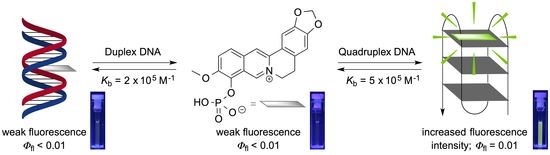
Berberrubine Phosphate: A Selective Fluorescent Probe for Quadruplex DNA
Abstract
1. Introduction
2. Results and Discussion
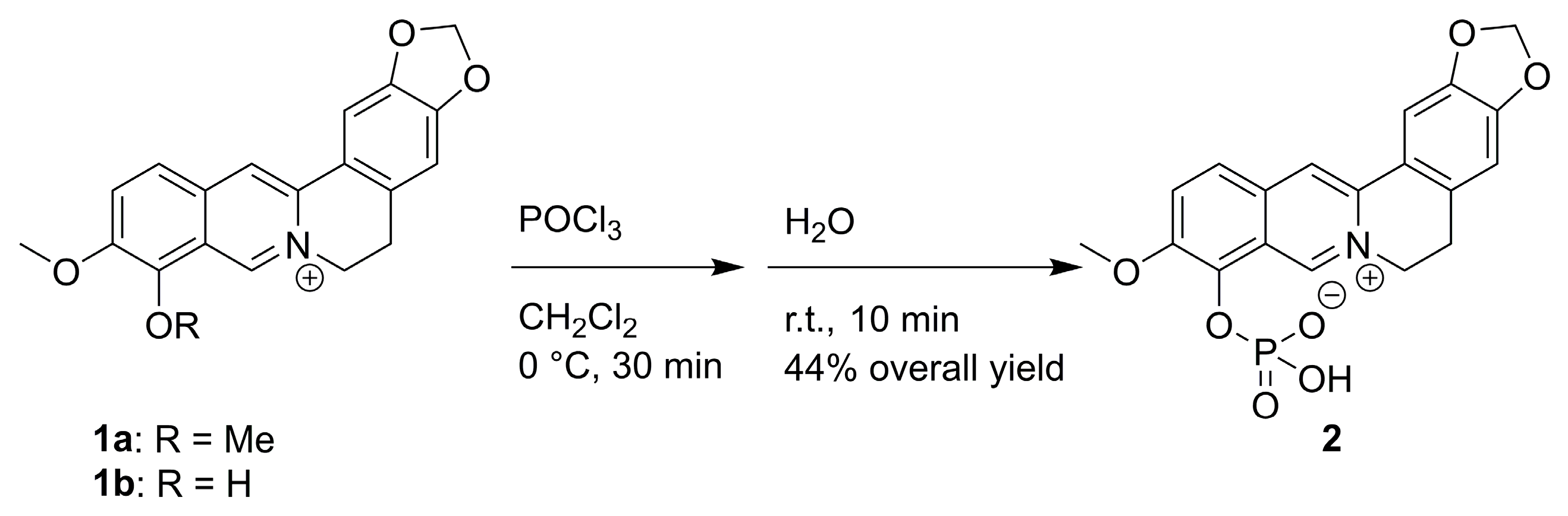
2.1. Synthesis
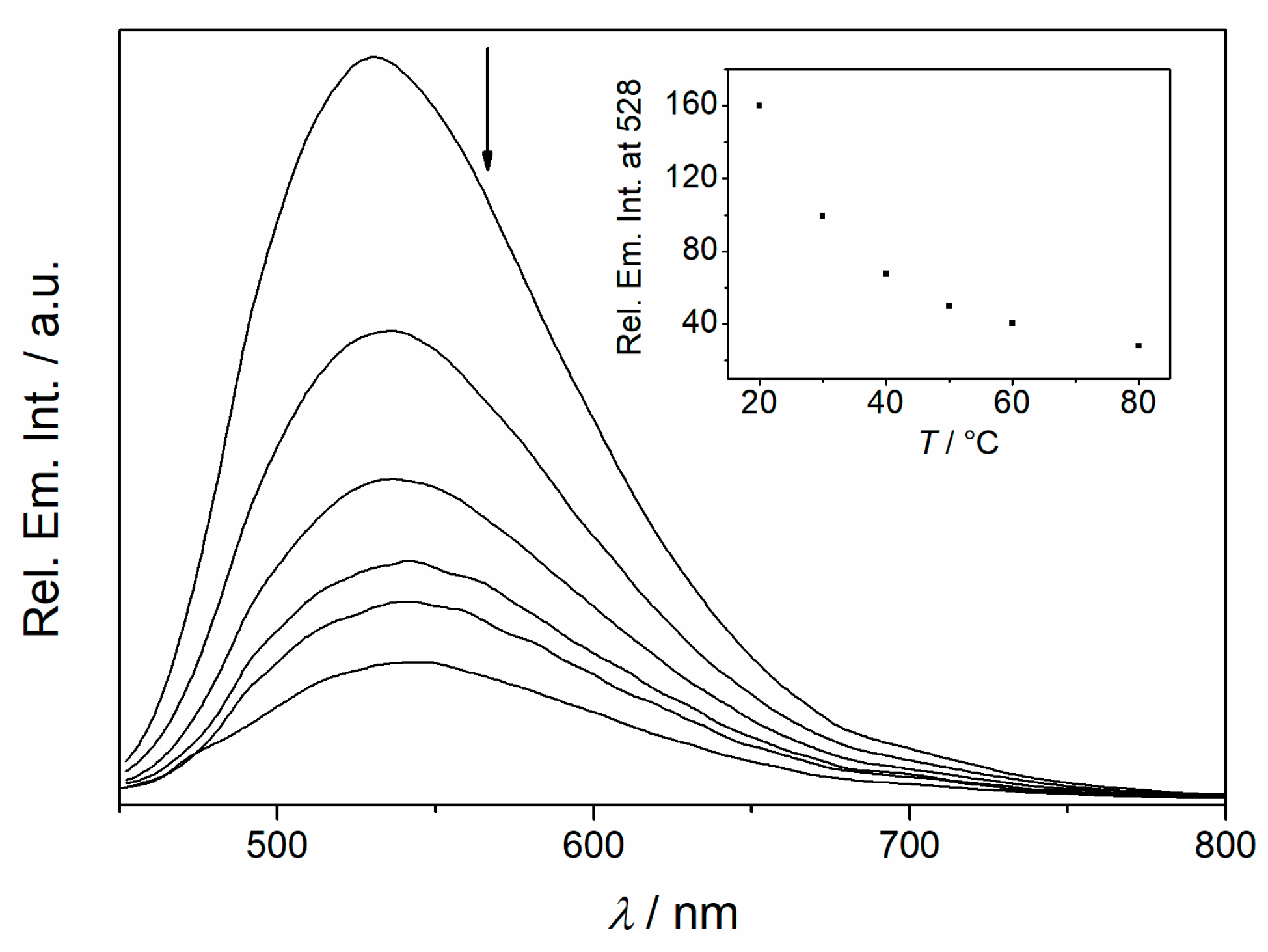
2.2. Absorption and Emission Properties
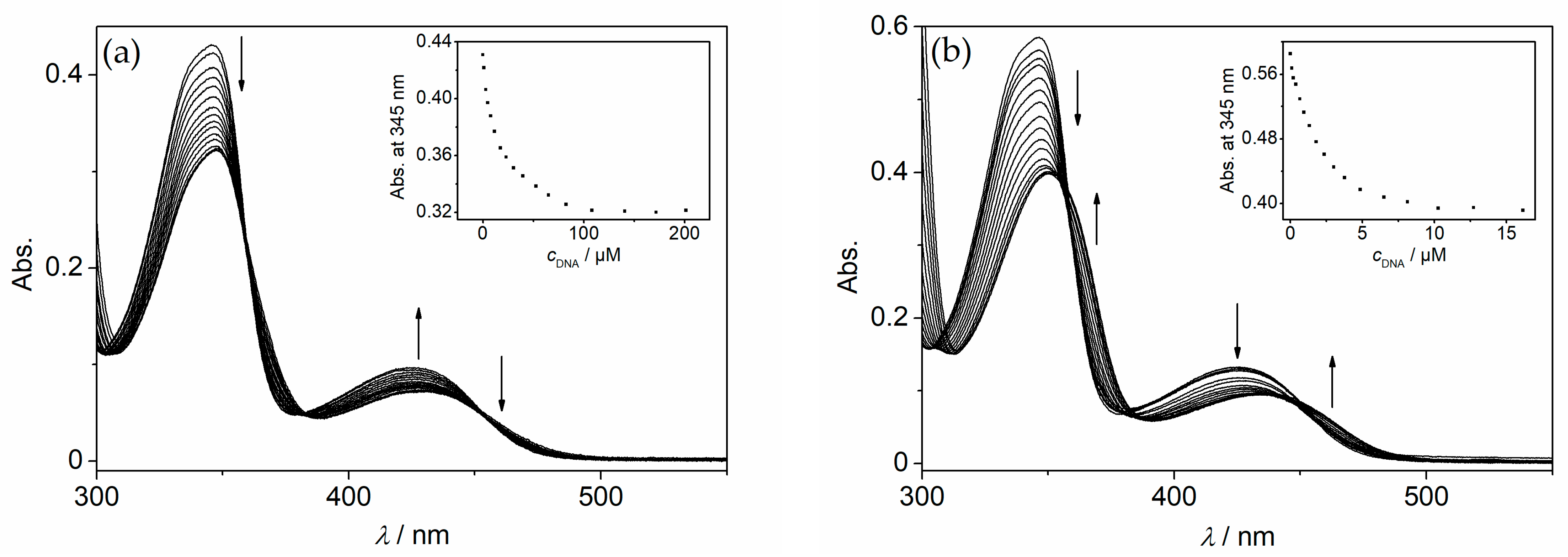
2.3. Photometric DNA Titrations
2.4. Thermal DNA Denaturation Experiments
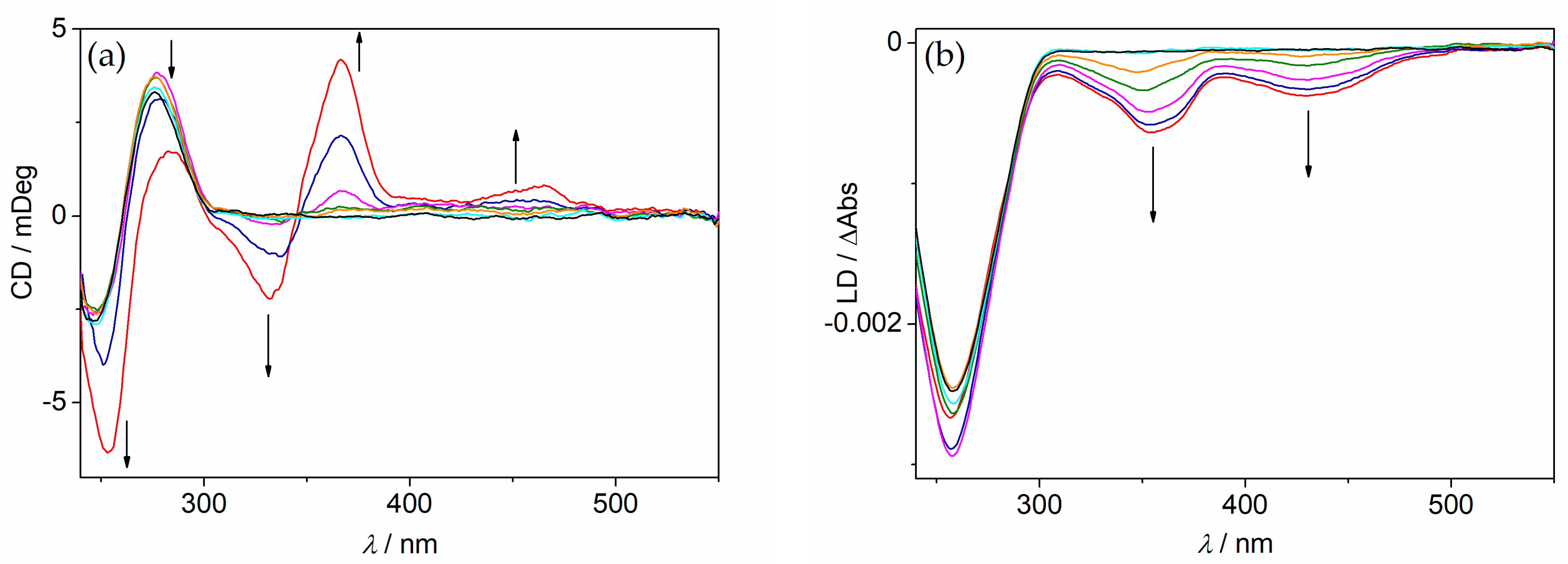
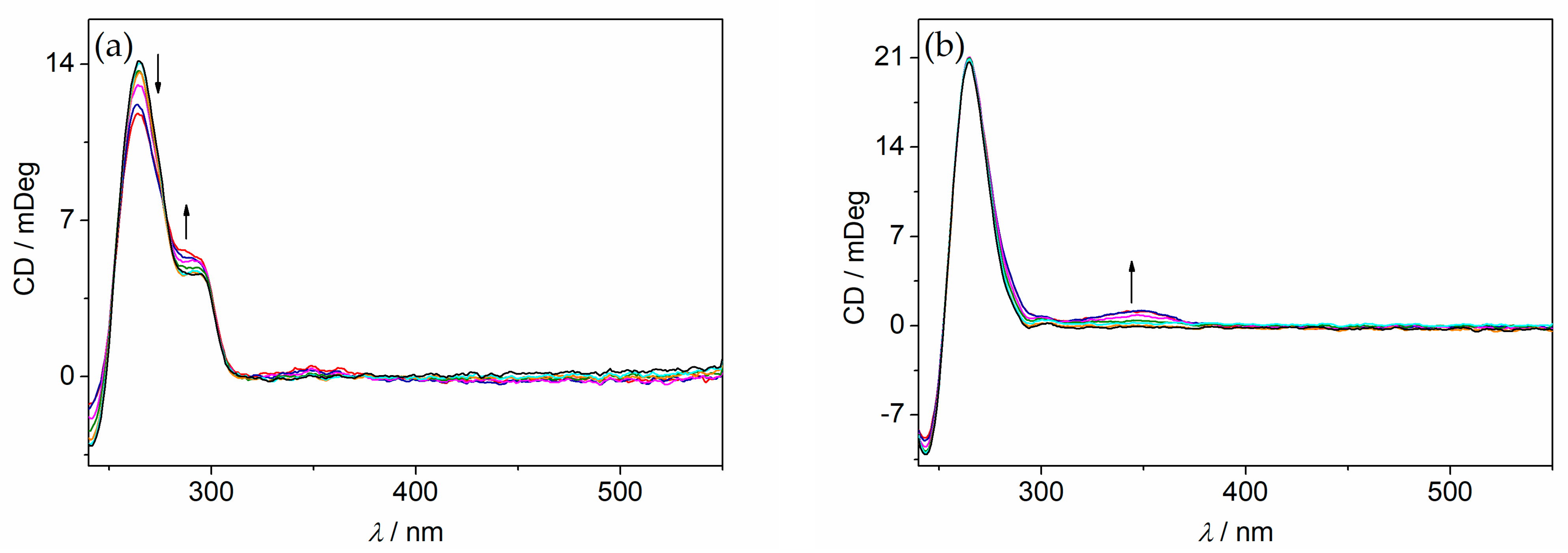
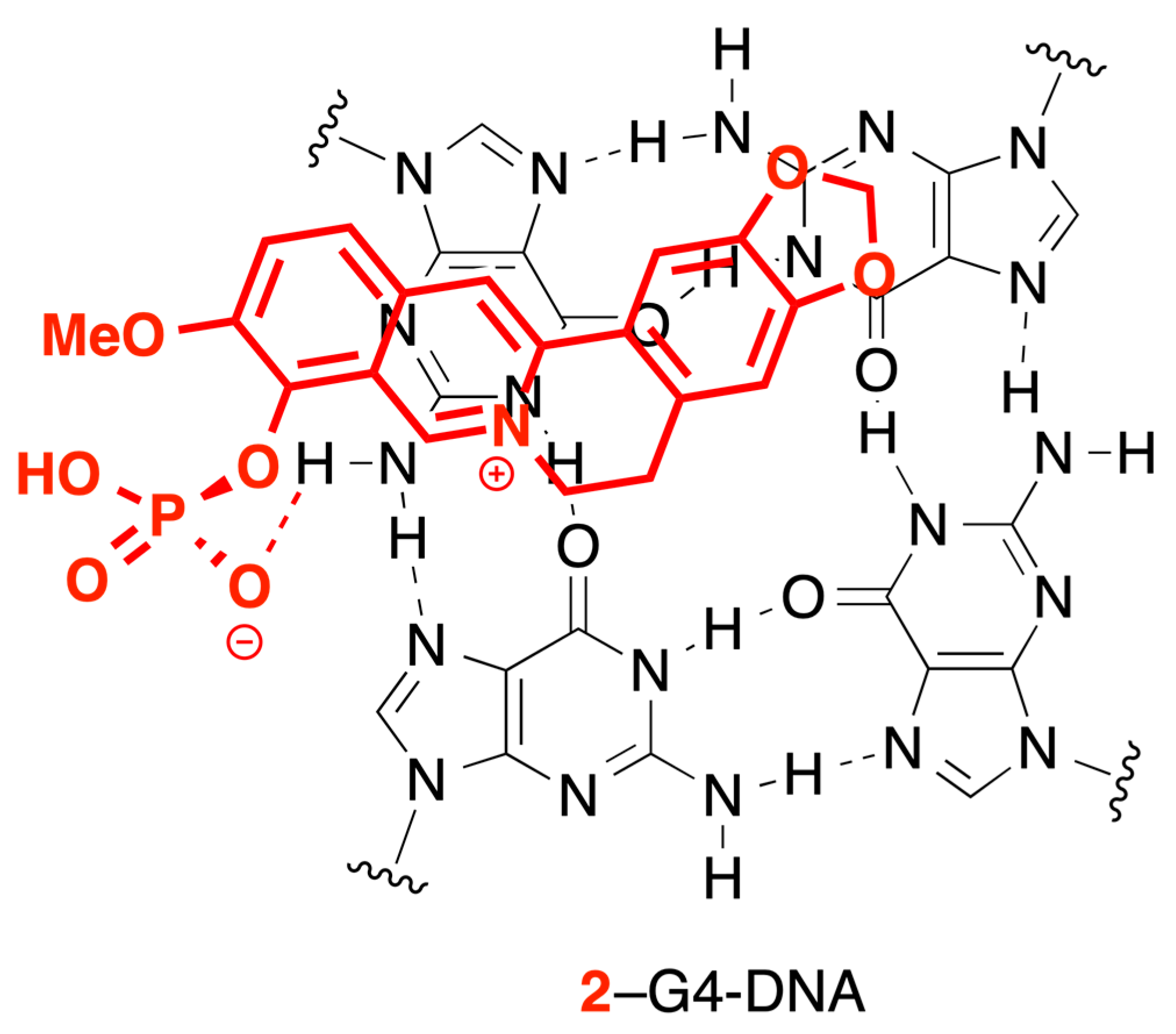
2.5. CD and LD Spectroscopy
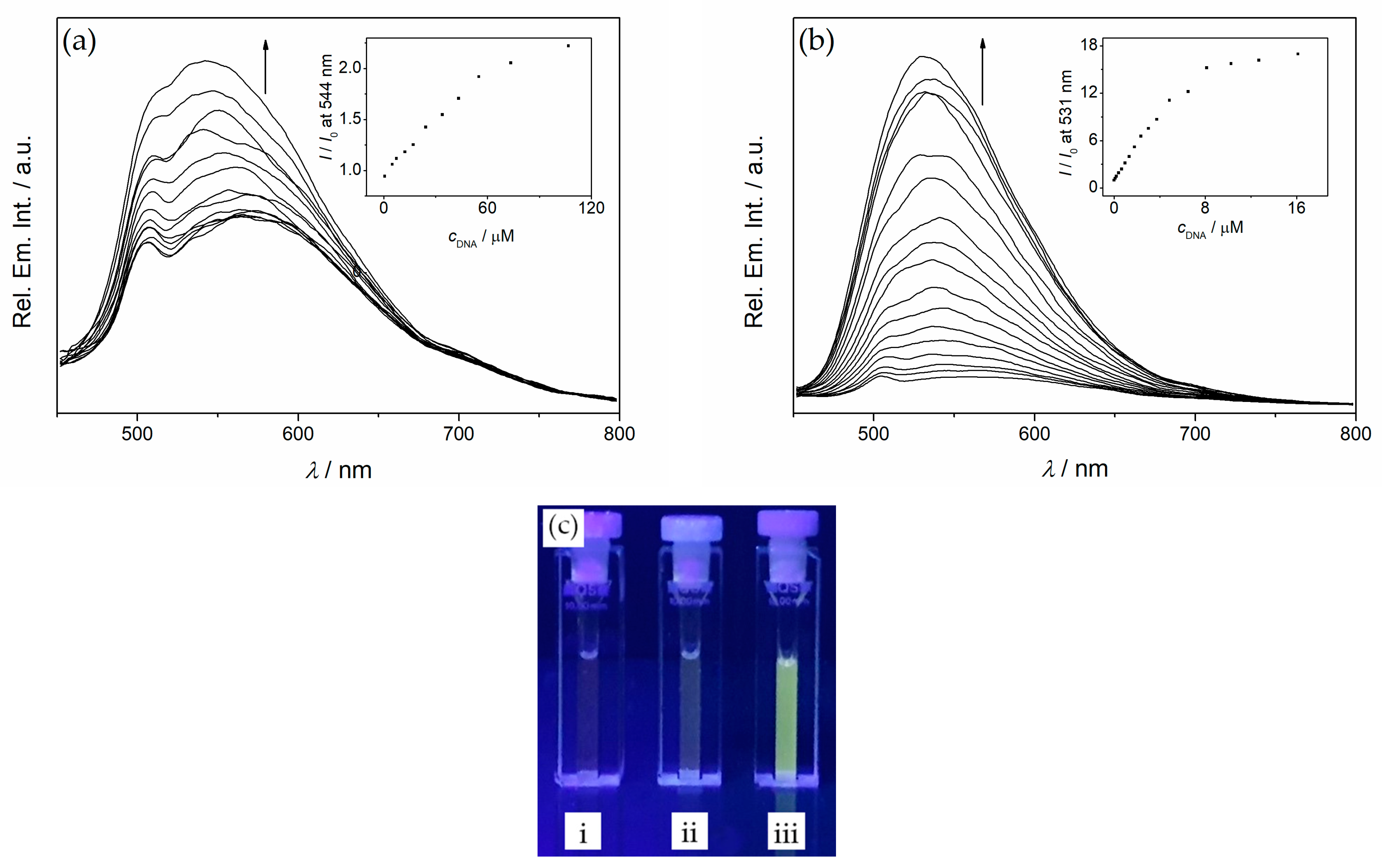
2.6. Fluorimetric Titrations
3. Materials and Methods
3.1. Equipment
3.2. Materials
3.3. Synthesis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Liu, H.; Lei, Y. A critical review: Recent advances in “digital” biomolecule detection with single copy sensitivity. Biosens. Bioelectron. 2021, 177, 112901. [Google Scholar] [CrossRef]
- Yang, X.; Lovell, J.F.; Murthy, N.; Zhang, Y. Topics in Medicinal Chemistry; Springer: Cham, Switzerland, 2020; Volume 34, pp. 33–53. [Google Scholar]
- Michel, B.Y.; Dziuba, D.; Benhida, R.; Demchenko, A.P.; Burger, A. Probing of nucleic acid structures, dynamics, and interactions with environment-sensitive fluorescent labels. Front. Chem. 2020, 8, 112. [Google Scholar] [CrossRef] [PubMed]
- Barnoy, E.A.; Popovtzer, R.; Fiixler, D. Fluorescence for biological logic gates. J. Biophotonics 2020, 13, e202000158. [Google Scholar] [CrossRef]
- Smith, S.J.; Nemr, C.R.; Kelley, S.O. Chemistry-driven approaches for ultrasensitive nucleic acid detection. J. Am. Chem. Soc. 2017, 139, 1020–1028. [Google Scholar] [CrossRef]
- Wiraja, C.; Yeo, D.C.; Lio, D.C.S.; Zheng, M.; Xu, C. Functional imaging with nucleic-acid-based sensors: Technology, application and future healthcare prospects. ChemBioChem 2019, 20, 437–450. [Google Scholar] [CrossRef]
- Yuan, J.-H.; Shao, W.; Chen, S.-B.; Huang, Z.-S.; Tan, J.-H. Recent advances in fluorescent probes for G-quadruplex nucleic acids. Biochem. Biophys. Res. Commun. 2020, 531, 18–24. [Google Scholar] [CrossRef]
- Suseela, Y.V.; Narayanaswamy, N.; Pratihar, S.; Govindaraju, T. Far-red fluorescent probes for canonical and non-canonical nucleic acid structures: Current progress and future implications. Chem. Soc. Rev. 2018, 47, 1098–1131. [Google Scholar] [CrossRef]
- Tateishi-Karimata, H.; Sugimoto, N. Chemical biology of non-canonical structures of nucleic acids for therapeutic applications. Chem. Commun. 2020, 56, 2379–2390. [Google Scholar] [CrossRef] [PubMed]
- Mathad, R.I.; Hatzakis, E.; Dai, J.; Yang, D. c-MYC promoter G-quadruplex formed at the 5′-end of NHE III1 element: Insights into biological relevance and parallel-stranded G-quadruplex stability. Nucleic Acids Res. 2011, 39, 9023–9033. [Google Scholar] [CrossRef]
- Rankin, S.; Reszka, A.P.; Huppert, J.; Zloh, M.; Parkinson, G.N.; Todd, A.K.; Ladame, S.; Balasubramanian, S.; Neidle, S. Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 2005, 127, 10584–10589. [Google Scholar] [CrossRef] [PubMed]
- Gatto, B.; Palumbo, M.; Sissi, C. Nucleic acid aptamers based on the G-quadruplex structure: Therapeutic and diagnostic potential. Curr. Med. Chem. 2009, 16, 1248–1265. [Google Scholar] [CrossRef]
- Rhodes, D.; Lipps, H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43, 8627–8637. [Google Scholar] [CrossRef]
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Kumari, B.; Yadav, A.; Pany, S.P.; Pradeepkumar, P.I.; Kanvah, S. Cationic red emitting fluorophore: A light up NIR fluorescent probe for G4-DNA. J. Photochem. Photobiol. B 2019, 190, 128–136. [Google Scholar] [CrossRef]
- Zhai, Q.; Gao, C.; Ding, J.; Zhang, Y.; Islam, B.; Lan, W.; Hou, H.; Deng, H.; Li, J.; Hu, Z.; et al. Selective recognition of c-MYC Pu22 G-quadruplex by a fluorescent probe. Nucleic Acids Res. 2019, 47, 2190–2204. [Google Scholar] [CrossRef]
- Karg, B.; Funke, A.; Ficht, A.; Sievers-Engler, A.; Lämmerhofer, M.; Weisz, K. Molecular recognition and visual detection of G-quadruplexes by a dicarbocyanine dye. Chem. Eur. J. 2015, 21, 13802–13811. [Google Scholar] [CrossRef]
- Yang, P.; De, C.A.; Teulade-Fichou, M.P.; Mergny, J.L.; Monchaud, D. Engineering bisquinolinium/thiazole orange conjugates for fluorescent sensing of G-quadruplex DNA. Angew. Chem. Int. Ed. 2009, 48, 2188–2191. [Google Scholar] [CrossRef]
- Xie, X.; Choi, B.; Largy, E.; Guillot, R.; Granzhan, A.; Teulade-Fichou, M.P. Asymmetric distyrylpyridinium dyes as red-emitting fluorescent probes for quadruplex DNA. Chem. Eur. J. 2013, 19, 1214–1226. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, X.; Lu, S.; Liu, J.; Zhong, S.; Wei, Y.; Bing, T.; Zhang, N.; Shangguan, D. Thiazole orange styryl derivatives as fluorescent probes for G-quadruplex DNA. ACS Appl. Bio Mater. 2020, 3, 2643–2650. [Google Scholar] [CrossRef]
- Das, A.K.; Druzhinin, S.I.; Ihmels, H.; Müller, M.; Schönherr, H. Colorimetric and fluorimetric dna detection with a hydroxystyryl–quinolizinium photoacid and its application for cell imaging. Chem. Eur. J. 2019, 25, 12703–12707. [Google Scholar] [CrossRef] [PubMed]
- Das, A.K.; Ihmels, H.; Kölsch, S. Diphenylaminostyryl-substituted quinolizinium derivatives as fluorescent light-up probes for duplex and quadruplex DNA. Photochem. Photobiol. Sci. 2019, 18, 1373–1381. [Google Scholar] [CrossRef]
- Bhadra, K.; Kumar, G.S. Interaction of berberine, palmatine, coralyne, and sanguinarine to quadruplex DNA: A comparative spectroscopic and calorimetric study. Biochim. Biophys. Acta. 2011, 1810, 485–496. [Google Scholar] [CrossRef]
- Umar, M.I.; Ji, D.; Chan, C.-Y.; Kwok, C.K. G-quadruplex-based fluorescent turn-on ligands and aptamers: From development to applications. Molecules 2019, 24, 2416. [Google Scholar] [CrossRef] [PubMed]
- Monchaud, D. Chapter Five–Quadruplex detection in human cells. Annu. Rep. Med. Chem. 2020, 54, 133–160. [Google Scholar]
- Di Antonio, M.; Ponjavic, A.; Radzevičius, A.; Ranasinghe, R.T.; Catalano, M.; Zhang, X.; Shen, J.; Needham, L.-M.; Lee, S.F.; Klenerman, D.; et al. Single-molecule visualization of DNA G-quadruplex formation in live cells. Nat. Chem. 2020, 12, 832–837. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Chan, M.K.; Kool, E.T. Fluorescent nucleobases as tools for studying DNA and RNA. Nature Chem. Rev. 2017, 9, 1043–1055. [Google Scholar] [CrossRef]
- Bhadra, K.; Kumar, G.S. Therapeutic potential of nucleic acid-binding isoquinoline alkaloids: Binding aspects and implications for drug design. Med. Res. Rev. 2011, 31, 821–862. [Google Scholar] [CrossRef]
- Becher, J.; Berdnikova, D.V.; Ihmels, H.; Stremmel, C. Synthesis and investigation of quadruplex-DNA-binding, 9-O-substituted berberine derivatives. Beilstein J. Org. Chem. 2020, 16, 2795–2806. [Google Scholar] [CrossRef]
- Pithan, P.M.; Decker, D.; Druzhinin, S.I.; Ihmels, H.; Schönherr, H.; Voß, Y. 8-Styryl-substituted coralyne derivatives as DNA binding fluorescent probes. RSC Adv. 2017, 7, 10660–10667. [Google Scholar] [CrossRef]
- Xu, L.; Hong, S.; Sun, N.; Wang, K.; Zhou, L.; Ji, L.; Pei, R. Berberine as a novel light-up i-motif fluorescence ligand and its application in designing molecular logic systems. Chem. Commun. 2016, 52, 179–182. [Google Scholar] [CrossRef]
- Zhou, C.-Q.; Yang, J.-W.; Dong, C.; Wang, Y.-M.; Sun, B.; Chen, J.-X.; Xu, Y.-S.; Chen, W.-H. Highly selective, sensitive and fluorescent sensing of dimeric G-quadruplexes by a dimeric berberine. Org. Biomol. Chem. 2016, 14, 191–197. [Google Scholar] [CrossRef]
- Duval, R.; Duplais, C. Fluorescent natural products as probes and tracersin biology. Nat. Prod. Rep. 2017, 34, 161–193. [Google Scholar] [CrossRef]
- Pang, B.; Zhao, L.-H.; Zhou, Q.; Zhao, T.-Y.; Wang, H.; Gu, C.-J.; Tong, X.-L.; Lee, H.C. Application of berberine on treating type 2 diabetes mellitus. Int. J. Endocrinol. 2015, 905749. [Google Scholar] [CrossRef] [PubMed]
- Gentry, E.J.; Jampani, H.B.; Keshavarz-Shokri, A.; Morton, M.D.; Velde, D.V.; Telikepalli, H.; Mitscher, L.; Shawar, R.; Humble, D.; Baker, W. Antitubercular natural products: Berberine from the roots of commercial Hydrastis canadensis powder. Isolation of inactive 8-oxotetrahydrothalifendine, canadine, beta-hydrastine, and two new quinic acid esters, hycandinic acid esters-1 and -2. J. Nat. Prod. 1998, 61, 1187–1193. [Google Scholar] [CrossRef]
- Yu, M.; Jin, X.; Liang, C.; Bu, F.; Pan, D.; He, Q.; Ming, Y.; Little, P.; Du, H.; Liang, S.; et al. Berberine for diarrhea in children and adults: A systematic review and meta-analysis. Ther. Adv. Gastroenterol. 2020, 13, 175628482096129. [Google Scholar] [CrossRef]
- Feng, X.; Sureda, A.; Jafari, S.; Memariani, Z.; Tewari, D.; Annunziata, G.; Barrea, L.; Hassan, S.T.S.; Šmejkal, K.; Malaník, M.; et al. Berberine in cardiovascular and metabolic diseases: From mechanisms to therapeutics. Theranostics 2019, 9, 1923–1951. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.M.; Basu, A.; Hossain, M.; Sureshkumar, G.; Hotha, S.; Kumar, G.S. Enhanced DNA binding of 9-ω-amino alkyl ether analogs from the plant alkaloid berberine. DNA Cell Biol. 2011, 30, 123–133. [Google Scholar] [CrossRef]
- Gumenyuk, V.G.; Bashmakova, N.V.; Kutovyy, S.Y.; Yashchuk, V.M.; Zaika, L.A. Binding parameters of alkaloids berberine and sanguinarine with DNA. Ukr. J. Phys. 2011, 56, 524–533. [Google Scholar]
- Kundu, N.; Roy, A.; Banik, D.; Sarkar, N. Unveiling the mode of interaction of berberine alkaloid in different supramolecular confined environments: Interplay of surface charge between nano-confined charged layer and DNA. J. Phys. Chem. B 2016, 120, 1106–1120. [Google Scholar] [CrossRef]
- Díaz, M.S.; Freile, M.L.; Gutiérrez, M.I. Solvent effect on the UV/Vis absorption and fluorescence spectroscopic properties of berberine. Photochem. Photobiol. Sci. 2009, 8, 970–974. [Google Scholar] [CrossRef]
- Kostjukova, L.O.; Kostjukov, V.V. The electronic states and vibronic absorption spectrum of berberine in aqueous solution. Int. J. Quantum. Chem. 2020, 121, e26537. [Google Scholar] [CrossRef]
- Auffinger, P.; Bielecki, L.; Westhof, E. Anion binding to nucleic acids. Structure 2004, 12, 379–388. [Google Scholar] [CrossRef]
- Rypniewski, W.; Adamiak, D.A.; Milecki, J.; Adamiak, R.W. Noncanonical G(syn)–G(anti) base pairs stabilized by sulphate anions in two X-ray structures of the (GUGGUCUGAUGAGGCC) RNA duplex. RNA 2008, 14, 1845–1851. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Van der Wijst, T.; Guerra, C.F.; Swart, M.; Bickelhaupt, F.M.; Lippert, B. A Ditopic ion-pair receptor based on stacked nucleobase quartets. Angew. Chem. Int. Ed. 2009, 48, 3285–3287. [Google Scholar] [CrossRef]
- van der Wijst, T.; Lippert, B.; Swart, M.; Guerra, C.F.; Bickelhaupt, F.M. Differential stabilization of adenine quartets by anions and cations. J. Biol. Inorg. Chem. 2010, 15, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Kawai, K.; Osakada, Y.; Fujitsuka, M.; Majima, T. Hole transfer in DNA and photosensitized DNA damage: Importance of adenine oxidation. J. Phys. Chem. B 2007, 111, 2322–2326. [Google Scholar] [CrossRef]
- Takada, T.; Kawai, K.; Tojo, S.; Majima, T. Effects of interaction of photosensitizer with DNA and stacked G bases on photosensitized one-electron oxidation of DNA. J. Phys. Chem. B 2004, 108, 761–766. [Google Scholar] [CrossRef]
- Kumar, G.S.; He, Q.-H.; Behr-Ventura, D.; Tomasz, M. Binding of 2,7-diaminomitosene to DNA: Model for the recognition of DNA by activated mitomycin C. Biochemistry 1995, 34, 2662–2671. [Google Scholar] [CrossRef]
- Takenaka, S.; Ihara, T.; Hamano, M.; Takagi, M. A zwitterionic anthraquinone derivative: First zwitterionic DNA binding ligand. J. Chem. Soc. Chem. Commun. 1990, 18, 1271–1273. [Google Scholar] [CrossRef]
- Nicoludis, J.M.; Miller, S.T.; Jeffrey, P.D.; Barrett, S.P.; Rablen, P.R.; Lawton, T.J.; Yatsunyk, L.A. Optimized end-stacking provides specificity of N-Methyl mesoporphyrin IX for human telomeric G-quadruplex DNA. J. Am. Chem. Soc. 2012, 134, 20446–20456. [Google Scholar] [CrossRef]
- Nasab, M.G.; Hassani, L.; Nejad, S.M.; Norouzi, D. Interaction of hemin with quadruplex DNA. J. Biol. Phys. 2017, 43, 5–14. [Google Scholar] [CrossRef]
- Yaku, H.; Murashima, T.; Miyoshi, D.; Sugimoto, N. Anionic phthalocyanines targeting G-quadruplexes and inhibiting telomerase activity in the presence of excessive DNA duplexes. Chem. Commun. 2010, 46, 5740–5742. [Google Scholar] [CrossRef]
- Yaku, H.; Murashima, T.; Miyoshi, D.; Sugimoto, N. Specific binding of anionic porphyrin and phthalocyanine to the G-quadruplex with a variety of in vitro and in vivo applications. Molecules 2012, 17, 10586–10613. [Google Scholar] [CrossRef]
- Luo, X.-J.; Qin, Q.-P.; Huang, K.-B.; Liu, Y.-C.; Liang, H. Spectroscopic studies on the interaction of hemin with human telomeric G-quadruplex DNA. Asian J. Chem. 2013, 25, 6396–6400. [Google Scholar] [CrossRef]
- Uchiyama, M.; Momotake, A.; Kobayashi, N.; Yamamoto, Y. Specific binding of an anionic phthalocyanine derivative to G-quadruplex DNAs. Chem. Lett. 2020, 49, 530–533. [Google Scholar] [CrossRef]
- Grabowski, Z.R.; Rotkiewicz, K.; Rettig, W. Structural changes accompanying intramolecular electron transfer: Focus on twisted intramolecular charge-transfer states and structures. Chem. Rev. 2003, 10, 3899–4032. [Google Scholar] [CrossRef]
- Mataga, N.; Kaifu, Y.; Koizumi, M. Solvent effects upon fluorescence spectra and the dipolemoments of excited molecules. Bull. Chem. Soc. Jpn. 1956, 29, 465–470. [Google Scholar] [CrossRef]
- Rurack, K.; Spieles, M. Fluorescence quantum yields of a series of red and near-infrared dyes emitting at 600−1000 nm. Anal. Chem. 2011, 83, 1232–1242. [Google Scholar] [CrossRef] [PubMed]
- Andrussow, L.; Schramm, B. Eigenschaften der Materie in ihren Aggregatzuständen; Transportphänomene I (Viskosität und Diffusion); Schäfer, K., Ed.; Springer: Berlin/Heidelberg, Germany, 1969. [Google Scholar]
- Paez, S.; Contreras, M. Densities and viscosities of binary mixtures of 1-propanol and 2-propanol with acetonitrile. J. Chem. Eng. Data 1989, 34, 455–459. [Google Scholar] [CrossRef]
- Stootman, F.H.; Fisher, D.M.; Rodger, A.; Aldrich-Wright, J.R. Improved curve fitting procedures to determine equilibrium binding constants. Analyst 2006, 131, 1145–1151. [Google Scholar] [CrossRef]
- De Cian, A.; Guittat, L.; Kaiser, M.; Saccà, B.; Amrane, S.; Bourdoncle, A.; Alberti, P.; Teulade-Fichou, M.-P.; Lacroix, L.; Mergny, J.-L. Fluorescence-based melting assays for studying quadruplex ligands. Methods 2007, 42, 183–195. [Google Scholar] [CrossRef]
- Nordén, B.; Rodger, A.; Dafforn, T. Linear Dichroism and Circular Dichroism. A Textbook on Polarized-Light Spectroscopy; RSC: Cambridge, UK, 2010. [Google Scholar]
- Šmidlehner, T.; Piantanida, I.; Pescitelli, G. Polarization spectroscopy methods in the determination of interactions of small molecules with nucleic acids–Tutorial. Beilstein J. Org. Chem. 2018, 14, 84–105. [Google Scholar] [CrossRef] [PubMed]
- Nordén, B.; Kurucsev, T. Analysing DNA complexes by circular and linear dichroism. J. Mol. Recognit. 1994, 7, 141–155. [Google Scholar] [CrossRef]
- Ferraroni, M.; Bazzicalupi, C.; Bilia, A.R.; Gratter, P. X-Ray diffraction analyses of the natural isoquinoline alkaloids Berberine and Sanguinarine complexed with double helix DNA d(CGTACG). Chem. Comm. 2011, 47, 4917–4919. [Google Scholar] [CrossRef] [PubMed]
- Nandi, R.; Debnath, D.; Maiti, M. Interactions of berberine with poly(A) and tRNA. Biochim. Biophys. Acta 1990, 1049, 339–342. [Google Scholar] [CrossRef]
- Dzubiel, D.; Ihmels, H.; Mahmoud, M.M.A.; Thomas, L. A comparative study of the interactions of cationic hetarenes with quadruplex-DNA forming oligonucleotide sequences of the insulin-linked polymorphic region (ILPR). Beilstein J. Org. Chem. 2014, 10, 2963–2974. [Google Scholar] [CrossRef] [PubMed]
- Vorlíčková, M.; Kejnovská, I.; Sagi, J.; Renčiuk, D.; Bednářová, K.; Motlová, J.; Kypr, J. Circular dichroism and guanine quadruplexes. Methods 2012, 57, 64–75. [Google Scholar] [CrossRef]
- Ihmels, H.; Mahmoud, M.M.A.; Patrick, B.O. Optical differentiation between quadruplex DNA and duplex DNA with a [2.2.2]heptamethinecyanine dye. J. Phys. Org. Chem. 2017, 30, e3736. [Google Scholar] [CrossRef]
- Jain, A.K.; Bhattacharya, S. Interaction of G-quadruplexes with nonintercalating duplex-DNA minor groove binding ligands. Bioconjug. Chem. 2011, 22, 2355–2368. [Google Scholar] [CrossRef]
- Yamashita, T.; Uno, T.; Ishikawa, Y. Stabilization of guanine quadruplex DNA by the binding of porphyrins with cationic side arms. Bioorg. Med. Chem. 2005, 13, 2423–2430. [Google Scholar] [CrossRef]
- Sun, H.; Tang, Y.; Xiang, J.; Xu, G.; Zhang, Y.; Zhang, H.; Xu, L. Spectroscopic studies of the interaction between quercetin and G-quadruplex DNA. Bioorg. Med. Chem. Lett. 2006, 16, 3586–3589. [Google Scholar] [CrossRef]
- Bazzicalupi, C.; Ferraroni, M.; Bilia, A.R.; Scheggi, F.; Gratteri, P. The crystal structure of human telomeric DNA complexed with berberine: An interesting case of stacked ligand to G-tetrad ratio higher than 1:1. Nucleic Acids Res. 2012, 41, 632–638. [Google Scholar] [CrossRef]
- Catasti, P.; Chen, X.; Moyzis, R.K.; Bradbury, E.M.; Gupta, G. Structure–function correlations of the insulin-linked polymorphic region. J. Mol. Biol. 1996, 264, 534–545. [Google Scholar] [CrossRef]
- Xu, Y.; Noguchi, Y.; Sugiyama, H. The new models of the human telomere d[AGGG(TTAGGG)3] in K+ solution. Bioorg. Med. Chem. 2006, 14, 5584–5591. [Google Scholar] [CrossRef]
- Phan, A.T.; Kuryavyi, V.; Burge, S.; Neidle, S.; Patel, D.J. Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J. Am. Chem. Soc. 2007, 129, 4386–4392. [Google Scholar] [CrossRef]
- Stump, S.; Mou, T.-C.; Sprang, S.R.; Natale, N.R.; Beall, H.D. Crystal structure of the major quadruplex formed in the promoter region of the human c-MYC oncogene. PLoS ONE 2018, 13, e0205584. [Google Scholar] [CrossRef] [PubMed]
- Cuervo, A.; Dans, P.D.; Carrascosa, J.L.; Orozco, M.; Gomila, G.; Fumagalli, L. Direct measurement of the dielectric polarization properties of DNA. Proc. Natl. Acad. Sci. USA 2014, 111, E3624–E3630. [Google Scholar] [CrossRef]
- Megyesi, M.; Biczók, L.; Jablonkai, I. Highly sensitive fluorescence response to inclusion complex formation of berberine alkaloid with cucurbit[7]uril. J. Phys. Chem. C 2008, 112, 3410–3416. [Google Scholar] [CrossRef]
- Le Bras, L.; Chaitou, K.; Aloïse, S.; Adamo, C.; Perrier, A. Aggregation-caused quenchingversuscrystallizationinduced emission in thiazolo[5,4-b]thieno[3,2-e]-pyridine (TTP) derivatives: Theoretical insights. Phys. Chem. Chem. Phys. 2018, 21, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Liang, Q.; Wang, R.; Hou, J.; Li, W.; Peng, Q.; Shuai, Z.; Qiao, J. Highly efficient thermally activated delayed fluorescence via J-aggregates with strong intermolecular charge transfer. Adv. Mater. 2019, 31, e1808242. [Google Scholar] [CrossRef]
- Jin, X.; Yan, L.; Li, H.-J.; Wang, R.-L.; Hu, Z.-L.; Jiang, Y.-Y.; Cao, Y.-B.; Yan, T.-H.; Sun, Q.-Y. Novel triazolyl berberine derivatives prepared via CuAAC click chemistry: Synthesis, anticancer activity and structure-activity relationships. Anti Cancer Agents Med. Chem. 2014, 15, 89–98. [Google Scholar] [CrossRef]
- Ren, J.; Chaires, J.B. Sequence and structural selectivity of nucleic acid binding ligands. Biochemistry 1999, 38, 16067–16075. [Google Scholar] [CrossRef]
- Bortolozzi, R.; Ihmels, H.; Thomas, L.; Tian, M.; Viola, G. 9-(4-Dimethylaminophenyl)-benzo[b]quinolizinium: A Near-Infrared Fluorophore for the Multicolor Analysis of Proteins and Nucleic Acids in Living Cells. Chem. Eur. J. 2013, 19, 8736–8741. [Google Scholar] [CrossRef]
- Duskova, K.; Lejault, P.; Benchimol, E.; Guillot, R.; Britton, S.; Granzhan, A.; Monchaud, D. DNA Junction Ligands Trigger DNA Damage and Are Synthetic Lethal with DNA Repair Inhibitors in Cancer Cells. J. Am. Chem. Soc. 2020, 142, 424–435. [Google Scholar] [CrossRef]
| Solvent | λabsa/nm | lg ε b | λflc/nm | νabs − νfl d/cm−1 | Φfle | ||||
|---|---|---|---|---|---|---|---|---|---|
| 2 | 1a f | 2 | 2 | 1a f | 2 | 1a f | 2 | 1a f | |
| MeOH | 431 | 429 | 3.98 | 558 | 540 | 5280 | 4790 | 0.01 | 0.01 |
| EtOH | 431 | 430 | 3.98 | 537 | 536 | 4580 | 4600 | 0.02 | 0.03 |
| 1-PrOH | 431 | 3.94 | 531 | 4369 | 0.04 | ||||
| MeCN | 431 | 430 | 3.91 | 537 | 537 | 4579 | 4630 | 0.03 | 0.02 |
| DMSO | 430 | 425 | 3.97 | 547 | 553 | 4974 | 5500 | 0.01 | <0.01 |
| Buffer | 424 | 3.92 | 535 | 4893 | <0.01 | ||||
| DNA | Kb/105 M−1 b | Φfl c | ∆Tm/°C d |
|---|---|---|---|
| ct DNA | 2.2 ± 0.2 | <0.01 | |
| 22AG/F21Ta | 5.0 ± 0.6 | 0.01 | 3.6 |
| c-kit/FkitTa | 6.9 ± 0.2 | 0.01 | 1.0 |
| a2/Fa2Ta | 6.7 ± 0.2 | 0.01 | 0.6 |
| c-myc/FmycT a | 5.9 ± 0.4 | 0.01 | 2.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wickhorst, P.J.; Ihmels, H. Berberrubine Phosphate: A Selective Fluorescent Probe for Quadruplex DNA. Molecules 2021, 26, 2566. https://doi.org/10.3390/molecules26092566
Wickhorst PJ, Ihmels H. Berberrubine Phosphate: A Selective Fluorescent Probe for Quadruplex DNA. Molecules. 2021; 26(9):2566. https://doi.org/10.3390/molecules26092566
Chicago/Turabian StyleWickhorst, Peter Jonas, and Heiko Ihmels. 2021. "Berberrubine Phosphate: A Selective Fluorescent Probe for Quadruplex DNA" Molecules 26, no. 9: 2566. https://doi.org/10.3390/molecules26092566
APA StyleWickhorst, P. J., & Ihmels, H. (2021). Berberrubine Phosphate: A Selective Fluorescent Probe for Quadruplex DNA. Molecules, 26(9), 2566. https://doi.org/10.3390/molecules26092566