Incorporation of 4J-HMBC and NOE Data into Computer-Assisted Structure Elucidation with WebCocon
Abstract
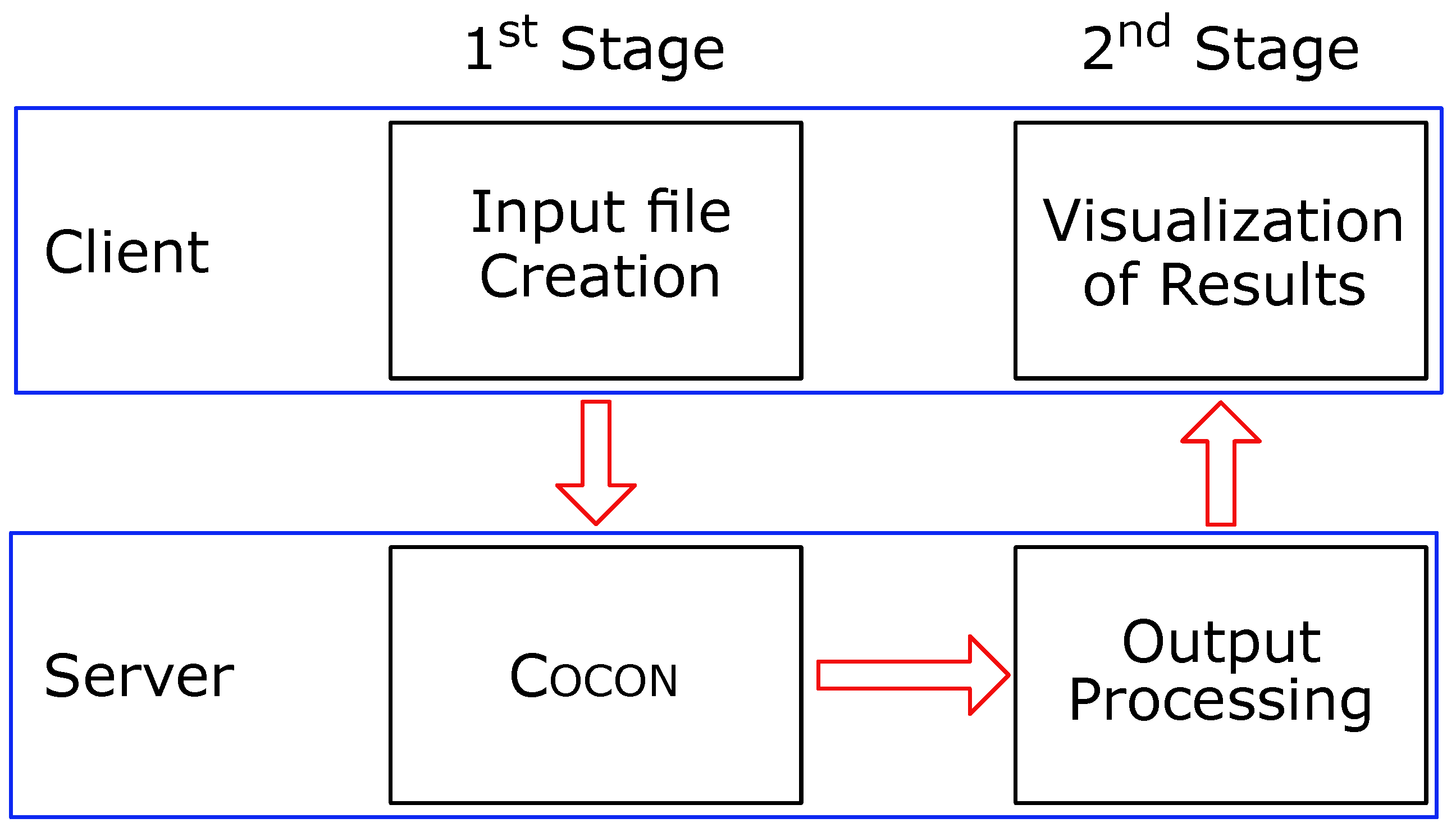
1. Introduction
2. Results
2.1. Reciprocity of Molecules and Correlation Data
2.2. Use of Correlation Data
- Allowing -HMBC correlations in the structural elucidation when there are none present in the input data increases the calculation time and possibly the number of results dramatically;
- The presence of -HMBC correlations in the input data without allowing the -HMBC interpretation during CASE makes the process fail;
- The best results are obtained when using no -HMBC correlation data or when the number of allowed -HMBC correlations in the CASE run matches the number of actually present -HMBC correlations.
2.3. Use of NOE Data in WebCocon’s Second-Stage Processing
3. Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
Abbreviations
| ADEQ | 1,1–ADEQUATE (“” equivalent) |
| CASE | Computer-Assisted Structure Elucidation [60] |
| calc. | calculated |
| COSY | H,H-Correlated Spectroscopy ( and ) |
| ⚠ | error ≫ 10 ppm |
| DFT | Density functional theory |
| DG | Distance geometry |
| exp. | experimental |
| HMBC | H,C-Heteronuclear Multiple Bond Correlation ( and ) |
| -HMBC | H,C-Heteronuclear Multiple Bond Correlation () |
| MD | Molecular Dynamics |
| NHMBC | H,N-Heteronuclear Multiple Bond Correlation ( and ) |
| NMR | Nuclear Magnetic Ressonance |
| NOE | Nuclear Overhauser Effect |
| sol. | number of solutions |
| theo. | theoretical |
References
- Ichi Sasaki, S.; Kudo, Y.; Ochiai, S.; Abe, H. Automated chemical structure analysis of organic compounds: An attempt to structure determination by the use of NMR. Mikrochim. Acta 1971, 59, 726–742. [Google Scholar] [CrossRef]
- Yamasaki, T.; Abe, H.; Kudo, Y.; Sasaki, S.I. CHEMICS: A Computer Program System for Structure Elucidation of Organic Compounds. In Computer-Assisted Structure Elucidation; American Chemical Society: Washington, DC, USA, 1977; Chapter 8; pp. 108–125. [Google Scholar] [CrossRef]
- Sasaki, S.I.; Abe, H.; Hirota, Y.; Ishida, Y.; Kudo, Y.; Ochiai, S.; Saito, K.; Yamasaki, T. CHEMICS-F: A Computer Program System for Structure Elucidation of Organic Compounds. J. Chem. Inf. Comput. Sci. 1978, 18, 211–222. [Google Scholar] [CrossRef]
- Funatsu, K.; Sasaki, S.I. Recent advances in the automated structure elucidation system, CHEMICS. Utilization of two-dimensional NMR spectral information and development of peripheral functions for examination of candidates. J. Chem. Inf. Comput. Sci. 1996, 36, 190–204. [Google Scholar] [CrossRef]
- Zlatina, L.A.; Elyashberg, M.E. Generation and pepresentation of stereoisomers of a molecular structure. J. Struct. Chem. 1992, 32, 528–533. [Google Scholar] [CrossRef]
- Pesek, M.; Juvan, A.; Jakoš, J.; Košmrlj, J.; Marolt, M.; Gazvoda, M. Database Independent Automated Structure Elucidation of Organic Molecules Based on IR, 1H NMR, 13C NMR, and MS Data. J. Chem. Inf. Model. 2021, 61, 756–763. [Google Scholar] [CrossRef]
- Gribov, L.A.; Elyashberg, M.E.; Raikhshtat, M.M. A new Approch to the Determination of Molecular Spatial Structures based on the use of Spectra and Computers. J. Mol. Struct. 1979, 53, 81–96. [Google Scholar] [CrossRef]
- Peng, C.; Yuan, S.; Zheng, C.; Hui, Y.; Wu, H.; Ma, K.; Han, X. Application of expert system CISOC-SES to the structure elucidation of complex natural products. J. Chem. Inf. Comput. Sci. 1993, 33, 814–819. [Google Scholar] [CrossRef]
- Elyashberg, M.E.; Blinov, K.A.; Williams, A.J.; Molodtsov, S.G.; Martin, G.E.; Martirosian, E.R. Structure elucidator: A versatile expert system for molecular structure elucidation from 1D and 2D NMR data and molecular fragments. J. Chem. Inf. Comput. Sci. 2004, 44, 771–792. [Google Scholar] [CrossRef]
- Christie, B.D.; Munk, M.E. Structure Generation by Reduction: A New Strategy for Computer-Assisted Structure Elucidation. J. Chem. Inf. Comput. Sci. 1988, 28, 87–93. [Google Scholar] [CrossRef]
- Nuzillard, J.M.; Georges, M. Logic for structure determination. Tetrahedron 1991, 47, 3655–3664. [Google Scholar] [CrossRef]
- Faulon, J.L. Stochastic Generator of Chemical Structure. 1. Application to the Structure Elucidation of Large Molecules. J. Chem. Inf. Comput. Sci. 1994, 34, 1204–1218. [Google Scholar] [CrossRef]
- Benecke, C.; Grund, R.; Hohberger, R.; Kerber, A.; Laue, R.; Wieland, T. MOLGEN+, a generator of connectivity isomers and stereoisomers for molecular structure elucidation. Anal. Chim. Acta 1995, 314, 141–147. [Google Scholar] [CrossRef]
- Benecke, C.; Grüner, T.; Kerber, A.; Laue, R.; Wieland, T. Molecular structure generation with MOLGEN, new features and future developments. Fresenius J. Anal. Chem. 1997, 359, 23–32. [Google Scholar] [CrossRef]
- Meringer, M.; Schymanski, E.L. Small molecule identification with MOLGEN and mass spectrometry. Metabolites 2013, 3, 440–462. [Google Scholar] [CrossRef] [PubMed]
- Gugisch, R.; Kerber, A.; Kohnert, A.; Laue, R.; Meringer, M.; Rücker, C.; Wassermann, A. MOLGEN 5.0, A Molecular Structure Generator. In Advances in Mathematical Chemistry and Applications: Revised Edition; Bentham Science Publishers: Sharjah, United Arab Emirates, 2015; Volume 1, Chapter 6; pp. 113–138. [Google Scholar] [CrossRef][Green Version]
- Kerber, A. MOLGEN, a generator for structural formulas. Match 2018, 80, 733–744. [Google Scholar]
- Will, M.; Fachinger, W.; Richert, J.R. Fully automated structure elucidation—A spectroscopist’s dream comes true. J. Chem. Inf. Comput. Sci. 1996, 36, 221–227. [Google Scholar] [CrossRef]
- Neudert, R.; Penk, M. Enhanced structure elucidation. J. Chem. Inf. Comput. Sci. 1996, 36, 244–248. [Google Scholar] [CrossRef]
- Lindel, T.; Junker, J.; Köck, M. Cocon: From NMR correlation data to molecular constitutions. J. Mol. Model. 1997, 3, 364–368. [Google Scholar] [CrossRef]
- Badertscher, M.; Korytko, A.; Schulz, K.P.; Madison, M.; Munk, M.E.; Portmann, P.; Junghans, M.; Fontana, P.; Pretsch, E. Assemble 2.0: A structure generator. Chemom. Intell. Lab. Syst. 2000, 51, 73–79. [Google Scholar] [CrossRef]
- Meiler, J.; Will, M. Automated Structure Elucidation of Organic Molecules from 13C NMR Spectra Using Genetic Algorithms and Neural Networks. J. Chem. Inf. Comput. Sci. 2001, 41, 1535–1546. [Google Scholar] [CrossRef]
- Meiler, J.; Will, M. Genius: A genetic algorithm for automated structure elucidation from 13C NMR spectra. J. Am. Chem. Soc. 2002, 124, 1868–1870. [Google Scholar] [CrossRef] [PubMed]
- Steinbeck, C. SENECA: A Platform-Independent, Distributed, and Parallel System for Computer-Assisted Structure Elucidation in Organic Chemistry. J. Chem. Inf. Comput. Sci. 2001, 41, 1500–1507. [Google Scholar] [CrossRef] [PubMed]
- Korytko, A.; Schulz, K.P.; Madison, M.S.; Munk, M.E. HOUDINI: A New Approach to Computer-Based Structure Generation. J. Chem. Inf. Comput. Sci. 2003, 43, 1434–1446. [Google Scholar] [CrossRef] [PubMed]
- Schulz, K.P.; Korytko, A.; Munk, M.E. Applications of a HOUDINI-Based Structure Elucidation System. J. Chem. Inf. Comput. Sci. 2003, 43, 1447–1456. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Steinbeck, C. Evolutionary-algorithm-based strategy for computer-assisted structure elucidation. J. Chem. Inf. Comput. Sci. 2004, 44, 489–498. [Google Scholar] [CrossRef]
- Elyashberg, M.E.; Blinov, K.A.; Molodtsov, S.G.; Williams, A.J.; Martin, G.E. Fuzzy structure generation: A new efficient tool for Computer-Aided Structure Elucidation (CASE). J. Chem. Inf. Model. 2007, 47, 1053–1066. [Google Scholar] [CrossRef]
- Elyashberg, M.; Blinov, K.; Williams, A. A systematic approach for the generation and verification of structural hypotheses. Magn. Reson. Chem. 2009, 47, 371–389. [Google Scholar] [CrossRef]
- Nicolaou, K.C.; Snyder, S.A. Chasing molecules that were never there: Misassigned natural products and the role of chemical synthesis in modern structure elucidation. Angew. Chem. Int. Ed. 2005, 44, 1012–1044. [Google Scholar] [CrossRef] [PubMed]
- Elyashberg, M.; Williams, A.J.; Blinov, K. Structural revisions of natural products by Computer-Assisted Structure Elucidation (CASE) systems. Nat. Prod. Rep. 2010, 27, 1296–1328. [Google Scholar] [CrossRef]
- Junker, J. Theoretical NMR correlations based structure discussion. J. Cheminform. 2011, 3, 27. [Google Scholar] [CrossRef]
- Elyashberg, M.; Blinov, K.; Molodtsov, S.; Williams, A. Elucidating ’undecipherable’ chemical structures using computer-assisted structure elucidation approaches. Magn. Reson. Chem. 2012, 50, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Marcarino, M.O.; Zanardi, M.M.; Sarotti, A.M. The Risks of Automation: A Study on DFT Energy Miscalculations and Its Consequences in NMR-based Structural Elucidation. Org. Lett. 2020, 22, 3561–3565. [Google Scholar] [CrossRef] [PubMed]
- Köck, M.; Junker, J.; Lindel, T. Impact of the 1H,15N-HMBC experiment on the constitutional analysis of alkaloids. Org. Lett. 1999, 1, 2041–2044. [Google Scholar] [CrossRef]
- Köck, M.; Junker, J.; Maier, W.; Will, M.; Lindel, T. A Cocon analysis of proton-poor heterocycles—Application of carbon chemical shift predictions for the evaluation of structural proposals. Eur. J. Org. Chem. 1999, 3, 579–586. [Google Scholar] [CrossRef]
- Junker, J.; Maier, W.; Lindel, T.; Köck, M. Computer-assisted constitutional assignment of large molecules: Cocon analysis of Ascomycin. Org. Lett. 1999, 1, 737–740. [Google Scholar] [CrossRef] [PubMed]
- Lindel, T.; Junker, J.; Köck, M. 2D-NMR-Guided Constitutional Analysis of Organic Compounds Employing the Computer Program Cocon. Eur. J. Org. Chem. 1999, 3, 573–577. [Google Scholar] [CrossRef]
- Martin, G.E.; Hadden, C.E. Long-Range 1H–15N Heteronuclear Shift Correlation at Natural Abundance. J. Nat. Prod. 2000, 63, 543–585. [Google Scholar] [CrossRef] [PubMed]
- Reif, B.; Köck, M.; Kerssebaum, R.; Kang, H.; Fenical, W.; Griesinger, C. ADEQUATE, a New Set of Experiments to Determine the Constitution of Small Molecules at Natural Abundance. J. Magn. Reson. Ser. A 1996, 118, 282–285. [Google Scholar] [CrossRef]
- Blinov, K.A.; Buevich, A.V.; Williamson, R.T.; Martin, G.E. The impact of LR-HSQMBC very long-range heteronuclear correlation data on computer-assisted structure elucidation. Org. Biomol. Chem. 2014, 12, 9505–9509. [Google Scholar] [CrossRef]
- Junker, J. Statistical filtering for NMR based structure generation. J. Cheminform. 2011, 3, 31. [Google Scholar] [CrossRef]
- Gilbert, K.; Guha, R. Simple 3D Conformer Generation with Smi23D. Depth-First, 12 December 2007. [Google Scholar]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Weininger, D. SMILES, a Chemical Language and Information System: 1: Introduction to Methodology and Encoding Rules. J. Chem. Inf. Model. 1988, 28, 31–36. [Google Scholar] [CrossRef]
- Weininger, D.; Weininger, A.; Weininger, J.L. SMILES. 2. Algorithm for Generation of Unique SMILES Notation. J. Chem. Inf. Model. 1989, 29, 97–101. [Google Scholar] [CrossRef]
- Pirhadi, S.; Sunseri, J.; Koes, D.R. Open source molecular modeling. J. Mol. Graph. Model. 2016, 69, 127–143. [Google Scholar] [CrossRef] [PubMed]
- Wikipedia. Category: Molecular Dynamics Software. Available online: https://en.wikipedia.org/wiki/Category:Molecular_dynamics_software (accessed on 1 April 2021).
- Wikipedia. Comparison of Software for Molecular Mechanics Modeling. Available online: https://en.wikipedia.org/wiki/Comparison_of_software_for_molecular_mechanics_modeling (accessed on 1 April 2021).
- Rackers, J.A.; Wang, Z.; Lu, C.; Laury, M.L.; Lagardère, L.; Schnieders, M.J.; Piquemal, J.P.; Ren, P.; Ponder, J.W. Tinker 8: Software Tools for Molecular Design. J. Chem. Theory Comput. 2018, 14, 5273–5289. [Google Scholar] [CrossRef] [PubMed]
- Grube, A.; Köck, M. Oxocyclostylidol, an intramolecular cyclized oroidin derivative from the marine sponge Stylissa caribica. J. Nat. Prod. 2006, 69, 1212–1214. [Google Scholar] [CrossRef]
- Al-Khdhairawi, A.A.Q.; Low, Y.Y.; Manshoor, N.; Arya, A.; Jelecki, M.; Alshawsh, M.A.; Kamran, S.; Suliman, R.S.; Low, A.; Shivanagere Nagojappa, N.B.; et al. Asperginols A and B, Diterpene Pyrones, from an Aspergillus sp. And the Structure Revision of Previously Reported Analogues. J. Nat. Prod. 2020, 83, 3564–3570. [Google Scholar] [CrossRef]
- Procházková, E.; Čechová, L.; Jansa, P.; Dračínský, M. Long-range heteronuclear coupling constants in 2,6-disubstituted purine derivatives. Magn. Reson. Chem. 2012, 50, 295–298. [Google Scholar] [CrossRef] [PubMed]
- Steinbeck, C.; Krause, S.; Kuhn, S. NMRShiftDB—Constructing a Free Chemical Information System with Open-Source Components. J. Chem. Inf. Comput. Sci. 2003, 43, 1733–1739. [Google Scholar] [CrossRef] [PubMed]
- Barca, G.M.J.; Bertoni, C.; Carrington, L.; Datta, D.; De Silva, N.; Deustua, J.E.; Fedorov, D.G.; Gour, J.R.; Gunina, A.O.; Guidez, E.; et al. Recent developments in the general atomic and molecular electronic structure system. J. Chem. Phys. 2020, 152, 154102. [Google Scholar] [CrossRef] [PubMed]
- Modgraph Consultants Ltd. NMRPredict v4.7.41. Available online: http://www.modgraph.co.uk/ (accessed on 1 April 2021).
- Pupier, M.; Nuzillard, J.M.; Wist, J.; Schlörer, N.E.; Kuhn, S.; Erdelyi, M.; Steinbeck, C.; Williams, A.J.; Butts, C.; Claridge, T.D.; et al. NMReDATA, a standard to report the NMR assignment and parameters of organic compounds. Magn. Reson. Chem. 2018, 56, 703–715. [Google Scholar] [CrossRef] [PubMed]
- Trevorrow, P.; Jeannerat, D. Reporting on the first NMReDATA Symposium, Porto, Portugal. Magn. Reson. Chem. 2020, 58, 218–222. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, S.; Wieske, L.H.E.; Trevorrow, P.; Schober, D.; Schlörer, N.E.; Nuzillard, J.; Kessler, P.; Junker, J.; Herráez, A.; Farès, C.; et al. NMReDATA: Tools and applications. Magn. Reson. Chem. 2021. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.H. Computer-Assisted Structure Elucidation; American Chemical Society Symposium Series; American Chemical Society: Washington, DC, USA, 1977. [Google Scholar] [CrossRef][Green Version]
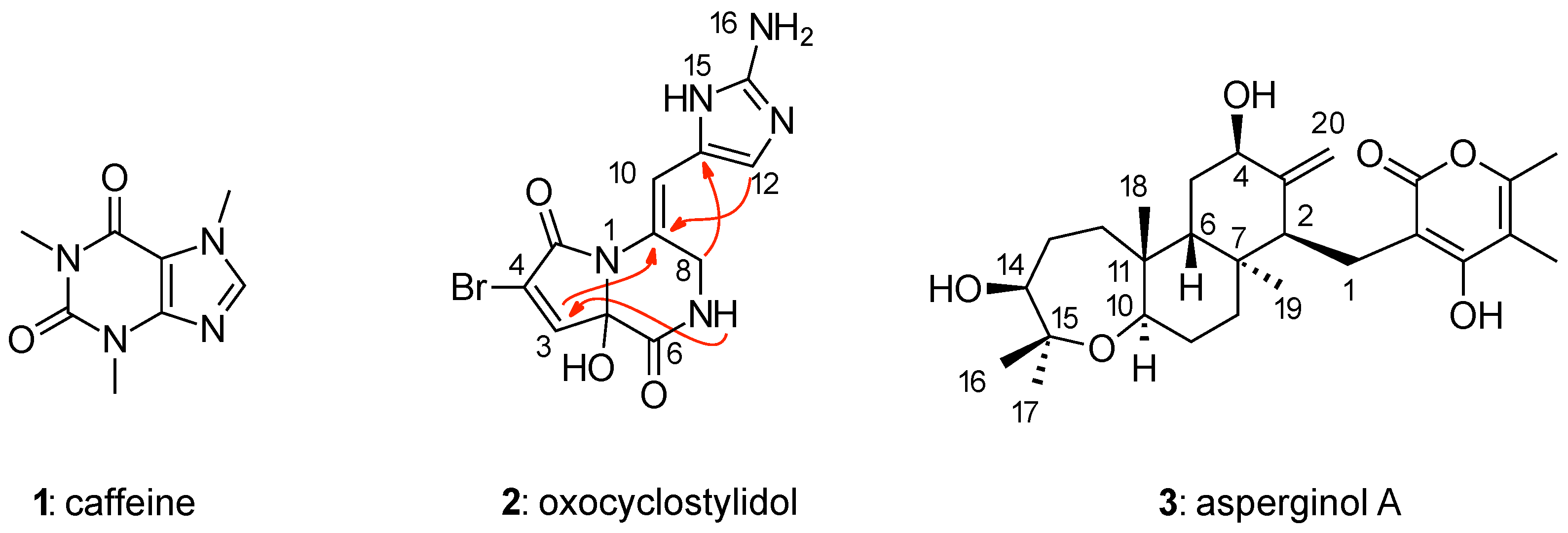


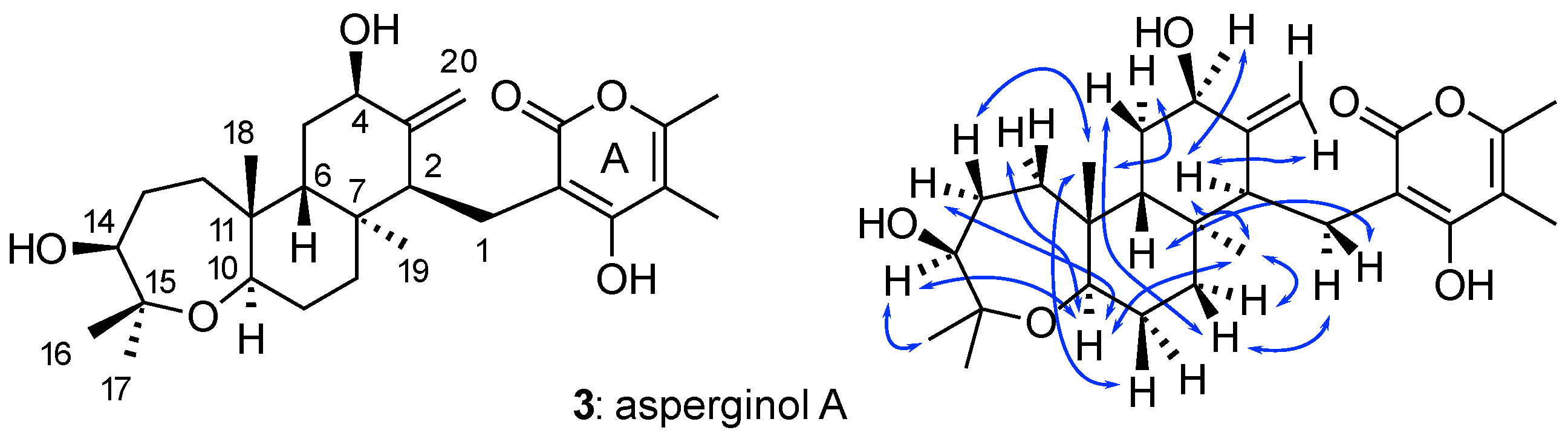
| Data | COSY | HMBC | -HMBC | ADEQ | NHMBC | NOE | |
|---|---|---|---|---|---|---|---|
| Caffeine (1) | theo. a | – | 8 | – | – | 5 | – |
| Oxocyclostylidol (2) | exp. | 1 | 25 | 4 | 6 | 9 | – |
| Asperginol A (3) | exp. | 18 | 38 | – | – | – | 15 |
| 1-1 | 1-2 | ||||||
|---|---|---|---|---|---|---|---|
| Atom | exp. | M-Ia | M-IIb | M-IIIc | M-Ia⚠ | M-IIb | M-IIIc |
| 2 | 148.5 | 150.7 | 149.5 | 151.4 | 151.1 | 152.4 | 150.8 |
| 4 | 151.5 | 149.0 | 153.3 | 147.0 | 157.4 | 152.0 | 149.7 |
| 5 | 107.4 | 107.3 | 104.5 | 111.5 | |||
| 6 | 155.2 | 155.3 | 154.7 | 154.3 | 149.2 | 149.0 | 149.4 |
| 7 | 60.1 | 115.9 | 117.1 | ||||
| 8 | 141.4 | 143.0 | 145.8 | 147.4 | 50.8 | 122.3 | 128.2 |
| 10 | 27.8 | 28.8 | 25.7 | 29.5 | 37.3 | 27.1 | 31.3 |
| 11 | 29.6 | 28.7 | 26.7 | 27.7 | 37.3 | 27.2 | 29.3 |
| 12 | 33.5 | 33.4 | 26.9 | 33.7 | 15.4 | 7.9 | 11.6 |
| 1.06 | 2.78 | 2.78 | 23.46 | 8.25 | 7.31 | ||
| Input | -HMBC | Cocon | |||||
|---|---|---|---|---|---|---|---|
| Data Set | 4J-Flag | H-3/C-9 | H-7/C-3 | H-8/C-11 | H-12/C-9 | sol. | Run Time [s] |
| A | 0 | - | - | - | - | 4 | 1 |
| 1 | - | - | - | - | 18 | 30 | |
| 2 | - | - | - | - | 107 | 42 | |
| 3 | - | - | - | - | 329 | 76 | |
| 4 | - | - | - | - | 889 | 153 | |
| −1 | - | - | - | - | 6045 | 907 | |
| B | 0 | X | - | - | - | 0 | 0 |
| 1 | X | - | - | - | 4 | 17 | |
| 2 | X | - | - | - | 19 | 20 | |
| 3 | X | - | - | - | 116 | 33 | |
| 4 | X | - | - | - | 330 | 66 | |
| −1 | X | - | - | - | 3974 | 525 | |
| C | 0 | - | X | - | - | 0 | 0 |
| 1 | - | X | - | - | 6 | 23 | |
| 2 | - | X | - | - | 32 | 27 | |
| 3 | - | X | - | - | 167 | 46 | |
| 4 | - | X | - | - | 529 | 98 | |
| −1 | - | X | - | - | 4664 | 592 | |
| D | 0 | - | - | X | - | 0 | 0 |
| 1 | - | - | X | - | 4 | 27 | |
| 2 | - | - | X | - | 18 | 30 | |
| 3 | - | - | X | - | 107 | 42 | |
| 4 | - | - | X | - | 329 | 74 | |
| −1 | - | - | X | - | 6045 | 788 | |
| E | 0 | - | - | - | X | 0 | 0 |
| 1 | - | - | - | X | 4 | 28 | |
| 2 | - | - | - | X | 18 | 31 | |
| 3 | - | - | - | X | 108 | 43 | |
| 4 | - | - | - | X | 346 | 79 | |
| −1 | - | - | - | X | 6045 | 791 | |
| F | 0 | X | X | - | - | 0 | 0 |
| 1 | X | X | - | - | 0 | 13 | |
| 2 | X | X | - | - | 6 | 14 | |
| 3 | X | X | - | - | 31 | 19 | |
| 4 | X | X | - | - | 172 | 39 | |
| −1 | X | X | - | - | 2910 | 402 | |
| G | 0 | X | X | X | - | 0 | 0 |
| 1 | X | X | X | - | 0 | 14 | |
| 2 | X | X | X | - | 0 | 14 | |
| 3 | X | X | X | - | 6 | 15 | |
| 4 | X | X | X | - | 31 | 18 | |
| −1 | X | X | X | - | 2910 | 400 | |
| H | 0 | X | X | X | X | 0 | 0 |
| 1 | X | X | X | X | 0 | 14 | |
| 2 | X | X | X | X | 0 | 14 | |
| 3 | X | X | X | X | 0 | 14 | |
| 4 | X | X | X | X | 6 | 14 | |
| −1 | X | X | X | X | 2910 | 401 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Köck, M.; Lindel, T.; Junker, J. Incorporation of 4J-HMBC and NOE Data into Computer-Assisted Structure Elucidation with WebCocon. Molecules 2021, 26, 4846. https://doi.org/10.3390/molecules26164846
Köck M, Lindel T, Junker J. Incorporation of 4J-HMBC and NOE Data into Computer-Assisted Structure Elucidation with WebCocon. Molecules. 2021; 26(16):4846. https://doi.org/10.3390/molecules26164846
Chicago/Turabian StyleKöck, Matthias, Thomas Lindel, and Jochen Junker. 2021. "Incorporation of 4J-HMBC and NOE Data into Computer-Assisted Structure Elucidation with WebCocon" Molecules 26, no. 16: 4846. https://doi.org/10.3390/molecules26164846
APA StyleKöck, M., Lindel, T., & Junker, J. (2021). Incorporation of 4J-HMBC and NOE Data into Computer-Assisted Structure Elucidation with WebCocon. Molecules, 26(16), 4846. https://doi.org/10.3390/molecules26164846






