Cellular NADH and NADPH Conformation as a Real-Time Fluorescence-Based Metabolic Indicator under Pressurized Conditions
Abstract
1. Introduction
2. Results
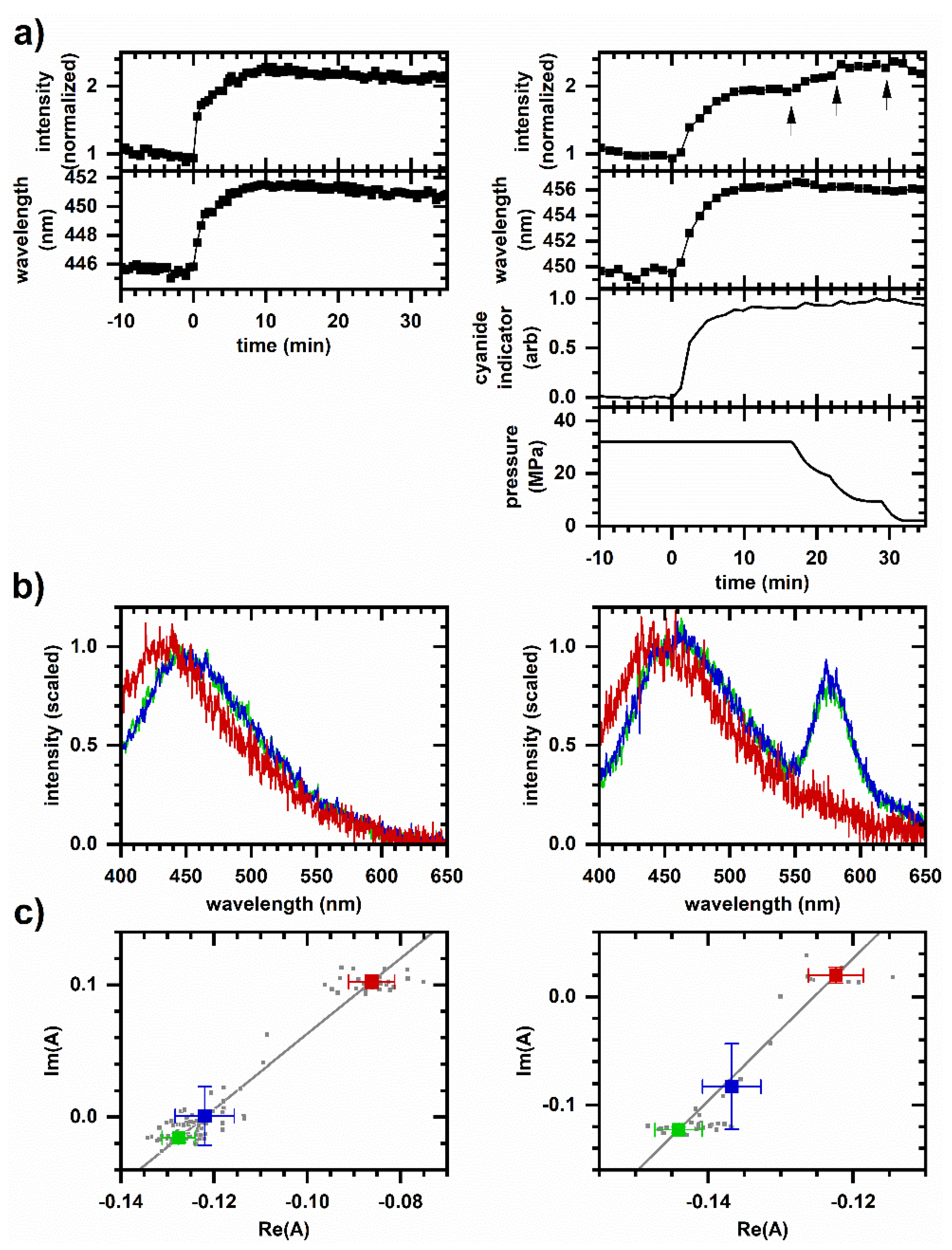
2.1. Cellular Autofluorescence Response to Cyanide
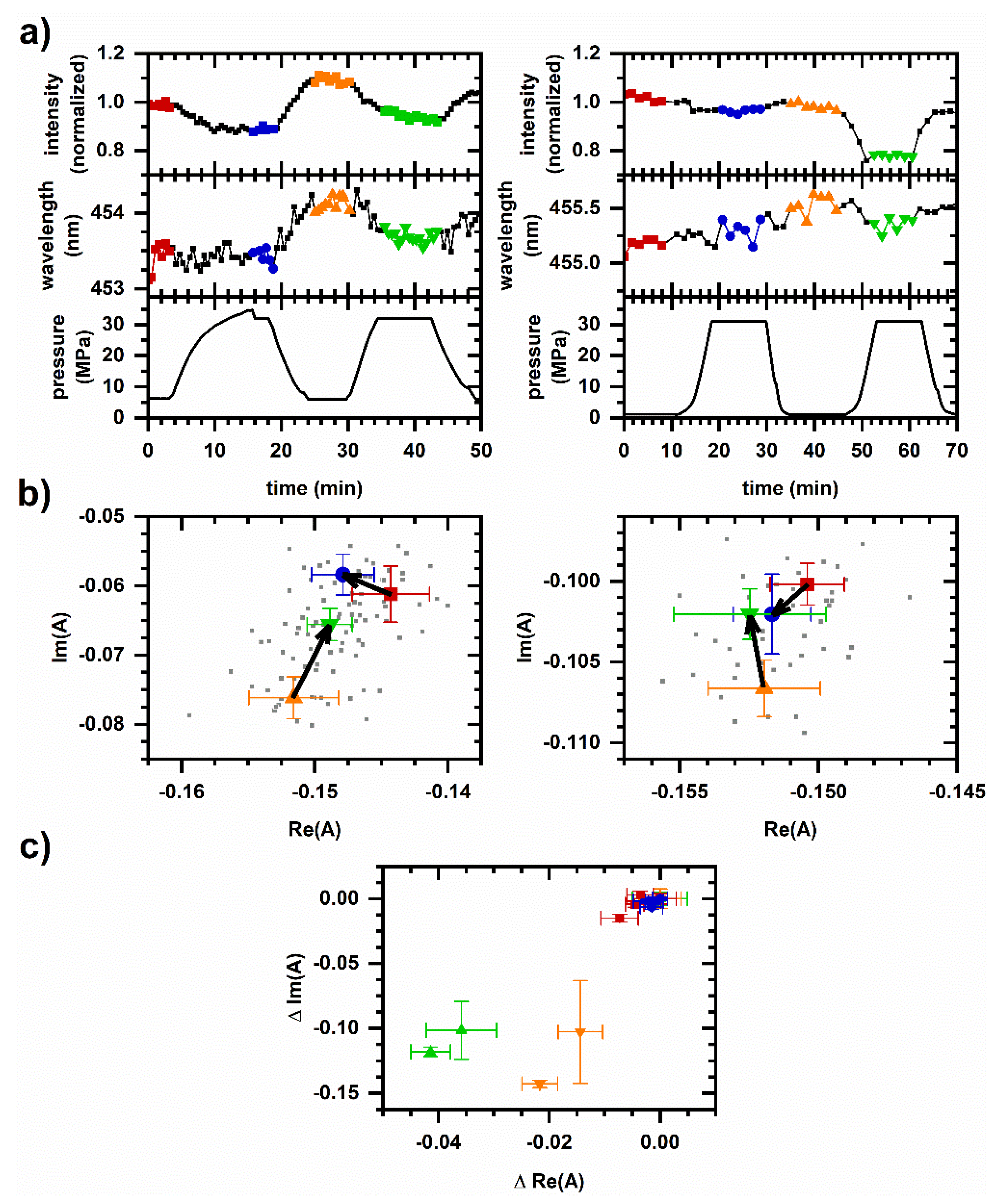
2.2. Cellular Autofluorescence Response to Pressure Cycling
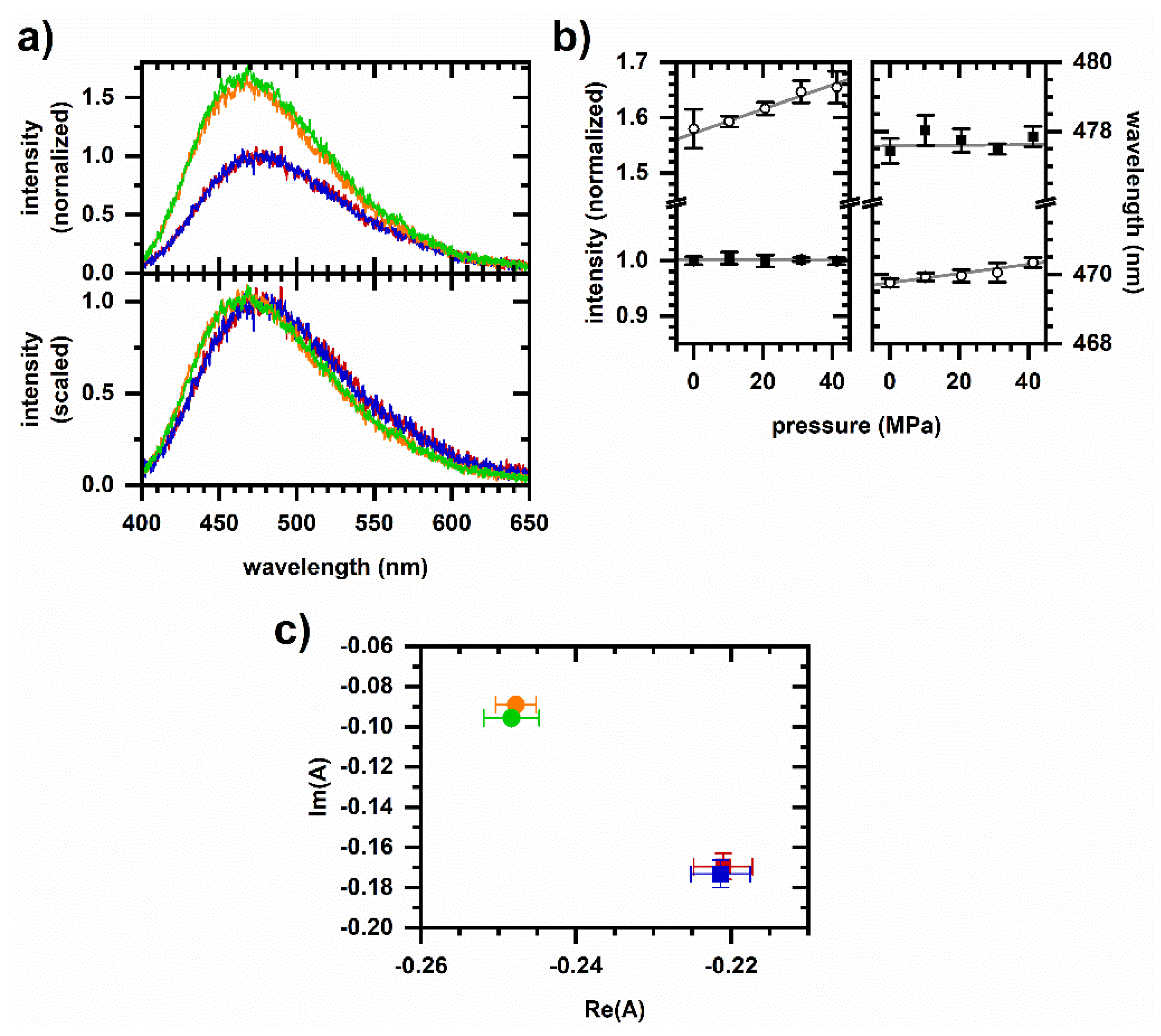
2.3. Excited-State Emission from NADH in Solution under Pressurized Conditions
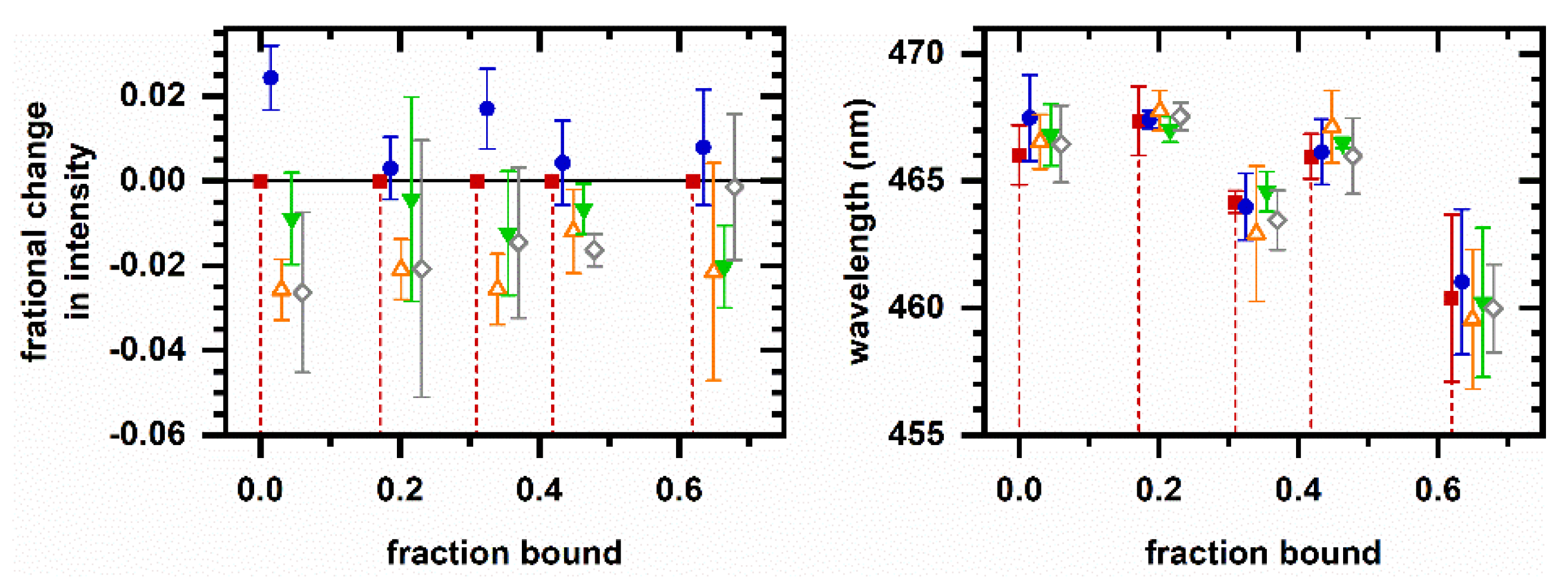
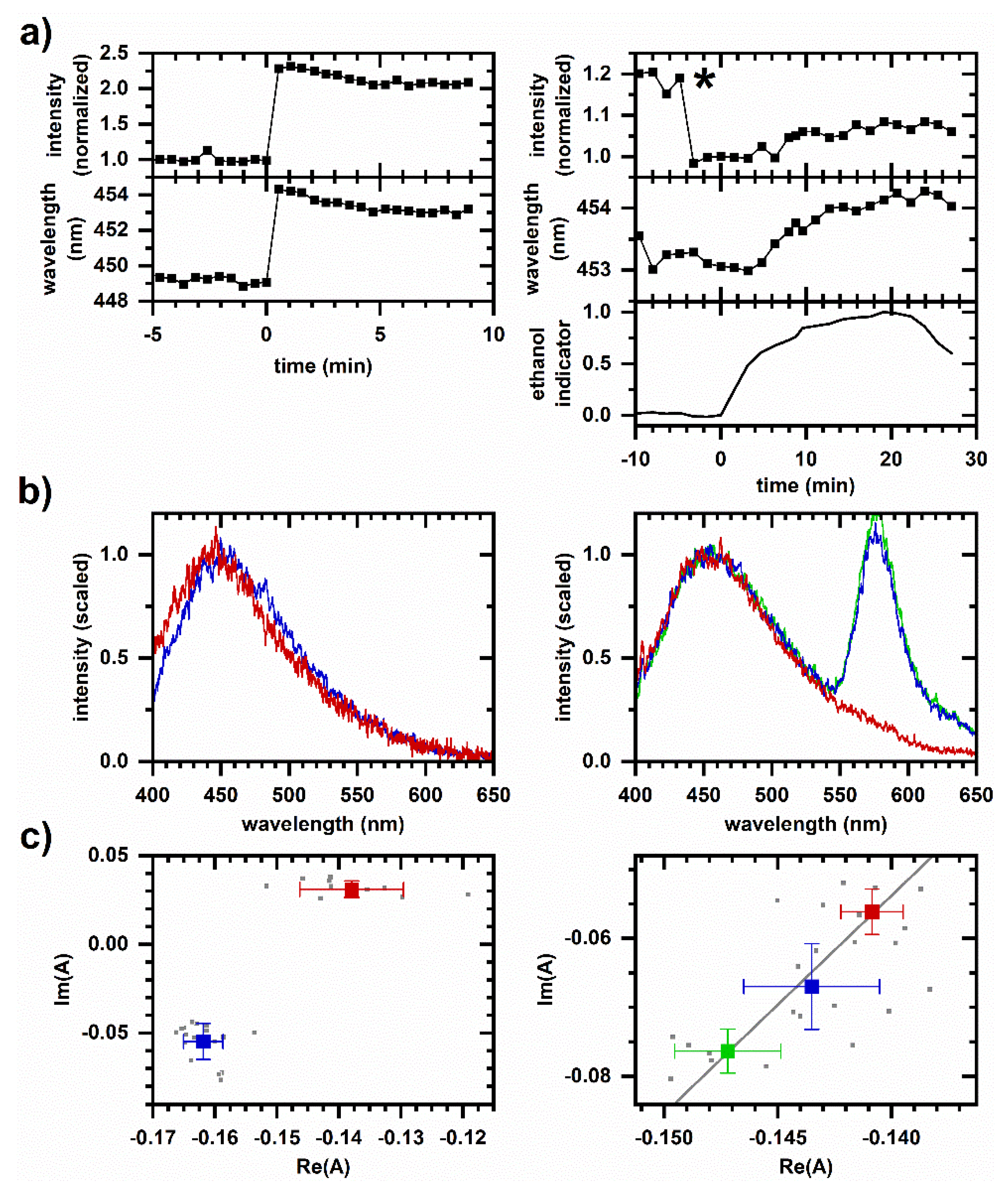
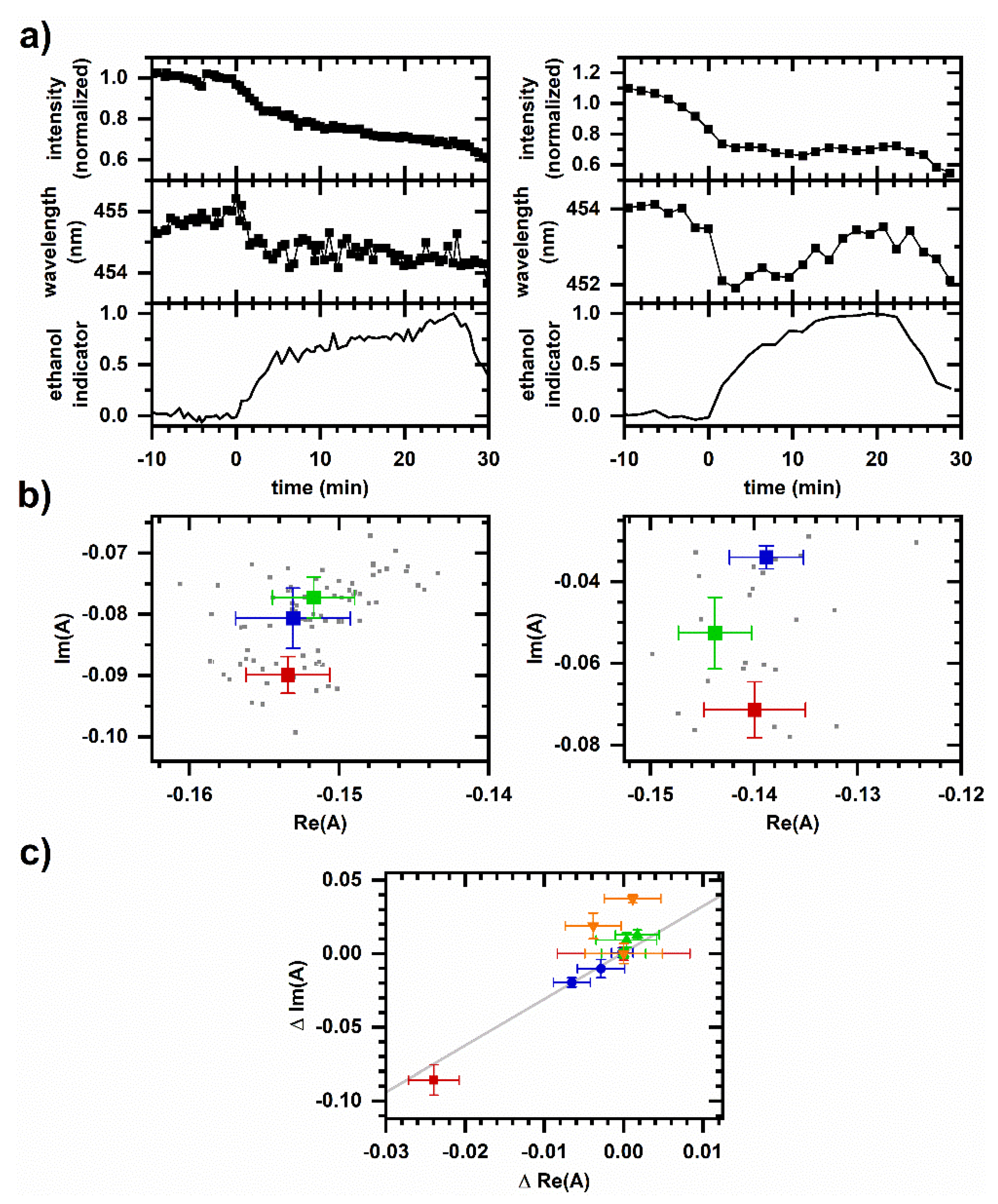
2.4. Cellular Autofluorescence Response to Ethanol
3. Discussion
4. Materials and Methods
4.1. Instrumentation
4.2. Sample Preparation and Data Acquisition
4.3. Spectral Phasor Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Blacker, T.S.; Duchen, M.R. Investigating mitochondrial redox state using NADH and NADPH autofluorescence. Free Radic. Biol. Med. 2016, 100, 53–65. [Google Scholar] [CrossRef]
- Ying, W.H. NAD+/NADH and NADP+/NADPH in cellular functions and cell death: Regulation and biological consequences. Antioxid. Redox Signal. 2008, 10, 179–206. [Google Scholar] [CrossRef] [PubMed]
- Croce, A.C.; Bottiroli, G. Autofluorescence spectroscopy and imaging: A tool for biomedical research and diagnosis. Eur. J. Histochem. 2014, 58, 2461. [Google Scholar] [CrossRef]
- Blacker, T.S.; Mann, Z.F.; Gale, J.E.; Ziegler, M.; Bain, A.J.; Szabadkai, G.; Duchen, M.R. Separating NADH and NADPH fluorescence in live cells and tissues using FLIM. Nat. Commun. 2014, 5, 3936. [Google Scholar] [CrossRef]
- Bartlett, D.H. Pressure effects on in vivo microbial processes. Biochim. Biophys. Acta 2002, 1595, 367–381. [Google Scholar] [CrossRef]
- Abe, F. Exploration of the effects of high hydrostatic pressure on microbial growth, physiology and survival: Perspectives from piezophysiology. Biosci. Biotechnol. Biochem. 2007, 71, 2347–2357. [Google Scholar] [CrossRef]
- Mota, M.J.; Lopes, R.P.; Delgadillo, I.; Saraiva, J.A. Microorganisms under high pressure—Adaptation, growth and biotechnological potential. Biotechnol. Adv. 2013, 31, 1426–1434. [Google Scholar] [CrossRef]
- Meersman, F.; McMillan, P.F. High hydrostatic pressure: A probing tool and a necessary parameter in biophysical chemistry. Chem. Commun. 2014, 50, 766–775. [Google Scholar] [CrossRef]
- Qureshi, M.H.; Kato, C.; Horikoshi, K. Purification of a ccb-type quinol oxidase specifically induced in a deep-sea barophilic bacterium, Shewanella sp. strain DB-172F. Extremophiles 1998, 2, 93–99. [Google Scholar] [CrossRef]
- Qureshi, M.H.; Kato, C.; Horikoshi, K. Purification of two pressure-regulated c-type cytochromes from a deep-sea barophilic bacterium, Shewanella sp. strain DB-172F. FEMS Microbiol. Lett. 1998, 161, 301–309. [Google Scholar] [CrossRef]
- Yamada, M.; Nakasone, K.; Tamegai, H.; Kato, C.; Usami, R.; Horikoshi, K. Pressure regulation of soluble cytochromes c in a deep-sea piezophilic bacterium, Shewanella violacea. J. Bacteriol. 2000, 182, 2945–2952. [Google Scholar] [CrossRef][Green Version]
- Tamegai, H.; Kawano, H.; Ishii, A.; Chikuma, S.; Nakasone, K.; Kato, C. Pressure-regulated biosynthesis of cytochrome bd in piezo- and psychrophilic deep-sea bacterium Shewanella violacea DSS12. Extremophiles 2005, 9, 247–253. [Google Scholar] [CrossRef]
- Vezzi, A.; Campanaro, S.; D’Angelo, M.; Simonato, F.; Vitulo, N.; Lauro, F.M.; Cestaro, A.; Malacrida, G.; Simionati, B.; Cannata, N.; et al. Life at depth: Photobacterium profundum genome sequence and expression analysis. Science 2005, 307, 1459–1461. [Google Scholar] [CrossRef] [PubMed]
- Tamegai, H.; Nishikawa, S.; Haga, M.; Bartlett, D.H. The respiratory system of the piezophile Photobacterium profundum SS9 grown under various pressures. Biosci. Biotechnol. Biochem. 2012, 76, 1506–1510. [Google Scholar] [CrossRef][Green Version]
- Stokes, M.D.; Somero, G.N. An optical oxygen sensor and reaction vessel for high-pressure applications. Limnol. Oceanogr. 1999, 44, 189–195. [Google Scholar] [CrossRef]
- Nagahama, T.; Hamamoto, M.; Nakase, T.; Horikoshi, K. Kluyveromyces nonfermentans sp. nov., a new yeast species isolated from the deep sea. Int. J. Syst. Bacteriol. 1999, 49, 1899–1905. [Google Scholar] [CrossRef] [PubMed]
- Hiraki, T.; Sekiguchi, T.; Kato, C.; Hatada, Y.; Maruyama, T.; Abe, F.; Konishi, M. New type of pressurized cultivation method providing oxygen for piezotolerant yeast. J. Biosci. Bioeng. 2012, 113, 220–223. [Google Scholar] [CrossRef]
- Ferreira, R.M.; Mota, M.J.; Lopes, R.P.; Sousa, S.; Gomes, A.M.; Delgadillo, I.; Saraiva, J.A. Adaptation of Saccharomyces cerevisiae to high pressure (15, 25 and 35 MPa) to enhance the production of bioethanol. Food Res. Int. 2019, 115, 352–359. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Niu, C.; Liu, C.; Li, Q. Unraveling the mechanisms for low-level acetaldehyde production during alcoholic fermentation in Saccharomyces pastorianus lager yeast. J. Agric. Food Chem. 2019, 67, 2020–2027. [Google Scholar] [CrossRef]
- Xu, X.; Song, Y.; Guo, L.; Cheng, W.; Niu, C.; Wang, J.; Liu, C.; Zheng, F.; Zhou, Y.; Li, X.; et al. Higher NADH availability of lager yeast increases the flavor stability of beer. J. Agric. Food Chem. 2020, 68, 584–590. [Google Scholar] [CrossRef]
- Frauenfelder, H.; McMahon, B.H.; Fenimore, P.W. Myoglobin: The hydrogen atom of biology and a paradigm of complexity. Proc. Natl. Acad. Sci. USA 2003, 100, 8615–8617. [Google Scholar] [CrossRef]
- Gruner, S.M. Soft materials and biomaterials under pressure. In High-Pressure Crystallography; Katrusiak, A., McMillan, P., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2004. [Google Scholar]
- Marchal, S.; Torrent, J.; Masson, P.; Kornblatt, J.M.; Tortora, P.; Fusi, P.; Lange, R.; Balny, C. The powerful high pressure tool for protein conformational studies. Braz. J. Med. Biol. Res. 2005, 38, 1175–1183. [Google Scholar] [CrossRef]
- Vishwasrao, H.D.; Heikal, A.A.; Kasischke, K.A.; Webb, W.W. Conformational dependence of intracellular NADH on metabolic state revealed by associated fluorescence anisotropy. J. Biol. Chem. 2005, 280, 25119–25126. [Google Scholar] [CrossRef]
- Vergen, J.; Hecht, C.; Zholudeva, L.V.; Marquardt, M.M.; Hallworth, R.; Nichols, M.G. Metabolic imaging using two-photon excited NADH intensity and fluorescence lifetime imaging. Microsc. Microanal. 2012, 18, 761–770. [Google Scholar] [CrossRef]
- Yaseen, M.A.; Sakadzic, S.; Wu, W.; Becker, W.; Kasischke, K.A.; Boas, D.A. In vivo imaging of cerebral energy metabolism with two-photon fluorescence lifetime microscopy of NADH. Biomed. Opt. Express 2013, 4, 307–321. [Google Scholar] [CrossRef]
- Williamson, D.H.; Lund, P.; Krebs, H.A. The redox state of free nicotinamide-adenine dinucleotide in the cytoplasm and mitochondria of rat liver. Biochem. J. 1967, 103, 514–527. [Google Scholar] [CrossRef]
- Palero, J.A.; Bader, A.N.; de Bruijn, H.S.; van den Heuvel, A.v.d.P.; Sterenborg, H.J.C.M.; Gerritsen, H.C. In vivo monitoring of protein-bound and free NADH during ischemia by nonlinear spectral imaging microscopy. Biomed. Opt. Express 2011, 2, 1030–1039. [Google Scholar] [CrossRef]
- Stringari, C.; Cinquin, A.; Cinquin, O.; Digman, M.A.; Donovan, P.J.; Gratton, E. Phasor approach to fluorescence lifetime microscopy distinguishes different metabolic states of germ cells in a live tissue. Proc. Natl. Acad. Sci. USA 2011, 108, 13582–13587. [Google Scholar] [CrossRef]
- Scott, T.G.; Spencer, R.D.; Leonard, N.J.; Weber, G. Emission properties of NADH: Studies of fluorescence lifetimes and quantum efficiencies of NADH, AcPyADH, and simplified synthetic models. J. Am. Chem. Soc. 1970, 92, 687–695. [Google Scholar] [CrossRef]
- Long, Z.; Maltas, J.; Zatt, M.C.; Cheng, J.; Alquist, E.J.; Brest, A.; Urayama, P. The real-time quantification of autofluorescence spectrum shape for the monitoring of mitochondrial metabolism. J. Biophotonics 2015, 8, 247–257. [Google Scholar] [CrossRef]
- Maltas, J.; Amer, L.; Long, Z.; Palo, D.; Oliva, A.; Folz, J.; Urayama, P. Autofluorescence from NADH conformations associated with different metabolic pathways monitored using nanosecond-gated spectroscopy and spectral phasor analysis. Anal. Chem. 2015, 87, 5117–5124. [Google Scholar] [CrossRef]
- Maltas, J.; Palo, D.; Wong, C.K.; Stefan, S.; O’Connor, J.; Al Aayedi, N.; Gaire, M.; Kinn, D.; Urayama, P. A metabolic interpretation for the response of cellular autofluorescence to chemical perturbations assessed using spectral phasor analysis. RSC Adv. 2018, 8, 41526–41535. [Google Scholar] [CrossRef]
- Short, A.H.; Aayedi, N.A.; Gaire, M.; Kreider, M.; Wong, C.K.; Urayama, P. Distinguishing chemically induced NADPH- and NADH-related metabolic responses using phasor analysis of UV-excited autofluorescence. RSC Adv. 2021, 11, 18757–18767. [Google Scholar] [CrossRef]
- Hull, R.V.; Conger, P.S.; Hoobler, R.J. Conformation of NADH studied by fluorescence excitation transfer spectroscopy. Biophys. Chem. 2001, 90, 9–16. [Google Scholar] [CrossRef]
- Gorbunova, I.A.; Sasin, M.E.; Rubayo-Soneira, J.; Smolin, A.G.; Vasyutinskii, O.S. Two-photon excited fluorescence dynamics in NADH in water-methanol solutions: The role of conformation states. J. Phys. Chem. B 2020, 124, 10682–10697. [Google Scholar] [CrossRef]
- Cadena-Caicedo, A.; Gonzalez-Cano, B.; Lopez-Arteaga, R.; Esturau-Escofet, N.; Peon, J. Ultrafast fluorescence signals from beta-dihydronicotinamide adenine dinucleotide: Resonant energy transfer in the folded and unfolded forms. J. Phys. Chem. B 2020, 124, 519–530. [Google Scholar] [CrossRef]
- Jones, R.P. Biological principles for the effects of ethanol. Enzym. Microb. Technol. 1989, 11, 130–153. [Google Scholar] [CrossRef]
- Carlsen, H.N.; Degn, H.; Lloyd, D. Effects of alcohols on the respiration and fermentation of aerated suspensions of baker’s yeast. J. Gen. Microbiol. 1991, 137, 2879–2883. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Griffiths, D.E.; Wharton, D.C. Studies of the electron transport system. XXXV. Purification and properties of cytochrome oxidase. J. Biol. Chem. 1961, 236, 1850–1856. [Google Scholar] [CrossRef]
- Siano, S.A.; Mutharasan, R. NADH and flavin fluorescence responses of starved yeast cultures to substrate additions. Biotechnol. Bioeng. 1989, 34, 660–670. [Google Scholar] [CrossRef] [PubMed]
- Bravim, F.; Mota, M.M.; Fernandes, A.A.R.; Fernandes, P.M.B. High hydrostatic pressure leads to free radicals accumulation in yeast cells triggering oxidative stress. Fems Yeast Res. 2016, 16. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Aertsen, A.; De Spiegeleer, P.; Vanoirbeek, K.; Lavilla, M.; Michiels, C.W. Induction of oxidative stress by high hydrostatic pressure in Escherichia coli. Appl. Environ. Microb. 2005, 71, 2226–2231. [Google Scholar] [CrossRef]
- Bravim, F.; da Silva, L.F.; Souza, D.T.; Lippman, S.I.; Broach, J.R.; Fernandes, A.A.R.; Fernandes, P.M.B. High hydrostatic pressure activates transcription factors involved in Saccharomyces cerevisiae stress tolerance. Curr. Pharm. Biotechnol. 2012, 13, 2712–2720. [Google Scholar] [CrossRef][Green Version]
- Charoenbhakdi, S.; Dokpikul, T.; Burphan, T.; Techo, T.; Auesukaree, C. Vacuolar H+- ATPase protects Saccharomyces cerevisiae cells against ethanol-induced oxidative and cell wall stresses. Appl. Environ. Microb. 2016, 82, 3121–3130. [Google Scholar] [CrossRef]
- Palhano, F.L.; Gomes, H.L.; Orlando, M.T.D.; Kurtenbach, E.; Fernandes, P.M.B. Pressure response in the yeast Saccharomyces cerevisiae: From cellular to molecular approaches. Cell. Mol. Biol. 2004, 50, 447–457. [Google Scholar]
- Mileykovskaya, E.; Penczek, P.A.; Fang, J.; Mallampalli, V.; Sparagna, G.C.; Dowhan, W. Arrangement of the respiratory chain complexes in Saccharomyces cerevisiae supercomplex III2IV2 revealed by single particle cryo-electron microscopy. J. Biol. Chem. 2012, 287, 23095–23103. [Google Scholar] [CrossRef] [PubMed]
- Raber, E.C.; Dudley, J.A.; Salerno, M.; Urayama, P. Capillary-based, high-pressure chamber for fluorescence microscopy imaging. Rev. Sci. Instrum. 2006, 77, 096106. [Google Scholar] [CrossRef]
- Maltas, J.; Long, Z.; Huff, A.; Maloney, R.; Ryan, J.; Urayama, P. A micro-perfusion system for use during real-time physiological studies under high pressure. Rev. Sci. Instrum. 2014, 85, 106106. [Google Scholar] [CrossRef] [PubMed]
- Thorne, C.J.R.; Kaplan, N.O. Physicochemical properties of pig and horse heart mitochondrial malate dehydrogenase. J. Biol. Chem. 1963, 238, 1861–1868. [Google Scholar] [CrossRef]
- Shore, J.D.; Evans, S.A.; Holbrook, J.J.; Parker, D.M. NADH binding to porcine mitochondrial malate dehydrogenase. J. Biological Chem. 1979, 254, 9059–9062. [Google Scholar] [CrossRef]
- Brown, S.W.; Oliver, S.G.; Harrison, D.E.F.; Righelato, R.C. Ethanol inhibition of yeast growth and fermentation: Differences in the magnitude and complexity of the effect. Eur. J. Appl. Microb. Biotechnol. 1981, 11, 151–155. [Google Scholar] [CrossRef]
- Fereidouni, F.; Bader, A.N.; Gerritsen, H.C. Spectral phasor analysis allows rapid and reliable unmixing of fluorescence microscopy spectral images. Opt. Express 2012, 20, 12729–12741. [Google Scholar] [CrossRef] [PubMed]
- Kreider, M.; Rodriguez, A.; Vishwanath, K.; Urayama, P. Spectral phasor analysis of autofluorescence responses from cells embedded in turbid media containing collagen. In Label-Free Biomedical Imaging and Sensing; SPIE: San Francisco, CA, USA, 2020. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heidelman, M.; Dhakal, B.; Gikunda, M.; Silva, K.P.T.; Risal, L.; Rodriguez, A.I.; Abe, F.; Urayama, P. Cellular NADH and NADPH Conformation as a Real-Time Fluorescence-Based Metabolic Indicator under Pressurized Conditions. Molecules 2021, 26, 5020. https://doi.org/10.3390/molecules26165020
Heidelman M, Dhakal B, Gikunda M, Silva KPT, Risal L, Rodriguez AI, Abe F, Urayama P. Cellular NADH and NADPH Conformation as a Real-Time Fluorescence-Based Metabolic Indicator under Pressurized Conditions. Molecules. 2021; 26(16):5020. https://doi.org/10.3390/molecules26165020
Chicago/Turabian StyleHeidelman, Martin, Bibek Dhakal, Millicent Gikunda, Kalinga Pavan Thushara Silva, Laxmi Risal, Andrew I. Rodriguez, Fumiyoshi Abe, and Paul Urayama. 2021. "Cellular NADH and NADPH Conformation as a Real-Time Fluorescence-Based Metabolic Indicator under Pressurized Conditions" Molecules 26, no. 16: 5020. https://doi.org/10.3390/molecules26165020
APA StyleHeidelman, M., Dhakal, B., Gikunda, M., Silva, K. P. T., Risal, L., Rodriguez, A. I., Abe, F., & Urayama, P. (2021). Cellular NADH and NADPH Conformation as a Real-Time Fluorescence-Based Metabolic Indicator under Pressurized Conditions. Molecules, 26(16), 5020. https://doi.org/10.3390/molecules26165020





