Abstract
This review provides information on available methods for engineering glycan-binding proteins (GBP). Glycans are involved in a variety of physiological functions and are found in all domains of life and viruses. Due to their wide range of functions, GBPs have been developed with diagnostic, therapeutic, and biotechnological applications. The development of GBPs has traditionally been hindered by a lack of available glycan targets and sensitive and selective protein scaffolds; however, recent advances in glycobiology have largely overcome these challenges. Here we provide information on how to approach the design of novel “designer” GBPs, starting from the protein scaffold to the mutagenesis methods, selection, and characterization of the GBPs.
1. Introduction
Glycans—a broad term describing carbohydrates, including oligosaccharides and polysaccharides—are the third class of important biological macromolecules following nucleic acids and proteins [1]. Glycans are found in all domains of life and in viruses. They can exist as free sugars, but are more commonly found as glycoconjugates, including proteoglycans, glycoproteins, and glycolipids. Glycans are involved in a wide variety of physiological functions and have implications in numerous infectious and non-infectious diseases, making them diagnostic and therapeutic targets [2]. Additionally, glycans are targeted in various biotechnological and industrial applications. The broad applications of glycans have spurred interest in the development of glycan binding proteins (GBPs).
GBPs include lectins, antibodies, pseudoenzymes, and carbohydrate-binding modules (CBMs). Lectins are non-immunoglobulin proteins containing at least one non-catalytic domain that exhibits reversible carbohydrate binding. CBMs are similar to lectins, but are small binding domains typically found in lectins or carbohydrate-active enzymes (CAZymes). CAZymes can be further classified into glycoside hydrolases, glycosyltransferases, polysaccharide lyases, and carbohydrate esterases—detailed information on these enzymes is available through the Carbohydrate Active Enzymes (CAZy) database [3]. Over time, some CAZymes have evolved into pseudoenzymes that have lost their catalytic activity but retain their glycan binding properties; these have been categorized separately from lectins as their overall structure is distinct. Antibodies against glycans are also found in nature, however glycans are generally poorly immunogenic, leading to low binding affinities and specificity of anti-glycan antibodies [4]. Despite this, some high-affinity anti-glycan antibodies have been developed [5,6]. The aforementioned protein categories have been used to create a variety of GBPs and their use in GBP engineering is discussed in Section 2.
One application for GBP engineering is towards the development of diagnostics and therapeutics. Glycan recognition is involved in a variety of bacterial and viral infections, which has led to the production of several diagnostic and therapeutic GBPs. For example, the glycan epitopes displayed on the envelope spike protein of the human immunodeficiency virus type-1 (HIV-1) are involved in immune system evasion [7]. More than a dozen lectins have been identified to exhibit anti-HIV properties and some are currently being further explored for therapeutic potential [8]. Similar to HIV-1, glycans are involved in immune system evasion in various other viral and microbial diseases—an excellent review has been published on the therapeutic value of GBPs in microbial infections [9].
GBPs also have the potential to target glycans that are involved in a variety of non-infectious diseases, such as diabetes [10], arthritis [11], and cancer—with glycans of the latter being the most well studied as biomarkers. Aberrant glycan profiles are a hallmark of malignant tumor transformations and are commonly targeted for cancer diagnostics and therapeutics [12]. The exact glycosylation patterns of tumors varies greatly, but include glycan epitopes that can be categorized as T-antigens, poly-N-acetyllactosamines (PLAs), Lewis antigens, and glycosaminoglycans, among others [13]. Cancer glycan epitopes have been recently reviewed elsewhere, and therefore will not be discussed in greater detail here [13].
GBP applications are not limited to the medical field and have seen broad applications in industrial settings and in biotechnologies. Lectins have been used as biological insecticides in genetically engineered crops and it has been suggested that this approach may reduce the use of synthetic insecticides—reducing risk to human health and the environment [14]. On another note, the CBM category of GBPs have applications in bioprocessing methods for affinity purification of biomolecules, using them in an immobilized selection step, or as affinity tags [15]. CBMs have also been used in textile industries to increase the efficiency of polysaccharide degrading enzymes—allowing for more efficient dyeing and printing on fabrics [16,17].
We have provided a summary of applications for engineered GBPs, but this list is by no means extensive—recent advances in glycomic techniques are expanding the known glycans that can be targeted for biotechnological, industrial, or medicinal purposes. In particular, development in laboratory techniques like lectin-arrays are leading to faster discovery of glycan biomarkers in human disease [18]. The rapidly growing literature on glycan targets has created a need for the development of novel diagnostic and therapeutic GBPs.
GBPs can be modified chemically or by using protein engineering techniques—here we will focus on the latter. The applications of GBPs range from diagnostics, therapeutics, and industrial settings to biotechnologies; however, the lack of known GBPs for specific glycans has created a need for GBPs with novel or altered binding specificities. Here we provide information that serves as a starting point for GBP engineering, with a focus on high-throughput directed evolution approaches. We address the topics of scaffold choice (Section 2), mutagenesis methods (Section 3), library screening and selection (Section 4), and characterization (Section 5), with a focus on how these methods have been used, and have yet to be used, for glycan binding protein engineering.
2. Glycan-Binding Protein Scaffolds
Choosing the right protein scaffold is a crucial aspect of engineering a GBP with improved or novel binding properties. In this section we discuss examples of several protein scaffolds available for GBP engineering, including lectins, carbohydrate binding modules (CBMs), pseudoenzymes, carbohydrate-active enzymes (CAZymes), and antibody-based scaffolds (Figure 1). The definition of lectins has changed over the years [19,20], but can generally be defined as proteins that bind carbohydrates. Hence, most GBPs can be categorized as a lectin; however, for the purpose of this review, we have categorized certain GBPs separately from lectins due to their distinct characteristic folds and properties. A summary of the scaffolds discussed in this section, along with example scaffolds that have structural and binding data available, is available in Table 1.
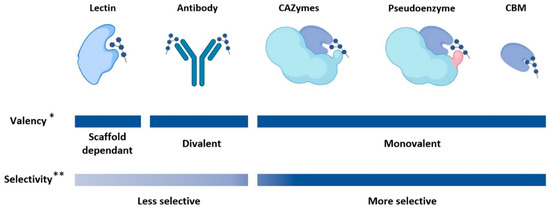
Figure 1.
Valency and selectivity of protein scaffolds with glycan binding sites. The valency of lectins and antibody-based scaffold varies, as some lectins contain tandem repeat units and antibody-based scaffolds can be designed to contain only a single, or multiple variable fragments. Similarly, CBMs can be designed in tandem to increase valency. The selectivity of antibody-based scaffold is affected by the poor immunogenicity of carbohydrates. * It should be noted that the valency of lectins, antibodies, and CBMs can be altered with protein engineering. ** There are exceptions to this trend and the selectivity can be affected by valency.
2.1. Lectins
Lectins are carbohydrate binding proteins that are placed into sub-categories based on their folds and function: P-type, I-type, L-type, R-type, C-type, and galectins. Lectins display a wide variety of physiological functions and have biotechnological and biomedical applications—lectins have already been used in the detection and targeted treatments of human diseases such as cancer [21,22]. Here we provide a brief overview of lectins and some examples in GBP engineering. An excellent resource for detailed information on the various sub-categories of lectins can be found in the comprehensive text, Essentials of Glycobiology (specifically chapters 28 to 38) [1].
Generally, lectins have relatively low affinities for their glycan targets, with dissociation constants in the micromolar range [23,24]. This may be explained by the shallow binding interface that is observed in most lectins, causing more competitive solvent interactions. The shallow binding interface may also explain the promiscuous binding observed in lectins—glycans with similar structures often bind similar lectins. In nature, the low affinity problem is overcome by oligomerization and multivalency; in biological settings lectins tend to assemble into oligomeric structures containing multiple binding sites, allowing for higher affinities to be reached. The relatively low affinities and promiscuity of lectins in the monomeric state must be considered when selecting scaffolds for GBP engineering; however, lectins with improved binding specificity and affinity have been developed [25]. One advantage of using lectins over other protein scaffolds is that databases like UniLectin3D are available that can search for lectin scaffolds based on the glycan target [26].
2.2. Carbohydrate Binding Modules
Carbohydrate binding modules (CBMs) [27], also known as carbohydrate binding domains (CBDs), are non-catalytic protein domains generally found on carbohydrate-active enzymes (CAZymes). There is low sequence identity between CBMs [27], but there are conserved tertiary folds that are categorized based on their binding site topology as types A, B, or C [28]. The topologies of CBMs are characterized in type A by a planar hydrophobic surface, in type B by an extended binding cavity, and in type C by a short binding pocket—for more information on the structures of CBM types please see the extensive review by Armenta et al. [27]. For the purposes of GBP engineering, type A CBMs are suitable for binding insoluble, crystalline carbohydrates, due to the exposed planar binding interface [29]. In contrast, type B CBMs bind oligosaccharides [30], and type C CBMs bind mono and di-saccharides [31]. One attractive aspect of CBMs as GBP scaffolds is their modularity; due to their small size CBMs can be designed in tandem to increase specificity or allow for multiple binding targets. Additionally, there is a variety of well characterized CBMs that can be used as scaffolds—not surprisingly, CBMs have been used to engineer a variety of GBPs with altered binding characteristics [30,32,33,34].
2.3. Pseudoenzymes
In nature, a number of GBPs have evolved from enzymes through the loss of catalytic activity while retaining binding function. These can be defined as pseudoenzymes, which are catalytically inactive proteins related to ancestral enzymes [35]. Pseudoglycosidases are a type of pseudoenzyme that evolved from glycosidases (glycoside hydrolases). These proteins, which bind glycans but cannot hydrolyze glycosidic linkages, can also be characterized as lectins since glycan-binding is their primary function. A few notable examples of pseudoglycosidases that act as GBPs have been observed in nature. In animals, chitinase-like proteins such as the human YKL-39 are pseudoglycosidases (GH18 homologues) with enigmatic biological functions that have been shown to bind to chitooligosaccharides as part of their apparent role in modulating the innate immune response [36,37]. Another example in animals is found in α- and β-klotho proteins, which each make up part of a receptor complex responsive to fibroblast growth factors (FGFs), wherein catalytically inactive GH1-like tandem repeats of the klotho proteins bind to “sugar-mimicking motifs” of FGF19 and FGF21 [38]. In protozoans, the CyRPA protein of Plasmodium falciparum—part of the invasion complex that allows the malaria-causing parasite to bind and enter red blood cells—appears to be a catalytically inactive pseudoglycosidase related to GH33 sialidases [39,40,41]. Pseudoenzymes evolved from other types of enzymes can also bind to glycans. For example, PgaB in E. coli is a deacetylase that is involved in the formation of the partially deacetylated poly-1,6-N-acetylglucosamine component of the bacterium’s biofilm coat, and the protein consists of two tandem domains related to carbohydrate esterase family 4 (CE4), with the C-terminal domain being a catalytically inactive pseudoesterase involved in binding poly-1,6-N-acetylglucosamine [42].
Although pseudoenzymes can, in theory, be used as glycan binding scaffolds, there are no published works on engineering pseudoenzyme scaffolds into novel GBPs as of 2020. This may be due to a lack of known pseudoenzymes scaffolds but may also be due to the prevalence of mutagenesis techniques that allow for inactivation of enzymes. The use of enzymes as GBP scaffolds is discussed in greater detail in the following section.
2.4. Carbohydrate-Active Enzymes (CAZymes)
Carbohydrate-active enzymes (CAZymes) catalyze reactions that break down, assemble, or modify saccharides, and they are categorized based on their activities and further subdivided into families based on sequences. Categories include glycosyltransferase, glycoside hydrolase, polysaccharide lyase, carbohydrate esterase, and auxiliary activity families. The examples of pseudoenzymes from nature demonstrate that inactivation of CAZymes can result in proteins that bind to glycans but do not catalytically turn them over. Naturally, this suggests that inactivating CAZymes through artificial mutations may be an effective method to engineer novel GBPs. Generating a GBP from a CAZyme requires the inactivation of catalytic residues, which sometimes only requires the mutation of a single amino acid. This may make CAZymes an attractive scaffold for GBP engineering. An example of a nanomolar affinity GBP engineered by inactivation of a CAZyme can be seen in the site-specific mutation of a glycoside hydrolase from E. coli K1 bacteriophages. The GH58 endosialidase, Endo-NF, was mutated to generate a catalytically inactive GBP that still binds to polysialic acid with a dissociation constant (KD) of 191 nM [43,44]. This engineered GBP has been applied as a very sensitive tool for detecting polysialic acid [45,46]. In another example, mutation of a CE2 carbohydrate esterase from Clostridium thermocellum has also been shown to produce a catalytically inactive GBP with micromolar affinity [47]. A single amino acid replacement of the CtCE2 enzyme not only abolished esterase activity, but increased the affinity to cellooligosaccharides nearly 8-fold, with the mutant binding to cellohexaose with a KD of 4.1 μM.
The strategy of engineering GBPs by inactivating CAZymes has been developed and commercialized most notably by the biotech company Lectenz Bio, who have produced a variety of catalytically inactivated CAZymes, which they have dubbed “Lectenz®” (lectins engineered from enzymes) [48]. The company has produced several Lectenz® through site-directed mutagenesis and computationally guided directed evolution. One advantage of using CAZyme scaffolds is that carbohydrate-processing enzymes tend to be more specific for their ligands than lectins, although this will vary between proteins.
2.5. Antibody-Based Scaffolds
Antibody-based scaffolds consist of immunoglobulin or immunoglobulin-like protein folds. A variety of antibody-based scaffolds are found in animals, but the most commonly used for developing antigen binding proteins are immunoglobulin G (IgG), and more recently, camelid antibodies [49]. The production of naturally occurring antibodies is time consuming and costly as it requires the immunization of an animal; however, antibody-based scaffolds have been engineered that circumvent the use of animals. These include, but are not limited to, antigen binding fragments (Fab) [50], single chain variable fragments (ScFvs) [51], diabodies [52], monobodies, and nanobodies [53]. There has been a concerted effort to produce antibodies against tumor-associated carbohydrate antigens (TACAs)—in total, antibodies have been designed for about 250 distinct glycan targets [54]. Antibody scaffolds offer certain advantages over lectins, including a larger binding interface for longer glycan epitopes, and generally more selective binding due to the complementary determining regions. However, glycans are poorly immunogenic and producing an anti-glycan antibody can be costly, labour intensive, and time consuming. Additionally, anti-glycan antibodies generally have lower affinities (KD in the micromolar range) than protein-targeting antibodies (KD in the nanomolar range). Within the last decade, phage display has provided methods for overcoming some of these limitations, resulting in antibodies with higher affinity for their glycan targets [55]. However, this approach still requires an initial scaffold obtained from immunization to be used as the base scaffold for improving affinity and selectivity.
2.6. Summary on Available GBP Scaffolds
Here we discussed the available protein scaffolds and some of their respective challenges and considerations when applied to GBP engineering. The scaffold that is chosen for GBP engineering will influence which mutagenesis techniques and selection methods are most appropriate. This brief overview provides a resource for glycobiologists who aim to design novel GBPs for specific glycan targets. Table 1 is by no means a complete scaffold list; it serves as a list of example scaffolds that are available and characteristics that need to be taken into consideration. Finding scaffolds ideal for a glycan of choice can be challenging and we recommend using UniLectin3D or equivalent GBP databases as starting point for finding potential scaffolds [26].

Table 1.
Carbohydrate binding proteins (CBPs) with available structural and ligand binding information.
Table 1.
Carbohydrate binding proteins (CBPs) with available structural and ligand binding information.
| Scaffold Category | Scaffold Sub-Category | Description | Origin | Example Protein (PE) | PE Length | PE Ligand | PE Oligomeric State | PE Multivalency |
|---|---|---|---|---|---|---|---|---|
| Lectins | P-type | Lectin that binds to mannose 6-phosphate | Animal | Bovine CD-MPR binding domain [56] | 154 aa | Mannose 6-Phosphate | Dimer | Monovalent |
| I-type | Protein that is homologous to the immunoglobulin superfamily (IgSF) | Vertebrata | hCD22 domains 1-3 [57] | 324 aa | Sialoglycans | Monomer | Monovalent | |
| L-type | Proteins that are structurally similar to lectins found in the seeds of leguminous plants | All domains of life and viruses | Concanavalin A [58,59] | 237 aa | Trimannoside containing-oligosaccharides [59] | Oligomer | Divalent | |
| R-type | Proteins that are structurally similar to the carbohydrate recognition domain (CRD) in ricin | All domains of life and viruses | Ricin [60] | 267 aa | β1,4 galactose, N-acetylgalactosamine | Dimer | Divalent | |
| C-type | Ca2+ dependant proteins that share a primary and secondary homology in their CRDs | Animal | C-type domain of murine DCIR2 [61] | 129 aa | N-glycans | Monomer | Monovalent | |
| Galectin | Globular proteins that share primary structural homology in their CRDs | Animal | hGalectin-3 [62] | 146 aa | N-acetyllactosamine | Monomer | Monovalent | |
| Carbohydrate Binding Modules (CBMs) | Type A | Protein domain that binds to crystalline surfaces of cellulose and chitin | All domains of life and viruses | CBM from Cel7A [63] | 36 aa | Cellulose | Monomer | Monovalent |
| Type B | Protein domain that binds endo-glycan chains | All domains of life and viruses | CBM4-2 from xylanase [33] | 150 aa | Xylans, β-glucans | Monomer | Monovalent | |
| Type C | Protein domain that binds exo-type glycan chains | All domains of life and viruses | Cp-CBM 32 of hexosaminidase [64] | 150 aa | N-acetyllactosamine | Monomer | Monovalent | |
| Pseudoenzymes | Pseudoglycosidase | Carbohydrate binding proteins that evolved from glycosidases but are no longer catalytically active | Possibly all domains of life* | hYKL-39 [36] | 365 aa | Chitooligo-saccharides | Monomer | Monovalent |
| Pseudoesterase | Carbohydrate binding proteins that evolved from carbohydrate esterases but are no longer catalytically active | Possibly all domains of life * | C-terminal domain of PgaB [42] | 367 aa | Poly-1,6-N-acetylgluco-samine | Monomer | Monovalent | |
| Carbohydrate- Active Enzymes (CAZymes) | Glycoside hydrolase | Enzymes that cleave glycosidic linkages | All domains of life and viruses | Endo-NF (GH58) [44] | 811 aa | Polysialic acid | Trimer | Multivalent |
| Carbohydrate esterase | Enzymes that hydrolyze ester linkages of acyl groups attached to carbohydrates | All domains of life and viruses | CtCE2 [47] | 333 aa | Cellooligo-saccharides | Monomer | Monovalent | |
| Other CAZymes (glycosyltransferase, polysaccharide lyase, auxiliary activities) | Enzymes involved in the assembly, break-down, and modification of carbohydrates | All domains of life and viruses | − | − | − | − | − | |
| Antibodies | N/A | Naturally or synthetically produced proteins with an immunoglobulin, or derived from an immunoglobulin-like structure | Vertebrata | hu3S193 [65] | LC: 219 aa HC: 222 aa | LewisY | Dimeric | Divalent |
* Ancestral enzymes found in all domains of life.
3. Mutagenesis Methods for Library Generation
When a suitable protein scaffold is chosen it can be engineered into a novel glycan binding protein (GBP) using random, rational, or semi-rational mutagenesis. Random mutagenesis can generate large libraries for directed evolution approaches without structural information, whereas semi-rational and rational mutagenesis require structural data. There are various mutagenesis methods that fall into random, semi-rational, and rational mutagenesis, some of which can be used in combination. Here we provide a brief overview of these methods and examples of their uses for engineering GBPs.
3.1. Sequence Agnostic Random Mutagenesis
Random mutagenesis techniques that do not require any structural information and allow for mutation at any position within the protein-coding region of a gene can be considered “sequence agnostic”. Sequence (and structure) agnostic mutagenesis approaches are the methods of choice for library generation when no structural data is available. The mutant libraries generated in this way can be used for directed evolution, in combination with high-throughput screening or selection techniques (Section 4). Methods for random mutagenesis include error prone PCR (epPCR) [66], DNA shuffling [67], in vivo mutagenesis using mutator strains [68], and external mutagens [69] (Table 2).

Table 2.
Overview of random mutagenesis methods.
A powerful and versatile yet straightforward technique, epPCR is the most common method for creating mutant libraries of a single gene. In epPCR, conditions are chosen to allow for a relatively high mutation rate by the DNA polymerase (i.e., low fidelity of replication). This can typically be achieved by adjusting the concentration of DNA polymerase and MgCl2, adding MnCl2, and adjusting the ratio of dNTPs, or by using an engineered DNA polymerase mutant with reduced fidelity [66]. As it is the most common mutagenesis technique, it comes as no surprise that epPCR has been applied to engineer novel GBPs. In one example from 2007, Yabe et al. cloned an earthworm galactose-binding lectin, EW29Ch, as the starting point for directed evolution wherein variants from successive generations were selected from mutant libraries generated by epPCR. This approach produced an engineered GBP specific for α2,6-sialic acid, a ligand not recognized by the parent protein [70].
In other examples, epPCR can also be combined with other mutagenesis techniques, such as DNA shuffling. DNA shuffling involves recombination of a population of homologous genes that have diverged either naturally or through laboratory mutagenesis of a parent (e.g., by epPCR). In this technique, random fragmentation of genes in a library (e.g., by DNase I digestion) is followed by PCR-based reassembly of overlapping fragments with sufficient homology, which effectively recombines mutations within the gene library [71]. Examples of DNA shuffling in combination with epPCR are seen in protein engineering efforts that have introduced mutations to the CBM of cyclodextrin glucanotransferase from Bacillus sp., and the glycan-binding regions of N-oligosaccharyltransferase from Campylobacter jejuni resulting in increased specificity and efficiency of those enzymes [72,73].
Alternative to the in vitro mutagenesis methods described above, one can perform in vivo mutagenesis on a target gene. These in vivo methods involve manipulating the DNA replication and repair machinery of the organism in which the target gene is cloned. For example, in mutator strains like E. coli XL1-red, which is deficient in three of the primary DNA repair pathways (carrying mutations mutS, mutD, and mutT), imperfect replication of DNA results in the accumulation of mutations in the cloned gene (along with the vector) [68]. In an example from 2011, Mendonça and Marana used in vivo mutagenesis in E.coli XL1-Red to alter the specificity of a β-glycosidase, SfβGly, from Spodoptera frugiperda [74]. Mutants from a library generated in the mutator strain were screened for their specificity towards fucosides vs. glucosides, and several variants were identified that differed from the parent enzyme in their substrate preference. Given that glycosidases can serve as scaffolds for GBPs through their catalytic inactivation (Section 2.4), this can be a useful strategy for engineering novel GBPs. The advantage of mutator strains is that their use involves simple protocols, generally involving transformation of the mutator strain by a plasmid followed by propagation and plasmid recovery. However, mutator strains get progressively sick as they divide due to the deficiencies in their DNA repair mechanisms, and consequently this mutagenesis method often requires frequent re-transformations. Other in vivo mutagenesis methods use external mutagens such as UV radiation or mutagenic chemicals (e.g., ethyl methanesulfonate) which can avoid some of the challenges of maintaining mutator strains.
Regardless of the sequence agnostic random mutagenesis techniques used, one disadvantage is that the produced libraries only cover a small fraction of the possible mutations and require considerable effort to screen. Rational and semi-rational mutagenesis can be more efficient at producing effective mutations based on the structural and functional information when it is available.
3.2. Rational and Semi-Rational Mutagenesis
Rational and semi-rational mutagenesis can produce smaller libraries than sequence agnostic random mutagenesis techniques, while simultaneously focusing on mutations that are more likely to impact protein function in a desirable way. Rational mutagenesis, as defined herein, involves making precise amino acid substitutions to a protein scaffold based on its structural data, whereas semi-rational mutagenesis uses the structural data to target specific sites that are then randomized. Some degree of rationality is always used to limit the sequence space that can be covered in mutagenesis, making it difficult to clearly distinguish between semi-rational and rational design—therefore, we have grouped these techniques together. Both techniques make use of various site-directed mutagenesis (SDM) methods (Table 3).
SDM methods are applied to engineering binding proteins when detailed structural information is known about the binding site, or binding-determining regions of the scaffold protein. The most common and efficient way to target specific sites is by PCR—desired single point mutations are included in the primers that amplify the gene of interest. PCR based SDM is one of the most common SDM methods applied in protein engineering and variations of PCR site-directed mutagenesis have been successfully applied in engineering GBPs in several examples including altering the binding specificity of a fucose-binding lectin PA-IIL [75] and increasing the affinity of a tail spike protein Sf6 to the glycans comprising bacterial O-antigens [76]. SDM has also been used to inactivate a streptococcal endo-N-acetylglucosaminidase (EndoS) [77] and an E. coli K1 bacteriophage endosialidase (EndoNF) to dissociate binding from hydrolytic activity [43,44] and produce novel GBPs for specific glycan detection [45,46].
One drawback of standard SDM approaches is that they only sample a few defined mutations that may or may not result in the desired protein function; hence a sub-category of SDM, site-saturation mutagenesis (SSM) is often used to increase the sample space. Instead of a straightforward replacement of one amino acid for another, SSM randomizes a specific codon, or short sequence of codons, to produce libraries of mutants with all possible amino acid substitutions (or a subset of possible substitutions) at the targeted positions [78]. Screening of such libraries allows for the identification of ideal amino acid replacements for those positions [79]. Although there are various SSM techniques, they all rely on site-directed mutagenesis PCR using degenerate codons. SSM has been successfully used to alter the glycan-binding specificity of a galectin towards α(2,3)-linked sialic acid in a single mutagenesis step [80]. This demonstrates the efficiency of SSM when compared to sequence agnostic random mutagenesis techniques that often require multiple cycles of mutagenesis to produce a desired mutant.
While directed mutagenesis can result in site-specific replacement of codons for targeted amino acid positions, it can also be employed to replace longer sequences of DNA for motif- or domain-swapping mutagenesis (substituting long strings of amino acids). Replacement DNA cassettes can be introduced by restriction digestion and ligation, or by overlap extension PCR to substitute a parental DNA fragment. In one example, motif-swapping has been used to alter the binding specificity of galactose-specific Bauhinia purpurea lectin by switching nine amino acids in the binding region with the corresponding nine residues present in the mannose-specific Lens culinaris lectin, generating a chimeric lectin that has a unique carbohydrate-binding specificity not observed in the parental proteins [81].

Table 3.
Overview of site-directed mutagenesis techniques.
Table 3.
Overview of site-directed mutagenesis techniques.
| Method | Definition | Pros | Cons | References |
|---|---|---|---|---|
| PCR site-directed mutagenesis | Primers containing the desired mutation(s) are used to alter the original gene | Not limited by the availability of nearby restriction enzyme cut sites | Primer design can be complicated when introducing multiple mutations | [43,44,45,46,75,76,77,82] |
| Site-saturation mutagenesis | A set of codons is substituted with every amino acid using degenerate codons | Allows for the screening of ideal amino acids at different positions | Mutation bias from the degenerate codon | [78,80] |
| Motif- and domain-swapping | Uses a “cassette” DNA fragment containing the mutations, which replaces the unmutated segment in the original gene | High mutation efficiency | Is limited by the domains/motifs that are used. | [81,83] |
3.3. Computational Tools for Rational and Semi-Rational Mutagenesis
In rational and semi-rational approaches, application of the aforementioned mutagenesis methods is guided by various computational approaches to identify positions for directed mutagenesis and predict beneficial mutations. These include homology modeling, molecular dynamics, deep learning, and tools for designing focused libraries (Table 4). Here we provide a brief overview of these computational methods and their applications in GBP engineering.
The 3D structure of a protein provides valuable information to guide its engineering, allowing one to identify residues that interact (directly or indirectly) with a ligand and to focus mutations at positions where they may be most effective. For proteins without solved structures, a homology model may be used where possible as an imperfect substitute. Homology modelling is one of the most common computational methods used for protein engineering, as it allows for the construction of an atomic resolution model for a target protein of unknown structure, provided that the structure of a sequence-homologous protein has been solved. The model is based on the structural data of homologous proteins and is generally considered reliable if there is more than 50% sequence identity between the target and homologue. In certain cases, homology models allow for precise editing of a protein. Using a homology model, Lienenmann et al. were able to identify and inactivate the catalytic residue in the Trichoderma harzianum chitinase, Chit42—turning the enzyme into a GBP [84]. However, X-ray crystal structures and homology models only provide a snapshot of the protein—unlike molecular dynamics (MD) simulations.
Molecular dynamics simulations generate a set of possible conformations based on the protein structure. One advantage of MD simulations is that they can identify flexible regions that may alter the protein activity. There are plenty of MD program packages available to use, with some of the most common being YASSA, MOE, Enlighten2, and GROMACS. In recent work on GBP engineering, Kunstmann et al. used MD simulations generated with the GROMACS 4.5.5 package to accurately link mutations in the tail-spike protein of bacteriophage Sf6 to varying affinities towards glycans of the O-antigens on Shigella flexneri (which is host to the phage) [76]. Yet being able to make such predictions accurately is rare and it is more common to identify beneficial mutations through screening focused libraries.
The insights gathered from analysis of protein structures, homology models, and MD simulations can be used in designing focused libraries of mutants. Focused libraries—in which specific positions are targeted for mutation (e.g., by SSM)—can be created with the assistance of computer tools for identifying amino acid sequences that are more likely to be involved in stability, catalytic activity, or specificity. These enriched libraries improve the efficiency of directed evolution by reducing the library size and have been used to design proteins with increased selectivity [85,86] and specificity [87]. Multiple tools are available for focused library design, such as CASTER and HotSpot Wizard 2.0, with the latter being a more accessible web-based tool that requires less bioinformatical knowledge.
One method of creating focused libraries that has been advancing rapidly is deep learning. Deep learning algorithms are a form of machine learning that trains artificial neural networks to recognize patterns in complex datasets such as sequencing data to predict properties of novel or uncharacterized proteins. Although deep learning is a fairly new technique, it has already been applied to lectin engineering. In 2012 Stephen et al. used a deep learning algorithm called CPred to produce a starch-binding domain with altered selectivity by predicting the effects of circular permutation [88,89].
Here we have only briefly covered some computational methods that are available for protein engineering of GBP—for more details on computational approaches to protein design, Kuhlman and Bradley provide an excellent review [90].

Table 4.
Example of computational approaches used in protein engineering.
Table 4.
Example of computational approaches used in protein engineering.
| Computational Approach | Definition | Examples | Utility in Protein Engineering | References |
|---|---|---|---|---|
| Homology model | Constructs an atomic resolution model of a protein based on available structural data of related homologous proteins. | Phyre2, ROBETTA, SWISS-MODEL | Predicted structures can be used in other computational methods, such as docking and molecular dynamics simulations. | [84] |
| Molecular Dynamics | Predicts the conformational energy landscape available to a protein based on the structure. | YASARA, Enlighten2, MOE, GROMACS | Dynamics can indicate how certain mutations can affect the behavior of protein such as folding, stability, ligand interaction and enzymatic activity. | [76,91,92] |
| Deep learning | Uses known protein sequences and properties to predict the properties of uncharacterized or novel proteins. | CPred, UniRep | Allows for the generation of more efficiency mutant libraries, by focusing on the most promising candidates. | [88,89,93] |
4. Library Selection and Screening Methods for GBPs
Library display techniques such as phage, yeast, ribosome, and mRNA display allow for the high-throughput selection of specific binding proteins from libraries of mutant variants. Regardless of the method used, there are three general steps involved: (1) library generation, (2) biopanning, and (3) characterization of variants. This section will focus on biopanning, since the library generation was previously described (Section 3), and characterization is dependent on the target protein (Section 5).
4.1. Phage Display
Phage display is a high throughput selection technique for large libraries of mutant proteins that links the protein library to the coat proteins of bacteriophages (most commonly M13 filamentous phage, T4, T7, and λ phage). This method displays protein libraries on the surface of bacteriophages by encoding the library into a phage-derived circular DNA vector—a phagemid. The phagemid encodes a coat protein fused to a mutant protein of interest, which links the phenotype of the protein (binding ability, enzymatic activity etc.) to its genotype. Typical elements of phagemids include bacterial and phage origins of replication, a selection marker, and the gene of the displayed protein appended to a phage protein-coding gene, which may also encode a periplasmic localization signal depending on the phage protein (Figure 2A). Displayed proteins can be selected for desired function then genotyped using high-throughput selection methods followed by sequencing. This section provides a brief overview of phage display; an extensive review is published in Chapter 3, volume 580 of Methods of Enzymology [94].
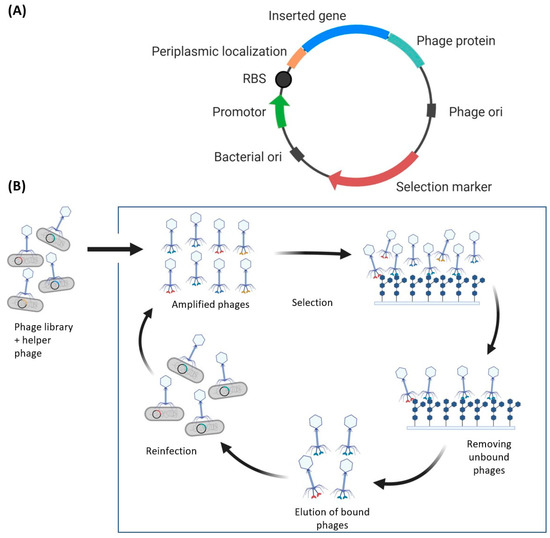
Figure 2.
(A) Example phagemid design for phage display. The phage plasmid requires two origins of replications, one for E. coli (the host) and one for the phage. Additionally, attaching a periplasmic localization signal to the target gene may improve the display, although this may depend on the appended phage protein. The phage protein is drawn attached to the C-terminus; however, it may be required to append it by the N-terminus—linkage is dependent on the phage protein. (B) Simplified schematic of phage display. The protein library is transformed into E. coli and amplified using helper phages, resulting in phages displaying the protein library. The phages can then be tested for antigen binding, using surfaces or beads that contain the antigen. A washing step removes unbound phages, and positive binders can be eluted and used for reinfection and amplification of the selected phages. This bio-panning cycle is repeated for multiple rounds and remaining phages are characterized and recombinantly expressed.
A phagemid library encoding a pool of mutants produced by one or more mutagenesis methods (Section 3) is used to transfect E. coli—the transformation efficiency limits the protein library diversity between 107 to 109 unique protein sequences [95]. Typically, the phagemid does not encode viral proteins that are needed for viral propagation. Hence, a helper phage that expresses viral proteins essential for efficient viral propagation is used to ensure high copy numbers of the phage library. Methods have been developed that eliminate the need for helper phages, but they are not as commonly used [96]. Regardless of whether helper phages are used, the amplification of the phage library is the beginning of the biopanning cycle that involves binding, washing, elution, reinfection, and phage amplification steps (Figure 2B). The phage library can then be used to test for binding to the target of interest and successful binders may be used to repeat the cycle using phage reinfection and amplification.
With regards to glycan binding proteins, phage display has been used to produce a single chain variable fragment (ScFv) against T-antigens—a glycan marker of many adenocarcinomas [97]. The anti T-antigen ScFv was selected from a library of human ScFvs and successfully bound T-antigens with micromolar affinities. Likewise CBM4-2 of Rhodothermus marinus xylanase Xyn10A (18 kDa) has been used as a scaffold in phage display to engineer new binding properties towards xylan, and the glycoprotein IgG4 [33]. An advantage of phage display over the other display methods is that it has been well optimized as it is the oldest and most common display technique. However, phage display does not allow for simple incorporation of many post-translational modifications (e.g., protein glycosylation) without chemical manipulation of the phage after amplification [98]—in contrast to yeast display.
4.2. Yeast Display
In yeast display, the proteins of interest are fused to cell surface proteins, most commonly α-agglutinin (Agα1), a-agglutinin (Aga1p-Aga2p), or flocculin (Figure 3). Depending on the anchor protein, the target protein can be fused to the N- or C-terminus, resulting in display of up to 100,000 copies [99]. The target protein is generally tethered to the anchor protein at the terminus farthest from the functional region, or binding region of the protein, to avoid interference. Additionally, the target protein can be flanked by protein tags for simplified purification and detection methods.
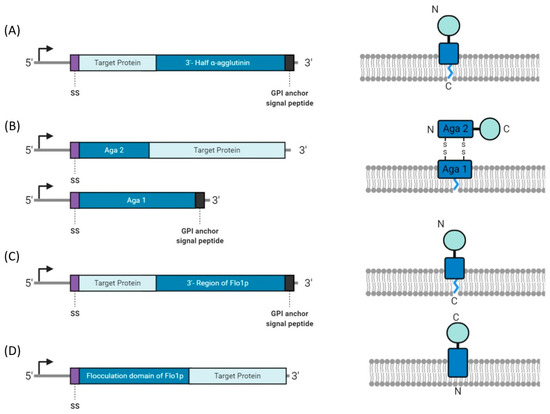
Figure 3.
Construct designs for yeast display systems. (A) α-agglutinin is fused to the C-terminus of the target protein and anchored to the cell membrane by a glycosylphosphatidylinositol (GPI) anchor. (B) a-agglutinin (Aga) is a dimer, with Aga2 linked to the N-terminus of the target protein. Aga1 is membrane bound by a GPI anchor and forms disulfide bonds with Aga2. (C) The C-terminal domain of Flo1p is linked to the C-terminal region of the target protein. (D) The N-terminal domain of the Flo1p is linked to the N-terminal region of the target protein.
Yeast display is commonly used for antibody-like proteins, such as ScFvs, Fabs, and monobodies. In 2013, Hong et al. used yeast display to produce monoclonal lamprey antibodies (lambodies) against several tumor associated carbohydrate antigens, Lewis antigens, and several glycoproteins [100]. More recently, a fully human ScFv was produced that could bind an epithelial tumor associated N-glycoform of periostin, further demonstrating the value of yeast display in engineering GBPs [101]. One advantage of yeast display over phage and in vitro display methods is the ability to include native (or similar to native) post-translational modifications, which can impact the protein’s fold and function. Additionally, yeast display can take advantage of fluorescence-activated cell sorting (FACS) for efficient library screening (after the cells bind to a fluorescently tagged target ligand). However, yeast display is more time intensive than in vitro display methods, and the mutant library size is restricted to 107 unique mutants—the smallest out of all display methods discussed in this review.
4.3. Ribosome Display
Unlike yeast display and phage display, ribosome display is performed in a cell-free process that involves in vitro transcription and translation, and it is not limited by transformation efficiencies. In ribosome display the target protein is linked to its mRNA in a ribosome-mRNA-target protein complex. This complex is established during in vitro translation, as the mRNA of the target protein lacks a stop codon—preventing the dissociation of the mRNA and protein from the ribosome. The design of the DNA construct, including the gene coding for the target protein, requires an upstream ribosomal binding site (RBS) and a downstream spacer (Figure 4A). The DNA construct is transcribed in vitro to mRNA, and the spacer sequence on the mRNA allows the protein of interest to sit outside of the ribosome tunnel, and fold properly, while remaining bound to the ribosome after it is translated in vitro. It is recommended that the construct include regions that will be transcribed into stemloops upstream of the RBS and downstream of the spacer—the secondary structure of the mRNA stemloops can prevent mRNA degradation and increase ribosome efficiency up to 15-fold [102]. Figure 4B provides an overview of the entire mRNA display cycle.
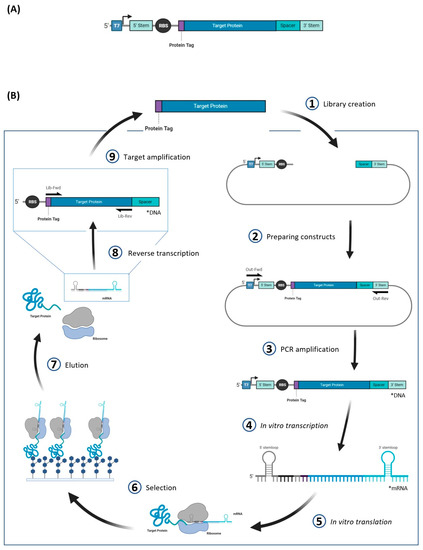
Figure 4.
Overview of the ribosome display method. (A) Construct design for ribosome display. The sequence that is transcribed (4) during the ribosome display contains 5′ and 3′ stem loop regions that help prevent degradation of the subsequent mRNA. (B) Cyclic representation of ribosome display. The plasmid used for ribosome display typically includes the promoter, stem loop regions, RBS, and spacer region.
In one example of ribosome display applied to GBP engineering, a sialic acid-binding lectin for use in analytical microarrays has been engineered from a galactose-binding lectin using ribosome display in combination with epPCR [70]. One advantage of cell-free display methods like ribosome display is the ease with which PCR-based mutagenesis methods can be implemented after each selection round—allowing for multiple generations of mutants to be evolved through iterative cycles of randomization and selection, mimicking the natural evolutionary process. However, during ribosome display the protein of interest is bound to an enormous 2.7 MDa ribosome complex, which may interfere with the target protein’s characteristics. A cell-free display technique that does not link the target protein to a macromolecule and may have less interference is mRNA display.
4.4. mRNA Display
mRNA display is a cell-free display method in which the protein of interest is covalently linked to the 3′ end of its own mRNA. DNA constructs for mRNA display require a promoter (commonly the T7 promoter for an E. coli-derived system, but may vary depending on the cell-free expression system) to recruit RNA polymerase for in vitro transcription, and a ribosomal binding site in the 5′ UTR, allowing for in vitro translation of the protein. The mRNA is transcribed in vitro, then enzymatically or photochemically ligated to a DNA-puromycin linker [103,104]. During in vitro translation, the puromycin mimics an aminoacyl-tRNA and forms a bond with the nascent peptide. The resulting protein-mRNA complex can then be used in selection methods to identify binding interactions—an overview of the mRNA display process is provided in Figure 5.
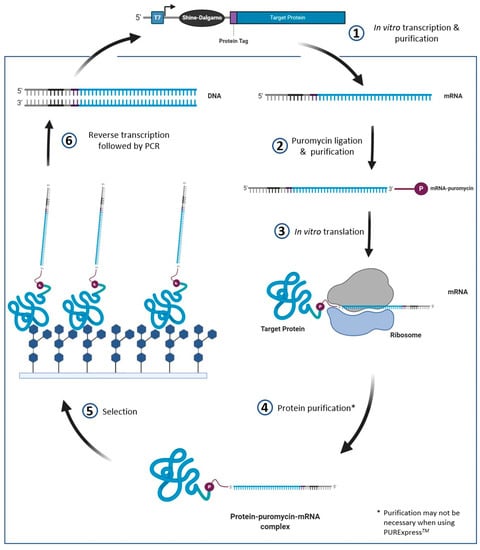
Figure 5.
Overview of the mRNA display method. The initial construct design has to contain a promoter (e.g., T7) and RBS (e.g., Shine Dalgarno) specific to the cell free expression system used. The puromycin mimics an aminoacyl tRNA which causes the mRNA to be linked to the nascent protein during in vitro translation. Once translated, the mRNA-protein complex is screened for glycan binding. The mRNA attached to the bound proteins is reverse transcribed and PCR amplified—allowing for simplified sequence detection.
The mRNA-protein linkage allows for simplified sequence detection following selection—the mRNA is reverse transcribed allowing for double-stranded cDNA to then be amplified by PCR. The resulting DNA can then be sent for next generation sequencing (NGS)—the sequencing data can be used to determine which proteins are enriched. As previously mentioned, one advantage of mRNA display over ribosome display is that it does not require the ribosome complex to be bound to the protein; hence interference is less likely. A review on mRNA display has recently been published, and discusses the topic in much greater detail than we will cover here [105]. However, we do note that as of 2020, no reports of mRNA display applied to the engineering novel GBPs have been published. It should also be noted that cell-free expression systems, like mRNA display and ribosome display, do not typically include protein folding chaperones, which may decrease the yield of properly folded proteins. Chaperones such as the E. coli proteins DnaK and GroEL can be added to cell free expression systems to increase the yield of functional proteins; however, they may not act as chaperones for every protein product and optimization would be required [106].
4.5. Glycan Immobilization Strategies for GBP Selection
Common to the range of selection methods to identify binding proteins, which involve different display techniques, is the need for an immobilized target. Various glycan immobilization strategies may be used for the selection and purification of GBPs. Glycan targets can be bound to a variety of solid supports including agarose resin [70], polymer-coated superparamagnetic particles (e.g., Dynabeads) [107], or the wells of standard microtiter plates [45,108]. This can be done using a number of chemical approaches such as amine coupling and click-chemistry.
Glycoproteins may serve as a convenient source of target glycans which can be fixed to a solid support. The entire protein along with its glycan modifications may be immobilized by covalent attachment to a chromatographic resin such as agarose. Glycoproteins such as bovine fetuin are readily available materials for cell biology and can be linked by amine coupling to resins that have been functionalized with, for example, aldehyde groups, cyanate esters, or N-hydroxysuccinimide esters. Fetuin-agarose produced in this fashion has been applied in the selection cycle for the directed evolution of a novel sialic acid-binding protein in a process involving ribosome display (Section 4.3) [70].
Coated magnetic particles, such as Dynabeads, can serve as another sort of solid support for target glycans. These versatile beads are used (often with the aid of automated magnetic particle handling robots) in the pull-down of proteins and nucleic acids. The use of glycan-coated Dynabeads has been demonstrated for specific pull-down of GBPs. Hence, Dynabead-linked carbohydrates can be used in a selection process for engineered GBPs. In a study by Liebroth et al. [107], LewisX and N-acetyllactosamine were immobilized to Dynabeads through a bovine serum albumin carrier, and these coated beads were then used to test the binding of TAG-1, Contactin, and NCAM120 (three lectin-like neuronal receptors). In pull-down assays carried out on a mixture of cellular proteins, it was determined that TAG-1 makes specific interactions with LewisX (but not N-acetyllactosamine), while the other two proteins do not. In a similar fashion, glycan-functionalized Dynabeads could be used to pull down binders to a target glycan from a library of GBPs, removing non-binders as part of a selection process.
A useful approach to link target glycans to solid support resins or surfaces involves “click” chemistry. Click chemistry describes a set of water-compatible, biocompatible reactions that can link two appropriately tagged reagents together in high yield (e.g., using azide-alkyne cycloaddition). Here we will focus on azide-alkyne based click chemistry, since it is a widely adapted tool in glycobiology research, seeing broad uses in applications including in vivo glycoengineering [109,110] and glycan labelling [108]. Notably, click chemistry has been used to immobilize functionalized glycans onto the surface of wells in microtitre plates. This has enabled high-throughput plate-based assays for glycosyltransferases exhibiting, for example, fucosyltransferase [111] or polysialyltransferase activity [45] on immobilized oligosaccaccharide substrates, detected through specific binding of the enzyme-product by tagged GBPs. Solid supports coated with immobilized glycans such as these could also be used for selection of GBPs.
5. Binding Characterization of GBPs
The most common binding characterization tools used for engineered GBPs are surface plasmon resonance (SPR) and titration calorimetry. SPR and titration calorimetry are both non-destructive method for determining binding characteristics; however, SPR requires the binding partner to be immobilized on a surface whereas titration calorimetry is done in solution. Specific applications of affinity chromatography using immobilized ligands have also proven useful in measuring GBP binding affinity, although this method is less accurate.
5.1. Frontal Affinity Chromatography
Using chromatographic resins functionalized with immobilized ligands, frontal affinity chromatography is an analytical technique that can be used to measure the binding interactions between molecular species. Application of this technique can be used to determine the binding constants of binding proteins of a wide range of equilibrium constants [112]. In particular, it has proven a useful tool in measuring protein-glycan interactions [113]. As a solution containing a GBP flows through a column packed with a glycan-functionalized resin, the degree to which the GBP is slowed by its interaction with the immobilized glycan can be used to measure binding interactions. Custom affinity resins with specific glycans can be generated, for example, as described in this review (Section 4.5). This approach has been demonstrated by Yabe et al. to characterize the binding of an engineered lectin to the sialic acid ligand to which it was tailored [70]. One advantage of affinity chromatography is that it can also be used to select and purify a protein target, yet the binding kinetics determined are not as accurate as SPR or titration calorimetry.
5.2. Surface Plasmon Resonance
Surface plasmon resonance (SPR) techniques measure the frequency of the electromagnetic oscillations on a metal surface, by exciting the surface electrons using a light source. The angle of the reflected light is influenced by the mass at the surface, hence mass changes on the surface can be measured based on the change in the reflected angle [114]. Glycan-labelled metal surfaces can therefore be measured for protein binding based on the change of the reflected angle (Figure 6). Moreover, SPR can measure binding interactions in real time—allowing for the determination of association and dissociation rate constants.
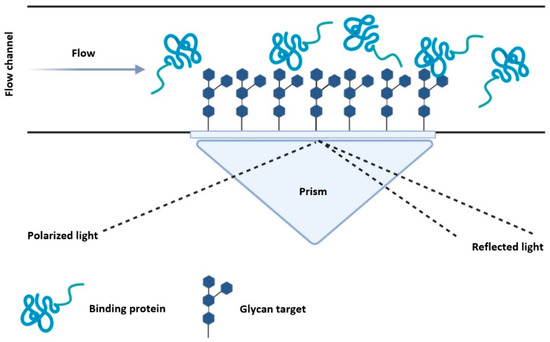
Figure 6.
Overview of SPR with glycan labelled surfaces. Glycans are bound to a metallic surface inside a flow chamber and a GBP solution is introduced allowing the protein to bind. A light source excites the metallic surface, and the angle of the reflected light is relative to the mass at the surface, which enables binding kinetics to be determined.
SPR has already been applied in glycobiology research to screen GBPs for glycan binding [115], produce a mannose biosensor capable of detecting nM concentrations of lectins [116], and for comparing glycan binding of lectin mutants [84]. The main advantage of SPR over other analytical techniques is the ability to provide real-time kinetic data for glycan-protein interactions, without the need for labelling methods. However, the equipment and specialized knowledge needed to apply SPR methods can prohibit the use of these techniques. One of the main challenges is attaching the glycans or GBPs to the surface. For glycans, protocols have been developed to attach phenoxy-derived sugars [117]. Immobilization of GBPs to the surface can be done in various approaches, including amine coupling, nickel affinity for His-tagged proteins, and streptavidin-binding for biotinylated proteins. Here we do not cover the various SPR techniques in detail—for a more detailed review on SPR and available surface labelling methods please refer to the Handbook of Surface Plasmon Resonance [114].
5.3. Titration Calorimetry
Isothermal titration calorimetry (ITC) can be used to screen GBPs for glycan binding by measuring changes in heat that occur during binding. Based on the heat changes, the enthalpy, entropy, stoichiometry, and binding constants can be determined. ITC has been used to screen the altered binding affinities of a mutated fucose lectin PA-IIL—providing evidence for the location of the specificity binding loop of the protein [75]. Additionally, ITC was used to determine the binding affinities of a mutated starch binding domain (CP90) for longer carbon chain starches [89]. Overall, this technique provides a non-destructive way to determine binding affinities; however, ITC is not suitable with high-throughput approaches since each mutant requires a separate ITC chamber.
6. Current Limitations in Lectin Engineering
Despite the available techniques for GBPs engineering, there are several factors that bottleneck the process, including the availability of glycans, the glycan binding interactions with the scaffolds, and the lack of available glycosylation profiles for cells and proteins.
6.1. Glycan Availability
One limiting factor is the availability of glycans—screening and selection methods to engineer a GBP for a specific glycan require the glycan to be available in high purity and quantity. Due to the large variety and complexity of glycans, only some glycan epitopes are available for purchase through vendors. Unlike oligopeptides and oligonucleotides synthesis, there are no general protocols for the synthesis of complex glycans—the development of chemical synthesis protocol for a single glycan can be time consuming and expensive. However, recent advances in enzymatic and chemoenzymatic synthesis of glycans using recombinant enzymes are providing a path for affordable and efficient production of glycans. Chemoenzymatic synthesis of glycans requires synthetic precursors, a series of glycosyltransferases (GTs), and protection and de-protection steps [118]—this approach has been shown to be more affordable than purely chemical approaches [119]. Additionally, the expanding database of characterized glycosyltransferases opens the possibilities for more glycan structures to be produced enzymatically. Enzymatic synthesis may expand on the available glycan targets available for GBP engineering, yet another bottleneck is in the interactions that a GBP scaffold needs to make with a glycan target.
6.2. Glycan Binding Protein Scaffolds
Glycans contain subtle stereochemical and regiochemical differences between isomers that would need to be distinguished by the binding protein. Additionally, glycans form extensive hydrogen bonding networks in water that would have to be broken by the GBP, making the interactions less favorable. In nature, lectins are used to bind complex carbohydrate structures with high specificity, however lectins with higher avidity for their targets are generally multivalent. The multivalency allows for stronger binding with its target but results in larger protein complexes that are more difficult to produce and hence less ideal for biotechnology applications. Unlike multivalent lectins, peptide aptamers and cyclic peptides are smaller and easier to produce, but do not provide large enough interfaces for complex glycan interactions, making them less ideal for glycan targets. Hence, there is a need in GBP engineering for protein scaffolds with large binding interfaces that are adaptable for biotechnological applications.
As high-throughput protein engineering methods become more common, future efforts may provide protein scaffolds for GBP engineering that bind glycans with higher affinity and avidity than what is currently available. Recently, designed ankyrin repeat proteins (DARPins) have been produced as highly stable, small molecular weight, modular scaffolds [120]. DARPins are based on ankyrin proteins, some of which have glycan binding pockets [121]; however, as of 2020, DARPins have yet to be used for GBP engineering. Due to their modular design that allows for multiple binding interfaces, these proteins may be ideal for glycan targets.
7. Conclusions
Engineered GBPs have broad applications in a wide variety of fields, including diagnostic, therapeutics, and biotechnology. GBPs that have been engineered so far have already been used in all the previously mentioned fields and as the field of glycobiology advances we expect more applications to become apparent. Although current methods are available to engineering GBPs, we are still limited by the number of available glycan structures that can be produced in pure quantities, and in the specificity and selectivity of the scaffolds. Additional research is needed to focus on glycan synthesis and on engineering novel scaffolds for glycan binding that provide larger binding interfaces to increase the specificity and selectivity of GBPs.
Author Contributions
Conceptualization, D.H.K., R.W.; funding acquisition, D.H.K.; writing—original draft preparation, R.W.; writing—review and editing, D.H.K., R.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by an NSERC Discovery Grant (RGPIN-2016-05464) awarded to D.H.K. and by the NSERC-CREATE (511601-2018)-supported SynBioApps program at Concordia University, which awarded a fellowship to R.W., and by PROTEO (Quebec Network for Research on Protein Function, Structure, and Engineering) who awarded an Undergraduate Summer Studentship to R.W.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
R.W. acknowledges the infrastructure and support available through the SynBioApps program at Concordia University. Figures were created with BioRender.com.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the writing of the manuscript, or in the decision to publish.
References
- Varki, A.; Cummings, R.D.; Esko, J.D.; Stanley, P.; Hart, G.W.; Aebi, M.; Darvill, A.G.; Kinoshita, T.; Packer, N.H.; Prestegard, J.H.; et al. Essentials of Glycobiology, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2015. [Google Scholar]
- Ohtsubo, K.; Marth, J.D. Glycosylation in Cellular Mechanisms of Health and Disease. Cell 2006, 126, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Ramulu, H.G.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Heimburg-Molinaro, J.; Lum, M.; Vijay, G.; Jain, M.; Almogren, A.; Rittenhouse-Olson, K. Cancer vaccines and carbohydrate epitopes. Vaccine 2011, 29, 8802–8826. [Google Scholar] [CrossRef] [PubMed]
- Polonskaya, Z.; Deng, S.; Sarkar, A.; Kain, L.; Comellas-Aragones, M.; McKay, C.S.; Kaczanowska, K.; Holt, M.; McBride, R.; Palomo, V.; et al. T cells control the generation of nanomolar-affinity anti-glycan antibodies. J. Clin. Investig. 2017, 127, 1491–1504. [Google Scholar] [CrossRef]
- Power, B.E.; Caine, J.M.; Burns, J.E.; Shapira, D.R.; Hattarki, M.K.; Tahtis, K.; Lee, F.-T.; Smyth, F.E.; Scott, A.M.; Kortt, A.A.; et al. Construction, expression and characterisation of a single-chain diabody derived from a humanised anti-LewisY cancer targeting antibody using a heat-inducible bacterial secretion vector. Cancer Immunol. Immunother. 2001, 50, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Balzarini, J. Targeting the glycans of gp120: A novel approach aimed at the Achilles heel of HIV. Lancet Infect. Dis. 2005, 5, 726–731. [Google Scholar] [CrossRef]
- Akkouh, O.; Ming, I.D.T.; Singh, S.S.; Yin, C.M.; Dan, X.; Chan, Y.S.; Pan, W.; Cheung, R.C.F. Lectins with Anti-HIV Activity: A Review. Molecules 2015, 20, 648–668. [Google Scholar] [CrossRef]
- Jandú, J.J.B.; Neto, R.N.M.; Zagmignan, A.; De Sousa, E.M.; Brelaz-De-Castro, M.C.A.; Correia, M.T.D.S.; Da Silva, L.C.N. Targeting the Immune System with Plant Lectins to Combat Microbial Infections. Front. Pharmacol. 2017, 8, 8. [Google Scholar] [CrossRef]
- Wang, Z.; Park, K.; Comer, F.; Hsieh-Wilson, L.C.; Saudek, C.D.; Hart, G.W. Site-Specific GlcNAcylation of Human Erythrocyte Proteins: Potential Biomarker(s) for Diabetes. Diabetes 2008, 58, 309–317. [Google Scholar] [CrossRef]
- Ercan, A.; Cui, J.; Hazen, M.M.; Batliwalla, F.; Royle, L.; Rudd, P.M.; Coblyn, J.S.; Shadick, N.A.; Weinblatt, M.; Gregersen, P.K.; et al. Hypogalactosylation of serum N-glycans fails to predict clinical response to methotrexate and TNF inhibition in rheumatoid arthritis. Arthritis Res. Ther. 2012, 14, R43. [Google Scholar] [CrossRef]
- Kuno, A.; Miyoshi, E.; Nakayama, J.; Ohyama, C.; Togayachi, A. Glycan Biomarkers for Cancer and Various Disease. In Glycoscience: Basic Science to Applications: Insights from the Japan Consortium for Glycobiology and Glycotechnology (JCGG); Taniguchi, N., Endo, T., Hirabayashi, J., Nishihara, S., Kadomatsu, K., Akiyoshi, K., Aoki-Kinoshita, K.F., Eds.; Springer: Singapore, 2019; pp. 297–309. ISBN 9789811358562. [Google Scholar]
- Pearce, O.M. Cancer glycan epitopes: Biosynthesis, structure and function. Glycobiology 2018, 28, 670–696. [Google Scholar] [CrossRef] [PubMed]
- Velkov, V.V.; Medvinsky, A.B.; Sokolov, M.S.; Marchenko, A.I. Will transgenic plants adversely affect the environment? J. Biosci. 2005, 30, 515–548. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, C.C.M.; Carvalho, V.; Domingues, L.; Gama, M. Recombinant CBM-fusion technology—Applications overview. Biotechnol. Adv. 2015, 33, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Kalum, L.; Andersen, B.G. Enzymatic Treatment of Denim. U.S. Patent 6,146,428, 14 November 2000. Available online: https://patents.google.com/patent/US6146428A/en (accessed on 3 November 2020).
- Von der Osten, C.; Bjornvad, M.E.; Vind, J.; Rasmussen, M.D. Process and Composition for Desizing Cellulosic Fabric with an Enzyme Hybrid. U.S. Patent 6,017,751, 25 January 2000. [Google Scholar]
- Dang, K.; Zhang, W.; Jiang, S.; Lin, X.; Qian, A. Application of Lectin Microarrays for Biomarker Discovery. ChemistryOpen 2020, 9, 285–300. [Google Scholar] [CrossRef]
- Barondes, S.H. Bifunctional properties of lectins: Lectins redefined. Trends Biochem. Sci. 1988, 13, 480–482. [Google Scholar] [CrossRef]
- Kocourek, J.; Horejsi, V. Defining a lectin. Nat. Cell Biol. 1981, 290, 188. [Google Scholar] [CrossRef]
- Feng, Y.; Guo, Y.; Li, Y.; Tao, J.; Ding, L.; Wu, J.; Ju, H. Lectin-mediated in situ rolling circle amplification on exosomes for probing cancer-related glycan pattern. Anal. Chim. Acta 2018, 1039, 108–115. [Google Scholar] [CrossRef]
- Hashim, O.H.; Jayapalan, J.J.; Lee, C.-S. Lectins: An effective tool for screening of potential cancer biomarkers. PeerJ 2017, 5, e3784. [Google Scholar] [CrossRef]
- Bertozzi, C.R. Chemical Glycobiology. Science 2001, 291, 2357–2364. [Google Scholar] [CrossRef]
- Liener, I.E.; Sharon, N.; Goldstein, I.J. The Lectins: Properties, Functions, and Applications in Biology and Medicine; Academic Press: Cambridge, MA, USA, 1986. [Google Scholar]
- Hu, D.; Tateno, H.; Hirabayashi, J. Lectin Engineering, a Molecular Evolutionary Approach to Expanding the Lectin Utilities. Molecules 2015, 20, 7637–7656. [Google Scholar] [CrossRef]
- Bonnardel, F.; Mariethoz, J.; Salentin, S.; Robin, X.; Schroeder, M.; Perez, S.; Lisacek, F.; Imberty, A. UniLectin3D, a database of carbohydrate binding proteins with curated information on 3D structures and interacting ligands. Nucleic Acids Res. 2019, 47, D1236–D1244. [Google Scholar] [CrossRef] [PubMed]
- Armenta, S.; Moreno-Mendieta, S.; Sánchez-Cuapio, Z.; Sanchez, S.; Rodríguez-Sanoja, R. Advances in molecular engineering of carbohydrate-binding modules. Proteins Struct. Funct. Bioinform. 2017, 85, 1602–1617. [Google Scholar] [CrossRef] [PubMed]
- Guillén, D.; Sanchez, S.; Rodríguez-Sanoja, R. Carbohydrate-binding domains: Multiplicity of biological roles. Appl. Microbiol. Biotechnol. 2009, 85, 1241–1249. [Google Scholar] [CrossRef] [PubMed]
- Simpson, H.D.; Barras, F. Functional Analysis of the Carbohydrate-Binding Domains of Erwinia chrysanthemi Cel5 (Endoglucanase Z) and an Escherichia coli Putative Chitinase. J. Bacteriol. 1999, 181, 4611–4616. [Google Scholar] [CrossRef]
- Simpson, P.J.; Jamieson, S.J.; Hachem, M.A.; Karlsson, E.N.; Gilbert, H.J.; Holst, O.; Williamson, M.P. The Solution Structure of the CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase. Biochemistry 2002, 41, 5712–5719. [Google Scholar] [CrossRef]
- Boraston, A.B.; Creagh, A.L.; Alam, M.; Kormos, J.M.; Tomme, P.; Haynes, C.A.; Warren, R.A.J.; Kilburn, U.G. Binding specificity and thermodynamics of a family 9 carbohydrate-binding module from Thermotoga maritima xylanase 10A. Biochemistry 2001, 40, 6240–6247. [Google Scholar] [CrossRef]
- Furtado, G.P.; Lourenzoni, M.R.; Fuzo, C.A.; Fonseca-Maldonado, R.; Guazzaroni, M.-E.; Ribeiro, L.F.; Ward, R.J. Engineering the affinity of a family 11 carbohydrate binding module to improve binding of branched over unbranched polysaccharides. Int. J. Biol. Macromol. 2018, 120, 2509–2516. [Google Scholar] [CrossRef]
- Gunnarsson, L.C.; Karlsson, E.N.; Albrekt, A.; Andersson, M.; Holst, O.; Ohlin, M. A carbohydrate binding module as a diversity-carrying scaffold. Protein Eng. Des. Sel. 2004, 17, 213–221. [Google Scholar] [CrossRef]
- Sakata, T.; Takakura, J.; Miyakubo, H.; Osada, Y.; Wada, R.; Takahashi, H.; Yatsunami, R.; Fukui, T.; Nakamura, S. Improvement of binding activity of xylan-binding domain by amino acid substitution. Nucleic Acids Symp. Ser. 2006, 50, 253–254. [Google Scholar] [CrossRef]
- Eyers, P.A.; Murphy, J.M. The evolving world of pseudoenzymes: Proteins, prejudice and zombies. BMC Biol. 2016, 14, 1–6. [Google Scholar] [CrossRef]
- Schimpl, M.; Rush, C.L.; Betou, M.; Eggleston, I.M.; Recklies, A.D.; Van Aalten, D.M. Human YKL-39 is a pseudo-chitinase with retained chitooligosaccharide-binding properties. Biochem. J. 2012, 446, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.G.; Da Silva, C.A.; Cruz, C.S.D.; Ahangari, F.; Ma, B.; Kang, M.-J.; He, C.-H.; Takyar, S.; Elias, J.A. Role of Chitin and Chitinase/Chitinase-Like Proteins in Inflammation, Tissue Remodeling, and Injury. Annu. Rev. Physiol. 2011, 73, 479–501. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Choi, J.; Mohanty, J.; Sousa, L.P.; Tome, F.; Pardon, E.; Steyaert, J.; Lemmon, M.A.; Lax, I.; Schlessinger, J. Structures of β-klotho reveal a ‘zip code’-like mechanism for endocrine FGF signalling. Nat. Cell Biol. 2018, 553, 501–505. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Xu, Y.; Wong, W.; Thompson, J.K.; Healer, J.; Goddard-Borger, E.D.; Lawrence, M.C.; Cowman, A.F. Structural basis for inhibition of erythrocyte invasion by antibodies to Plasmodium falciparum protein CyRPA. eLife 2017, 6, 213. [Google Scholar] [CrossRef] [PubMed]
- Favuzza, P.; Guffart, E.; Tamborrini, M.; Scherer, B.; Dreyer, A.M.; Rufer, A.C.; Erny, J.; Hoernschemeyer, J.; Thoma, R.; Schmid, G.; et al. Structure of the malaria vaccine candidate antigen CyRPA and its complex with a parasite invasion inhibitory antibody. eLife 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Wong, W.; Huang, R.; Menant, S.; Hong, C.; Sandow, J.J.; Birkinshaw, R.W.; Healer, J.; Hodder, A.N.; Kanjee, U.; Tonkin, C.J.; et al. Structure of Plasmodium falciparum Rh5–CyRPA–Ripr invasion complex. Nat. Cell Biol. 2018, 565, 118–121. [Google Scholar] [CrossRef]
- Little, D.J.; Li, G.; Ing, C.; DiFrancesco, B.R.; Bamford, N.C.; Robinson, H.; Nitz, M.; Pomès, R.; Howell, P.L. Modification and periplasmic translocation of the biofilm exopolysaccharide poly-1,6-N-acetyl-D-glucosamine. Proc. Natl. Acad. Sci. USA 2014, 111, 11013–11018. [Google Scholar] [CrossRef]
- Stummeyer, K.; Dickmanns, A.; Mühlenhoff, M.; Gerardy-Schahn, R.; Ficner, R. Crystal structure of the polysialic acid–degrading endosialidase of bacteriophage K1F. Nat. Struct. Mol. Biol. 2004, 12, 90–96. [Google Scholar] [CrossRef]
- Jakobsson, E.; Jokilammi, A.; Aalto, J.; Ollikka, P.; Lehtonen, J.V.; Hirvonen, H.; Finne, J. Identification of amino acid residues at the active site of endosialidase that dissociate the polysialic acid binding and cleaving activities in Escherichia coli K1 bacteriophages. Biochem. J. 2007, 405, 465–472. [Google Scholar] [CrossRef]
- Yu, C.-C.; Hill, T.; Kwan, D.; Chen, H.-M.; Lin, C.-C.; Wakarchuk, W.; Withers, S.G. A plate-based high-throughput activity assay for polysialyltransferase from Neisseria meningitidis. Anal. Biochem. 2014, 444, 67–74. [Google Scholar] [CrossRef]
- Yu, C.-C.; Huang, L.-D.; Kwan, D.; Wakarchuk, W.; Withers, S.G.; Lin, C.-C. A glyco-gold nanoparticle based assay for α-2,8-polysialyltransferase from Neisseria meningitidis. Chem. Commun. 2013, 49, 10166–10168. [Google Scholar] [CrossRef] [PubMed]
- Montanier, C.; Money, V.A.; Pires, V.M.R.; Flint, J.E.; Pinheiro, B.A.; Goyal, A.; Prates, J.A.M.; Izumi, A.; Stålbrand, H.; Morland, C.; et al. The Active Site of a Carbohydrate Esterase Displays Divergent Catalytic and Noncatalytic Binding Functions. PLoS Biol. 2009, 7, e1000071. [Google Scholar] [CrossRef] [PubMed]
- Woods, R.J.; Yang, L. Glycan-Specific Analytical Tools. U.S. Patent 9,926,612, 27 March 2018. [Google Scholar]
- Arbabi-Ghahroudi, M. Camelid Single-Domain Antibodies: Historical Perspective and Future Outlook. Front. Immunol. 2017, 8, 1589. [Google Scholar] [CrossRef] [PubMed]
- Nelson, A.L. Antibody fragments. mAbs 2010, 2, 77–83. [Google Scholar] [CrossRef]
- Ahmad, Z.A.; Yeap, S.K.; Ali, A.M.; Ho, W.Y.; Alitheen, N.B.M.; Hamid, M. ScFv Antibody: Principles and Clinical Application. J. Immunol. Res. 2012. Available online: https://www.hindawi.com/journals/jir/2012/980250/ (accessed on 2 January 2021). [CrossRef]
- Holliger, P.; Prospero, T.; Winter, G. Diabodies: Small bivalent and bispecific antibody fragments. Proc. Natl. Acad. Sci. USA 1993, 90, 6444–6448. [Google Scholar] [CrossRef]
- Sha, F.; Salzman, G.; Gupta, A.; Koide, S. Monobodies and other synthetic binding proteins for expanding protein science. Protein Sci. 2017, 26, 910–924. [Google Scholar] [CrossRef]
- Sterner, E.; Flanagan, N.; Gildersleeve, J.C. Perspectives on Anti-Glycan Antibodies Gleaned from Development of a Community Resource Database. ACS Chem. Biol. 2016, 11, 1773–1783. [Google Scholar] [CrossRef]
- Stewart, A.; Liu, Y.; Lai, J.R. A strategy for phage display selection of functional domain-exchanged immunoglobulin scaffolds with high affinity for glycan targets. J. Immunol. Methods 2012, 376, 150–155. [Google Scholar] [CrossRef]
- Olson, L.J.; Hindsgaul, O.; Dahms, N.M.; Kim, J.-J.P. Structural Insights into the Mechanism of pH-dependent Ligand Binding and Release by the Cation-dependent Mannose 6-Phosphate Receptor. J. Biol. Chem. 2008, 283, 10124–10134. [Google Scholar] [CrossRef]
- Ereño-Orbea, J.; Sicard, T.; Cui, H.; Mazhab-Jafari, M.T.; Benlekbir, S.; Guarné, A.; Rubinstein, J.L.; Julien, J.-P. Molecular basis of human CD22 function and therapeutic targeting. Nat. Commun. 2017, 8, 1–11. [Google Scholar] [CrossRef]
- Parkin, S.; Rupp, B.; Hope, H. Atomic Resolution Structure of Concanavalin A at 120 K. Acta Crystallogr. Sect. D Biol. Crystallogr. 1996, 52, 1161–1168. [Google Scholar] [CrossRef] [PubMed]
- Mandal, D.K.; Kishore, N.; Brewer, C.F. Thermodynamics of Lectin-Carbohydrate Interactions. Titration Microcalorimetry Measurements of the Binding of N-Linked Carbohydrates and Ovalbumin to Concanavalin A. Biochemistry 1994, 33, 1149–1156. [Google Scholar] [CrossRef] [PubMed]
- Rutenber, E.; Katzin, B.J.; Ernst, S.; Collins, E.J.; Mlsna, D.; Ready, M.P.; Robertus, J.D. Crystallographic refinement of ricin to 2.5 Å. Proteins Struct. Funct. Bioinform. 1991, 10, 240–250. [Google Scholar] [CrossRef] [PubMed]
- Nagae, M.; Yamanaka, K.; Hanashima, S.; Ikeda, A.; Morita-Matsumoto, K.; Satoh, T.; Matsumoto, N.; Yamamoto, K.; Yamaguchi, Y. Recognition of Bisecting N-Acetylglucosamine. J. Biol. Chem. 2013, 288, 33598–33610. [Google Scholar] [CrossRef]
- Sörme, P.; Arnoux, P.; Kahl-Knutsson, B.; Leffler, H.; Rini, J.M.; Nilsson, U.J. Structural and Thermodynamic Studies on Cation−Π Interactions in Lectin−Ligand Complexes: High-Affinity Galectin-3 Inhibitors through Fine-Tuning of an Arginine−Arene Interaction. J. Am. Chem. Soc. 2005, 127, 1737–1743. [Google Scholar] [CrossRef] [PubMed]
- Werneérus, H.; Lehtiö, J.; Teeri, T.; Nygren, P.-Å.; Ståahl, S. Generation of Metal-Binding Staphylococci through Surface Display of Combinatorially Engineered Cellulose-Binding Domains. Appl. Environ. Microbiol. 2001, 67, 4678–4684. [Google Scholar] [CrossRef]
- Ficko-Blean, E.; Boraston, A.B. The Interaction of a Carbohydrate-binding Module from aClostridium perfringens N-Acetyl-β-hexosaminidase with Its Carbohydrate Receptor. J. Biol. Chem. 2006, 281, 37748–37757. [Google Scholar] [CrossRef]
- Ramsland, P.A.; Farrugia, W.; Bradford, T.M.; Hogarth, P.M.; Scott, A.M. Structural Convergence of Antibody Binding of Carbohydrate Determinants in Lewis Y Tumor Antigens. J. Mol. Biol. 2004, 340, 809–818. [Google Scholar] [CrossRef]
- McCullum, E.O.; Williams, B.A.R.; Zhang, J.; Chaput, J.C. Random Mutagenesis by Error-Prone PCR. In In Vitro Mutagenesis Protocols, 3rd ed.; Braman, J., Ed.; Humana Press: Totowa, NJ, USA, 2010; pp. 103–109. ISBN 978-1-60761-652-8. [Google Scholar]
- Coco, W.M.; Levinson, W.E.; Crist, M.J.; Hektor, H.J.; Darzins, A.; Pienkos, P.T.; Squires, C.H.; Monticello, D.J. DNA shuffling method for generating highly recombined genes and evolved enzymes. Nat. Biotechnol. 2001, 19, 354–359. [Google Scholar] [CrossRef]
- Muteeb, G.; Sen, R. Random Mutagenesis Using a Mutator Strain. In In Vitro Mutagenesis Protocols, 3rd ed.; Braman, J., Ed.; Humana Press: Totowa, NJ, USA, 2010; pp. 411–419. ISBN 978-1-60761-652-8. [Google Scholar]
- Poluri, K.M.; Gulati, K. Protein Engineering Techniques: Gateways to Synthetic Protein Universe; Springer: Singapore, 2017; ISBN 978-981-10-2731-4. [Google Scholar]
- Yabe, R.; Suzuki, R.; Kuno, A.; Fujimoto, Z.; Jigami, Y.; Hirabayashi, J. Tailoring a Novel Sialic Acid-Binding Lectin from a Ricin-B Chain-like Galactose-Binding Protein by Natural Evolution-Mimicry. J. Biochem. 2006, 141, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Stemmer, W.P.C. Rapid evolution of a protein in vitro by DNA shuffling. Nat. Cell Biol. 1994, 370, 389–391. [Google Scholar] [CrossRef] [PubMed]
- Melzer, S.; Sonnendecker, C.; Föllner, C.; Zimmermann, W. Stepwise error-prone PCR and DNA shuffling changed the pH activity range and product specificity of the cyclodextrin glucanotransferase from an alkaliphilicBacillussp. FEBS Open Bio 2015, 5, 528–534. [Google Scholar] [CrossRef] [PubMed]
- Ihssen, J.; Haas, J.; Kowarik, M.; Wiesli, L.; Wacker, M.; Schwede, T.; Thöny-Meyer, L. Increased efficiency of Campylobacter jejuni N -oligosaccharyltransferase PglB by structure-guided engineering. Open Biol. 2015, 5, 140227. [Google Scholar] [CrossRef] [PubMed]
- Mendonça, L.M.; Marana, S.R. Single mutations outside the active site affect the substrate specificity in a β-glycosidase. Biochim. Biophys. Acta Proteins Proteom. 2011, 1814, 1616–1623. [Google Scholar] [CrossRef] [PubMed]
- Adam, J.; Pokorná, M.; Sabin, C.; Mitchell, E.; Imberty, A.; Wimmerová, M. Engineering of PA-IIL lectin from Pseudomonas aeruginosa—Unravelling the role of the specificity loop for sugar preference. BMC Struct. Biol. 2007, 7, 36. [Google Scholar] [CrossRef]
- Kunstmann, S.; Engström, O.; Wehle, M.; Widmalm, G.; Santer, M.; Barbirz, S. Increasing the Affinity of an O-Antigen Polysaccharide Binding Site in Shigella flexneri Bacteriophage Sf6 Tailspike Protein. Chem. A Eur. J. 2020, 26, 7263–7273. [Google Scholar] [CrossRef]
- Allhorn, M.; Olin, A.I.; Nimmerjahn, F.; Collin, M. Human IgG/FcγR Interactions Are Modulated by Streptococcal IgG Glycan Hydrolysis. PLoS ONE 2008, 3, e1413. [Google Scholar] [CrossRef]
- O’Donohue, M.J.; Kneale, G.G. Site-Directed and Site-Saturation Mutagenesis Using Oligonucleotide Primers. In DNA-Protein Interactions: Principles and Protocols; Kneale, G.G., Ed.; Humana Press: Totowa, NJ, USA, 1994; pp. 211–225. ISBN 978-1-59259-517-4. [Google Scholar]
- Hutchison, C.A.; Phillips, S.; Edgell, M.H.; Gillam, S.; Jahnke, P.; Smith, M. Mutagenesis at a specific position in a DNA sequence. J. Biol. Chem. 1978, 253, 6551–6560. [Google Scholar] [CrossRef]
- Imamura, K.; Takeuchi, H.; Yabe, R.; Tateno, H.; Hirabayashi, J. Engineering of the glycan-binding specificity of Agrocybe cylindracea galectin towards α(2,3)-linked sialic acid by saturation mutagenesis. J. Biochem. 2011, 150, 545–552. [Google Scholar] [CrossRef]
- Yamamoto, K.; Konami, Y.; Osawa, T. A chimeric lectin formed from Bauhinia purpurea lectin and Lens culinaris lectin recognizes a unique carbohydrate structure. J. Biochem. 2000, 127, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Naismith, J.H. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 2008, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Nandwani, N.; Surana, P.; Negi, H.; Mascarenhas, N.M.; Udgaonkar, J.B.; Das, R.; Gosavi, S. A five-residue motif for the design of domain swapping in proteins. Nat. Commun. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Lienemann, M.; Boer, H.; Paananen, A.; Cottaz, S.; Koivula, A. Toward understanding of carbohydrate binding and substrate specificity of a glycosyl hydrolase 18 family (GH-18) chitinase from Trichoderma harzianum. Glycobiology 2009, 19, 1116–1126. [Google Scholar] [CrossRef] [PubMed]
- Seifert, A.; Pleiss, J. Identification of selectivity-determining residues in cytochrome P450 monooxygenases: A systematic analysis of the substrate recognition site 5. Proteins Struct. Funct. Bioinform. 2009, 74, 1028–1035. [Google Scholar] [CrossRef]
- Cheriyan, M.; Toone, E.J.; Fierke, C.A. Mutagenesis of the phosphate-binding pocket of KDPG aldolase enhances selectivity for hydrophobic substrates. Protein Sci. 2007, 16, 2368–2377. [Google Scholar] [CrossRef]
- Schneider, S.; Gutiérrez, M.; Sandalova, T.; Schneider, G.; Clapés, P.; Sprenger, G.A.; Samland, A.K. Redesigning the Active Site of Transaldolase TalB from Escherichia coli: New Variants with Improved Affinity towards Nonphosphorylated Substrates. ChemBioChem 2010, 11, 681–690. [Google Scholar] [CrossRef]
- Lo, W.-C.; Wang, L.-F.; Liu, Y.-Y.; Dai, T.; Hwang, J.-K.; Lyu, P.-C. CPred: A web server for predicting viable circular permutations in proteins. Nucleic Acids Res. 2012, 40, W232–W237. [Google Scholar] [CrossRef]
- Stephen, P.; Tseng, K.-L.; Liu, Y.-N.; Lyu, P.-C. Circular permutation of the starch-binding domain: Inversion of ligand selectivity with increased affinity. Chem. Commun. 2012, 48, 2612–2614. [Google Scholar] [CrossRef]
- Kuhlman, B.; Bradley, P. Advances in protein structure prediction and design. Nat. Rev. Mol. Cell Biol. 2019, 20, 681–697. [Google Scholar] [CrossRef]
- Zinovjev, K.; Van der Kamp, M.W. Enlighten2: Molecular Dynamics Simulations of Protein-Ligand Systems Made Accessible. chemRxiv 2020. [Google Scholar] [CrossRef]
- Karplus, M.; Kuriyan, J. Molecular dynamics and protein function. Proc. Natl. Acad. Sci. USA 2005, 102, 6679–6685. [Google Scholar] [CrossRef] [PubMed]
- Alley, E.C.; Khimulya, G.; Biswas, S.; AlQuraishi, M.; Church, G.M. Unified rational protein engineering with sequence-based deep representation learning. Nat. Methods 2019, 16, 1315–1322. [Google Scholar] [CrossRef] [PubMed]
- Frei, J.; Lai, J. Protein and Antibody Engineering by Phage Display. Methods Enzym. 2016, 580, 45–87. [Google Scholar] [CrossRef]
- Peltomaa, R.; Benito-Peña, E.; Barderas, R.; Bondi, M.C.C.M. Phage Display in the Quest for New Selective Recognition Elements for Biosensors. ACS Omega 2019, 4, 11569–11580. [Google Scholar] [CrossRef]
- Chasteen, L.; Ayriss, J.; Pavlik, P.; Bradbury, A. Eliminating helper phage from phage display. Nucleic Acids Res. 2006, 34, e145. [Google Scholar] [CrossRef]
- Yuasa, N.; Koyama, T.; Fujita-Yamaguchi, Y. Purification and refolding of anti-T-antigen single chain antibodies (scFvs) expressed in Escherichia coli as inclusion bodies. Biosci. Trends 2014, 8, 24–31. [Google Scholar] [CrossRef]
- Ng, S.; Tjhung, K.F.; Paschal, B.M.; Noren, C.J.; Derda, R. Chemical Posttranslational Modification of Phage-Displayed Peptides. In Peptide Libraries: Methods and Protocols; Derda, R., Ed.; Springer: New York, NY, USA, 2015; pp. 155–172. ISBN 978-1-4939-2020-4. [Google Scholar]
- Boder, E.T.; Wittrup, K.D. Yeast Surface Display for Directed Evolution of Protein Expression, Affinity, and Stability. Methods Enzymol. 2000, 328, 430–444. [Google Scholar] [CrossRef]
- Hong, X.; Ma, M.Z.; Gildersleeve, J.C.; Chowdhury, S.; Barchi, J.J.; Mariuzza, R.A.; Murphy, M.B.; Mao, L.; Pancer, Z. Sugar-Binding Proteins from Fish: Selection of High Affinity Lambodies That Recognize Biomedically Relevant Glycans. ACS Chem. Biol. 2012, 8, 152–160. [Google Scholar] [CrossRef]
- Lu, Z.; Kamat, K.; Johnson, B.P.; Yin, C.C.; Scholler, N.; Abbott, K.L. Generation of a Fully Human scFv that binds Tumor-Specific Glycoforms. Sci. Rep. 2019, 9, 5101. [Google Scholar] [CrossRef]
- Hanes, J.; Plückthun, A. In vitro selection and evolution of functional proteins by using ribosome display. Proc. Natl. Acad. Sci. USA 1997, 94, 4937–4942. [Google Scholar] [CrossRef]
- Kurz, M.; Gu, K.; Lohse, P.A. Psoralen Photo-Crosslinked MRNA–Puromycin Conjugates: A Novel Template for the Rapid and Facile Preparation of MRNA–Protein Fusions. Nucleic Acids Res. 2000, 28, e83. [Google Scholar] [CrossRef]
- Nemoto, N.; Miyamoto-Sato, E.; Husimi, Y.; Yanagawa, H. In vitro virus: Bonding of mRNA bearing puromycin at the 3′-terminal end to the C-terminal end of its encoded protein on the ribosome in vitro. FEBS Lett. 1997, 414, 405–408. [Google Scholar] [CrossRef]
- Newton, M.S.; Cabezas-Perusse, Y.; Tong, C.L.; Seelig, B. In Vitro Selection of Peptides and Proteins—Advantages of mRNA Display. ACS Synth. Biol. 2019, 9, 181–190. [Google Scholar] [CrossRef]
- Niwa, T.; Kanamori, T.; Ueda, T.; Taguchi, H. Global analysis of chaperone effects using a reconstituted cell-free translation system. Proc. Natl. Acad. Sci. USA 2012, 109, 8937–8942. [Google Scholar] [CrossRef]
- Lieberoth, A.; Splittstoesser, F.; Katagihallimath, N.; Jakovcevski, I.; Loers, G.; Ranscht, B.; Karagogeos, D.; Schachner, M.; Kleene, R. Lewisx and 2,3-Sialyl Glycans and Their Receptors TAG-1, Contactin, and L1 Mediate CD24-Dependent Neurite Outgrowth. J. Neurosci. 2009, 29, 6677–6690. [Google Scholar] [CrossRef]
- Sprung, R.; Nandi, A.; Chen, Y.; Kim, S.C.; Barma, D.; Falck, J.R.; Zhao, Y. Tagging-via-Substrate Strategy for Probing O-GlcNAc Modified Proteins. J. Proteome Res. 2005, 4, 950–957. [Google Scholar] [CrossRef]
- Mahal, L.K.; Yarema, K.J.; Bertozzi, C.R. Engineering Chemical Reactivity on Cell Surfaces Through Oligosaccharide Biosynthesis. Science 1997, 276, 1125–1128. [Google Scholar] [CrossRef]
- Luchansky, S.J.; Argade, S.; Hayes, B.K.; Bertozzi, C.R. Metabolic Functionalization of Recombinant Glycoproteins. Biochemistry 2004, 43, 12358–12366. [Google Scholar] [CrossRef]
- Bryan, M.C.; Lee, L.V.; Wong, C.-H. High-throughput identification of fucosyltransferase inhibitors using carbohydrate microarrays. Bioorg. Med. Chem. Lett. 2004, 14, 3185–3188. [Google Scholar] [CrossRef]
- Winzor, D.J. Determination of binding constants by affinity chromatography. J. Chromatogr. A 2004, 1037, 351–367. [Google Scholar] [CrossRef]
- Kasai, K. Frontal Affinity Chromatography: A Unique Research Tool for Biospecific Interaction That Promotes Glycobiology. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2014, 90, 215–234. [Google Scholar] [CrossRef] [PubMed]
- Schasfoort, R.B.M. Chapter 1: Introduction to Surface Plasmon Resonance. In Handbook of Surface Plasmon Resonance; Royal Society of Chemistry: Cambridge, UK, 2017; pp. 1–26. [Google Scholar]
- Gutiérrez-Gallego, R.; Haseley, S.R.; Van Miegem, V.F.L.; Vliegenthart, J.F.; Kamerling, J.P. Identification of carbohydrates binding to lectins by using surface plasmon resonance in combination with HPLC profiling. Glycobiology 2004, 14, 373–386. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bellapadrona, G.; Tesler, A.B.; Grünstein, D.; Hossain, L.H.; Kikkeri, R.; Gilmore, K.; Vaskevich, A.; Rubinstein, I. Optimization of Localized Surface Plasmon Resonance Transducers for Studying Carbohydrate–Protein Interactions. Anal. Chem. 2011, 84, 232–240. [Google Scholar] [CrossRef]
- Huang, C.-F.; Yao, G.-H.; Liang, R.-P.; Qiu, J.-D. Graphene oxide and dextran capped gold nanoparticles based surface plasmon resonance sensor for sensitive detection of concanavalin A. Biosens. Bioelectron. 2013, 50, 305–310. [Google Scholar] [CrossRef]
- Liu, L.; Prudden, A.R.; Capicciotti, C.J.; Bosman, G.P.; Yang, J.-Y.; Chapla, D.G.; Moremen, K.W.; Boons, G.-J. Streamlining the chemoenzymatic synthesis of complex N-glycans by a stop and go strategy. Nat. Chem. 2019, 11, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Palcic, M.M. Glycosyltransferases as biocatalysts. Curr. Opin. Chem. Biol. 2011, 15, 226–233. [Google Scholar] [CrossRef]
- Shilova, O.N.; Deyev, S.M. DARPins: Promising Scaffolds for Theranostics. Acta Nat. 2019, 11, 42–53. [Google Scholar] [CrossRef]
- Hofmeister, D.L.; Thoden, J.B.; Holden, H.M. Investigation of a sugar N-formyltransferase from the plant pathogen Pantoea ananatis. Protein Sci. 2019, 28, 707–716. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).