Inhibition of Cytosolic Phospholipase A2α Induces Apoptosis in Multiple Myeloma Cells
Abstract
1. Introduction
2. Results
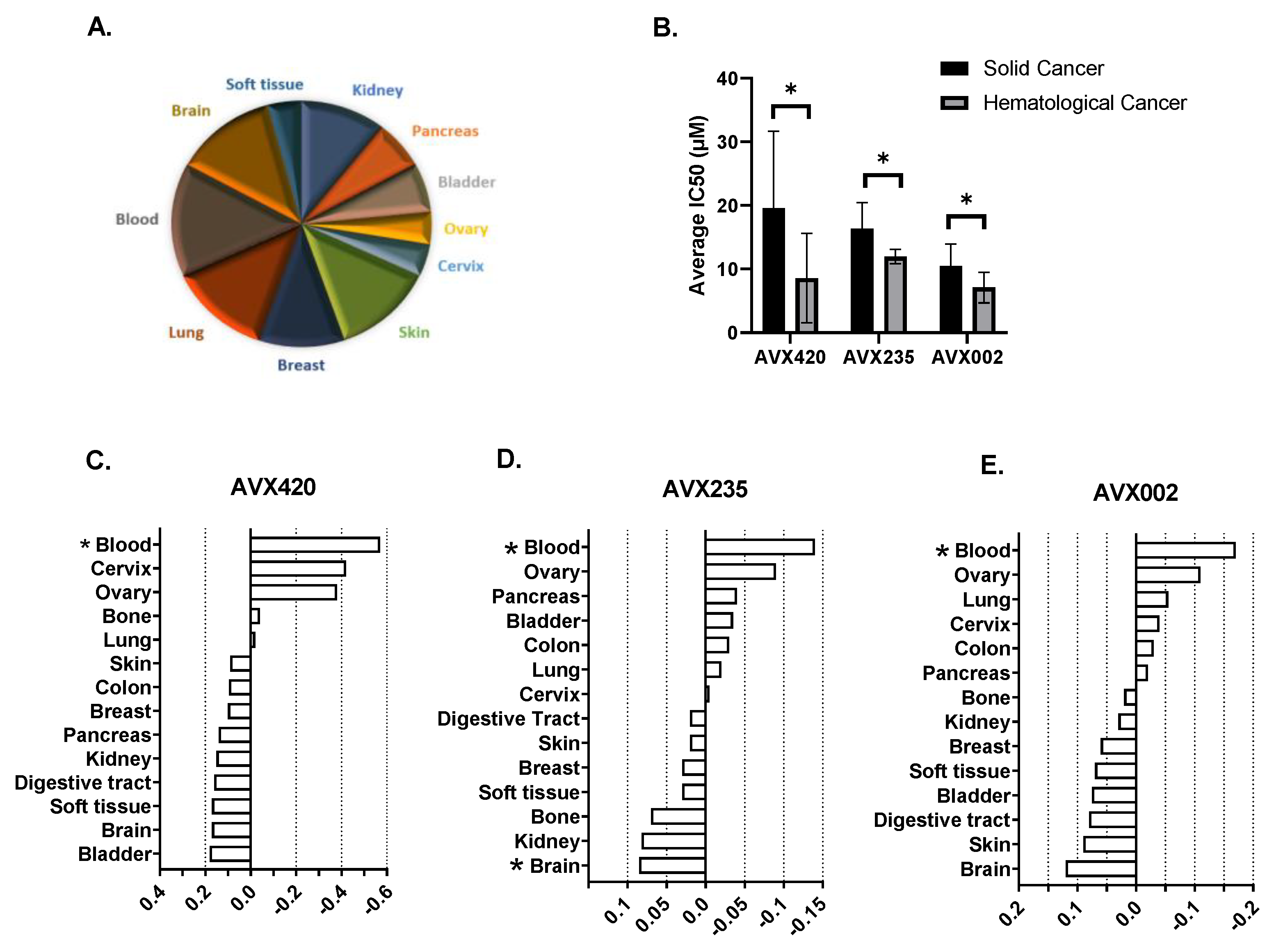
2.1. cPLA2α Is Overexpressed in Cancers and Cancer Cell Lines from Different Tissue Origins
2.2. Hematological Cancer Cells Are Sensitive to cPLA2α Inhibition
2.3. cPLA2α Is Overexpressed in Hematological Cancers including Multiple Myeloma
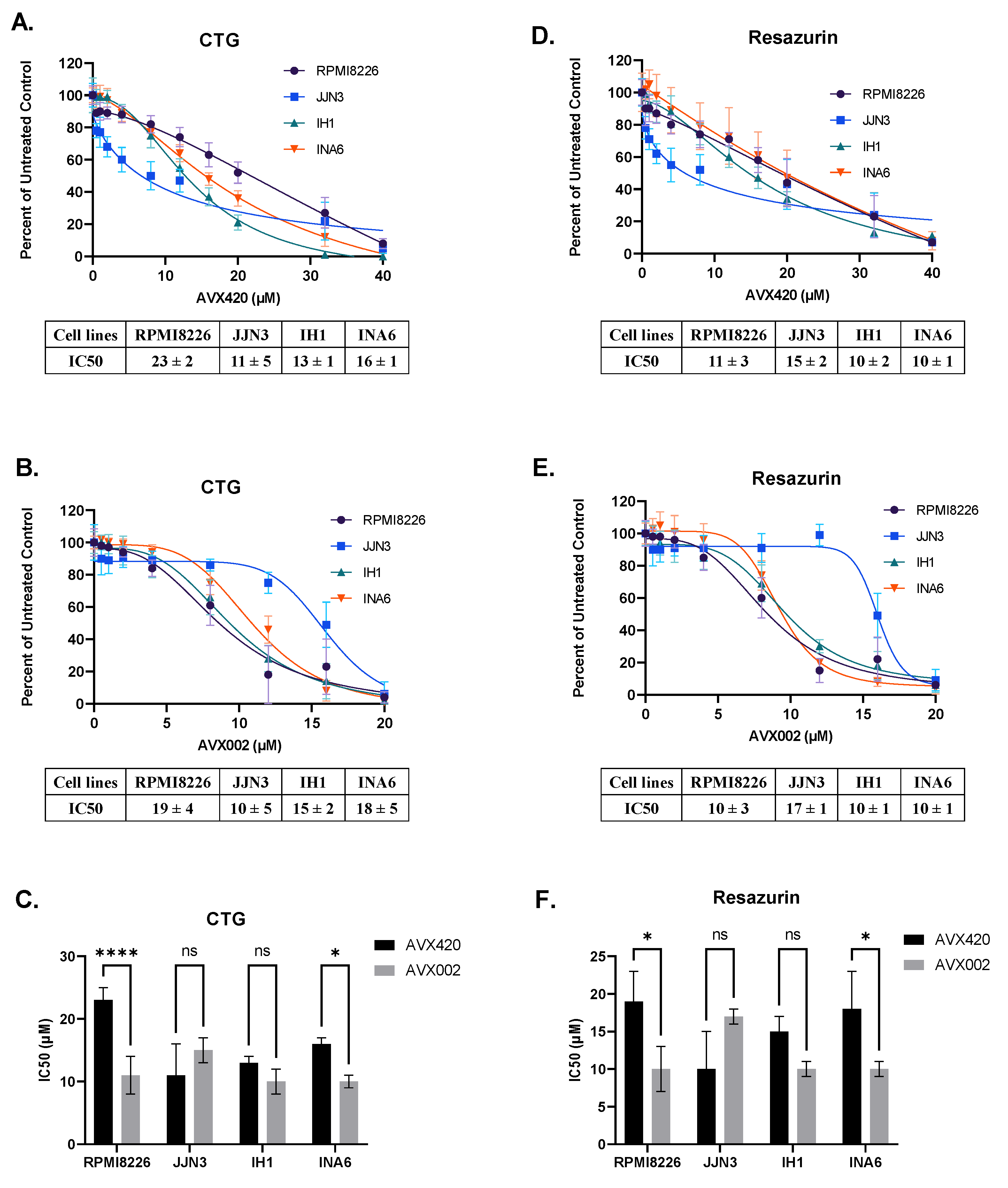
2.4. Inhibition of cPLA2α Reduces Cell Viability of Multiple Myeloma Cells
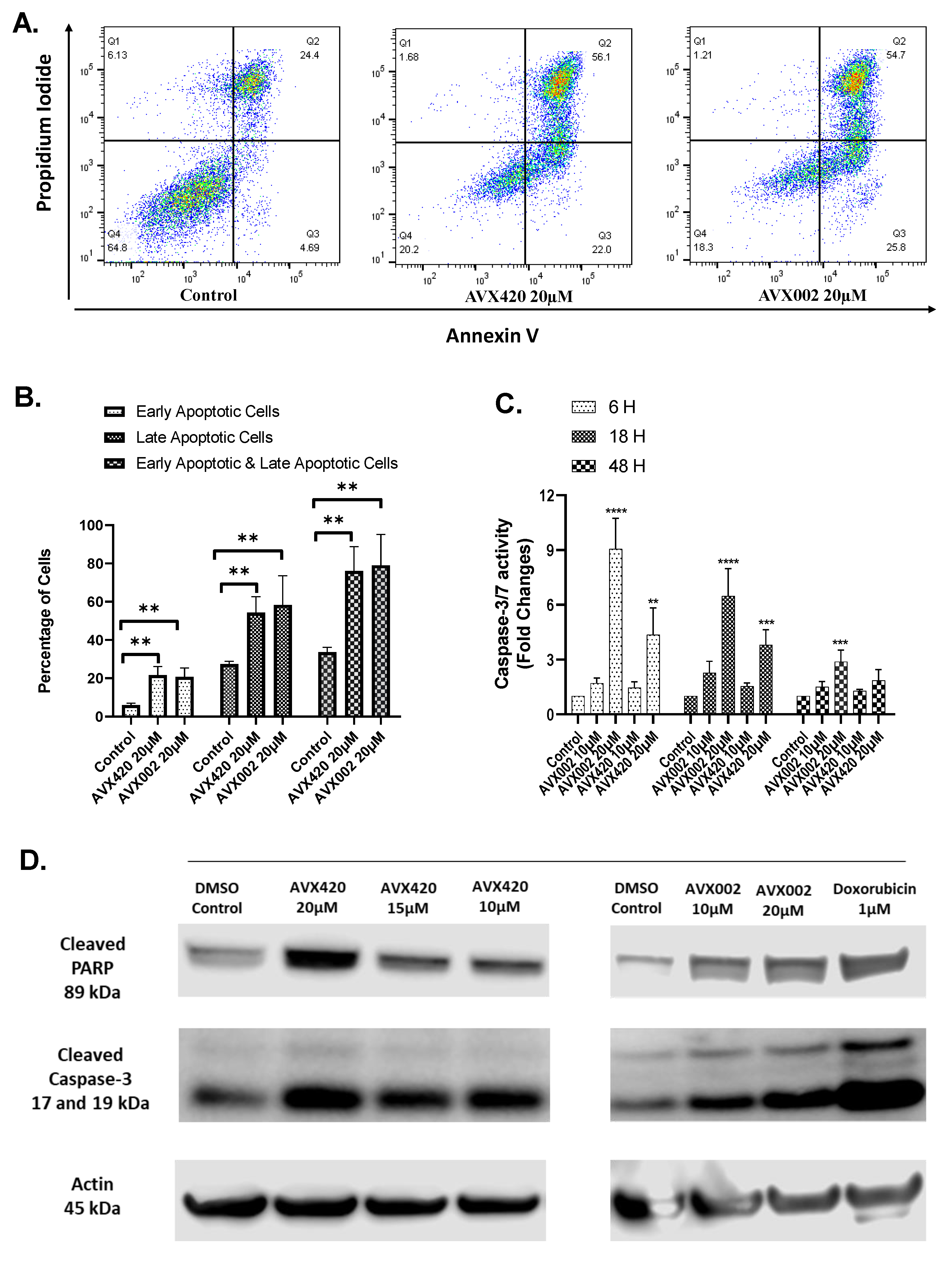
2.5. cPLA2α Inhibitors Induce Apoptosis in Caspase-3 Dependent Pathways
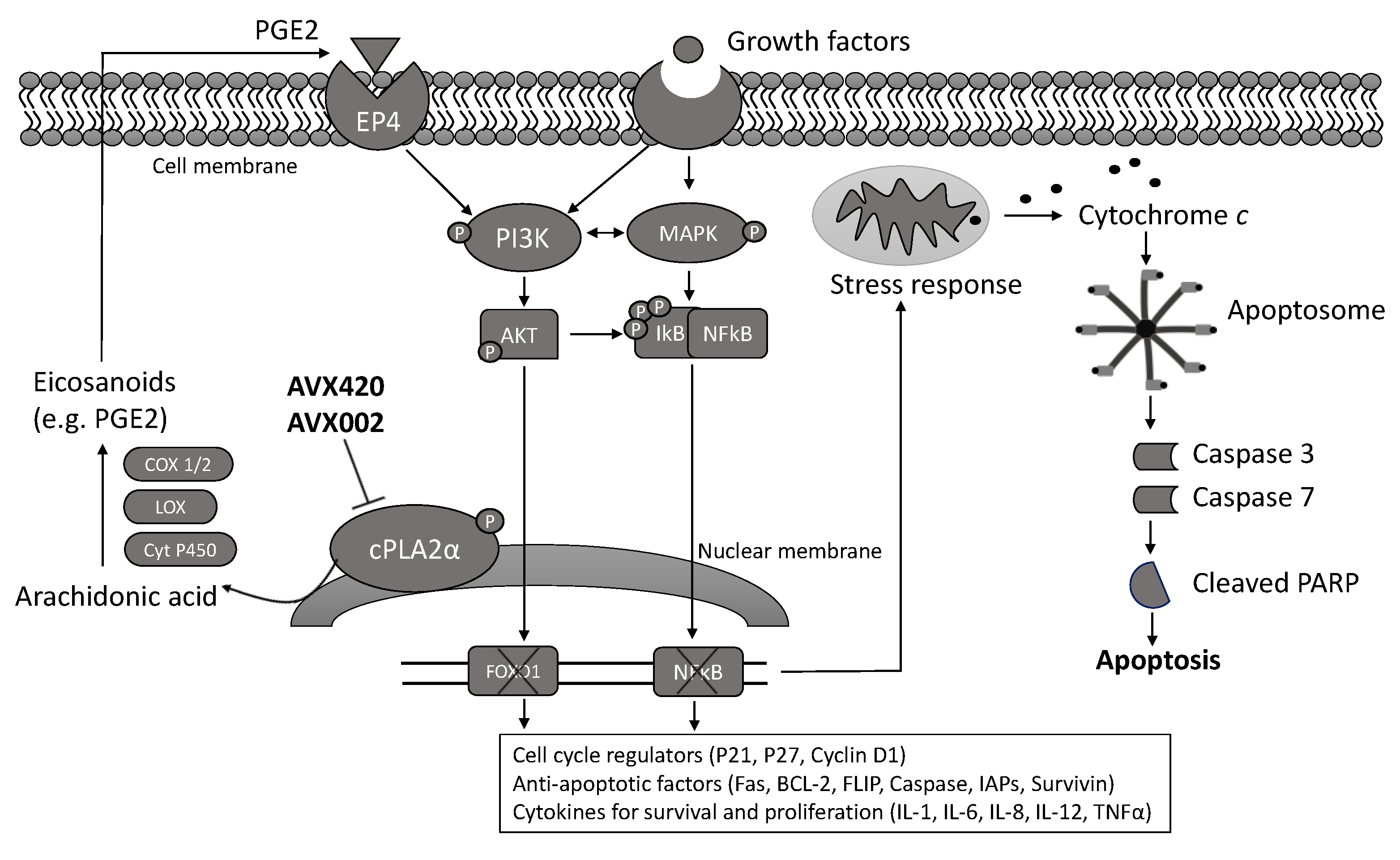
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Maintenance of Multiple Myeloma Cell Lines
4.3. Viability Screening of Oncolines Cancer Cell Panel
4.4. Cell Viability Assays
4.5. Annexin V-FITC and Propidium Iodide Apoptosis Assay
4.6. Caspase-3/7 Activity Assay
4.7. Western Blot
4.8. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Rodriguez-Abreu, D.; Bordoni, A.; Zucca, E. Epidemiology of hematological malignancies. Ann. Oncol. 2007, 18 (Suppl. 1), i3–i8. [Google Scholar] [CrossRef]
- Kyle, R.A.; Rajkumar, S.V. Multiple myeloma. Blood 2008, 111, 2962–2972. [Google Scholar] [CrossRef]
- Hideshima, T.; Mitsiades, C.; Tonon, G.; Richardson, P.G.; Anderson, K.C. Understanding multiple myeloma pathogenesis in the bone marrow to identify new therapeutic targets. Nat. Rev. Cancer 2007, 7, 585–598. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, A.; Anderson, K. Multiple myeloma. N. Engl. J. Med. 2011, 364, 1046–1060. [Google Scholar] [CrossRef] [PubMed]
- Eslick, R.; Talaulikar, D. Multiple myeloma: From diagnosis to treatment. Aust. Fam. Phys. 2013, 42, 684–688. [Google Scholar]
- Firth, J. Haematology: Multiple myeloma. Clin. Med. (Lond.) 2019, 19, 58–60. [Google Scholar] [CrossRef]
- Wang, D.; Dubois, R.N. Eicosanoids and cancer. Nat. Rev. Cancer 2010, 10, 181–193. [Google Scholar] [CrossRef]
- Dennis, E.A.; Cao, J.; Hsu, Y.H.; Magrioti, V.; Kokotos, G. Phospholipase A2 enzymes: Physical structure, biological function, disease implication, chemical inhibition, and therapeutic intervention. Chem. Rev. 2011, 111, 6130–6185. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, M.; Tucker, D.E.; Burchett, S.A.; Leslie, C.C. Properties of the Group IV phospholipase A2 family. Prog. Lipid Res. 2006, 45, 487–510. [Google Scholar] [CrossRef]
- Chen, L.; Fu, H.; Luo, Y.; Chen, L.; Cheng, R.; Zhang, N.; Guo, H. cPLA2α mediates TGF-β-induced epithelial-mesenchymal transition in breast cancer through PI3k/Akt signaling. Cell Death Dis. 2017, 8, e2728. [Google Scholar] [CrossRef]
- Fu, H.; He, Y.; Qi, L.; Chen, L.; Luo, Y.; Chen, L.; Li, Y.; Zhang, N.; Guo, H. cPLA2α activates PI3K/AKT and inhibits Smad2/3 during epithelial-mesenchymal transition of hepatocellular carcinoma cells. Cancer Lett. 2017, 403, 260–270. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Sun, Y.; Zeng, L.; Li, Y.; Hu, S.; He, S.; Chen, H.; Zou, Q.; Luo, B. Inhibition of cytosolic phospholipase A2 alpha increases chemosensitivity in cervical carcinoma through suppressing β-catenin signaling. Cancer Biol. Ther. 2019, 20, 912–921. [Google Scholar] [CrossRef]
- Patel, M.I.; Singh, J.; Niknami, M.; Kurek, C.; Yao, M.; Lu, S.; Maclean, F.; King, N.J.; Gelb, M.H.; Scott, K.F.; et al. Cytosolic phospholipase A2-alpha: A potential therapeutic target for prostate cancer. Clin. Cancer Res. 2008, 14, 8070–8079. [Google Scholar] [CrossRef] [PubMed]
- Xin, C.; Chu, L.; Zhang, L.; Geng, D.; Wang, Y.; Sun, D.; Sui, P.; Zhao, X.; Gong, Z.; Sui, M.; et al. Expression of Cytosolic Phospholipase A2 (cPLA2)-Arachidonic Acid (AA)-Cyclooxygenase-2 (COX-2) Pathway Factors in Lung Cancer Patients and Its Implication in Lung Cancer Early Detection and Prognosis. Med. Sci. Monit. 2019, 25, 5543–5551. [Google Scholar] [CrossRef]
- Bai, H.; Zhou, M.; Zeng, M.; Han, L. PLA2G4A Is a Potential Biomarker Predicting Shorter Overall Survival in Patients with Non-M3/NPM1 Wildtype Acute Myeloid Leukemia. DNA Cell Biol. 2020, 39, 700–708. [Google Scholar] [CrossRef]
- Runarsson, G.; Feltenmark, S.; Forsell, P.K.; Sjöberg, J.; Björkholm, M.; Claesson, H.E. The expression of cytosolic phospholipase A2 and biosynthesis of leukotriene B4 in acute myeloid leukemia cells. Eur. J. Haematol. 2007, 79, 468–476. [Google Scholar] [CrossRef] [PubMed]
- Guriec, N.; Le Jossic-Corcos, C.; Simon, B.; Ianotto, J.C.; Tempescul, A.; Dréano, Y.; Salaün, J.P.; Berthou, C.; Corcos, L. The arachidonic acid-LTB4-BLT2 pathway enhances human B-CLL aggressiveness. Biochim. Biophys. Acta 2014, 1842, 2096–2105. [Google Scholar] [CrossRef] [PubMed]
- Chitale, D.; Gong, Y.; Taylor, B.S.; Broderick, S.; Brennan, C.; Somwar, R.; Golas, B.; Wang, L.; Motoi, N.; Szoke, J.; et al. An integrated genomic analysis of lung cancer reveals loss of DUSP4 in EGFR-mutant tumors. Oncogene 2009, 28, 2773–2783. [Google Scholar] [CrossRef]
- Gravendeel, L.A.; Kouwenhoven, M.C.; Gevaert, O.; de Rooi, J.J.; Stubbs, A.P.; Duijm, J.E.; Daemen, A.; Bleeker, F.E.; Bralten, L.B.; Kloosterhof, N.K.; et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009, 69, 9065–9072. [Google Scholar] [CrossRef] [PubMed]
- Clarke, C.; Madden, S.F.; Doolan, P.; Aherne, S.T.; Joyce, H.; O’Driscoll, L.; Gallagher, W.M.; Hennessy, B.T.; Moriarty, M.; Crown, J.; et al. Correlating transcriptional networks to breast cancer survival: A large-scale coexpression analysis. Carcinogenesis 2013, 34, 2300–2308. [Google Scholar] [CrossRef] [PubMed]
- Zhan, F.; Huang, Y.; Colla, S.; Stewart, J.P.; Hanamura, I.; Gupta, S.; Epstein, J.; Yaccoby, S.; Sawyer, J.; Burington, B.; et al. The molecular classification of multiple myeloma. Blood 2006, 108, 2020–2028. [Google Scholar] [CrossRef] [PubMed]
- Minn, A.J.; Gupta, G.P.; Padua, D.; Bos, P.; Nguyen, D.X.; Nuyten, D.; Kreike, B.; Zhang, Y.; Wang, Y.; Ishwaran, H.; et al. Lung metastasis genes couple breast tumor size and metastatic spread. Proc. Natl. Acad. Sci. USA 2007, 104, 6740–6745. [Google Scholar] [CrossRef]
- Magrioti, V.; Kokotos, G. Phospholipase A2 inhibitors for the treatment of inflammatory diseases: A patent review (2010--present). Expert Opin. Ther. Pat. 2013, 23, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Linkous, A.; Geng, L.; Lyshchik, A.; Hallahan, D.E.; Yazlovitskaya, E.M. Cytosolic phospholipase A2: Targeting cancer through the tumor vasculature. Clin. Cancer Res. 2009, 15, 1635–1644. [Google Scholar] [CrossRef]
- Linkous, A.G.; Yazlovitskaya, E.M.; Hallahan, D.E. Cytosolic phospholipase A2 and lysophospholipids in tumor angiogenesis. J. Natl. Cancer Inst. 2010, 102, 1398–1412. [Google Scholar] [CrossRef] [PubMed]
- Thotala, D.; Craft, J.M.; Ferraro, D.J.; Kotipatruni, R.P.; Bhave, S.R.; Jaboin, J.J.; Hallahan, D.E. Cytosolic phospholipaseA2 inhibition with PLA-695 radiosensitizes tumors in lung cancer animal models. PLoS ONE 2013, 8, e69688. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, X.; Zhang, L.; Geng, D.; Wang, Y.; Sun, D.; Sui, P.; Zhao, X.; Xin, C.; Jiang, J.; et al. Inhibition of PLA2G4A Reduces the Expression of Lung Cancer-Related Cytokines. DNA Cell Biol. 2018, 37, 1076–1081. [Google Scholar] [CrossRef] [PubMed]
- Ashcroft, F.J.; Mahammad, N.; Midtun Flatekvål, H.; Jullumstrø Feuerherm, A.; Johansen, B. cPLA(2)α Enzyme Inhibition Attenuates Inflammation and Keratinocyte Proliferation. Biomolecules 2020, 10, 1402. [Google Scholar] [CrossRef]
- Huwiler, A.; Feuerherm, A.J.; Sakem, B.; Pastukhov, O.; Filipenko, I.; Nguyen, T.; Johansen, B. The ω3-polyunsaturated fatty acid derivatives AVX001 and AVX002 directly inhibit cytosolic phospholipase A(2) and suppress PGE(2) formation in mesangial cells. Br. J. Pharmacol. 2012, 167, 1691–1701. [Google Scholar] [CrossRef]
- Sommerfelt, R.M.; Feuerherm, A.J.; Skuland, T.; Johansen, B. Cytosolic phospholipase A2 modulates TLR2 signaling in synoviocytes. PLoS ONE 2015, 10, e0119088. [Google Scholar] [CrossRef] [PubMed]
- Sommerfelt, R.M.; Feuerherm, A.J.; Jones, K.; Johansen, B. Cytosolic phospholipase A2 regulates TNF-induced production of joint destructive effectors in synoviocytes. PLoS ONE 2013, 8, e83555. [Google Scholar] [CrossRef] [PubMed]
- Omland, S.H.; Habicht, A.; Damsbo, P.; Wilms, J.; Johansen, B.; Gniadecki, R. A randomized, double-blind, placebo-controlled, dose-escalation first-in-man study (phase 0) to assess the safety and efficacy of topical cytosolic phospholipase A2 inhibitor, AVX001, in patients with mild to moderate plaque psoriasis. J. Eur. Acad. Dermatol. Venereol. 2017, 31, 1161–1167. [Google Scholar] [CrossRef]
- Feuerherm, A.J.; Dennis, E.A.; Johansen, B. Cytosolic group IVA phospholipase A2 inhibitors, AVX001 and AVX002, ameliorate collagen-induced arthritis. Arthritis Res. Ther. 2019, 21, 29. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Tunset, H.M.; Cebulla, J.; Vettukattil, R.; Helgesen, H.; Feuerherm, A.J.; Engebråten, O.; Mælandsmo, G.M.; Johansen, B.; Moestue, S.A. Anti-vascular effects of the cytosolic phospholipase A2 inhibitor AVX235 in a patient-derived basal-like breast cancer model. BMC Cancer 2016, 16, 191. [Google Scholar] [CrossRef] [PubMed]
- Tunset, H.M.; Feuerherm, A.J.; Selvik, L.M.; Johansen, B.; Moestue, S.A. Cytosolic Phospholipase A2 Alpha Regulates TLR Signaling and Migration in Metastatic 4T1 Cells. Int. J. Mol. Sci. 2019, 20, 4800. [Google Scholar] [CrossRef] [PubMed]
- Johansen, B.; Sanderberg, M.; Aukrust, I.R.; Kokotos, G.; Barbayianni, E. Antiiflammatory and antitumor 2-oxothiazoles and 2-oxothiophenes compounds. US Patent Application No. 14/764,509, 31 December 2015. [Google Scholar]
- Mahammad, N.; Ashcroft, F.J.; Feuerherm, A.J.; Kokotos, G.; Gjertsen, B.T.; Dennis, E.A.; Johansen, B. AVX420, a novel cPLA2α inhibitor targeting hematological cancers. Manuscript in preparation.
- Zimmermann, P.; Hennig, L.; Gruissem, W. Gene-expression analysis and network discovery using Genevestigator. Trends Plant Sci. 2005, 10, 407–409. [Google Scholar] [CrossRef] [PubMed]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, D.R.; Yu, J.; Shanker, K.; Deshpande, N.; Varambally, R.; Ghosh, D.; Barrette, T.; Pandey, A.; Chinnaiyan, A.M. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia 2004, 6, 1–6. [Google Scholar] [CrossRef]
- Chaudhry, P.; Singh, M.; Parent, S.; Asselin, E. Prostate apoptosis response 4 (Par-4), a novel substrate of caspase-3 during apoptosis activation. Mol. Cell Biol. 2012, 32, 826–839. [Google Scholar] [CrossRef]
- Elsaadi, S.; Steiro, I.; Abdollahi, P.; Vandsemb, E.N.; Yang, R.; Slørdahl, T.S.; Rø, T.B.; Menu, E.; Sponaas, A.M.; Børset, M. Targeting phosphoglycerate dehydrogenase in multiple myeloma. Exp Hematol. Oncol. 2021, 10, 3. [Google Scholar] [CrossRef] [PubMed]
- Sayan, B.S.; Sayan, A.E.; Yang, A.L.; Aqeilan, R.I.; Candi, E.; Cohen, G.M.; Knight, R.A.; Croce, C.M.; Melino, G. Cleavage of the transactivation-inhibitory domain of p63 by caspases enhances apoptosis. Proc. Natl. Acad. Sci. USA 2007, 104, 10871–10876. [Google Scholar] [CrossRef] [PubMed]
- El-Hamid, E.S.A.; Gamal-Eldeen, A.M.; Sharaf Eldeen, A.M. Liposome-coated nano doxorubicin induces apoptosis on oral squamous cell carcinoma CAL-27 cells. Arch Oral. Biol. 2019, 103, 47–54. [Google Scholar] [CrossRef]
- Tacar, O.; Sriamornsak, P.; Dass, C.R. Doxorubicin: An update on anticancer molecular action, toxicity and novel drug delivery systems. J. Pharm. Pharmacol. 2013, 65, 157–170. [Google Scholar] [CrossRef]
- Vu, M.; Kassouf, N.; Ofili, R.; Lund, T.; Bell, C.; Appiah, S. Doxorubicin selectively induces apoptosis through the inhibition of a novel isoform of Bcl-2 in acute myeloid leukaemia MOLM-13 cells with reduced Beclin 1 expression. Int. J. Oncol. 2020, 57, 113–121. [Google Scholar] [CrossRef]
- Heasley, L.E.; Thaler, S.; Nicks, M.; Price, B.; Skorecki, K.; Nemenoff, R.A. Induction of cytosolic phospholipase A2 by oncogenic Ras in human non-small cell lung cancer. J. Biol. Chem. 1997, 272, 14501–14504. [Google Scholar] [CrossRef]
- Wu, T.; Han, C.; Lunz, J.G., 3rd; Michalopoulos, G.; Shelhamer, J.H.; Demetris, A.J. Involvement of 85-kd cytosolic phospholipase A(2) and cyclooxygenase-2 in the proliferation of human cholangiocarcinoma cells. Hepatology 2002, 36, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Soydan, A.S.; Tavares, I.A.; Weech, P.K.; Temblay, N.M.; Bennett, A. High molecular weight phospholipase A2 and fatty acids in human colon tumours and associated normal tissue. Eur. J. Cancer 1996, 32a, 1781–1787. [Google Scholar] [CrossRef]
- Naini, S.M.; Choukroun, G.J.; Ryan, J.R.; Hentschel, D.M.; Shah, J.V.; Bonventre, J.V. Cytosolic phospholipase A2α regulates G1 progression through modulating FOXO1 activity. Faseb J. 2016, 30, 1155–1170. [Google Scholar] [CrossRef]
- Kisslov, L.; Hadad, N.; Rosengraten, M.; Levy, R. HT-29 human colon cancer cell proliferation is regulated by cytosolic phospholipase A(2)α dependent PGE(2)via both PKA and PKB pathways. Biochim. Biophys. Acta 2012, 1821, 1224–1234. [Google Scholar] [CrossRef]
- Han, C.; Bowen, W.C.; Li, G.; Demetris, A.J.; Michalopoulos, G.K.; Wu, T. Cytosolic phospholipase A2alpha and peroxisome proliferator-activated receptor gamma signaling pathway counteracts transforming growth factor beta-mediated inhibition of primary and transformed hepatocyte growth. Hepatology 2010, 52, 644–655. [Google Scholar] [CrossRef]
- Goodwin, A.M. In vitro assays of angiogenesis for assessment of angiogenic and anti-angiogenic agents. Microvasc Res. 2007, 74, 172–183. [Google Scholar] [CrossRef]
- Zeeshan, R.; Mutahir, Z. Cancer metastasis—Tricks of the trade. Bosn J. Basic Med. Sci. 2017, 17, 172–182. [Google Scholar] [CrossRef][Green Version]
- Clodi, K.; Kliche, K.O.; Zhao, S.; Weidner, D.; Schenk, T.; Consoli, U.; Jiang, S.; Snell, V.; Andreeff, M. Cell-surface exposure of phosphatidylserine correlates with the stage of fludarabine-induced apoptosis in chronic lymphocytic leukemia and expression of apoptosis-regulating genes. Cytometry 2000, 40, 19–25. [Google Scholar] [CrossRef]
- Nagata, S.; Suzuki, J.; Segawa, K.; Fujii, T. Exposure of phosphatidylserine on the cell surface. Cell Death Differ. 2016, 23, 952–961. [Google Scholar] [CrossRef] [PubMed]
- van den Eijnde, S.M.; Boshart, L.; Baehrecke, E.H.; De Zeeuw, C.I.; Reutelingsperger, C.P.; Vermeij-Keers, C. Cell surface exposure of phosphatidylserine during apoptosis is phylogenetically conserved. Apoptosis 1998, 3, 9–16. [Google Scholar] [CrossRef]
- Duarte, R.A.; Mello, E.R.; Araki, C.; Bolzani Vda, S.; Siqueira e Silva, D.H.; Regasini, L.O.; Silva, T.G.; de Morais, M.C.; Ximenes, V.F.; Soares, C.P. Alkaloids extracted from Pterogyne nitens induce apoptosis in malignant breast cell line. Tumour. Biol. 2010, 31, 513–522. [Google Scholar] [CrossRef]
- Habiba, K.; Encarnacion-Rosado, J.; Garcia-Pabon, K.; Villalobos-Santos, J.C.; Makarov, V.I.; Avalos, J.A.; Weiner, B.R.; Morell, G. Improving cytotoxicity against cancer cells by chemo-photodynamic combined modalities using silver-graphene quantum dots nanocomposites. Int. J. Nanomed. 2016, 11, 107–119. [Google Scholar] [CrossRef]
- Sačková, V.; Kuliková, L.; Kello, M.; Uhrinová, I.; Fedoročko, P. Enhanced antiproliferative and apoptotic response of HT-29 adenocarcinoma cells to combination of photoactivated hypericin and farnesyltransferase inhibitor manumycin A. Int. J. Mol. Sci. 2011, 12, 8388–8405. [Google Scholar] [CrossRef]
- Sundquist, T.; Moravec, R.; Niles, A.; O’Brien, M.; Riss, T. Timing Your Apoptosis Assays. Cell Notes 2016, 16, 18–21. [Google Scholar]
- Anthonsen, M.W.; Solhaug, A.; Johansen, B. Functional coupling between secretory and cytosolic phospholipase A2 modulates tumor necrosis factor-alpha- and interleukin-1beta-induced NF-kappa B activation. J. Biol. Chem. 2001, 276, 30527–30536. [Google Scholar] [CrossRef] [PubMed]
- George, R.J.; Sturmoski, M.A.; Anant, S.; Houchen, C.W. EP4 mediates PGE2 dependent cell survival through the PI3 kinase/AKT pathway. Prostaglandins Other Lipid Mediat. 2007, 83, 112–120. [Google Scholar] [CrossRef]
- Holmeide, A.K.; Skattebol, L. Syntheses of some polyunsaturated trifluoromethyl ketones as potential phospholipase A(2) inhibitors. J. Chem. Soc. Perkin. Trans. 2000, 1, 2271–2276. [Google Scholar] [CrossRef]
- Kokotos, G.; Feuerherm, A.J.; Barbayianni, E.; Shah, I.; Sæther, M.; Magrioti, V.; Nguyen, T.; Constantinou-Kokotou, V.; Dennis, E.A.; Johansen, B. Inhibition of group IVA cytosolic phospholipase A2 by thiazolyl ketones in vitro, ex vivo, and in vivo. J. Med. Chem. 2014, 57, 7523–7535. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahammad, N.; Ashcroft, F.J.; Feuerherm, A.J.; Elsaadi, S.; Vandsemb, E.N.; Børset, M.; Johansen, B. Inhibition of Cytosolic Phospholipase A2α Induces Apoptosis in Multiple Myeloma Cells. Molecules 2021, 26, 7447. https://doi.org/10.3390/molecules26247447
Mahammad N, Ashcroft FJ, Feuerherm AJ, Elsaadi S, Vandsemb EN, Børset M, Johansen B. Inhibition of Cytosolic Phospholipase A2α Induces Apoptosis in Multiple Myeloma Cells. Molecules. 2021; 26(24):7447. https://doi.org/10.3390/molecules26247447
Chicago/Turabian StyleMahammad, Nur, Felicity J. Ashcroft, Astrid J. Feuerherm, Samah Elsaadi, Esten N. Vandsemb, Magne Børset, and Berit Johansen. 2021. "Inhibition of Cytosolic Phospholipase A2α Induces Apoptosis in Multiple Myeloma Cells" Molecules 26, no. 24: 7447. https://doi.org/10.3390/molecules26247447
APA StyleMahammad, N., Ashcroft, F. J., Feuerherm, A. J., Elsaadi, S., Vandsemb, E. N., Børset, M., & Johansen, B. (2021). Inhibition of Cytosolic Phospholipase A2α Induces Apoptosis in Multiple Myeloma Cells. Molecules, 26(24), 7447. https://doi.org/10.3390/molecules26247447






