 2020, 12, 12308–12316. [Google Scholar] [CrossRef] [PubMed]
- Pinotsi, D.; Grisanti, L.; Mahou, P.; Gebauer, R.; Kaminski, C.F.; Hassanali, A.; Kaminski Schierle, G.S. Proton Transfer and Structure-Specific Fluorescence in Hydrogen Bond-Rich Protein Structures. J. Am. Chem. Soc. 2016, 138, 3046–3057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
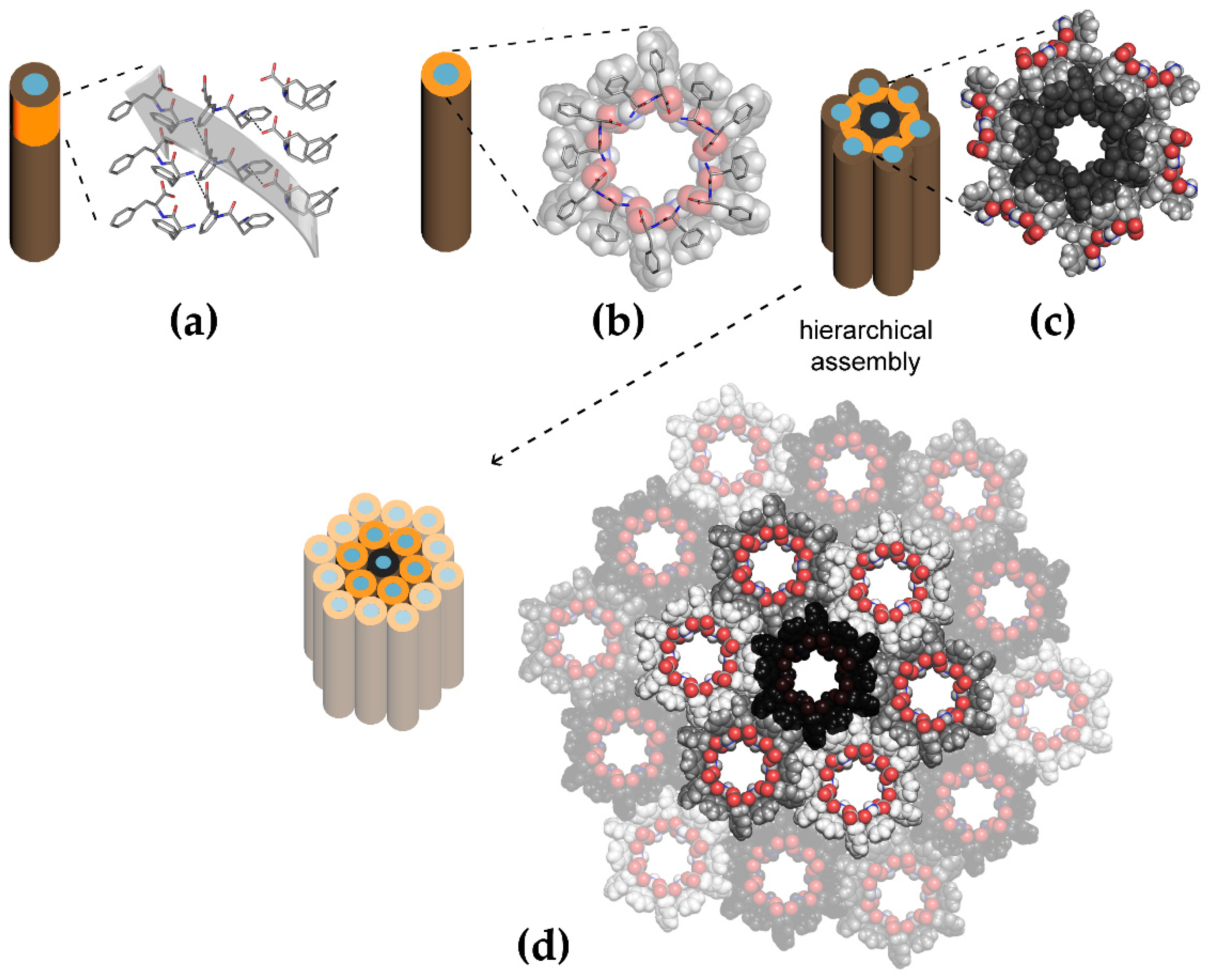


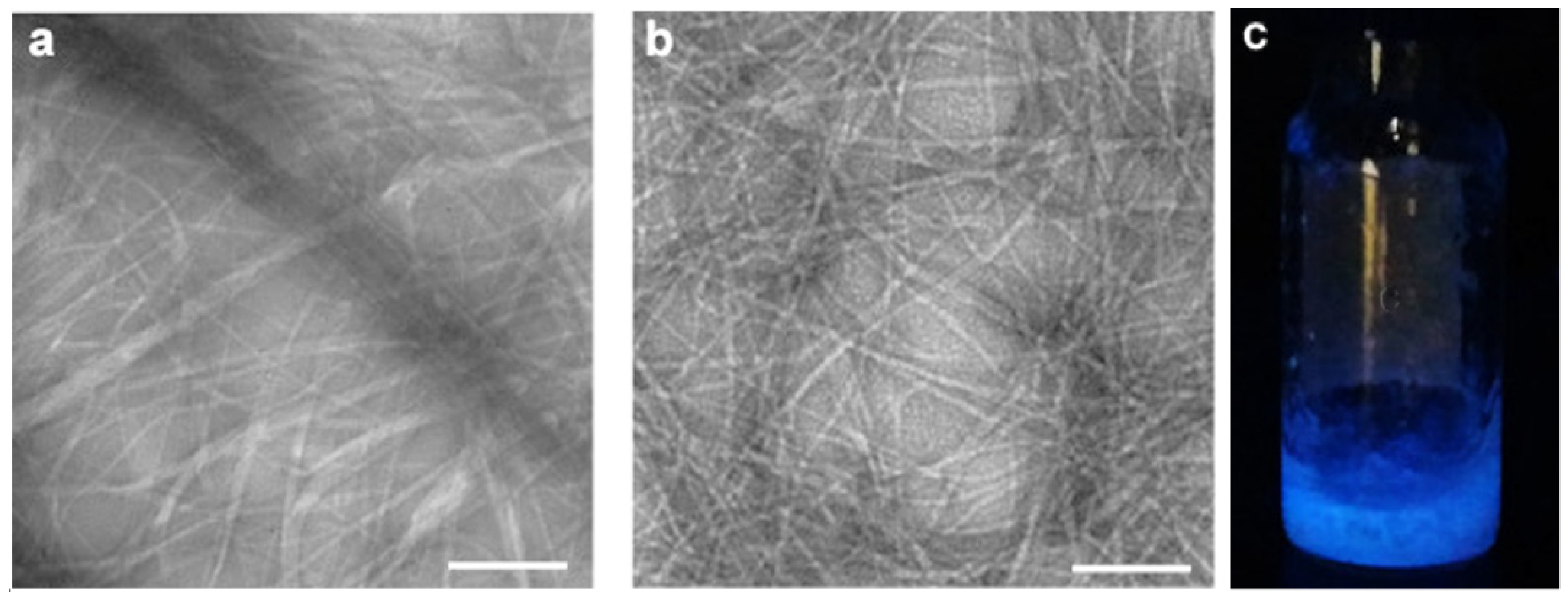
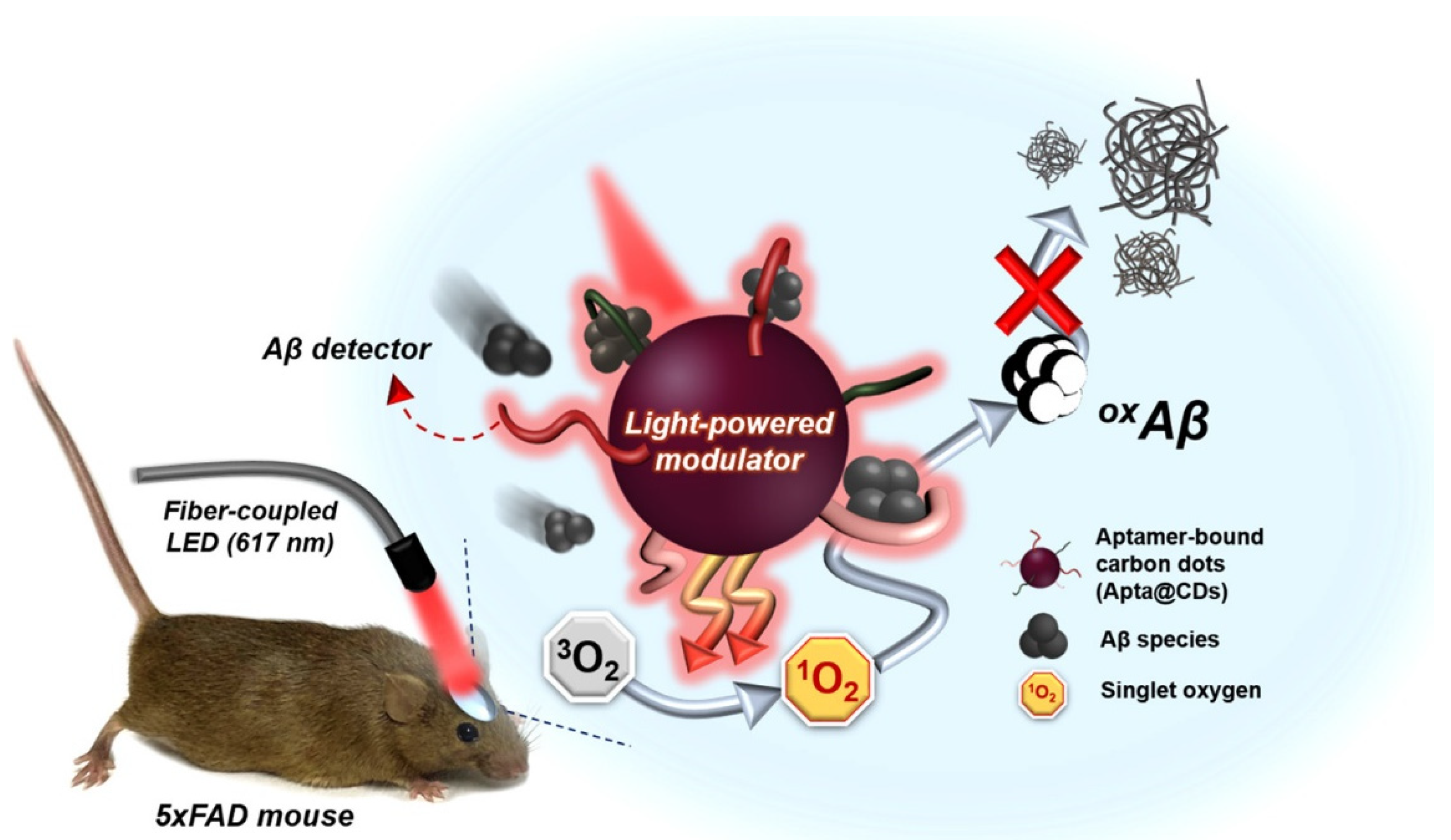
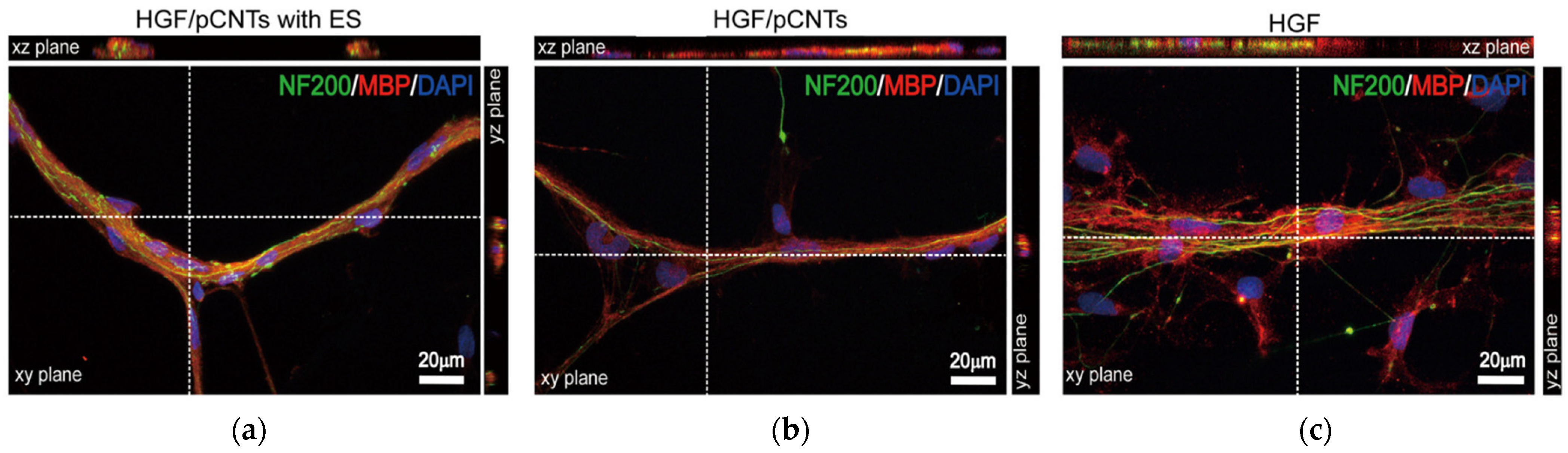

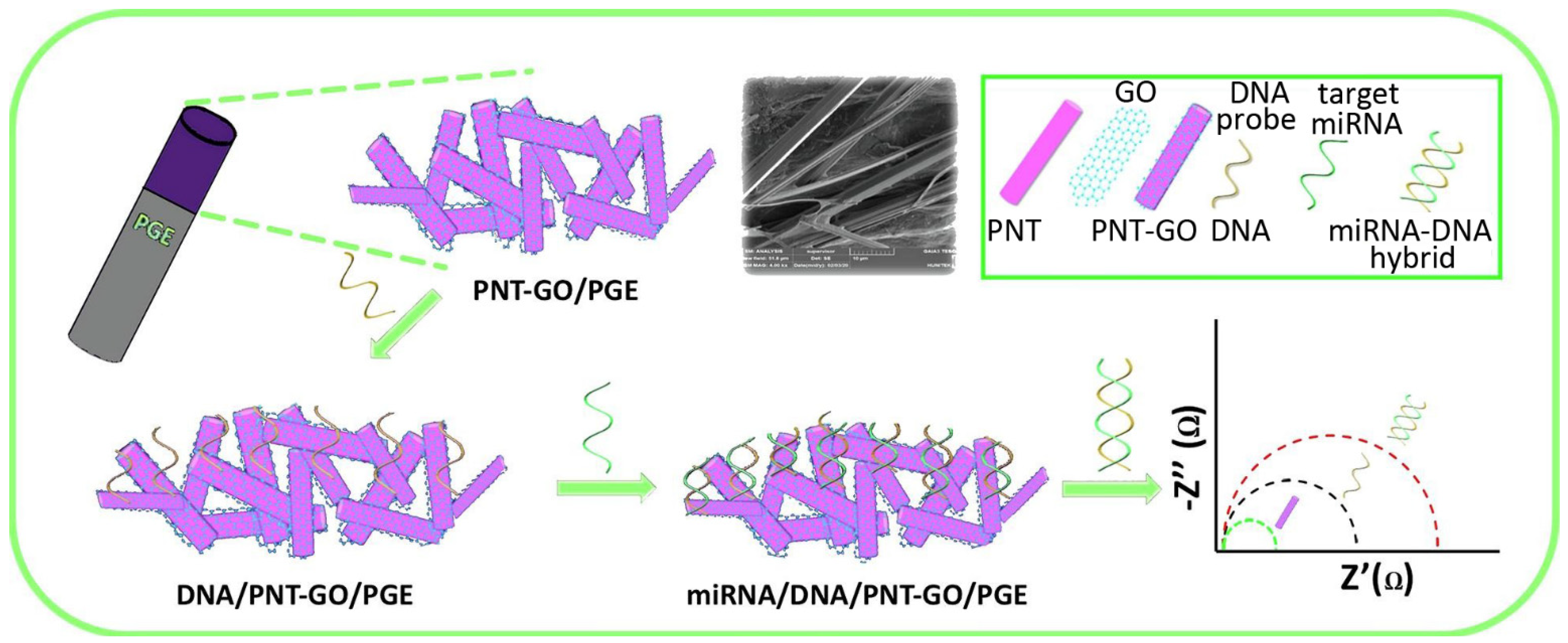
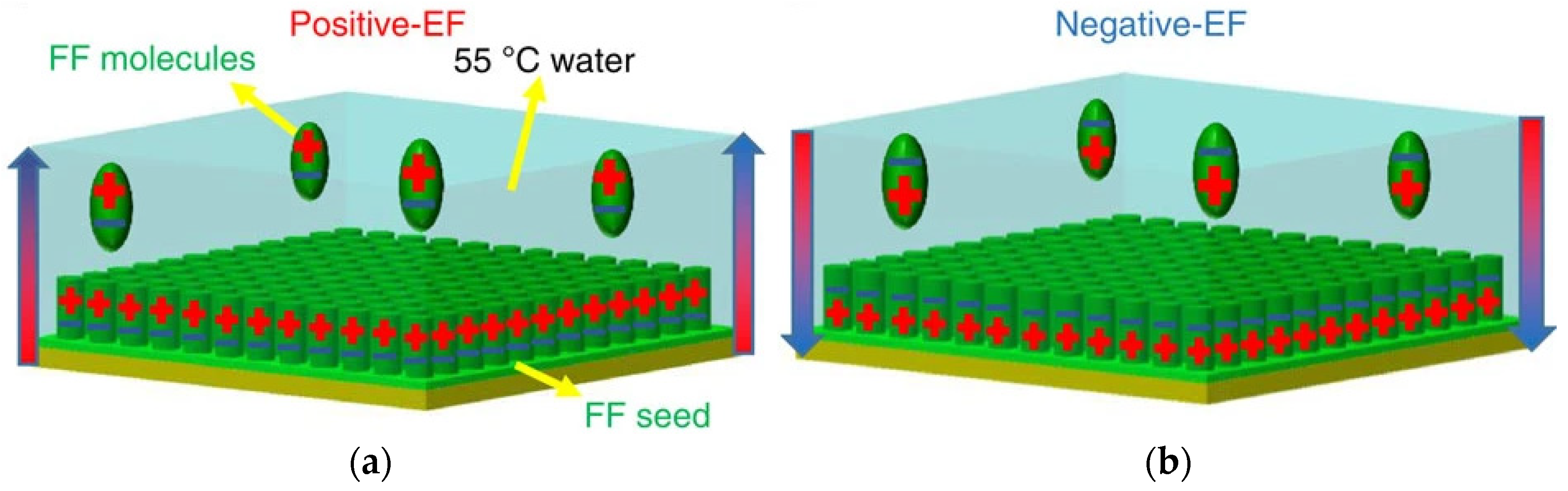
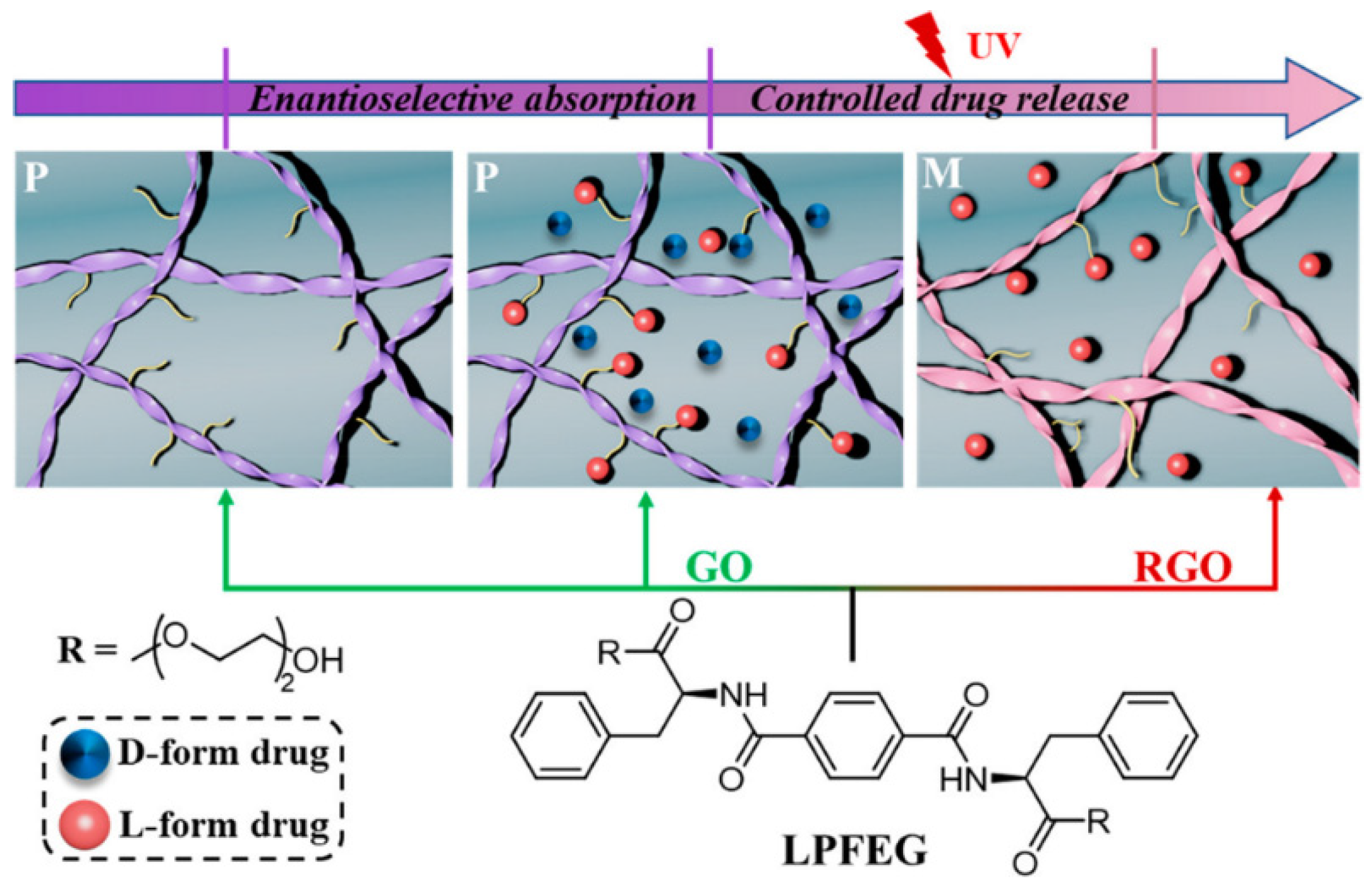
| Carbon Nanostructure | Peptide | Material | Application | Ref. |
|---|---|---|---|---|
| Fullerene | Aβ(1—40), Aβ(1—42) | Solution | Amyloidosis inhibition | [112] |
| Fullerene | Aβ(1—42) | Solution | Amyloidosis inhibition | [113] |
| Fullerene | Cyclopeptide nanotube | Solution | Bioelectronics | [114] |
| Fullerene | Fmoc-Phe-Phe | Hydrogel | Antibacterials | [115] |
| Carbon dots | D-Leu-L-Phe-L-Phe | Hydrogel | Biomaterial, biosensing | [116] |
| Carbon dots | Aβ(1—42) | Solution | Amyloidosis inhibition | [117,118] |
| Carbon dots | Aβ(1—42) | Solution | Biosensing | [119] |
| Carbon dots | Biofilm amyloids | Solution | Antibacterials | [120] |
| Carbon dots | RGDAEAKAEAKYWYAFAEAKAEAKRGD | Solution | Theranostics | [121] |
| Carbon dots, GO 1 | AEAKAEAKYWYAFAEAKAEAK | Solution | Biosensing | [111] |
| Carbon dots, CNTs, GO 1 | Aβ33—42 | Solution | Amyloidosis inhibition | [67] |
| CNHs, CNTs, GO 1 | L-Leu-D-Phe-D-Phe | Hydrogel | Biomaterial, drug delivery | [66] |
| CNTs | EFK8 | Hydrogel | Tissue regeneration | [98] |
| CNTs | RADA16-I | Hydrogel | Tissue regeneration | [122] |
| CNTs | Boc-β3(R)Phe-β3(R)Phe-OH Boc-γ4(R)-Phe-γ4(R)Phe-OH | Fibrils | Tissue regeneration | [123] |
| CNTs, graphene | Trp, Phe | Solution | Bioelectronics | [124] |
| CNTs, GO 1 | C8H16(-CH2-NH-Gly5)2·2HCl | Film | Tissue regeneration | [100] |
| CNTs, GO 1 | Fmoc-Tyr-OH, Fmoc-Tyr(Bzl)-OH | Hydrogel | Drug delivery | [125] |
| Graphene | Phe-Phe | Solution | Biosensing | [126] |
| Graphene | IMVTESSDYSSY | Film | Biosensing | [127] |
| Graphene | HSSYWYAFNNKT IMVTESSDYSSY | Film | Biosensing | [128] |
| Graphene | YGAGAGAY, EGAGAGAE, RGAGAGAR | Solution | Biosensing | [129] |
| Graphene | LLVFGAKMLPHHGA | Scaffold | Tissue regeneration | [130] |
| GO 1 | Phe-Phe | Solution | Biosensing | [131] |
| GO 1 | Phe-Phe, Tyr-Tyr | Solution | Theranostics | [132] |
| GO 1 | CLVPRGSC, CRGC | Solution | Biosensing | [133] |
| GO 1 | C16CO-KKFF | Membrane | Antibacterials | [134] |
| GO 1 | FEFKFEFK | Hydrogel | Tissue regeneration | [135] |
| GO 1 | AEAKAEAKYWYAFAEAKAEAK | Solution | Tissue regeneration | [136] |
| GO 1 | N3-KKPPPPKGPLGVRGC-CONH2 N3-KKPPPPKGPLGVRGA-CONH2 N3-KKPPPPKAAPFC-CONH2 | Solution | Biosensing | [137] |
| GO 1 | Block copolymer polypeptide PBLG-b-PDMS-b-PBLG | Gel | Tissue regeneration | [138] |
| GO 1, rGO 2 | VEVKVEVK, FEFKFEFK, FEFEFKFE | Hydrogel | Tissue regeneration | [139] |
| rGO 2 | Boc-Trp-PABA-OMe, Boc-Phe-PABA-OMe, Boc-Phg-PABA-OMe | Hydrogel | Tissue regeneration | [140] |
| rGO 2 | Polylysine, polyglutamate | Dispersion | Tissue regeneration | [141] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rozhin, P.; Charitidis, C.; Marchesan, S. Self-Assembling Peptides and Carbon Nanomaterials Join Forces for Innovative Biomedical Applications. Molecules 2021, 26, 4084. https://doi.org/10.3390/molecules26134084
Rozhin P, Charitidis C, Marchesan S. Self-Assembling Peptides and Carbon Nanomaterials Join Forces for Innovative Biomedical Applications. Molecules. 2021; 26(13):4084. https://doi.org/10.3390/molecules26134084
Chicago/Turabian StyleRozhin, Petr, Costas Charitidis, and Silvia Marchesan. 2021. "Self-Assembling Peptides and Carbon Nanomaterials Join Forces for Innovative Biomedical Applications" Molecules 26, no. 13: 4084. https://doi.org/10.3390/molecules26134084
APA StyleRozhin, P., Charitidis, C., & Marchesan, S. (2021). Self-Assembling Peptides and Carbon Nanomaterials Join Forces for Innovative Biomedical Applications. Molecules, 26(13), 4084. https://doi.org/10.3390/molecules26134084








