Exploration of Anti-HIV Phytocompounds against SARS-CoV-2 Main Protease: Structure-Based Screening, Molecular Simulation, ADME Analysis and Conceptual DFT Studies
Abstract
1. Introduction
2. Results
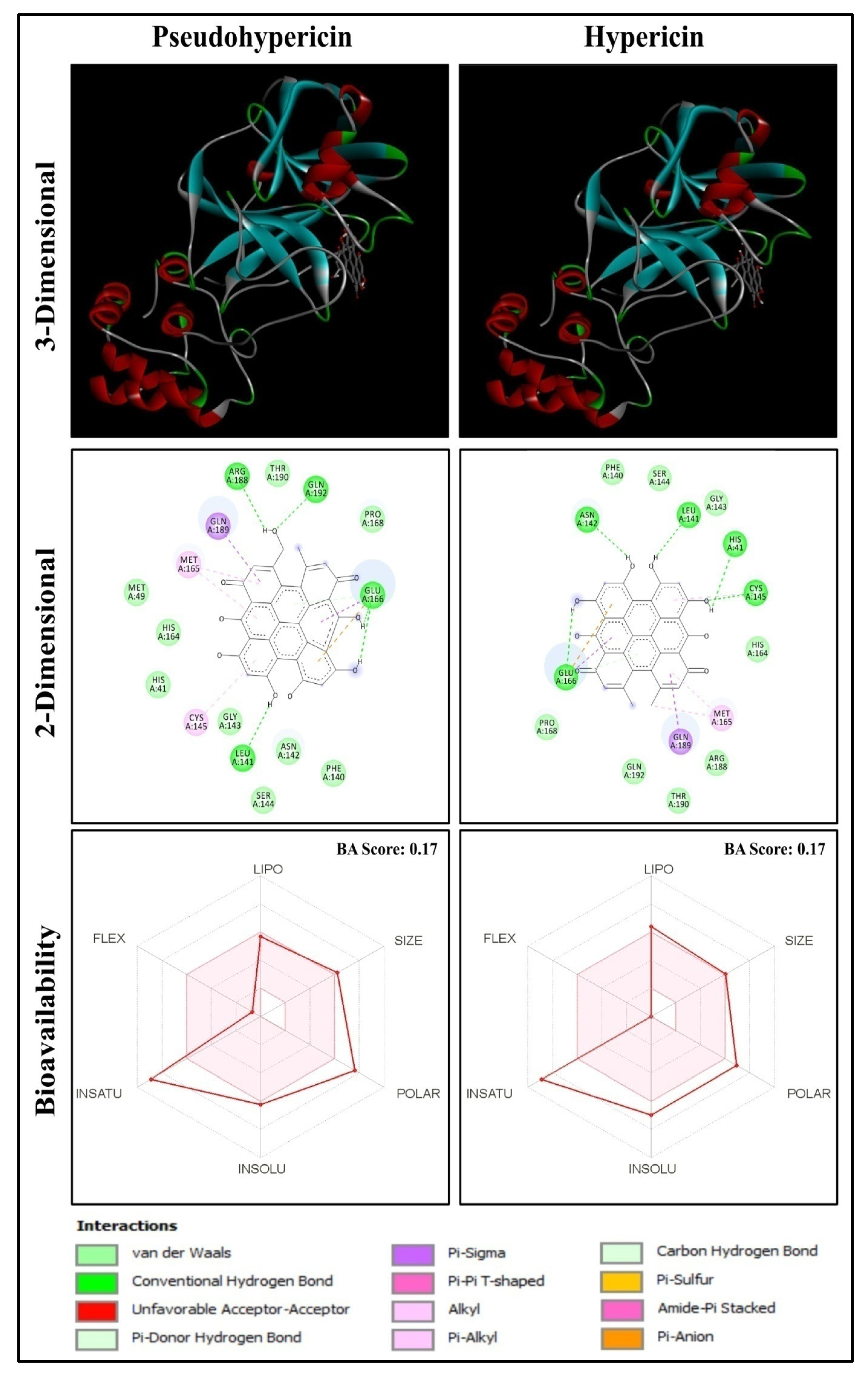
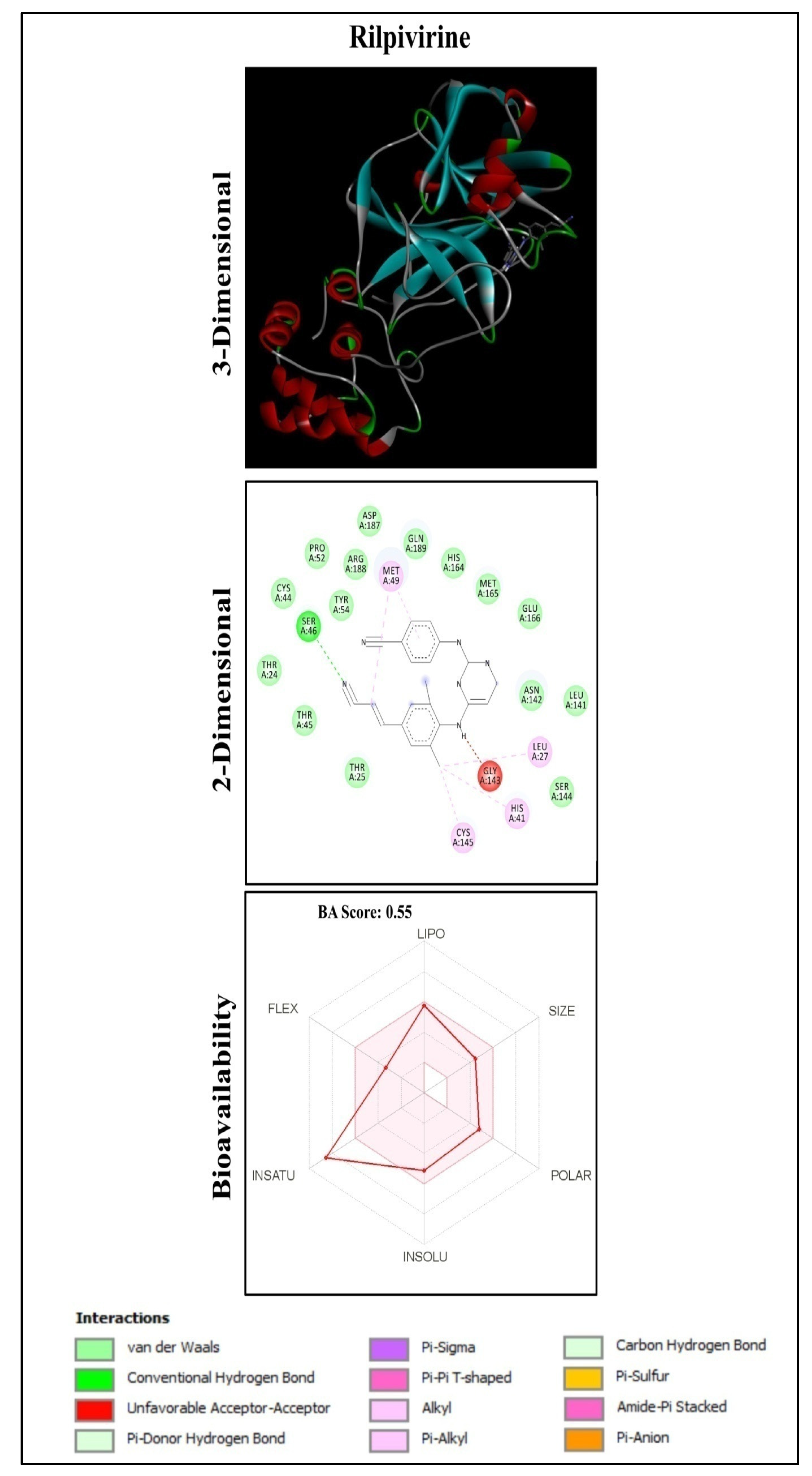
2.1. Molecular Docking Analysis
2.2. Functional Group Analysis
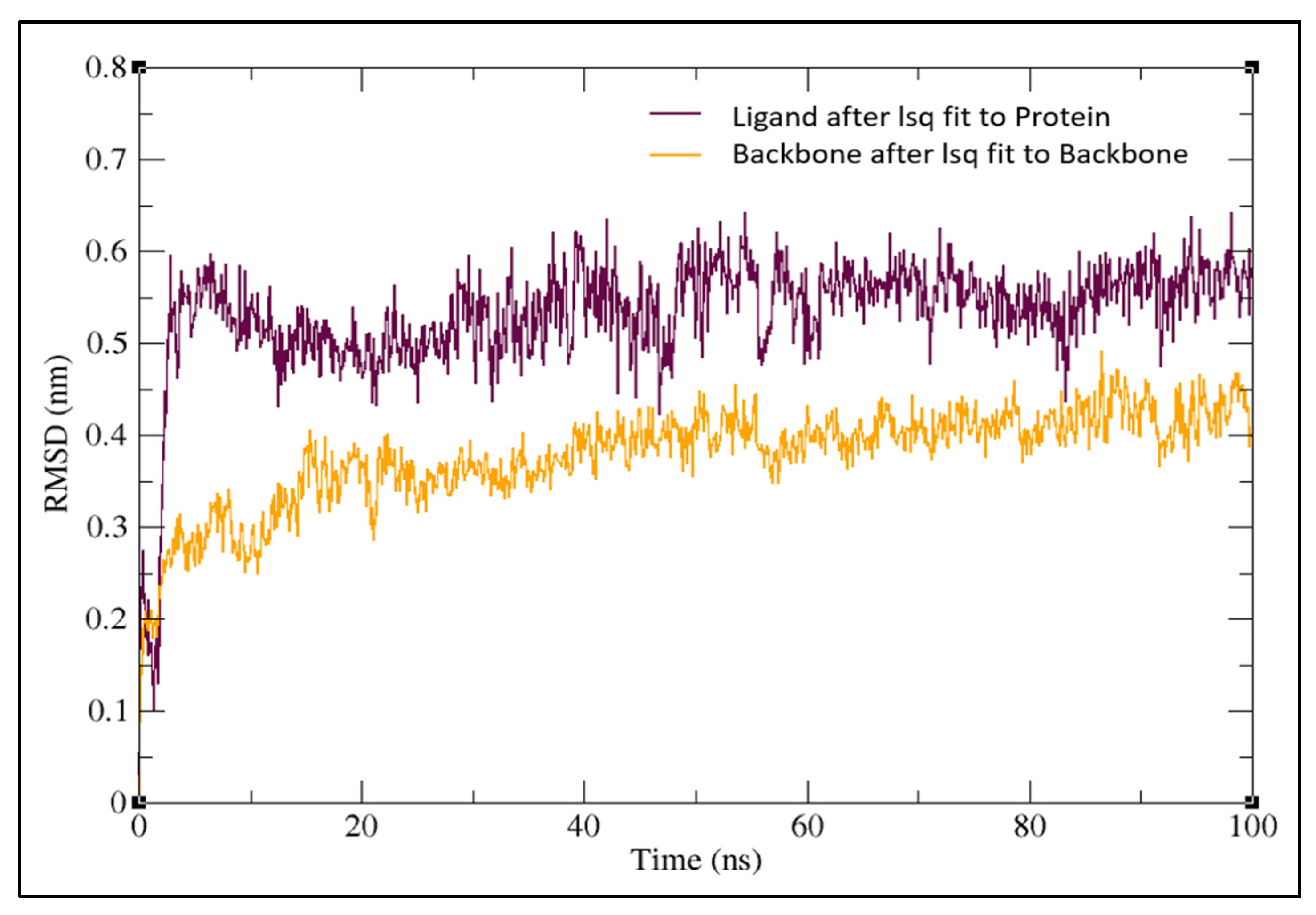
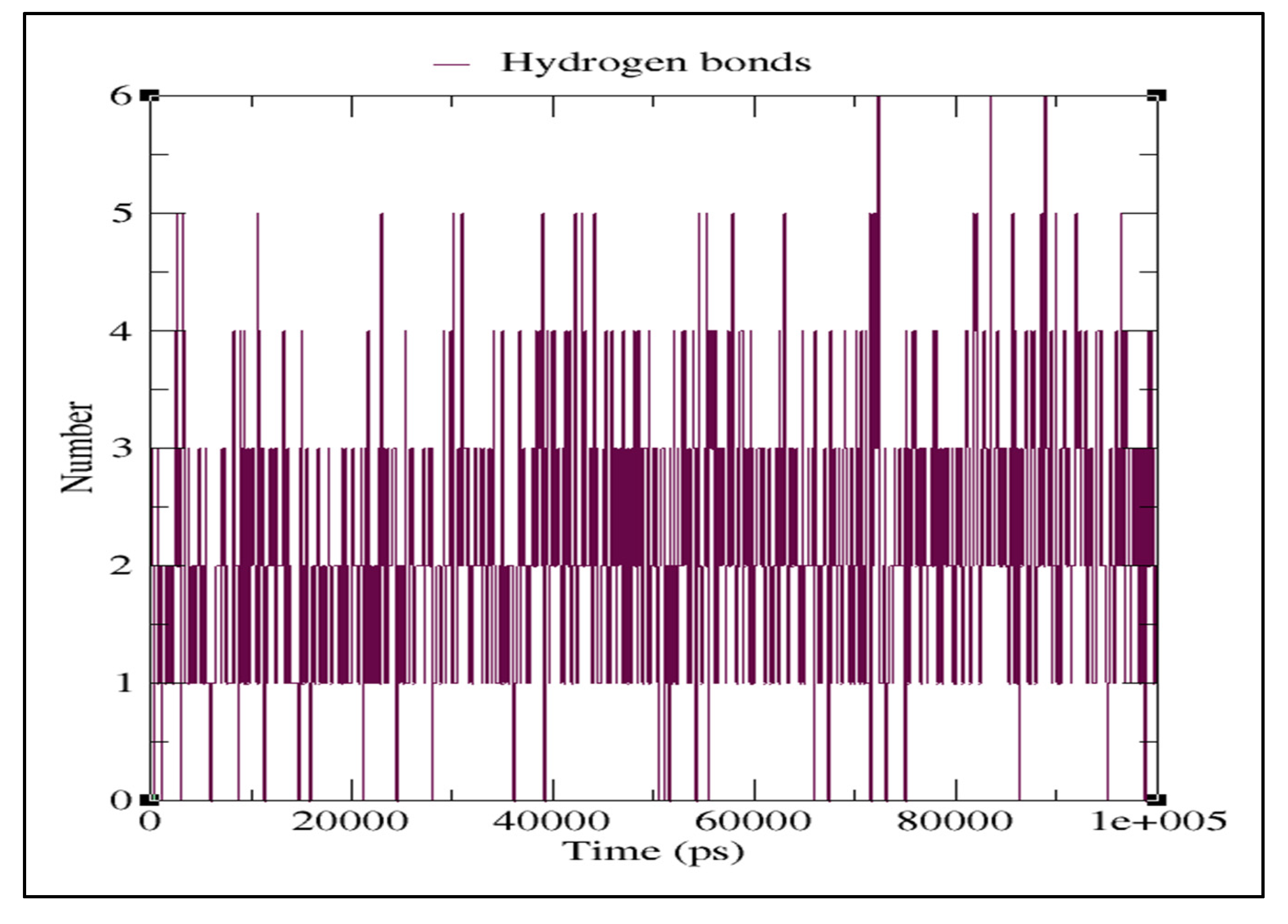
2.3. Molecular Dynamics Simulation
2.4. ADME Properties of Ligands
2.5. Conceptual DFT Studies
3. Discussion
4. Materials and Methods
4.1. Ligand Preparation
4.2. Preparation of the Target Protein
4.3. Protein Structure Validation
4.4. Molecular Docking
4.5. Molecular Dynamics (MD) Simulations
4.6. ADME (Absorption, Distribution, Metabolism and Excretion) Test
4.7. Conceptual DFT Studies
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abrahão, J.S.; de Arruda, L.B. Special Issue Emerging Viruses: Surveillance, Prevention, Evolution, and Control. Viruses 2020, 12, 306. [Google Scholar] [CrossRef] [PubMed]
- Ryu, W.S. New Emerging Viruses. In Molecular Virology of Human Pathogenic Viruses; Elsevier: Amsterdam, The Netherlands, 2017; pp. 289–302. [Google Scholar]
- Jiang, S. Don’t Rush to Deploy COVID-19 Vaccines and Drugs Without Sufficient Safety Guarantees. Nature 2020, 579, 321. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Lin, B.; Chu, V.; Hu, Z.; Hu, X.; Xiao, J.; Wang, A.Q.; Schweitzer, C.J.; Li, Q.; Imamura, M.; et al. Repurposing of the Antihistamine Chlorcyclizine and Related Compounds for Treatment of Hepatitis C Virus Infection. Sci. Transl. Med. 2015, 7, 282ra49. [Google Scholar] [CrossRef] [PubMed]
- Barrows, N.J.; Campos, R.K.; Powell, S.T.; Prasanth, K.R.; Schott-Lerner, G.; Soto-Acosta, R.; Galarza-Muñoz, G.; McGrath, E.L.; Urrabaz-Garza, R.; Gao, J.; et al. A Screen of FDA-Approved Drugs for Inhibitors of Zika Virus Infection. Cell Host Microbe 2016, 20, 259–270. [Google Scholar] [CrossRef]
- Cao, B.; Wang, Y.; Wen, D.; Liu, W.; Wang, J.; Fan, G.; Ruan, L.; Song, B.; Cai, Y.; Wei, M.; et al. A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe COVID-19. New Engl. J. Med. 2020, 382, 1787–1799. [Google Scholar] [CrossRef] [PubMed]
- Hui, D.S.; Azhar, E.I.; Madani, T.A.; Ntoumi, F.; Kock, R.; Dar, O.; Ippolito, G.; Mchugh, T.D.; Memish, Z.A.; Drosten, C.; et al. The Continuing 2019-nCoV Epidemic Threat of Novel Coronaviruses to Global Health—The Latest 2019 Novel Coronavirus Outbreak in Wuhan, China. Int. J. Infect. Dis. 2020, 91, 264–266. [Google Scholar] [CrossRef]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and Clinical Characteristics of 99 Cases of 2019 Novel Coronavirus Pneumonia in Wuhan, China: A Descriptive Study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Kumar, Y.; Singh, H.; Patel, C.N. In silico Prediction of Potential Inhibitors for the Main Protease of SARS-CoV-2 Using MolecularDocking and Dynamics Simulation Based Drug-Repurposing. J. Infect. Public Health 2020, 13, 1210–1223. [Google Scholar] [CrossRef]
- Wu, Y.; Chang, K.Y.; Lou, L.; Edwards, L.G.; Doma, B.K.; Xie, Z.R. In Silico Identification of Drug Candidates Against COVID-19. Inform. Med. Unlocked 2020, 21, 100461. [Google Scholar] [CrossRef]
- Yang, Y.M.; Gupta, S.K.; Kim, K.J.; Powers, B.E.; Cerqueira, A.; Wainger, B.J.; Ngo, H.D.; Rosowski, K.A.; Schein, P.A.; Ackeifi, C.A.; et al. A Small Molecule Screen in Stem-Cell-Derived Motor Neurons Identifies a Kinase Inhibitor as a Candidate Therapeutic for ALS. Cell Stem Cell 2013, 12, 713–726. [Google Scholar] [CrossRef]
- Hilgenfeld, R. From SARS to MERS: Crystallographic Studies on Coronaviral Proteases Enable Antiviral Drug Design. FEBSJ 2014, 281, 4085–4096. [Google Scholar] [CrossRef] [PubMed]
- Anand, K. Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of Anti-SARS Drugs. Science 2003, 300, 1763–1767. [Google Scholar] [CrossRef] [PubMed]
- Shereen, M.A.; Khan, S.; Kazmi, A.; Bashir, N.; Siddique, R. COVID-19 Infection: Emergence, Transmission, and Characteristics of Human Coronaviruses. J. Adv. Res. 2020, 24, 91–98. [Google Scholar] [CrossRef]
- Elzupir, A.O. Inhibition of SARS-CoV-2 Main Protease 3Clpro by Means of α-Ketoamide and Pyridone-containing Pharmaceuticals Using in silico Molecular Docking. J. Mol. Struct. 2020, 1222, 128878. [Google Scholar] [CrossRef] [PubMed]
- Pattnaik, B.; Patil, S.S.; Chandrashekar, S.; Amachawadi, R.G.; Dash, A.P.; Yadav, M.P.; Prasad, K.S.; Sushma, P.; Jain, A.S.; Shivamallu, C. COVID-19 Pandemic: A Systematic Review on the Coronaviruses of Animals and SARS-CoV-2. J. Exp. Biol. Agric. Sci. 2021, 9, 117–130. [Google Scholar] [CrossRef]
- Dharmashekara, C.; Pradeep, S.; Prasad, S.K.; Jain, A.S.; Syed, A.; Prasad, K.S.; Patil, S.S.; Beelagi, M.S.; Srinivasa, C.; Shivamallu, C. Virtual Screening of Potential Phyto Candidates as Therapeutic Leads Against SARS-CoV-2 Infection. Environ. Chall. 2021, 4, 100136. [Google Scholar] [CrossRef]
- Zhang, L.; Lin, D.; Sun, X.; Curth, U.; Drosten, C.; Sauerhering, L.; Becker, S.; Rox, K.; Hilgenfeld, R. Crystal Structure of SARS-CoV-2 Main Protease Provides a Basis for Design of Improved α-Ketoamide Inhibitors. Science 2020, 368, 409–412. [Google Scholar] [CrossRef]
- Anandan, S.; Mahadevamurthy, M.; Ansari, M.A.; Alzohairy, M.A.; Alomary, M.N.; Siraj, S.F.; Nagaraja, S.H.; Chikkamadaiah, M.; Ramachandrappa, L.T.; Krishnappa, H.K.N.; et al. Biosynthesized ZnO-NPs from Morus indica Attenuates Methylglyoxal-Induced Protein Glycation and RBC Damage: In-Vitro, In-Vivo and Molecular Docking Study. Biomolecules 2019, 9, 882. [Google Scholar] [CrossRef] [PubMed]
- Gowtham, H.G.; Murali, M.; Singh, S.B.; Shivamallu, C.; Pradeep, S.; Shivakumar, C.S.; Anandan, S.; Thampy, A.; Achar, R.R.; Silina, E.; et al. Phytoconstituents of Withaniasomnifera unveiled Ashwagandhanolide as a potential drug targeting breast cancer: Investigations through computational, molecular docking and conceptual DFT studies. PLoS ONE 2022, 17, e0275432. [Google Scholar] [CrossRef]
- Anandan, S.; Gowtham, H.G.; Shivakumara, C.S.; Thampy, A.; Brijesh Singh, S.; Murali, M.; Shivamallu, C.; Pradeep, S.; Shilpa, N.; Shati, A.A.; et al. Integrated approach for studying bioactive compounds from Cladosporium spp. against estrogen receptor alpha as breast cancer drug target. Sci. Rep. 2022, 22038. [Google Scholar] [CrossRef]
- Choudhary, M.I.; Shaikh, M.; tul-Wahab, A.; ur-Rahman, A. In silico Identification of Potential Inhibitors of KeySARS-CoV-2 3CL Hydrolase (Mpro) via Molecular Docking, MMGBSA Predictive Binding Energy Calculations, and Molecular Dynamics Simulation. PLoS ONE 2020, 15, e0235030. [Google Scholar] [CrossRef]
- Odar, H.A.; Ahiel, S.W.; Albeer, A.A.M.A.; Hashim, A.F.; Rayshan, A.M.; Humadi, S.S. Molecular Docking and Dynamics Simulation of FDA Approved Drugs with the Main Protease from 2019 Novel Coronavirus. Bioinformation 2020, 16, 236–244. [Google Scholar] [CrossRef] [PubMed]
- Murali, M.; Gowtham, H.G.; Ansari, M.A.; Alomary, M.N.; Alghamdi, S.; Almehmadi, M.; Singh, S.B.; Shilpa, N.; Aiyaz, M.; Kalegowda, N.; et al. Repositioning Therapeutics for SARS-CoV-2: Virtual Screening of Plant-based Anti-HIV Compounds as Possible Inhibitors against COVID-19 Viral RdRp. Curr. Pharm. Des. 2022, 28, 969–980. [Google Scholar] [CrossRef] [PubMed]
- Kurapati, K.R.V.; Atluri, V.S.; Samikkannu, T.; Garcia, G.; Nair, M.P.N. Natural Products as Anti-HIV Agents and Role in HIV-Associated Neurocognitive Disorders (HAND): A Brief Overview. Front. Microbiol. 2016, 6, 1444. [Google Scholar] [CrossRef] [PubMed]
- Laila, U.; Akram, M.; Shariati, M.A.; Hashmi, A.M.; Akhtar, N.; Tahir, I.M.; Ghauri, A.O.; Munir, N.; Riaz, M.; Akhter, N.; et al. Role of Medicinal Plants in HIV/AIDS Therapy. Clin. Exp. Pharmacol. Physiol. 2019, 46, 1063–1073. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—AVisualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 155–461. [Google Scholar] [CrossRef]
- Dallakyan, S.; Olson, A.J. Small-Molecule Library Screening by Docking with PyRx. In Methods in Molecular Biology; Springer: NewYork, NY, USA, 2014; pp. 243–250. [Google Scholar]
- Jain, A.S.; Sushma, P.; Dharmashekar, C.; Beelagi, M.S.; Prasad, S.K.; Shivamallu, C.; Prasad, A.; Syed, A.; Marraiki, N.; Prasad, K.S. In silico Evaluation of Flavonoids as Effective Antiviral Agents on the Spike Glycoprotein of SARS-CoV-2. Saudi J. Biol. Sci. 2021, 28, 1040–1051. [Google Scholar] [CrossRef]
- Nand, M.; Maiti, P.; Joshi, T.; Chandra, S.; Pande, V.; Kuniyal, J.C.; Ramakrishnan, M.A. Virtual screening of anti-HIV1 Compounds Against SARS-CoV-2: Machine Learning Modeling, Chemoinformatics and Molecular Dynamics Simulation Based Analysis. Sci. Rep. 2020, 10, 1–2. [Google Scholar] [CrossRef]
- Sang, P.; Tian, S.H.; Meng, Z.H.; Yang, L.Q. Anti-HIV Drug Repurposing Against SARS-CoV-2. RSC Advances. 2020, 10, 15775–15783. [Google Scholar] [CrossRef]
- Barros, R.O.; Junior, F.L.C.C.; Pereira, W.S.; Oliveira, N.M.N.; Ramos, R.M. Interaction of Drug Candidates with Various SARS-CoV-2 Receptors: An in silico Study to Combat COVID-19. J. Proteome Res. 2020, 19, 4567–4575. [Google Scholar] [CrossRef]
- Eleftheriou, P.; Amanatidou, D.; Petrou, A.; Geronikaki, A. In Silico Evaluation of the Effectivity of Approved Protease Inhibitors against the Main Protease of the Novel SARS-CoV-2 Virus. Molecules 2020, 25, 2529. [Google Scholar] [CrossRef]
- Shah, B.; Modi, P.; Sagar, S.R. In silico studies on therapeutic agents for COVID-19: Drug repurposing approach. Life Sci. 2020, 252, 117652. [Google Scholar] [CrossRef]
- Wu, R.; Wang, L.; Kuo, H.C.D.; Shannar, A.; Peter, R.; Chou, P.J.; Li, S.; Hudlikar, R.; Liu, X.; Liu, Z.; et al. An Update on Current Therapeutic Drugs Treating COVID-19. Curr. Pharmacol. Rep. 2020, 6, 56–70. [Google Scholar] [CrossRef]
- Sithisarn, P.; Michaelis, M.; Schubert-Zsilavecz, M.; Cinatl, J. Differential Antiviral and anti-Inflammatory Mechanisms of the Flavonoids Biochanin A and Baicalein in H5N1 Influenza A Virus-Infected Cells. Antivir. Res. 2013, 97, 41–48. [Google Scholar] [CrossRef]
- Carvalho, O.; Botelho, C.; Ferreira, C.; Ferreira, H.; Santos, M.; Diaz, M.; Oliveira, T.; Soares-Martins, J.; Almeida, M.; Júnior, A.S. In Vitro Inhibition of Canine Distemper Virus by Flavonoids and Phenolic Acids: Implications of Structural Differences for Antiviral Design. Res. Vet. Sci. 2013, 95, 717–724. [Google Scholar] [CrossRef]
- He, J.; Xu, L.; Yang, L.; Wang, X. Epigallocatechin Gallate Is the Most Effective Catechin Against Antioxidant Stress via Hydrogen Peroxide and Radical Scavenging Activity. Med. Sci. Monit. 2018, 24, 8198–8206. [Google Scholar] [CrossRef]
- Mahdi, M.; Mótyán, J.A.; Szojka, Z.I.; Golda, M.; Miczi, M.; Tőzsér, J. Analysis of the efficacy of HIV protease inhibitors against SARS-CoV-2′s main protease. Virol. J. 2020, 17, 190. [Google Scholar] [CrossRef]
- Pitsillou, E.; Liang, J.; Karagiannis, C.; Ververis, K.; Darmawan, K.K.; Ng, K.; Hung, A.; Karagiannis, T.C. Interaction of small molecules with the SARS-CoV-2 main protease in silico and in vitro validation of potential lead compounds using an enzyme-linked immunosorbent assay. Comput. Biol. Chem. 2020, 89, 107408. [Google Scholar] [CrossRef]
- Domingo, L.R.; Aurell, M.J.; Pérez, P.; Contreras, R. Quantitative Characterization of the Global Electrophilicity Power of Common diene/Dienophile Pairs in Diels-Alder Reactions. Tetrahedron 2002, 58, 4417–4423. [Google Scholar] [CrossRef]
- Swain, S.S.; Singh, S.R.; Sahoo, A.; Hussain, T.; Pati, S. Anti-HIV-drug and phyto-flavonoid combination against SARS-CoV-2: A molecular docking-simulation base assessment. J. Biomol. Struct. Dyn. 2022, 40, 6463–6476. [Google Scholar] [CrossRef] [PubMed]
- Ayan, A.K.; Çirak, C. Hypericin and Pseudohypericin Contents in Some Hypericum. Species Growing in Turkey. Pharm. Biol. 2008, 46, 288–291. [Google Scholar] [CrossRef]
- Brockmöller, J.; Reum, T.; Bauer, S.; Kerb, R.; Hübner, W.D.; Roots, I. Hypericin and Pseudohypericin: Pharmacokinetics and Effects on Photosensitivity in Humans. Pharmacopsychiatry 1997, 30, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Natesh, J.; Mondal, P.; Kaur, B.; Salam, A.A.A.; Kasilingam, S.; Meeran, S.M. Promising Phytochemicals of Traditional Himalayan Medicinal Plants Against Putative Replication and Transmission Targets of SARS-CoV-2 by Computational Investigation. Comput. Biol. Med. 2021, 133, 104383. [Google Scholar] [CrossRef] [PubMed]
- Domingo, L.R.; Ríos-Gutiérrez, M.; Pérez, P. Applications of the Conceptual Density Functional Theory Indices to Organic Chemistry Reactivity. Molecules 2016, 21, 748. [Google Scholar] [CrossRef]
- Jaramillo, P.; Domingo, L.R.; Chamorro, E.; Pérez, P. A Further Exploration of a Nucleophilicity Index Based on the Gas-Phase Ionization Potentials. J. Mol. Struct. Theochem. 2008, 865, 68–72. [Google Scholar] [CrossRef]
- Chakraborty, D.; Chattaraj, P.K. Conceptual Density Functional Theory Based Electronic Structure Principles. Chem. Sci. 2021, 12, 6264–6279. [Google Scholar] [CrossRef]
- Domingo, L.R.; Sáez, J.A. Understanding the Mechanism of Polar Diels-Alder Reactions. Org. Biomol. Chem. 2009, 7, 3576–3583. [Google Scholar] [CrossRef]
- Halgren, T.A. Merck Molecular Force Field. Basis, Form, Scope, Parameterization, and Performance of MMFF94. J. Comput. Chem. 1996, 17, 490–519. [Google Scholar] [CrossRef]
- Halgren, T.A. MMFF VI. MMFF94s Option for Energy Minimization Studies. J. Comput. Chem. 1999, 20, 720–729. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheesemanet, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian16 Revision, C.01; Gaussian Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Peveratia, R.; Truhlar, D.G. Screened-exchange density functionals with broad accuracy for chemistry and solid-state physics. Phys. Chem. Chem. Phys. 2012, 14, 16187–16191. [Google Scholar] [CrossRef]
- Flores-Holguín, N.; Frau, J.; Glossman-Mitnik, D. Conceptual DFT-Based Computational Peptidology of Marine Natural Com- pounds: Discodermins A–H. Molecules 2020, 25, 4158. [Google Scholar] [CrossRef]
- Marenich, A.V.; Cramer, C.J.; Truhlar, D.G. Universal Solvation Model Based on Solute Electron Density and a Continuum Model of the Solvent Defined by the Bulk Dielectric Constant and Atomic Surface Tensions. J. Phys. Chem. B 2009, 113, 6378–6396. [Google Scholar] [CrossRef]
- Geerlings, P.; Chamorro, E.; Chattaraj, P.K.; De Proft, F.; Gázquez, J.L.; Liu, S.; Morell, C.; Toro-Labbé, A.; Vela, A.; Ayers, P. Conceptual Density Functional Theory: Status, Prospects, Issues. Theor. Chem. Acc. 2020, 139, 1–8. [Google Scholar] [CrossRef]


| Sl. No. | Name of Compound | Name of Plants | Class | Binding Energy (kcal/mol) | Interaction of Residues with Hydrogen Bond |
|---|---|---|---|---|---|
| 1 | Pseudohypericin | Hypericum perforatum | Quinone | −10.2 | LEU141, GLU166, ARG188, GLN192 |
| 2 | Hypericin | Hypericum perforatum | Quinone | −10.1 | HIS41, LEU141, ASN142, CYS145, GLU166 |
| 3 | Robustaflavone | Rhus succedanea | Flavonoid | −9.7 | HIS163, HIS164 |
| 4 | Procyanidin B2 | Maytenus senegalensis | Flavonoid | −9.3 | THR26, HIS41, LEU141, GLU166 |
| 5 | (-)-Epicatechin-(4beta→8)-(-)-epigallocatechin | Maytenus senegalensis | Phenolic | −9.3 | LEU141, SER144, GLU166 |
| 6 | (-)-Epicatechin(4.beta.→8)(-)-4′-methylepigallocatechin | Maytenus senegalensis | Phenolic | −9.2 | THR26, SER144, GLU166 |
| 7 | Agathisflavone | Rhus succedanea | Flavonoid | −9.2 | GLU166 |
| 8 | Hinokiflavone | Rhus succedanea | Flavonoid | −9.2 | GLY109, GLN110 |
| 9 | Michellamine B | Ancistrocladuskorupensis | Alkaloid | −9.2 | LEU220, ARG222 |
| 10 | Rhusflavanone | Rhus succedanea | Flavonoid | −9.1 | GLN189 |
| 11 | GB-1a 7′-glucoside | Garcinia multiflora | Flavonoid | −9.1 | THR26, GLU166 |
| 12 | Quercetin 3-O-(2″galloyl)-alpha-L-arabinopyranoside | Acer okamotoanum | Flavonoid | −9.0 | ASN142, GLU166 |
| 13 | Wikstrol B | Wikstroemia indica | Flavonoid | −9.0 | HIS41, HIS163 |
| 14 | Morelloflavone | Garcinia multiflora | Flavonoid | −8.9 | PHE140, ASN142, GLN189, THR190 |
| 15 | Quercitrin | Acer okamotoanum | Flavonoid | −8.9 | LEU141, ASN142, HIS163, GLU166 |
| 16 | Actein | Cimicifuga racemosa | Terpene | −8.9 | ARG131, GLY195 |
| 17 | Rilpivirine | Drug | −8.9 | SER46 |
| Sl. No. | Homo | Lumo | Homo-Lumo Gap | χ | η | ω | S | N | ω− | ω+ | Δω± |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | −5.29 | −3.33 | 1.96 | 4.31 | 1.96 | 4.74 | 0.51 | 3.51 | 11.76 | 7.45 | 19.20 |
| 2 | −5.27 | −3.28 | 1.99 | 4.27 | 1.99 | 4.59 | 0.50 | 3.52 | 11.44 | 7.17 | 18.61 |
| 3 | −6.16 | −2.43 | 3.73 | 4.30 | 3.73 | 2.47 | 0.27 | 2.63 | 7.33 | 3.03 | 10.36 |
| 4 | −5.90 | −0.68 | 5.21 | 3.29 | 5.21 | 1.04 | 0.19 | 2.90 | 4.05 | 0.76 | 4.80 |
| 5 | −5.94 | −0.68 | 5.26 | 3.31 | 5.26 | 1.04 | 0.19 | 2.85 | 4.07 | 0.76 | 4.83 |
| 6 | −5.94 | −0.68 | 5.26 | 3.31 | 5.26 | 1.04 | 0.19 | 2.85 | 4.07 | 0.76 | 4.83 |
| 7 | −6.12 | −2.42 | 3.70 | 4.27 | 3.70 | 2.46 | 0.27 | 2.67 | 7.29 | 3.02 | 10.32 |
| 8 | −6.24 | −2.46 | 3.78 | 4.35 | 3.78 | 2.50 | 0.26 | 2.55 | 7.42 | 3.07 | 10.49 |
| 9 | −5.43 | −1.70 | 3.73 | 3.56 | 3.73 | 1.80 | 0.27 | 3.37 | 5.42 | 1.85 | 7.27 |
| 10 | −6.11 | −1.87 | 4.24 | 3.99 | 4.24 | 1.87 | 0.24 | 2.69 | 6.01 | 2.03 | 8.04 |
| 11 | −6.25 | −1.98 | 4.27 | 4.11 | 4.27 | 1.98 | 0.23 | 2.55 | 6.29 | 2.18 | 8.47 |
| 12 | −6.14 | −2.52 | 3.62 | 4.33 | 3.62 | 2.59 | 0.28 | 2.65 | 7.56 | 3.23 | 10.79 |
| 13 | −5.91 | −2.41 | 3.50 | 4.16 | 3.50 | 2.47 | 0.29 | 2.89 | 7.23 | 3.08 | 10.31 |
| 14 | −6.06 | −2.28 | 3.78 | 4.17 | 3.78 | 2.31 | 0.26 | 2.73 | 6.93 | 2.76 | 9.70 |
| 15 | −6.10 | −2.38 | 3.72 | 4.24 | 3.72 | 2.42 | 0.27 | 2.69 | 7.19 | 2.95 | 10.14 |
| 16 | −6.60 | −0.33 | 6.27 | 3.46 | 6.27 | 0.96 | 0.16 | 2.19 | 4.04 | 0.57 | 4.51 |
| 17 | −6.49 | −0.22 | 6.27 | 3.35 | 6.28 | 0.90 | 0.16 | 2.30 | 3.86 | 0.51 | 4.37 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murali, M.; Gowtham, H.G.; Shilpa, N.; Krishnappa, H.K.N.; Ledesma, A.E.; Jain, A.S.; Shati, A.A.; Alfaifi, M.Y.; Elbehairi, S.E.I.; Achar, R.R.; et al. Exploration of Anti-HIV Phytocompounds against SARS-CoV-2 Main Protease: Structure-Based Screening, Molecular Simulation, ADME Analysis and Conceptual DFT Studies. Molecules 2022, 27, 8288. https://doi.org/10.3390/molecules27238288
Murali M, Gowtham HG, Shilpa N, Krishnappa HKN, Ledesma AE, Jain AS, Shati AA, Alfaifi MY, Elbehairi SEI, Achar RR, et al. Exploration of Anti-HIV Phytocompounds against SARS-CoV-2 Main Protease: Structure-Based Screening, Molecular Simulation, ADME Analysis and Conceptual DFT Studies. Molecules. 2022; 27(23):8288. https://doi.org/10.3390/molecules27238288
Chicago/Turabian StyleMurali, Mahadevamurthy, Hittanahallikoppal Gajendramurthy Gowtham, Natarajamurthy Shilpa, Hemanth Kumar Naguvanahalli Krishnappa, Ana E. Ledesma, Anisha S. Jain, Ali A. Shati, Mohammad Y. Alfaifi, Serag Eldin I. Elbehairi, Raghu Ram Achar, and et al. 2022. "Exploration of Anti-HIV Phytocompounds against SARS-CoV-2 Main Protease: Structure-Based Screening, Molecular Simulation, ADME Analysis and Conceptual DFT Studies" Molecules 27, no. 23: 8288. https://doi.org/10.3390/molecules27238288
APA StyleMurali, M., Gowtham, H. G., Shilpa, N., Krishnappa, H. K. N., Ledesma, A. E., Jain, A. S., Shati, A. A., Alfaifi, M. Y., Elbehairi, S. E. I., Achar, R. R., Silina, E., Stupin, V., Ortega-Castro, J., Frau, J., Flores-Holguín, N., Amruthesh, K. N., Shivamallu, C., Kollur, S. P., & Glossman-Mitnik, D. (2022). Exploration of Anti-HIV Phytocompounds against SARS-CoV-2 Main Protease: Structure-Based Screening, Molecular Simulation, ADME Analysis and Conceptual DFT Studies. Molecules, 27(23), 8288. https://doi.org/10.3390/molecules27238288











