Potent Activity of a High Concentration of Chemical Ozone against Antibiotic-Resistant Bacteria
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Ozone Generating and Monitoring
2.3. Inoculation of the Test Surface
2.4. Ozone Treatment
2.5. Cell Viability
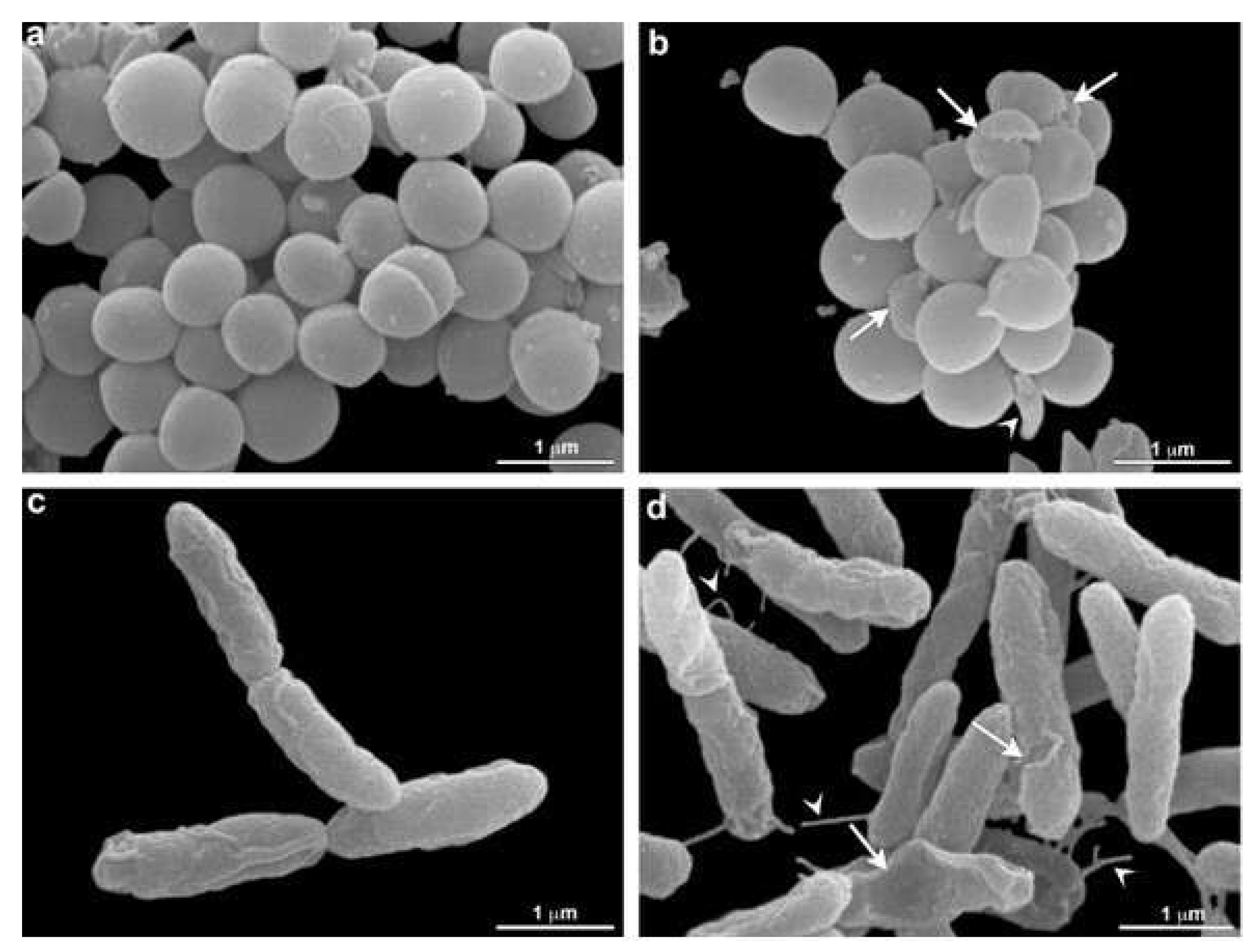
2.6. Scanning Electron Microscopy (SEM)
3. Results
3.1. Monitoring of Ozone Concentration
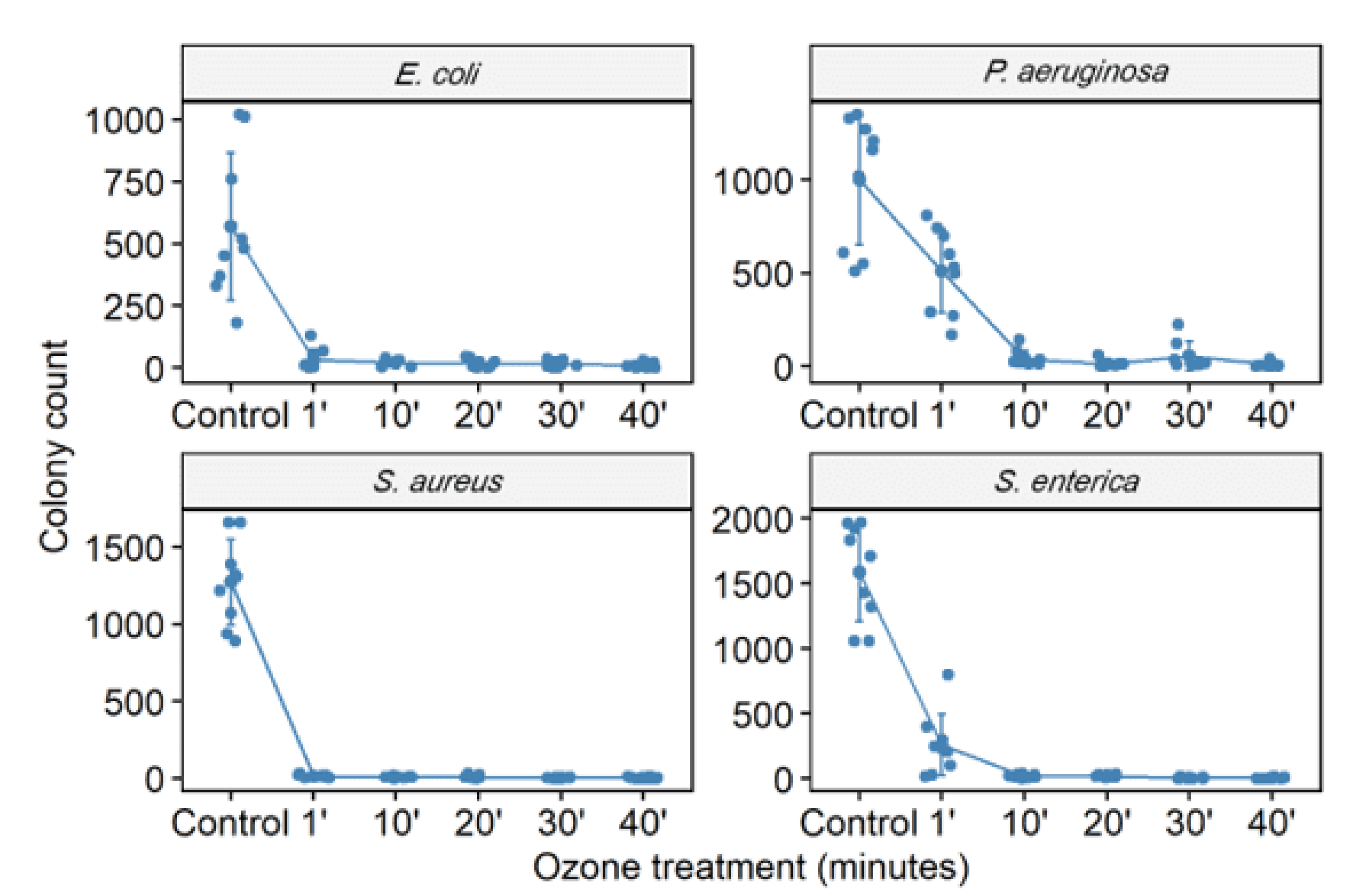
3.2. Ozone Treatment
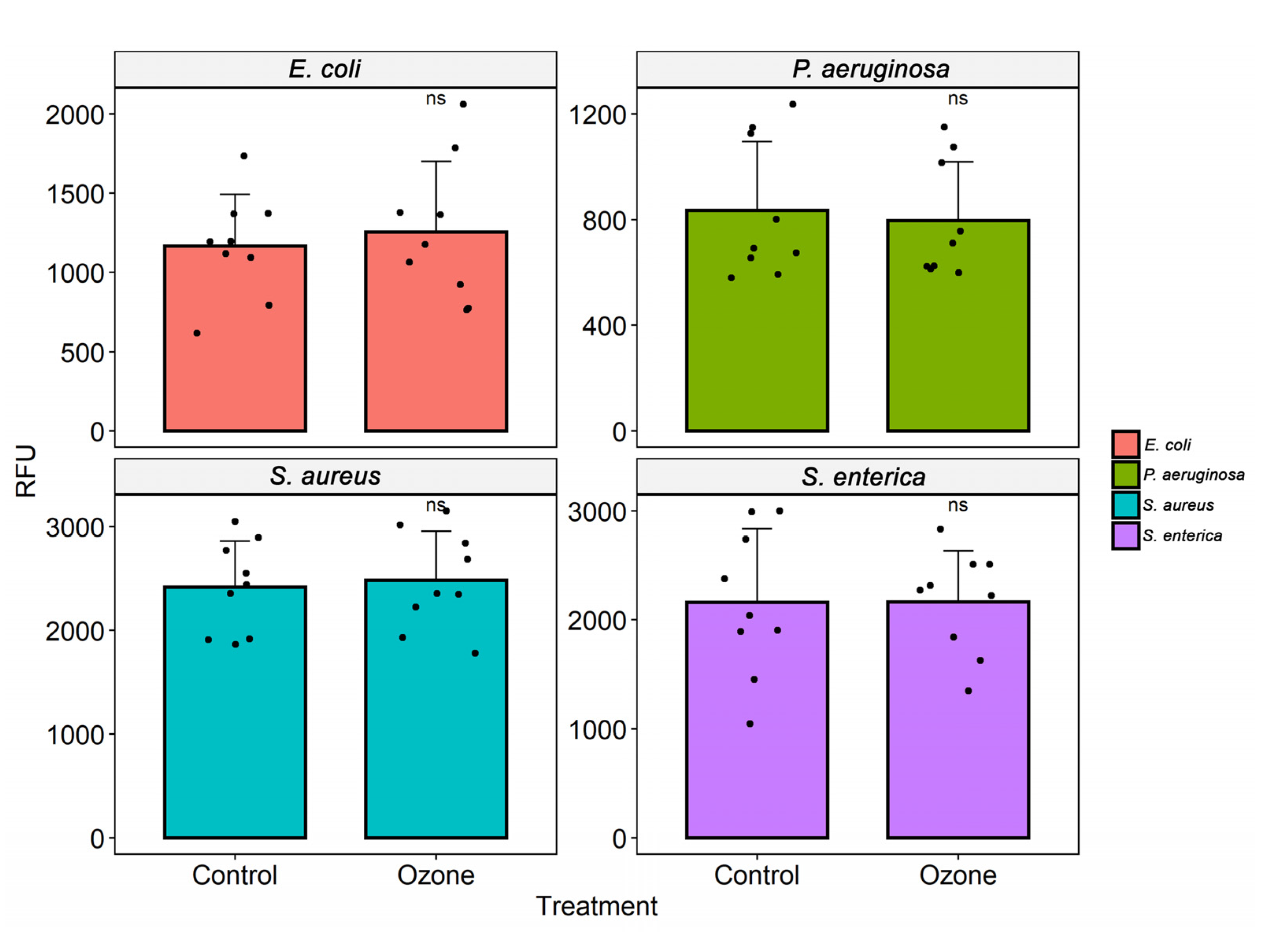
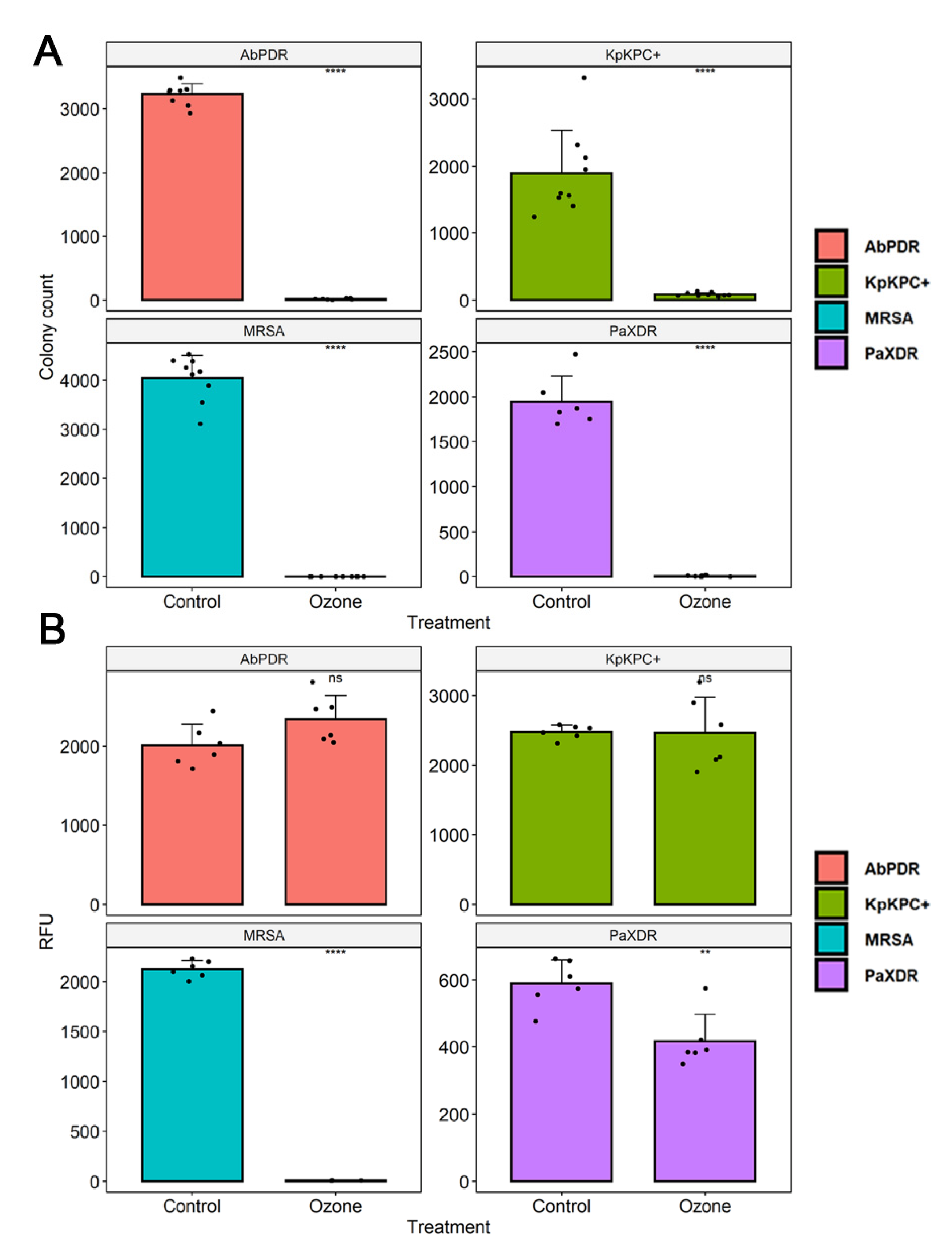
3.3. Cell Viability
3.4. Scanning Electron Microscopy (SEM)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Allegranzi, B.; Bagheri Nejad, S.; Combescure, C.; Graafmans, W.; Attar, H.; Donaldson, L.; Pittet, D. Burden of endemic health-care-associated infection in developing countries: Systematic review and meta-analysis. Lancet 2011, 337, 228–241. [Google Scholar] [CrossRef]
- ECDC. European Centre for Disease Prevention and Control Annual Epidemiological Report 2016- Healthcare-Associated Infections Acquired in Intensive Care Units. 2017. Available online: https://www.ecdc.Europa.eu/en/publications-data/healthcare-associated-infections-intensive-care-units-annualepidemiological-0 (accessed on 20 August 2021).
- Brasil. Boletim Segurança do Paciente e Qualidade em Serviços de Saúde n° 16: Avaliação dos indicadores nacionais das Infecções Relacionadas à Assistência à Saúde (IRAS) e Resistência microbiana do ano de 2016. 2017. Available online: https://www20.Anvisa.gov.br/segurancadopaciente/index.php/publicacoes/item/boletim-seguranca-do-paciente-e-qualidade-em-servicos-de-saude-n-16-avaliacaodos-indicadores-nacionais-das-infeccoes-relacionadas-a-assistencia-a-saudeiras-e-resistenciamicrobiana-do-ano-de-2016 (accessed on 20 August 2021).
- Jenks, J.; Duse, A.; Wattal, C.; Zaidi, A.K.; Wertheim, H.F.; Sumpradit, N.; Vlieghe, E.; Hara, G.L.; Gould, I.M.; Goossens, H.; et al. Antibiotic resistance needs global solutions. Lancet Infect. Dis. 2014, 14, 550. [Google Scholar] [CrossRef]
- WHO (World Health Organization). The Burden of Healthcare-Associated Infection Worldwide: A Summary; World Health Organization: Lisboa, Portugal, 2011; Volume 3, Available online: http://www.who.int/gpsc/countrywork/summary20100430em.pdf (accessed on 20 August 2021).
- Ali, S.; Birhane, M.; Bekele, S.; Kibru, G.; Teshager, L.; Yilma, Y.; Ahmed, Y.; Fentahun, N.; Assefa, H.; Gashaw, M.; et al. Healthcare-associated infection and its risk factors among patients admitted to a tertiary hospital in Ethiopia: A longitudinal study. Antimicrob. Resist. Infect. Control 2018, 7, 2. [Google Scholar] [CrossRef] [PubMed]
- Umscheid, C.; Mitchell, M.D.; Doshi, J.A.; Agarwal, R.; Williams, K.; Brennan, P.J. Estimating the proportion of healthcare-associated infections that are reasonably preventable and the associated mortality and costs. Infect. Control Hosp. Epidemiol. 2011, 32, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Gatt, Y.E.; Margalit, H. Common adaptive strategies underlie within-host evolution of bacterial pathogens. Mol. Biol. Evol. 2021, 38, 1101–1121. [Google Scholar] [CrossRef] [PubMed]
- Santajit, S.; Indrawattana, N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed. Res. Int. 2016, 2016, 2475067. [Google Scholar] [CrossRef] [PubMed]
- Georgescu, M.; Gheorghe, I.; Curutiu, C.; Lazar, V.; Bleotu, C.; Chifiriuc, M.C. Virulence and resistance features of Pseudomonas aeruginosa strains isolated from chronic leg ulcers. BMC Infect. Dis. 2016, 16, 92. [Google Scholar] [CrossRef] [PubMed]
- Coelho, F.; Coelho, M.; Diniz, A.; Vicente, G.; Vieira, C. Velhos Problemas, novos desafios. Rev. Technol. Hospitalar. 2011, 43, 30–32. [Google Scholar]
- Tadesse, B.T.; Ashley, E.A.; Ongarello, S.; Havumaki, J.; Wijegoonewardena, M.; Gonzalez, I.J.; Dittrich, S. Antimicrobial resistance in Africa: A systematic review. BMC Infect. Dis. 2017, 17, 616. [Google Scholar] [CrossRef] [PubMed]
- Gajdács, M.; Albericio, F. Antibiotic resistance: From the bench to patients. Antibiotics 2019, 8, 129. [Google Scholar] [CrossRef] [PubMed]
- Bocé, M.; Tassé, M.; Mallet-Ladeira, S.; Pillet, F.; Da Silva, C.; Vicendo, P.; Lacroix, P.G.; Malfant, I.; Rols, M.P. Effect of trans(NO, OH)-[RuFT(Cl)(OH)NO](PF(6)) ruthenium nitrosyl complex on methicillin-resistant Staphylococcus epidermidis. Sci. Rep. 2019, 9, 4867. [Google Scholar] [CrossRef] [PubMed]
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef]
- Park, S.E. Epidemiology, virology, and clinical features of severe acute respiratory syndrome -coronavirus-2 (SARS-CoV-2; Coronavirus Disease-19). Clin. Exp. Pediatr. 2020, 63, 119–124. [Google Scholar] [CrossRef]
- Cucinotta, D.; Vanelli, M. WHO Declares COVID-19 a Pandemic. Acta Biomed. 2020, 91, 157–160. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. China novel coronavirus investigating and research team. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Canto, N.R.; Ruiz-Garbajosa, P. Co-resistance: An opportunity for the bacteria and resistance genes. Curr. Opin. Pharmacol. 2011, 11, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Clancy, C.J.; Nguyen, M.H. Coroviruses disease 2019, superinfections, and antimicrobial development. What can we expect? Clin. Infect. Dis. 2020, 71, 2736–2743. [Google Scholar] [CrossRef] [PubMed]
- Bengoechea, J.Á.; Bamford, C.G.G. SARS-CoV-2, bacterial coinfections, and AMR: The deadly trio in COVID-19? EMBO Mol. Med. 2020, 26, e12560. [Google Scholar] [CrossRef]
- Fu, Y.; Yang, Q.; Xu, M.; Kong, H.; Chen, H.; Fu, Y.; Yao, Y.; Zhou, H.; Zhou, J. Secondary bacterial infections in critically ill patients with coronavirus disease 2019. Open Forum Infect. Dis. 2020, 5, ofaa220. [Google Scholar] [CrossRef] [PubMed]
- Rawson, T.M.; Moore, L.S.P.; Zhu, N.; Ranganathan, N.; Skolimowska, K.; Gilchrist, M.; Satta, G.; Cooke, G.; Holmes, A. Bacterial and fungal coinfection in individuals with coronavirus: A rapid review to support COVID-19 antimicrobial prescribing. Clin. Infect. Dis. 2020, 71, 2459–2468. [Google Scholar] [CrossRef] [PubMed]
- Chirumbolo, S.; Simonetti, V.; Franzini, M.; Valdenassi, L.; Bertossi, D.; Pandolfi, S. Estimating coronavirus disease 2019 (COVID19)-caused deaths in hospitals and healthcare units: Do hospital-acquired infections play a role? Comments with a proposal. Infect. Control. Hosp. Epidemiol. 2021, 19, 1–2. [Google Scholar] [CrossRef]
- Lizioli, A.; Privitera, G.; Alliata, E.; Banfi, E.M.A.; Boselli, L.; Panceri, M.L.; Perna, M.C.; Porretta, A.D.; Santini, M.G.; Carreri, V. Prevalence of nosocomial infections in Italy: Result from the Lombardy survey in 2000. J. Hosp. Infect. 2003, 54, 141–148. [Google Scholar] [CrossRef]
- Nicastri, E.; Petrosillo, N.; Martini, L.; Larosa, M.; Gesu, G.P.; Ippolito, G.; INF-NOS Study Group. Prevalence of nosocomial infections in 15 Italian hospitals: First point prevalence study for the INF-NOS project. Infection 2003, 71, 10–15. [Google Scholar] [PubMed]
- Zotti, C.M.; Ioli, G.M.; Charrier, L.; Arditi, G.; Argentero, P.A.; Biglino, A.; Farina, E.C.; Ruggenini, A.M.; Reale, R.; Romagnoli, S.; et al. Hospital-acquired infections in Italy: A region wide prevalence study. J. Hosp. Infect. 2004, 56, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging strategies to combat eskape pathogens in the era of antimicrobial resistance: A review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef]
- Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance. Microbiol. Spectr. 2016, 4, 10. [Google Scholar] [CrossRef]
- Miller, W.R.; Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance in enterococci. Expert. Rev. Anti. Infect. Ther. 2014, 12, 1221–1236. [Google Scholar] [CrossRef]
- WHO (World Health Organization). Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics; World Health Organization: Geneva, Switzerland, 2017; Available online: https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf (accessed on 15 August 2021).
- Fernandes, A.T.; Fernandes, M.O.V.; Ribeiro Filho, N.; Graziano, K.U.; Gabrielloni, M.C.; Cavalcante, N.J.F.; Lacerda, R.A. Infecção Hospitalar e Suas Interfaces na Área da Saúde, 1st ed.; Atheneu: São Paulo, Brazil, 2000; p. 1795. [Google Scholar]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pan drug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Oliveira, A.C. Infecções Hospitalares, Epidemiologia, Prevenção e Controle, 1st ed.; Guanabara Koogan—Grupo Gen: Rio de Janeiro, Brazil, 2005; p. 710. [Google Scholar]
- Stephanie, J.D. Controlling hospital-acquired infection: Focus on the role of the environment and new technologies for decontamination. Clin. Microbiol. Rev. 2014, 27, 665–690. [Google Scholar] [CrossRef]
- Moccia, G.; Motta, O.; Pironti, C.; Proto, A.; Capunzo, M.; De Caro, F. An alternative approach for the decontamination of hospital settings. J. Infect. Public Health 2020, 13, 2038–2044. [Google Scholar] [CrossRef] [PubMed]
- Pironti, C.; Motta, O.; Proto, A. Development of a new vapor phase methodology for textiles disinfection. Clean. Engin. Technol. 2021, 4, 100170. [Google Scholar] [CrossRef]
- Bayarri, B.; Cruz-Alcalde, A.; López-Vinent, N.; Micó, M.M.; Sans, C.J. Can ozone inactivate SARS-CoV-2? A review of mechanisms and performance on viruses. J. Hazard. Mater. 2021, 415, 125658. [Google Scholar] [CrossRef] [PubMed]
- Nascente, E.P.; Chagas, S.R.; Pessoa, A.V.C.; Matos, M.P.C.; Andrade, M.A.; Pascoal, L.M. Potencial antimicrobiano do ozônio: Aplicações e perspectivas em medicina veterinária. Pubvet 2019, 13, 130. [Google Scholar] [CrossRef]
- Bocci, V. Ozone: A New Medical Drug, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2010; p. 336. [Google Scholar] [CrossRef]
- Wickramanayake, G.B.; Sproul, O.J. Ozone concentration and temperature effects on disinfection kinetics. Ozone Sci. Eng. 1988, 10, 123–135. [Google Scholar] [CrossRef]
- Wolf, C.; Von Gunten, U.; Kohn, T. Kinetics of inactivation of waterborne enteric viruses by ozone. Environ. Sci. Technol. 2018, 52, 2170–2177. [Google Scholar] [CrossRef]
- Blanchard, E.L.; Lawrence, J.D.; Noble, J.A.; Xu, M.; Joo, T.; Ng, N.L.; Schmidt, B.E.; Santangelo, P.J.; Finn, M.G.G. Enveloped virus inactivation on personal protective equipment by exposure to ozone. medRxiv 2020. [Google Scholar] [CrossRef]
- Dubuis, M.E.E.; Dumont-Leblond, N.; Lalibert’e, C.; Veillette, M.; Turgeon, N.; Jean, J.; Duchaine, C. Ozone efficacy for the control of airborne viruses: Bacteriophage and norovirus models. PLoS ONE 2020, 15, e0231164. [Google Scholar] [CrossRef]
- Davies, A.; Pottage, T.; Bennett, A.; Walker, J. Gaseous and air decontamination technologies for Clostridium difficile in the healthcare environment. J. Hosp. Infect. 2011, 77, 199–203. [Google Scholar] [CrossRef]
- Moat, J.; Cargill, J.; Shone, J.; Upton, M. Application of a novel decontamination process using gaseous ozone. Canad. J. Microbiol. 2009, 55, 928–933. [Google Scholar] [CrossRef]
- Piletić, K.; Kovač, B.; Perčić, M.; Žigon, J.; Broznić, D.; Karleuša, L.; Blagojević, S.L.; Oder, M.; Gobin, I. Disinfectin action of gaseous ozone on OXA-48-producing Klebsiella pneumoniae biofilm in vitro. Int. J. Environ. Res. Public Health 2022, 19, 6177. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Hudson, J.B. Ozone gas is an effective and practical antibacterial agent. Am. J. Infect. Control 2008, 36, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Moccia, G.; De Caro, F.; Pironti, C.; Boccia, G.; Capunzo, M.; Borrelli, A.; Motta, O. Development and improvement of an effective method for air and surfaces disinfection with ozone gas as a decontaminating agent. Medicina 2020, 56, 578. [Google Scholar] [CrossRef] [PubMed]
- Rangel, K.; Cabral, F.O.; Lechuga, G.C.; Carvalho, J.P.R.S.C.; Villas-Bôas, M.H.S.; Midlej, V.; De-Simone, S.G. Detrimental effect of ozone on pathogenic bacteria. Microorganisms 2022, 10, 40. [Google Scholar] [CrossRef] [PubMed]
- Fontes, B.; Heimbecker, A.M.C.; de Souza Brito, G.; Costa, S.F.; van der Heijden, I.M.; Levin, A.S.; Rasslan, S. Effect of low-dose gaseous ozone on pathogenic bacteria. BMC Infect. Dis. 2012, 12, 358. [Google Scholar] [CrossRef]
- Merks, P.; Religioni, U.; Bilmin, K.; Bogusz, J.; Juszczyk, G.; Barańska, A.; Kuthan, R.; Drelich, E.; Jakubowska, M.; Świeczkowski, D.; et al. Ozone disinfection of community pharmacies during the COVID-19 pandemic as a possible preventive measure for infection spread. Med. Pr. 2021, 72, 529–534. [Google Scholar] [CrossRef] [PubMed]
- Franke, G.; Knobling, B.; Brill, F.H.; Becker, B.; Klupp, E.M.; Belmar Campos, C.; Pfefferle, S.; Lütgehetmann, M.; Knobloch, J.K. An automated room disinfection system using ozone is highly active against surrogates for SARS-CoV-2. J. Hosp. Infect. 2021, 112, 108–113. [Google Scholar] [CrossRef]
- Percivalle, E.; Clerici, M.; Cassaniti, I.; Nepita, E.V.; Marchese, P.; Olivati, D.; Catelli, C.; Berri, A.; Baldanti, F.; Marone, P.; et al. SARS-CoV-2 viability on different surfaces after gaseous ozone treatment: A preliminary evaluation. J. Hosp. Infect. 2021, 110, 33–36. [Google Scholar] [CrossRef]
- Radvar, S.; Karkon-Shayan, S.; Motamed-Sanaye, A.; Majidi, M.; Hajebrahimi, S.; Taleschian-Tabrizi, N.; Pashazadeh, F.; Sahebkar, A. Using ozone therapy as an option for treatment of COVID-19 Patients: A scoping review. Adv. Exp. Med. Biol. 2021, 1327, 151–160. [Google Scholar] [CrossRef]
- Brodowska, A.J.; Nowak, A.; Śmigielski, K. Ozone in the food industry: Principles of ozone treatment, mechanisms of action, and applications: An overview. Crit. Rev. Food Sci. Nutr. 2018, 58, 2176–2201. [Google Scholar] [CrossRef]
- Feng, L.; Zhang, K.; Gao, M.; Shi, C.; Ge, C.; Qu, D.; Zhu, J.; Shi, Y.; Han, J. Inactivation of Vibrio parahaemolyticus by aqueous ozone. J. Microbiol. Biotechnol. 2018, 28, 1233–1246. [Google Scholar] [CrossRef] [PubMed]
- Ersoy, Z.G.; Barisci, S.; Turkay, O. Mechanisms of the Escherichia coli and Enterococcus faecalis inactivation by ozone. Food Sci. Technol. 2019, 100, 306–313. [Google Scholar] [CrossRef]
- Zhang, Y.Q.; Wu, Q.P.; Zhang, J.M.; Yang, X.H. Effects of ozone on membrane permeability and ultrastructure in Pseudomonas aeruginosa. J. Appl. Microbiol. 2011, 111, 1006–1015. [Google Scholar] [CrossRef] [PubMed]
- Lall, N.; Henley-Smith, C.J.; Canha, M.N.; Oosthuizen, C.B.; Barrington, D. Viability reagent presto blue in comparison with other available reagents utilized in cytotoxicity and antimicrobial assays. Int. J. Microbiol. 2013, 2013, 420601. [Google Scholar] [CrossRef] [PubMed]
- Riss, T.; Moravec, R.; Niles, A. Development for cell viability and apoptosis for high-throughput screening. In A Practical Guide to Assay Development and High-Throughput Screening in Drug Discovery; Chen, T., Ed.; CRC Press: Boca Raton, FL, USA, 2010; pp. 109–110. [Google Scholar]
- Riss, T.; Moravec, R.A.; Niles, A.L.; Duellman, S.; Benink, H.A.; Worzella, T.J.; Minor, L.; Markossian, S.; Grossman, A.; Brimacombe, K.; et al. Cell Viability Assays. In The Assay Guidance Manual; Sittampalam, G., Ed.; Eli Lilly Co.: Indianapolis, IN, USA; National Center for Advancing Translational Sciences: Bethesda, MD, USA, 2016; pp. 10–12. [Google Scholar]
- Präbst, K.; Engelhardt, H.; Ringgeler, S.; Hübner, H. Basic colorimetric proliferation assays: MTT WST and resazurin. Methods Mol. Biol. 2017, 1601, 1–17. [Google Scholar] [CrossRef]
- Sousa, C.S.; Torres, L.M.; Azevedo, M.P.F.; Camargo, T.C.; Graziano, K.U.; Lacerda, R.A.; Turrini, R.N.T. Sterilization with ozone in health care: An integrative literature review. Rev. Esc. Enfer. USP 2011, 45, 1243–1249. [Google Scholar] [CrossRef][Green Version]
- Rubio-Romero, J.C.; Pardo-Ferreira, M.D.C.; Torrecilla-García, J.A.; Calero-Castro, S. Disposable masks: Disinfection and sterilization for reuse, and non-certified manufacturing, in the face of shortages during the COVID-19 pandemic. Saf. Sci. 2020, 129, 104830. [Google Scholar] [CrossRef]
- EPA. Environmental Protection Agency. Integrated Science Assessment for Ozone and Related Photochemical Oxidant (Final Report, April 2020). 2020. Available online: https://cfpub.epa.gov/ncea/isa/recordisplay.cfm?deid=348522 (accessed on 15 August 2021).
- Blanco, A.; Ojembarrena, F.B.; Clavo, B.; Negro, C. Ozone potential to fight against SAR-COV-2 pandemic: Facts and research needs. Environ. Sci. Pollut. Res. Int. 2021, 28, 16517–16531. [Google Scholar] [CrossRef]
- Grignani, E.; Mansi, A.; Cabella, R.; Castellano, P.; Tirabasso, A.; Sisto, R.; Spagnoli, M.; Fabrizi, G.; Frigerio, F.; Tranfo, G. Safe and effective use of ozone as air and surface disinfectant in the conjuncture of COVID-19. Gases 2021, 1, 19–32. [Google Scholar] [CrossRef]
- Hudson, J.B.; Sharma, M.; Vimalanathan, S. Development of a practical method for using ozone gas as a virus decontaminating agent. Ozone Sci. Eng. 2009, 31, 216–223. [Google Scholar] [CrossRef]
- McClurkin, J.D.; Maier, D.E.; Ileleji, K.E. Half-life time of ozone as a function of air movement and conditions in a sealed container. J. Stored Prod. Res. 2013, 55, 41–47. [Google Scholar] [CrossRef]
- Dennis, R.; Cashion, A.; Emanuel, S.; Hubbard, D. Ozone gas: Scientific justification and practical guidelines for improvised disinfection using consumer-grade ozone generators and plastic storage boxes. J. Sci. Med. 2020, 2, 1–15. [Google Scholar] [CrossRef]
- Tseng, C.; Li, C. Inactivation of surface viruses by gaseous ozone. J. Environ. Health 2008, 70, 56–63. [Google Scholar]
- Knobling, B.; Franke, G.; Klupp, E.M.; Belmar Campos, C.; Knobloch, J.K. Evaluation of the effectiveness of two automated room decontamination devices under real-life conditions. Front. Public Health 2021, 9, 618263. [Google Scholar] [CrossRef] [PubMed]
- Zoutman, D.; Shannon, M.; Mandel, A. Effectiveness of a novel ozone-based system for the rapid high-level disinfection of health care spaces and surfaces. Am. J. Infect. Control. 2011, 39, 873–879. [Google Scholar] [CrossRef]
- Xu, Y.; Ji, J.; Wu, H.; Pi, F.; Blaženović, I.; Zhang, Y.; Sun, X. Untargeted GC-TOFMS-based cellular metabolism analysis to evaluate ozone degradation effect of deoxynivalenol. Toxicon 2019, 168, 49–57. [Google Scholar] [CrossRef]
- Heß, S.; Gallert, C. Sensitivity of antibiotic-resistant and antibiotic susceptible Escherichia coli, Enterococcus and Staphylococcus, strains against ozone. J. Water Health 2015, 13, 1020–1028. [Google Scholar] [CrossRef]
- Schulz zur Wiesch, P.; Engelstadter, J.; Bonhoeffer, S. Compensation of fitness costs and reversibility of antibiotic resistance mutations. Antimicrob. Agents Chemother. 2010, 54, 2085–2095. [Google Scholar] [CrossRef]
- Von Gunten, U. Ozonation of drinking water: Part II. Disinfection and by-product formation in presence of bromide, iodide, or chlorine. Water Res. 2003, 37, 1469–1487. [Google Scholar] [CrossRef]
- Von Sonntag, C.; Von Gunten, U. Chemistry of Ozone in Water and Wastewater Treatment from Basic Principles to Applications; IWA Publishing: London, UK, 2012; p. 306. [Google Scholar] [CrossRef]
- Casolari, A. Microbial Death. In Physiological Models in Microbiology; Bazin, M.J., Prosser, J.I., Eds.; CRC Press: Boca Raton, FL, USA, 1988; Volume 2, pp. 1–44. [Google Scholar]
- Broadwater, W.T.; Hoehn, R.C.; King, P.H. Sensitivity of three selected bacterial species to ozone. Appl. Microbiol. 1973, 26, 391–393. [Google Scholar] [CrossRef]
- Patil, S.; Valdramidis, V.P.; Karatzas, K.A.; Cullen, P.J.; Bourke, P. Assessing the microbial oxidative stress mechanism of ozone treatment through the responses of Escherichia coli mutants. J. Appl. Microbiol. 2011, 111, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Janex, M.L.; Savoye, P.; Cockx, A.; Lazarova, V. Wastewater disinfection by ozone: Main parameters for process design. Water Res. 2002, 36, 1043–1055. [Google Scholar] [CrossRef]
- Patil, S.; Bourke, P.; Frias, J.M.; Tiwari, B.K. Inactivation of Escherichia coli in orange juice using ozone. Inn. Food Sci. Emerg. Technol. 2009, 10, 551–557. [Google Scholar] [CrossRef]
- Pak, G.; Salcedo, D.E.; Lee, H.; Oh, J.; Maeng, S.K.; Song, K.G.; Hong, S.W.; Kim, H.C.; Chandran, K.; Kim, S. Comparison of antibiotic resistance removal efficiencies using ozone disinfection under different pH and suspended solids and humic substance concentrations. Environ. Sci. Technol. 2016, 50, 7590–7600. [Google Scholar] [CrossRef] [PubMed]
- Adler, M.G.; Hill, G.R. The kinetics and mechanism of hydroxide iron-catalyzed ozone decomposition in aqueous solutions. J. Am. Chem. Soc. 1950, 72, 1884–1886. [Google Scholar] [CrossRef]
- Hoigne, J.; Bader, H. Ozonation of water: Role of hydroxyl radicals as oxidizing intermediates. Science 1975, 190, 782–784. [Google Scholar] [CrossRef]
- Kang, S.W. Superoxide dismutase 2 gene and cancer risk: Evidence from an updated meta-analysis. Int. J. Clinic. Exp. Med. 2015, 8, 14647–14655. [Google Scholar]
- Imlay, J.A. Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 2008, 77, 755–776. [Google Scholar] [CrossRef]
- Pomposiello, P.J.; Demple, B. Redox-operated genetic switches: The SoxR and OxyR transcription factors. Trends. Biotechnol. 2021, 19, 109–114. [Google Scholar] [CrossRef]
- Greenberg, J.T.; Monach, P.; Chou, J.H.; Josephy, P.D.; Demple, B. Positive control of a global antioxidant defense regulon activated by superoxide generating agents in Escherichia coli. Proc. Natl. Acad. Sci. USA 1990, 87, 6181–6185. [Google Scholar] [CrossRef]
- Delaney, J.M. Requirement of the Escherichia coli dnaK gene for thermotolerance and protection against H2O2. J. Gen. Microbiol. 1990, 136, 2113–2118. [Google Scholar] [CrossRef] [PubMed]
- Rockabrand, D.; Arthur, T.; Korinek, G.; Livers, K.; Blum, P. An essential role for the Escherichia coli DnaK protein in starvation-induced thermotolerance, H2O2 resistance, and reductive division. J. Bacteriol. 1995, 177, 3695–3703. [Google Scholar] [CrossRef] [PubMed]
- Loewen, P.; Hu, B.; Strutinsky, J.; Sparling, R. Regulation in the rpoS regulon of Escherichia coli. Can. J. Microbiol. 1998, 44, 707–717. [Google Scholar] [CrossRef] [PubMed]
- Chaffin, D.O.; Taylor, D.; Skerrett, S.J.; Rubens, C.E. Changes in the Staphylococcus aureus transcriptome during early adaptation to the lung. PLoS ONE 2012, 7, e41329. [Google Scholar] [CrossRef]
- Hamelin, C.; Chung, Y.S. Optimal conditions for mutagenesis by ozone in Escherichia coli K12. Mutat. Res. 1974, 24, 271–279. [Google Scholar] [CrossRef]
- Hamelin, C.; Sarhan, F.; Chung, Y.S. Ozone-induced DNA degradation in different DNA polymerase I mutants of Escherichia coli K12. Biochem. Biophys. Res. Commun. 1977, 77, 220–224. [Google Scholar] [CrossRef]
- Hamelin, C.; Sarhan, F.; Chung, Y.S. Induction of deoxyribonucleic acid degradation in Escherichia coli by ozone. Experientia 1978, 34, 1578–1579. [Google Scholar] [CrossRef]
- Dodd, M.C. Potential impacts of disinfection processes on eliminating and deactivating antibiotic resistance genes during water and wastewater treatment. J. Environ. Moni. 2012, 14, 1754–1771. [Google Scholar] [CrossRef]
- Scott, D.B.M.; Lesher, E.C. Effect of ozone on survival and permeability of Escherichia coli. J. Bacteriol. 1963, 85, 567–576. [Google Scholar] [CrossRef]
- Hunt, N.K.; Mariñas, B.J. Inactivation of Escherichia coli with ozone: Chemical and inactivation kinetics. Water Res. 1999, 33, 263–264. [Google Scholar] [CrossRef]
- Han, L.; Patil, S.; Boehm, D.; Milosavljević, V.; Cullen, P.J.; Bourke, P. Mechanisms of inactivation by high-voltage atmospheric cold plasma differ for Escherichia coli and Staphylococcus aureus. Appl. Environ. Microbiol. 2016, 82, 450–458. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Zhang, D.; Graham, N.J.D. Membrane fouling by extracellular polymeric substances after ozone pre-treatment: Variation of nano-particle size. Water Res. 2017, 120, 146–155. [Google Scholar] [CrossRef] [PubMed]
- Halder, S.; Yadav, K.K.; Sarkar, R.; Mukherjee, S.; Saha, P.; Haldar, S.; Karmakar, S.; Sen, T. Alteration of Zeta potential and membrane permeability in bacteria: A study with cationic agents. Springerplus 2015, 4, 672–686. [Google Scholar] [CrossRef] [PubMed]
- Nagayoshi, M.; Kitamura, C.; Fukuizumi, T.; Nishihara, T.; Terashita, M. Antimicrobial effect of ozonated water on bacteria invading dentinal tubules. J. Endod. 2004, 30, 778–781. [Google Scholar] [CrossRef] [PubMed]
- Russell, A.D. Similarities and differences in the responses of microorganisms to biocides. J. Antimicrob. Chemother. 2003, 52, 750–763. [Google Scholar] [CrossRef]

| Ozone Exposure Time (Minutes) | Evaluated Parameters | ||
|---|---|---|---|
| Temperature (°C) | Relative Humidity of Air (% RH) | Ozone Concentration (ppm) | |
| 1 | 22.5 | 77.2 | 71.7 |
| 10 | 22.8 | 74.6 | 48.8 |
| 20 | 23.7 | 75.5 | 47.2 |
| 30 | 23.9 | 71.4 | 21.1 |
| 40 | 24.3 | 72.4 | 30.8 |
| Mean | 23.4 | 74.2 | 43.9 |
| Bacterial Strains | Ozone Exposure Times | |||||
|---|---|---|---|---|---|---|
| C | 1′ | 10′ | 20′ | 30′ | 40′ | |
| Count Number CFU/% of Reduction | ||||||
| S. aureus (ATCC 6538) | 6287 | 123.1/98 | 36/99.4 | 31.9/99.5 | 27.2/99.6 | 20.4/99.7 |
| P. aeruginosa (ATCC 15442) | 3767 | 3109/17.5 | 256/93.2 | 105.2/97.2 | 65.33/98.3 | 57.8/98.5 |
| S. enterica (ATCC 10708) | 7391 | 711.3/90.4 | 360.8/95.1 | 69.7/99.1 | 63.11/99.2 | 53.9/99.3 |
| E. coli (ATCC 25922) | 3090 | 62/98 | 66.1/98 | 57.2/98.1 | 31.2/99 | 30.3/99 |
| S. aureus (MRSA) | 4041 | - | - | - | - | 0.1/99.99 |
| P. aeruginosa (XDR) | 1946 | - | - | - | - | 6.33/99.7 |
| A. baumannii (PDR) | 3228 | - | - | - | - | 16.6/99.5 |
| K. pneumoniae (KPC+) | 1894 | - | - | - | - | 86/95.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rangel, K.; Cabral, F.O.; Lechuga, G.C.; Carvalho, J.P.R.S.; Villas-Bôas, M.H.S.; Midlej, V.; De-Simone, S.G. Potent Activity of a High Concentration of Chemical Ozone against Antibiotic-Resistant Bacteria. Molecules 2022, 27, 3998. https://doi.org/10.3390/molecules27133998
Rangel K, Cabral FO, Lechuga GC, Carvalho JPRS, Villas-Bôas MHS, Midlej V, De-Simone SG. Potent Activity of a High Concentration of Chemical Ozone against Antibiotic-Resistant Bacteria. Molecules. 2022; 27(13):3998. https://doi.org/10.3390/molecules27133998
Chicago/Turabian StyleRangel, Karyne, Fellipe O. Cabral, Guilherme C. Lechuga, João P. R. S. Carvalho, Maria H. S. Villas-Bôas, Victor Midlej, and Salvatore G. De-Simone. 2022. "Potent Activity of a High Concentration of Chemical Ozone against Antibiotic-Resistant Bacteria" Molecules 27, no. 13: 3998. https://doi.org/10.3390/molecules27133998
APA StyleRangel, K., Cabral, F. O., Lechuga, G. C., Carvalho, J. P. R. S., Villas-Bôas, M. H. S., Midlej, V., & De-Simone, S. G. (2022). Potent Activity of a High Concentration of Chemical Ozone against Antibiotic-Resistant Bacteria. Molecules, 27(13), 3998. https://doi.org/10.3390/molecules27133998