Synthesis, Self-Assembly, and Cell Responses of Aromatic IKVAV Peptide Amphiphiles
Abstract
:1. Introduction
2. Results and Discussion
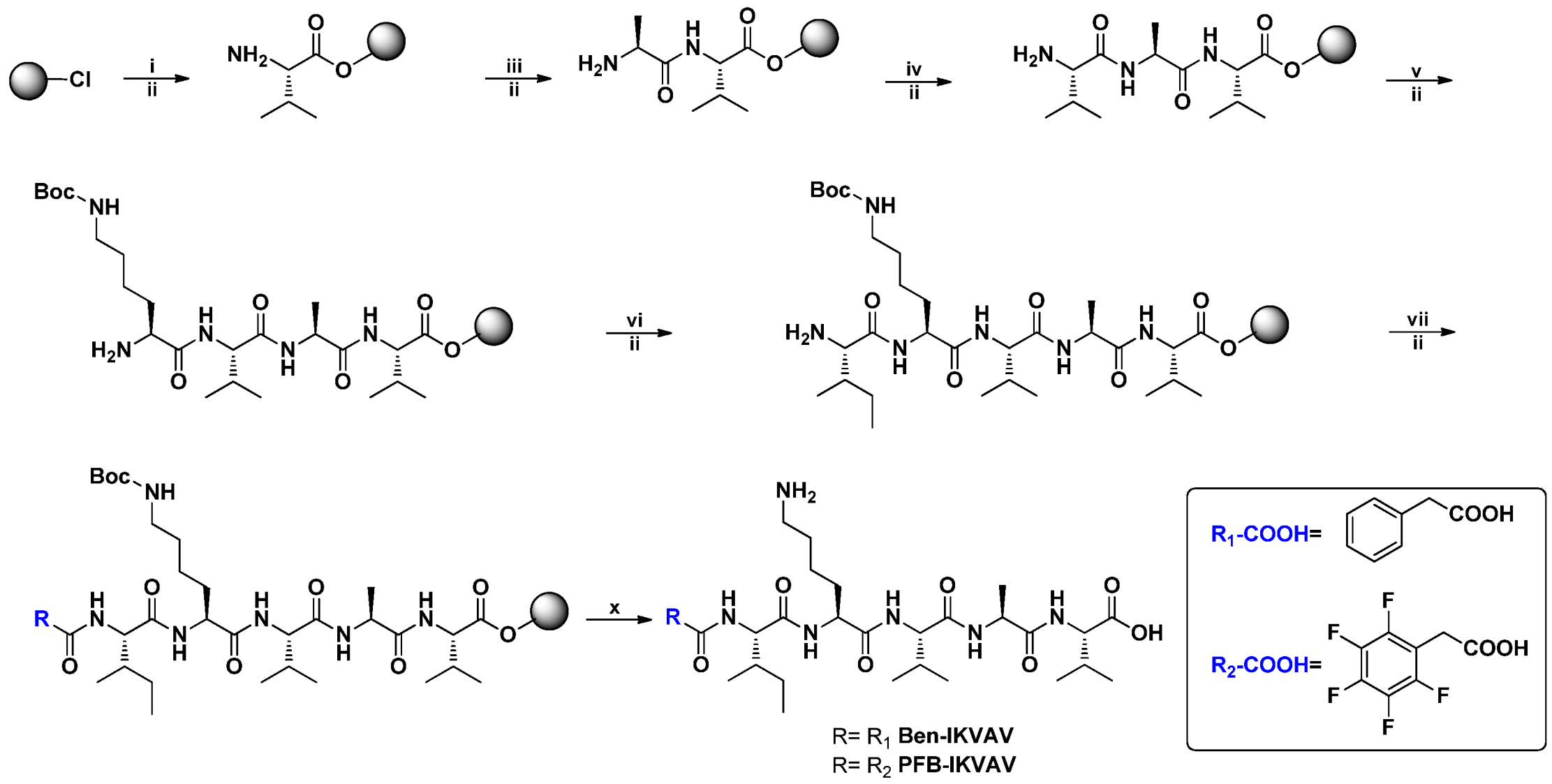
2.1. Molecular Design and Synthesis
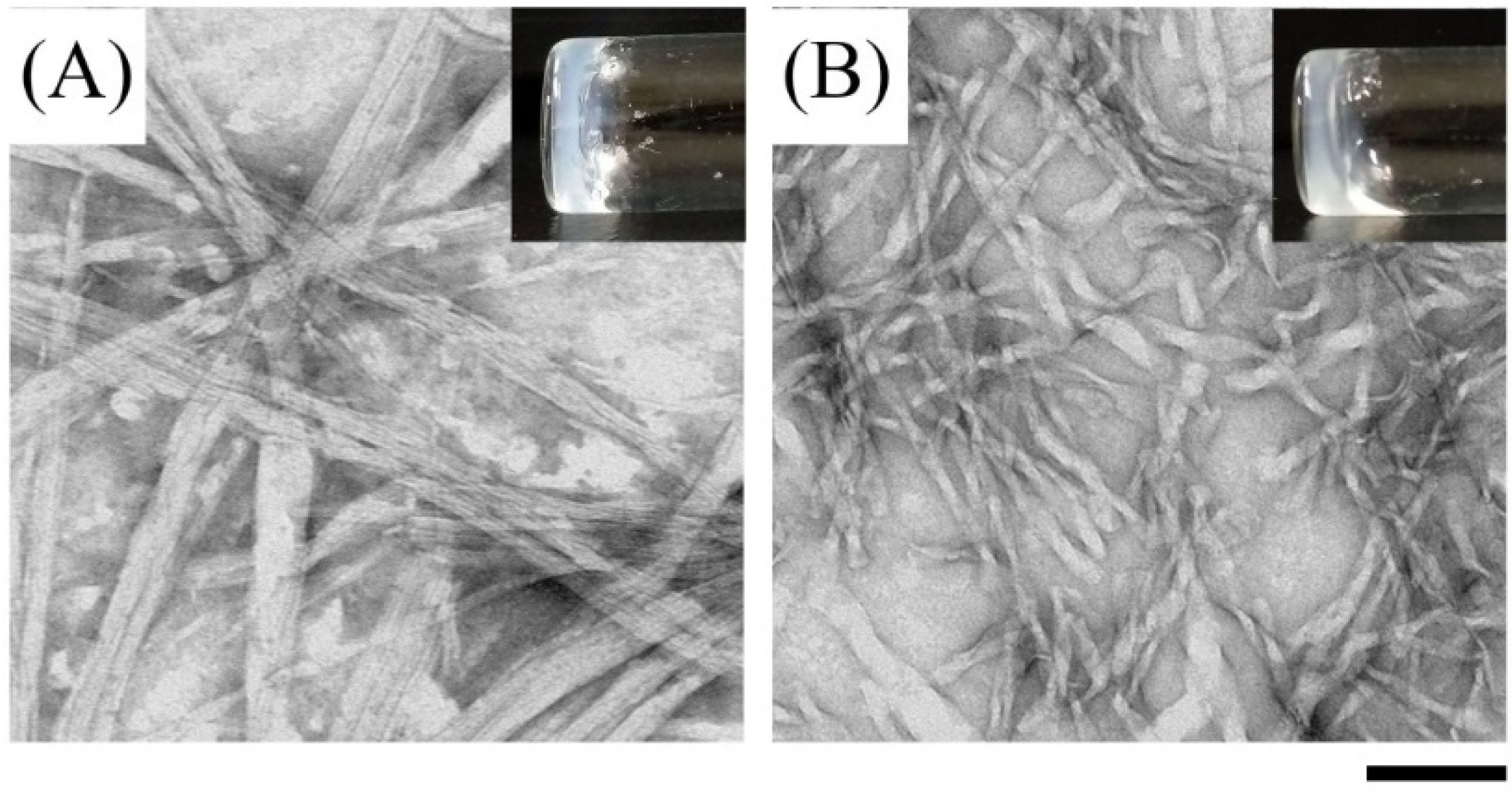
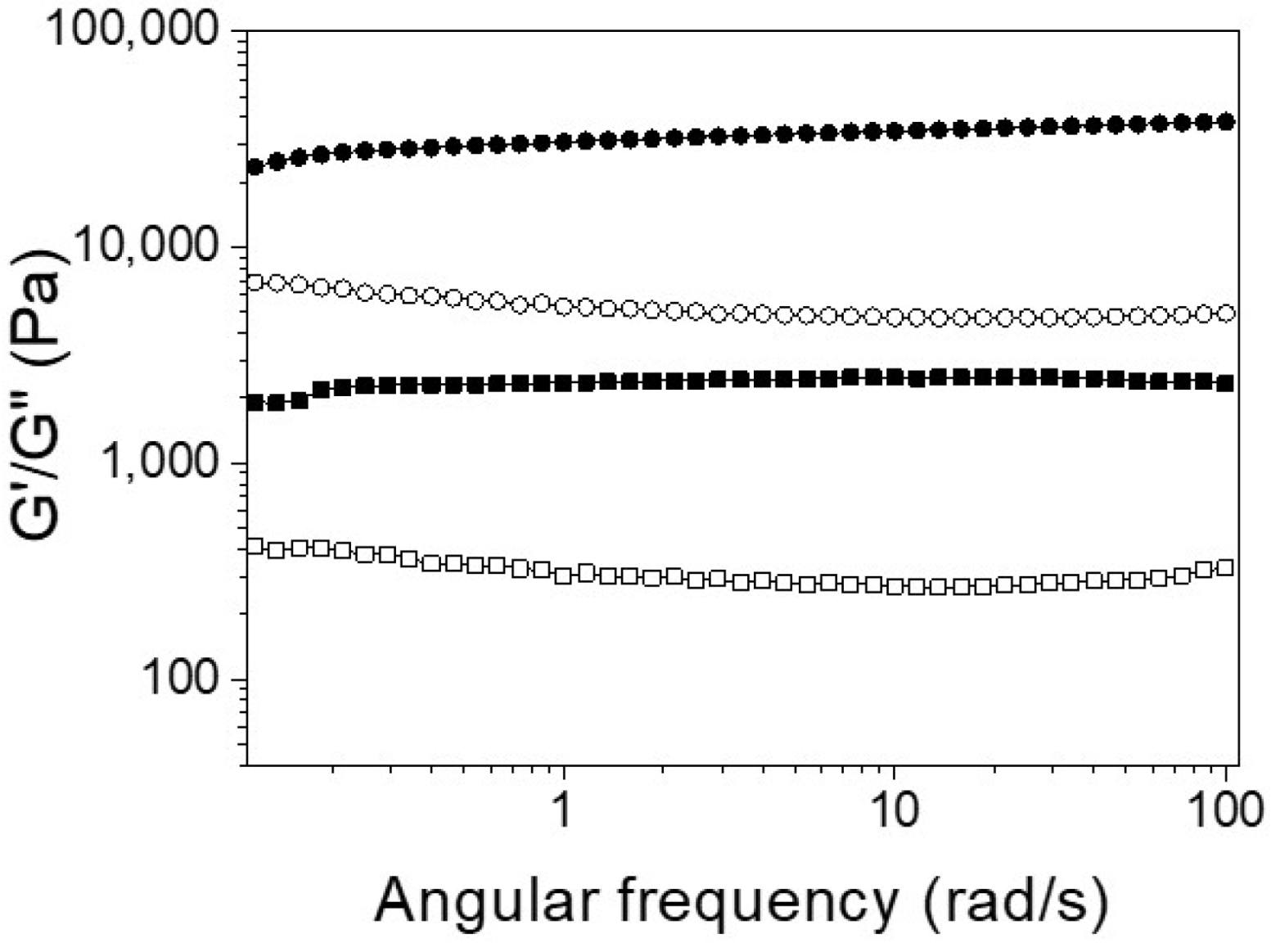
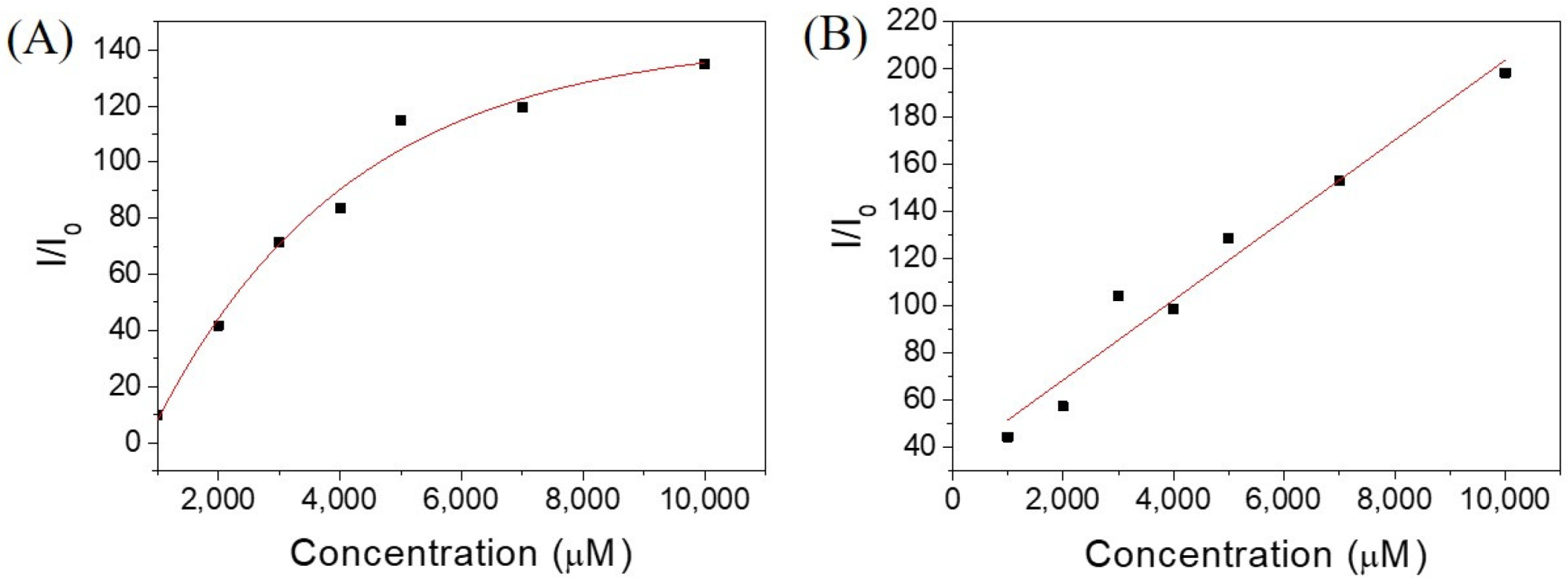
2.2. Investigation of Self-Assembly Properties
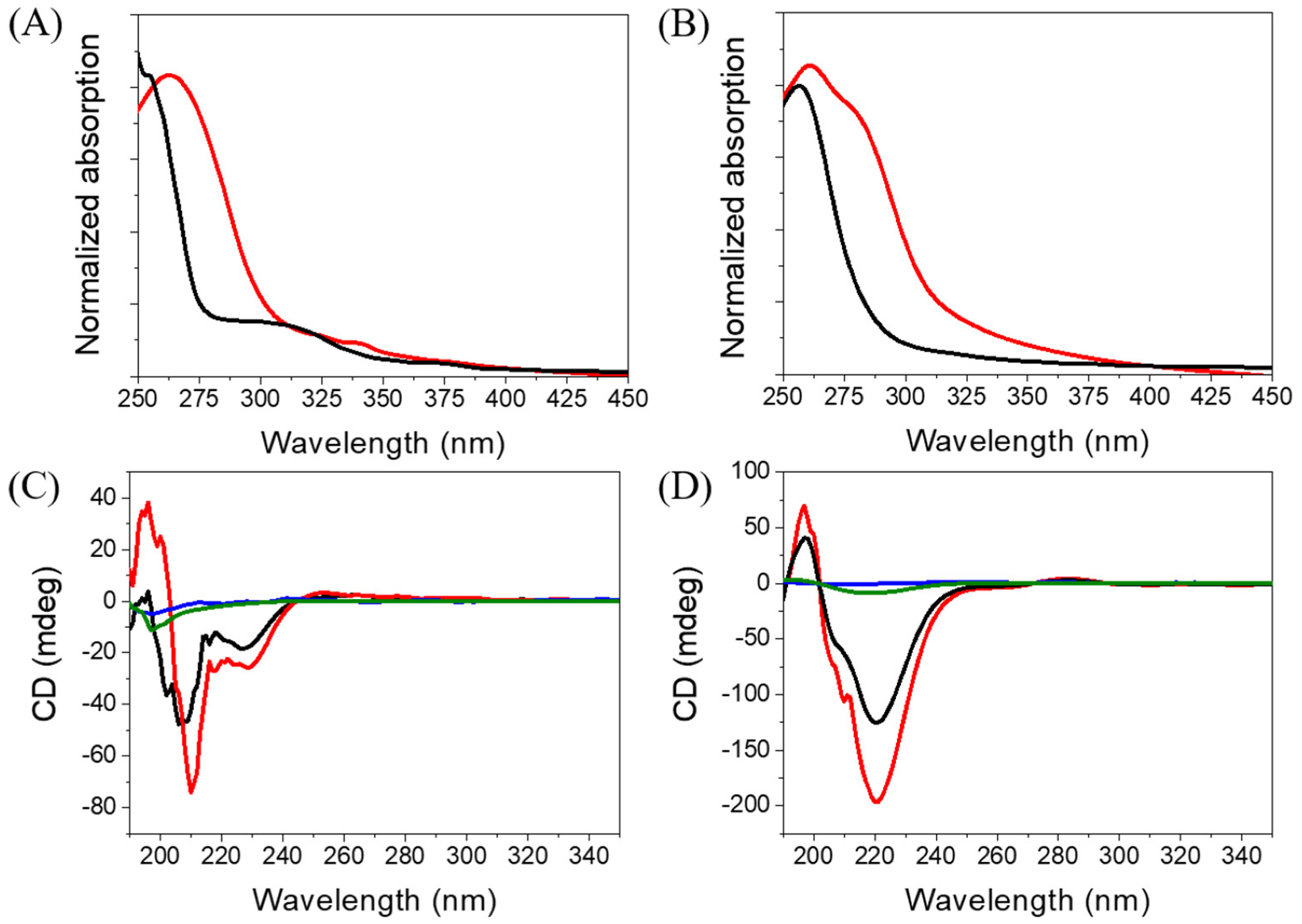
2.3. Spectroscopic Characterization
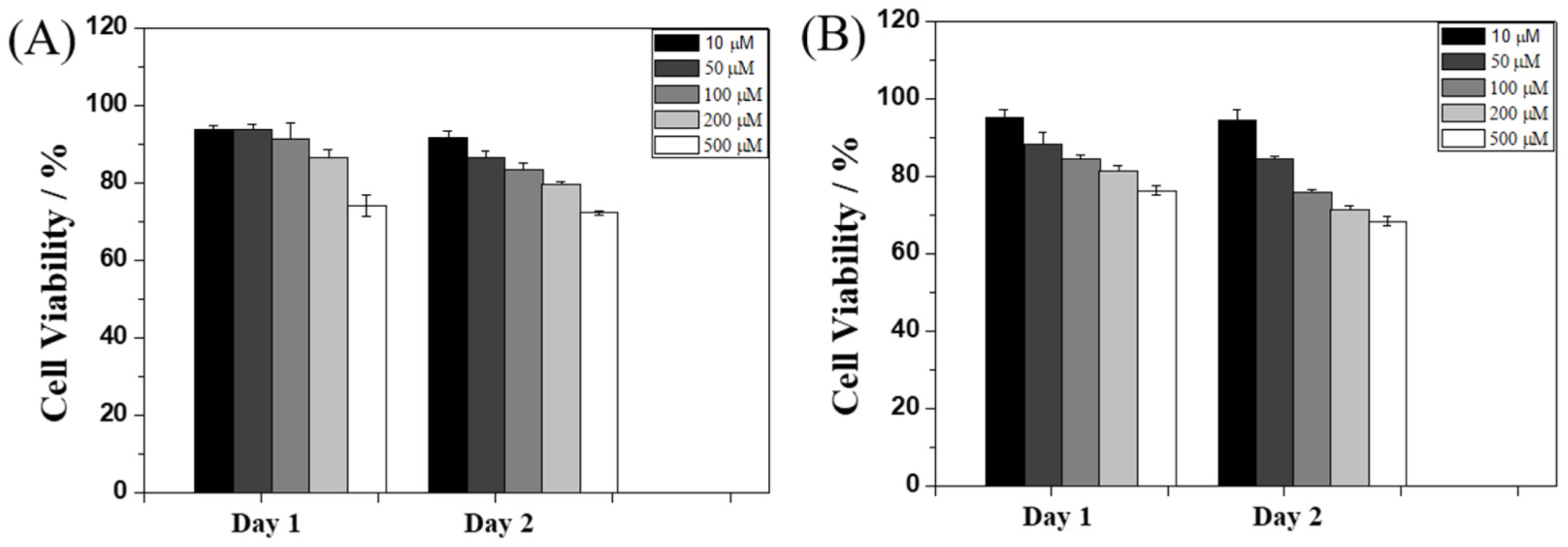
2.4. Cytotoxicity Test
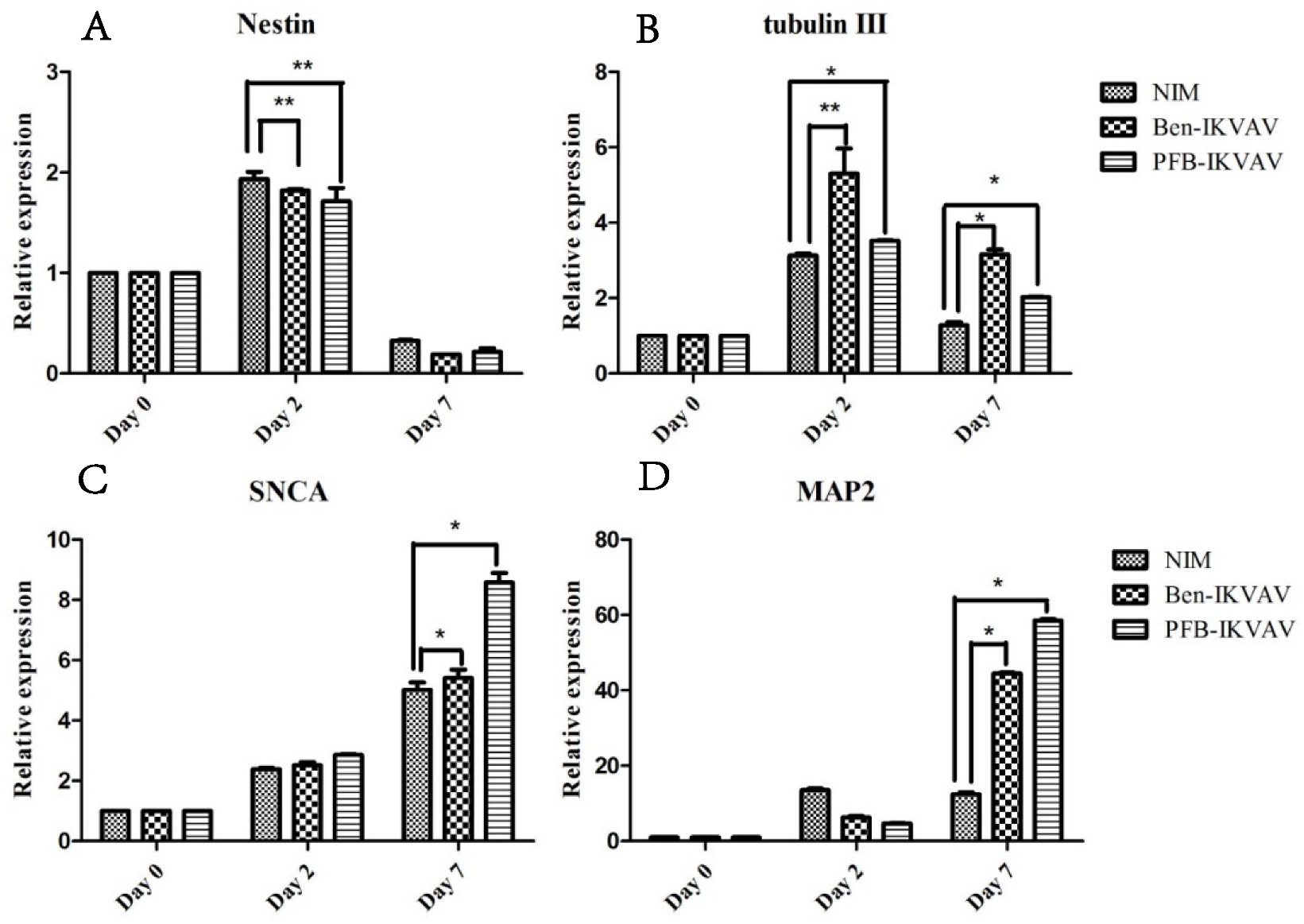
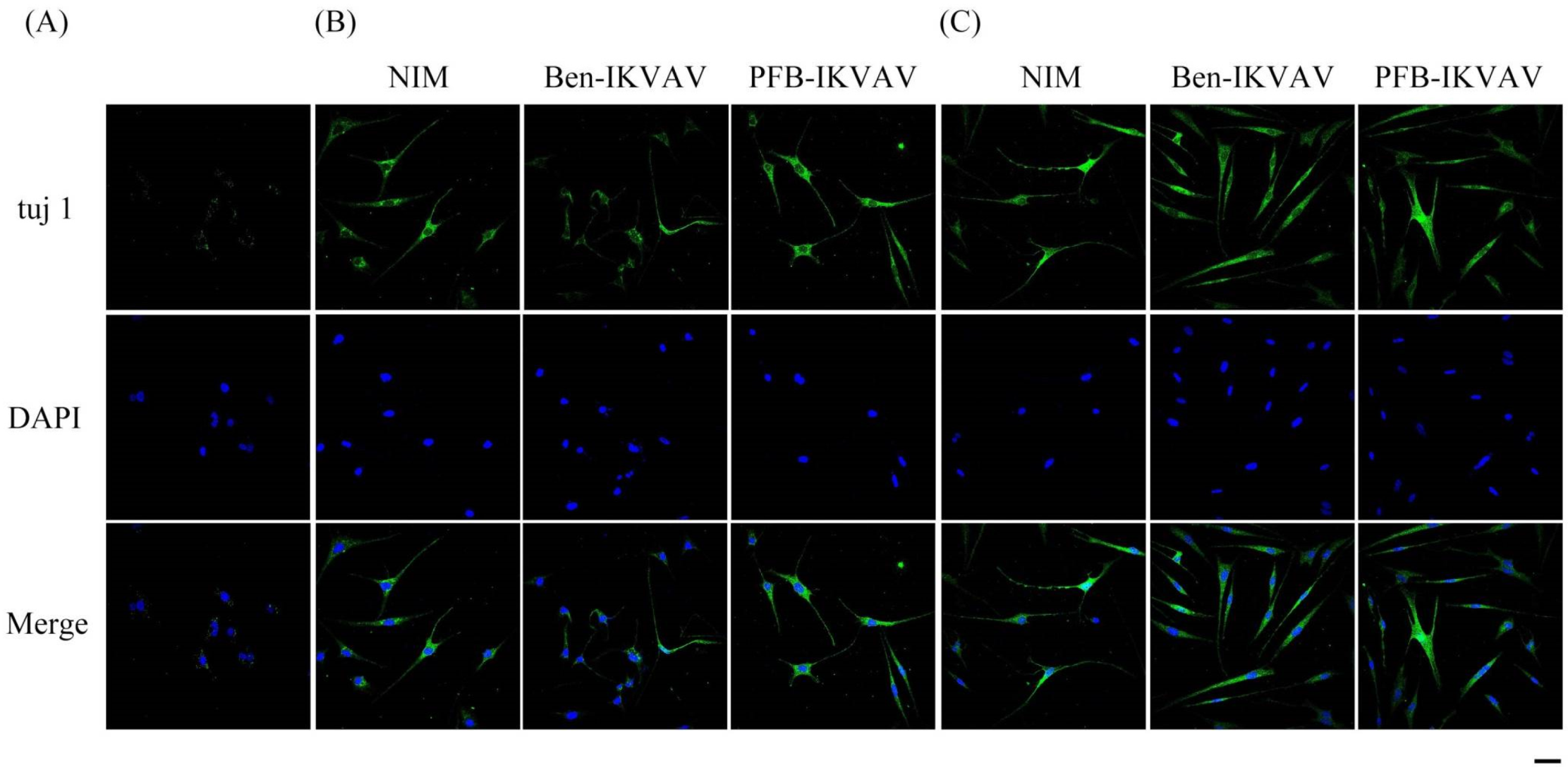
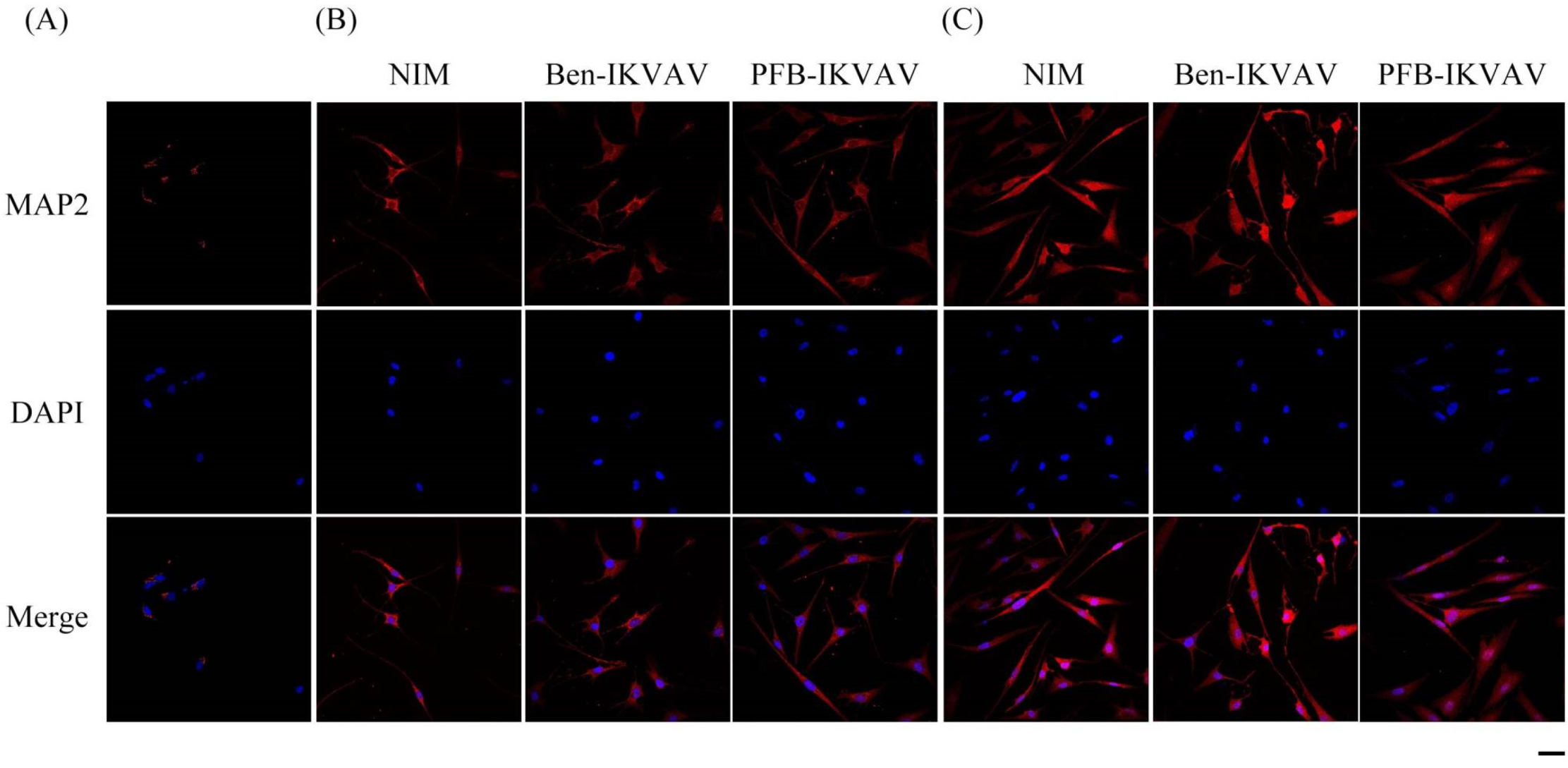
2.5. Cell Differentiation Study
3. Materials and Methods
3.1. Materials and Apparatus
3.2. Synthesis of Ben-IKVAV
3.3. Synthesis of PFB-IKVAV
3.4. Cell Viability Tests
3.5. Cell Differentiation
3.6. Real-Time Quantitative Polymerase Chain Reaction
3.7. Immunofluorescent Staining
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Manandhar, A.; Kang, M.; Chakraborty, K.; Tang, P.K.; Loverde, S.M. Molecular simulations of peptide amphiphiles. Org. Biomol. Chem. 2017, 15, 7993–8005. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Zhao, H.; Li, Y.; Lee, A.L.; Li, Z.; Fu, M.; Li, C.; Yang, Y.Y.; Yuan, P. Synthetic peptide hydrogels as 3D scaffolds for tissue engineering. Adv. Drug Deliv. Rev. 2020, 160, 78–104. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Q.; Yan, H.Y.; Li, J.; Zhang, L.Y.; Zhao, Y.R.; Xu, H. Artificial Metalloenzymes Based on Peptide Self-Assembly. Prog. Chem. 2018, 30, 1121–1132. [Google Scholar]
- Bertolani, A.; Pirrie, L.; Stefan, L.; Houbenov, N.; Haataja, J.S.; Catalano, L.; Terraneo, G.; Giancane, G.; Valli, L.; Milani, R.; et al. Supramolecular amplification of amyloid self-assembly by iodination. Nat. Commun. 2015, 6, 7574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; Wang, X. Biomimetic Self-Assembling Peptide Hydrogels for Tissue Engineering Applications. Adv. Exp. Med. Biol. 2018, 1064, 297–312. [Google Scholar]
- Tesauro, D.; Accardo, A.; Diaferia, C.; Milano, V.; Guillon, J.; Ronga, L.; Rossi, F. Peptide-based drug-delivery systems in biotechnological applications: Recent advances and perspectives. Molecules 2019, 24, 351. [Google Scholar] [CrossRef] [Green Version]
- Stupp, S.I.; Zha, R.H.; Palmer, L.C.; Cui, H.; Bitton, R. Self-Assembly of Biomolecular. Soft. Matter. Faraday Discuss. 2013, 166, 9–30. [Google Scholar] [CrossRef]
- Chen, J.; Zou, X. Self-assemble peptide biomaterials and their biomedical applications. Bioact. Mater. 2019, 4, 120–131. [Google Scholar] [CrossRef]
- Stephanopoulos, N.; Ortony, J.H.; Stupp, S.I. Self-Assembly for the Synthesis of Functional Biomaterials. Acta Mater. 2013, 61, 912–930. [Google Scholar] [CrossRef] [Green Version]
- Yokoi, H.; Kinoshita, T.; Zhang, S. Dynamic reassembly of peptide RADA16 nanofiber scaffold. Proc. Natl. Acad. Sci. USA 2005, 102, 8414–8419. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.Y.; Song, B.; Ao, Y.; Nowak, A.P.; Abelowitz, R.B.; Korsak, R.A.; Havton, L.A.; Deming, T.J.; Sofroniew, M.V. Biocompatibility of amphiphilic diblock copolypeptide hydrogels in the central nervous system. Biomaterials 2009, 30, 2881–2898. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhao, S.; Yu, Y.; Wang, H.; Yang, Y.; Liu, C. Biocompatibility profile and in vitro cellular uptake of self-assembled alginate nanoparticles. Molecules 2019, 24, 555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gazit, E. Self-assembled peptide nanostructures: The design of molecular building blocks and their technological utilization. Chem. Soc. Rev. 2007, 36, 1263–1269. [Google Scholar] [CrossRef] [PubMed]
- Ramakers, B.E.I.; Van Hest, J.C.M.; Löwik, D.W.P.M. Molecular tools for the construction of peptide-based materials. Chem. Soc. Rev. 2014, 43, 2743–2756. [Google Scholar] [CrossRef] [PubMed]
- Matsuurua, K. Rational design of self-assembled proteins and peptides for nano- and micro-sized architectures. RSC Adv. 2014, 4, 2942–2953. [Google Scholar] [CrossRef]
- Edwards-Gayle, C.J.C.; Hamley, I.W. Self-assembly of bioactive peptides, peptide conjugates, and peptide mimetic materials. Org. Biomol. Chem. 2017, 15, 5867–5876. [Google Scholar] [CrossRef] [Green Version]
- Matsuura, K. Synthetic approaches to construct viral capsid-like spherical nanomaterials. Chem. Commun. 2018, 54, 8944–8959. [Google Scholar] [CrossRef]
- Zhao, L.; Zou, Q.; Yan, X. Self-Assembling Peptide-Based Nanoarchitectonics. Bull. Chem. Soc. Jpn. 2018, 92, 70–79. [Google Scholar] [CrossRef]
- Lou, S.; Wang, X.; Yu, Z.; Shi, L. Peptide Tectonics: Encoded Structural Complementarity Dictates Programmable Self-Assembly. Adv. Sci. 2019, 6, 1802043. [Google Scholar] [CrossRef] [Green Version]
- Fukunaga, K.; Tsutsumi, H.; Mihara, H. Self-Assembling Peptides as Building Blocks of Functional Materials for Biomedical Applications. Bull. Chem. Soc. Jpn. 2018, 92, 391–399. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Trinh, T.H.T.; Yoo, M.; Shin, J.; Lee, H.; Kim, J.; Hwang, E.; Lim, Y.-b.; Ryou, C. Self-Assembling Peptides and Their Application in the Treatment of Diseases. Int. J. Mol. Sci. 2019, 20, 5850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levin, A.; Hakala, T.A.; Schnaider, L.; Bernardes, G.J.L.; Gazit, E.; Knowles, T.P.J. Biomimetic peptide self-assembly for functional materials. Nat. Rev. Chem. 2020, 4, 615–634. [Google Scholar] [CrossRef]
- Bertuzzi, S.; Gimeno, A.; Martinez-Castillo, A.; Lete, M.G.; Delgado, S.; Airoldi, C.; Rodrigues Tavares, M.; Bláhová, M.; Chytil, P.; Křen, V.; et al. Cross-Linking Effects Dictate the Preference of Galectins to Bind LacNAc-Decorated HPMA Copolymers. Int. J. Mol. Sci. 2021, 22, 12662. [Google Scholar] [CrossRef] [PubMed]
- Sinha, N.J.; Langenstein, M.G.; Pochan, D.J.; Kloxin, C.J.; Saven, J.G. Peptide Design and Self-assembly into Targeted Nanostructure and Functional Materials. Chem. Rev. 2021, 121, 13915–13935. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Feng, Z.; Xu, B. Assemblies of Peptides in a Complex Environment and their Applications. Angew. Chem. Int. Ed. 2019, 58, 10423–10432. [Google Scholar] [CrossRef]
- Lee, J.K.; Link, J.M.; Hu, J.C.Y.; Athanasiou, K.A. The Self-Assembling Process and Applications in Tissue Engineering. Cold. Spring Harb. Perspect. Med. 2017, 7, a025668. [Google Scholar] [CrossRef]
- Kyle, S.; Aggeli, A.; Ingham, E.; McPherson, M.J. Production of self-assembling biomaterials for tissue engineering. Trends Biotechnol. 2009, 27, 423–433. [Google Scholar] [CrossRef] [Green Version]
- Hosseinkhani, H.; Hong, P.-D.; Yu, D.-S. Self-Assembled Proteins and Peptides for Regenerative Medicine. Chem. Rev. 2013, 113, 4837–4861. [Google Scholar] [CrossRef]
- Verma, G.; Hassan, P.A. Self-assembled materials: Design strategies and drug delivery perspectives. Phys. Chem. Chem. Phys. 2013, 15, 17016–17028. [Google Scholar] [CrossRef]
- Hendricks, M.P.; Sato, K.; Palmer, L.C.; Stupp, S.I. Supramolecular Assembly of Peptide Amphiphiles. Acc. Chem. Res. 2017, 50, 2440–2448. [Google Scholar] [CrossRef]
- Song, Z.; Chen, X.; You, X.; Huang, K.; Dhinakar, A.; Gu, Z.; Wu, J. Self-assembly of peptide amphiphiles for drug delivery: The role of peptide primary and secondary structures. Biomater. Sci. 2017, 5, 2369–2380. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, A.; Das, D. Designer Peptide Amphiphiles: Self-Assembly to Applications. Langmuir 2019, 35, 10704–10724. [Google Scholar] [CrossRef] [PubMed]
- Lampel, A.; McPhee, S.A.; Park, H.A.; Scott, G.G.; Humagain, S.; Hekstra, D.R.; Yoo, B.; Frederix, P.; Li, T.D.; Abzalimov, R.R.; et al. Polymeric peptide pigments with sequence-encoded properties. Science 2017, 356, 1064–1068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, J.D.; Mura, C.; Lampe, K.J. Stimuli-Responsive, Pentapeptide, Nanofiber Hydrogel for Tissue Engineering. J. Am. Chem. Soc. 2019, 141, 4886–4899. [Google Scholar] [CrossRef] [PubMed]
- Kleinsmann, A.J.; Nachtsheim, B.J. Phenylalanine-containing cyclic dipeptides—The lowest molecular weight hydrogelators based on unmodified proteinogenic amino acids. Chem. Commun. 2013, 49, 7818–7820. [Google Scholar] [CrossRef]
- Smith, A.M.; Williams, R.J.; Tang, C.; Coppo, P.; Collins, R.F.; Turner, M.L.; Saiani, A.; Ulijn, R.V. Fmoc-Diphenylalanine Self Assembles to a Hydrogel via a Novel Architecture Based on π–π Interlocked β-Sheets. Adv. Mater. 2008, 20, 37. [Google Scholar] [CrossRef]
- Fleming, S.; Ulijn, R.V. Design of nanostructures based on aromatic peptide amphiphiles. Chem. Soc. Rev. 2014, 43, 8150–8177. [Google Scholar] [CrossRef]
- Li, J.; Du, X.; Hashim, S.; Shy, A.; Xu, B. Aromatic–Aromatic Interactions Enable α-Helix to β-Sheet Transition of Peptides to Form Supramolecular Hydrogels. J. Am. Chem. Soc. 2017, 139, 71–74. [Google Scholar] [CrossRef]
- Reches, M.; Gazit, E. Casting Metal Nanowires Within Discrete Self-Assembled Peptide Nanotubes. Science 2003, 300, 625–627. [Google Scholar] [CrossRef] [Green Version]
- Cai, Y.; Zheng, C.; Xiong, F.; Ran, W.; Zhai, Y.; Zhu, H.H.; Wang, H.; Li, Y.; Zhang, P. Recent Progress in the Design and Application of Supramolecular Peptide Hydrogels in Cancer Therapy. Adv. Healthc. Mater. 2021, 10, e2001239. [Google Scholar] [CrossRef]
- Patel, R.; Santhosh, M.; Dash, J.K.; Karpoormath, R.; Jha, A.; Kwak, J.; Patel, M.; Kim, J.H. Ile-Lys-Val-ala-Val (IKVAV) peptide for neuronal tissue engineering. Polym. Adv. Technol. 2019, 30, 4–12. [Google Scholar] [CrossRef] [Green Version]
- Tashiro, K.; Sephel, G.C.; Weeks, B.; Sasaki, M.; Martin, G.R.; Kleinman, H.K.; Yamada, Y. A synthetic peptide containing the IKVAV sequence from the A chain of laminin mediates cell attachment, migration, and neurite outgrowth. J. Biol. Chem. 1989, 264, 16174–16182. [Google Scholar] [CrossRef]
- Kubinová, Š.; Horák, D.; Kozubenko, N.; Vanecek, V.; Proks, V.; Price, J.; Cocks, G.; Sykova, E. The use of superporous AC-CGGASIKVAVS-OH-modified phema scaffolds to promote cell adhesion and the differentiation of human fetal neural precursors. Biomaterials 2010, 31, 5966–5975. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Wang, W.; Shao, Q.; Li, B.; Yu, D.; Zhou, X.; Parajuli, J.; Xu, H.; Qiu, T.; Yetisen, A.K.; et al. Pentapeptide IKVAV-engineered hydrogels for neural stem cell attachment. Biomater. Sci. 2021, 9, 2887–2892. [Google Scholar] [CrossRef] [PubMed]
- Silva, G.A.; Czeisler, C.; Niece, K.; Beniash, E.; Harrington, D.A.; Kessler, J.A.; Stupp, S.I. Selective Differentiation of Neural Progenitor Cells by High-Epitope Density Nanofibers. Science 2004, 303, 1352–1355. [Google Scholar] [CrossRef] [Green Version]
- Brun, P.; Zamuner, A.; Peretti, A.; Conti, J.; Messina, G.M.L.; Marletta, G.; Dettin, M. 3D Synthetic Peptide-based Architectures for the Engineering of the Enteric Nervous System. Sci. Rep. 2019, 9, 5583. [Google Scholar] [CrossRef]
- Farrukh, A.; Ortega, F.; Fan, W.; Marichal, N.; Paez, J.I.; Berninger, B.; Del Campo, A.; Salierno, M.J. Bifunctional Hydrogels Containing the Laminin Motif IKVAV Promote Neurogenesis. Stem Cell Rep. 2017, 9, 1432–1440. [Google Scholar] [CrossRef] [Green Version]
- Jain, R.; Roy, S. Controlling Neuronal Cell Growth through Composite Laminin Supramolecular Hydrogels. ACS Biomater. Sci. Eng. 2020, 6, 2832–2846. [Google Scholar] [CrossRef]
- Somaa, F.A.; Wang, T.-Y.; Niclis, J.C.; Bruggeman, K.F.; Kauhausen, J.A.; Guo, H.; McDougall, S.; Williams, R.J.; Nisbet, D.R.; Thomposon, L.H.; et al. Peptide-Based Scaffolds Support Human Cortical Progenitor Graft Integration to Reduce Atrophy and Promote Functional Repair in a Model of Stroke. Cell Rep. 2017, 20, 1964–1977. [Google Scholar] [CrossRef] [Green Version]
- Hsu, S.-M.; Lin, Y.-C.; Chang, J.-W.; Liu, Y.-H.; Lin, H.-C. Intramolecular Interactions of a Phenyl/Perfluorophenyl Pair in the Formation of Supramolecular Nanofibers and Hydrogels. Angew. Chem. Int. Ed. 2014, 53, 1921–1927. [Google Scholar] [CrossRef]
- Hsu, S.-M.; Wu, F.-Y.; Lai, T.-S.; Lin, Y.-C.; Lin, H.-C. Self-assembly and Hydrogelation from Multicomponent Coassembly of Pentafluorobenzyl-phenylalanine and Pentafluorobenzyl-diphenylalanine. RSC Adv. 2015, 5, 22943–22946. [Google Scholar] [CrossRef]
- Hsu, S.-M.; Chang, J.-W.; Wu, F.-Y.; Lin, Y.-C.; Lai, T.-S.; Hsun, C.; Lin, H.-C. A Supramolecular Hydrogel Self-assembled from Pentafluorobenzyl-dipeptide. RSC Adv. 2015, 5, 32431–32434. [Google Scholar] [CrossRef]
- Talloj, S.K.; Cheng, B.; Weng, J.-P.; Lin, H.-C. Glucosamine-Based Supramolecular Nanotubes for Human Mesenchymal Cell Therapy. ACS Appl. Mater. Interfaces 2018, 10, 15079–15087. [Google Scholar] [CrossRef] [PubMed]
- Saddik, A.; Chakravarthy, R.D.; Mohiuddin, M.; Lin, H.-C. Effects of fluoro substitutions and electrostatic interactions on self-assembled structures and hydrogelation of tripeptides: Tuning the mechanical properties of co-assembled hydrogels. Soft Matter 2020, 16, 10143–10150. [Google Scholar] [CrossRef]
- Merrifield, R.B. Solid phase peptide synthesis. I. The synthesis of a tetrapeptide. J. Am. Chem. Soc. 1963, 85, 2149–2154. [Google Scholar] [CrossRef]
- Yan, C.; Pochan, D. Rheological properties of peptide-based hydrogels for biomedical and other applications. Chem. Soc. Rev. 2010, 39, 3528–3540. [Google Scholar] [CrossRef] [Green Version]
- George, S.J.; De Bruijn, R.; Tomović, Z.; Averbeke, B.V.; Beljonne, D.; Lazzaroni, R.; Schenning, A.; Meijer, E.W. Asymmetric Noncovalent Synthesis of Self-Assembled One-Dimensional Stacks by a Chiral Supramolecular Auxiliary Approach. J. Am. Chem. Soc. 2012, 134, 17789–17796. [Google Scholar] [CrossRef]
- Yeh, M.-Y.; Huang, C.-T.; Lai, T.-S.; Chen, F.-Y.; Chu, N.-T.; Tseng, D.T.-H.; Hung, S.-C.; Lin, H.-C. Effect of Peptide Sequences on Supramolecular Interactions of Naphthaleneimide/Tripeptide Conjugates. Langmuir 2016, 32, 7630–7638. [Google Scholar] [CrossRef]
- Ma, M.; Kuang, Y.; Gao, Y.; Zhang, Y.; Gao, P.; Xu, B. Aromatic−Aromatic Interactions Induce the Self-Assembly of Pentapeptidic Derivatives in Water To Form Nanofibers and Supramolecular Hydrogels. J. Am. Chem. Soc. 2010, 132, 2719–2728. [Google Scholar] [CrossRef]
- Adochitei, A.; Drochioiu, G. Rapid Characterization Of Peptide Secondary Structure By FT-IR Spectroscopy. Rev. Roum. Chim. 2011, 56, 783–791. [Google Scholar]
- Biancalana, M.; Koide, S. Molecular mechanism of Thioflavin-T binding to amyloid fibrils. Biochim. Biophys. Acta Proteins Proteom. 2010, 1804, 1405–1412. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Peterson, D.A.; Kimura, H.; Schubert, D. Mechanism of cellular 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reduction. J. Neurochem. 1997, 69, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Jin, Z.; Liu, L.; Yan, Y.; Li, T.; Zhu, X.; Jing, N. Characterization and promoter analysis of the mouse nestin gene. FEBS Lett. 2004, 565, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Lewis, S.A.; Cowan, N.J.J. Complex regulation and functional versatility of mammalian alpha- and beta-tubulin isotypes during the differentiation of testis and muscle cells. J. Cell Biol. 1988, 106, 2023–2033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, L.J.; Mallory, M.; Xia, Y.; Veinbergs, I.; Hashimoto, M.; Yoshimoto, M.; Thal, L.J.; Saitoh, T.; Masliah, E. Expression pattern of synucleins (non-Abeta component of Alzheimer’s disease amyloid precursor protein/alpha-synuclein) during murine brain development. J. Neurochem. 1998, 71, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Goedert, M.; Crowther, R.; Garner, C. Molecular characterization of microtubule-associated proteins tau and MAP2. Trends Neurosci. 1991, 14, 193–199. [Google Scholar] [CrossRef]
- Caceres, A.; Mautino, J.; Kosik, K.S. Suppression of MAP2 in cultured cerebeller macroneurons inhibits minor neurite formation. Neuron 1992, 9, 607–618. [Google Scholar] [CrossRef]
- Subburaju, S.; Benes, F.M. Induction of the GABA Cell Phenotype: An In Vitro Model for Studying Neurodevelopmental Disorders. PLoS ONE 2012, 7, e33352. [Google Scholar] [CrossRef]
- Chen, W.; Liu, J.; Zhang, L.; Xu, H.; Guo, X.; Deng, S.; Liu, L.; Yu, D.; Chen, Y.; Li, Z. Generation of the SCN1A epilepsy mutation in hiPS cells using the TALEN technique. Sci. Rep. 2014, 4, 5404. [Google Scholar] [CrossRef] [Green Version]


| Gene | Primer Sequence | Product Size (bp) |
|---|---|---|
| Nestin | 5′-CTGGAGCAGGAGAAACAGG-3′ (forward) 5′-TGAAAGCTGAGGGAAGTCTTG-3′ (reverse) | 182 |
| β-tubulin | 5′-AGCAAGAACAGCAGCTACTTCGT -3′ (forward) 5′-GATGAAGGTGGAGGACATCTTGA -3′ (reverse) | 102 |
| α-synuclein (SNCA) | 5′-AGGACTTTCAAAGGCCAAGG-3′ (forward) 5′-TCC TCCAACATTTGTCACTTG-3′ (reverse) | 187 |
| MAP2 | 5′-CTTCAGCTTGTCTCTAACCGAG-3′ (forward) 5′-CCTTTGCTTCATCTTTCCGTTC-3′ (reverse) | 199 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, F.-Y.; Lin, H.-C. Synthesis, Self-Assembly, and Cell Responses of Aromatic IKVAV Peptide Amphiphiles. Molecules 2022, 27, 4115. https://doi.org/10.3390/molecules27134115
Wu F-Y, Lin H-C. Synthesis, Self-Assembly, and Cell Responses of Aromatic IKVAV Peptide Amphiphiles. Molecules. 2022; 27(13):4115. https://doi.org/10.3390/molecules27134115
Chicago/Turabian StyleWu, Fang-Yi, and Hsin-Chieh Lin. 2022. "Synthesis, Self-Assembly, and Cell Responses of Aromatic IKVAV Peptide Amphiphiles" Molecules 27, no. 13: 4115. https://doi.org/10.3390/molecules27134115
APA StyleWu, F.-Y., & Lin, H.-C. (2022). Synthesis, Self-Assembly, and Cell Responses of Aromatic IKVAV Peptide Amphiphiles. Molecules, 27(13), 4115. https://doi.org/10.3390/molecules27134115







