The Genus Cladosporium: A Rich Source of Diverse and Bioactive Natural Compounds
Abstract
1. Introduction
2. Fifty Years of Metabolomic Studies in Cladosporium
2.1. Alkaloids
2.2. Azaphilones
2.3. Benzofluorantheneones
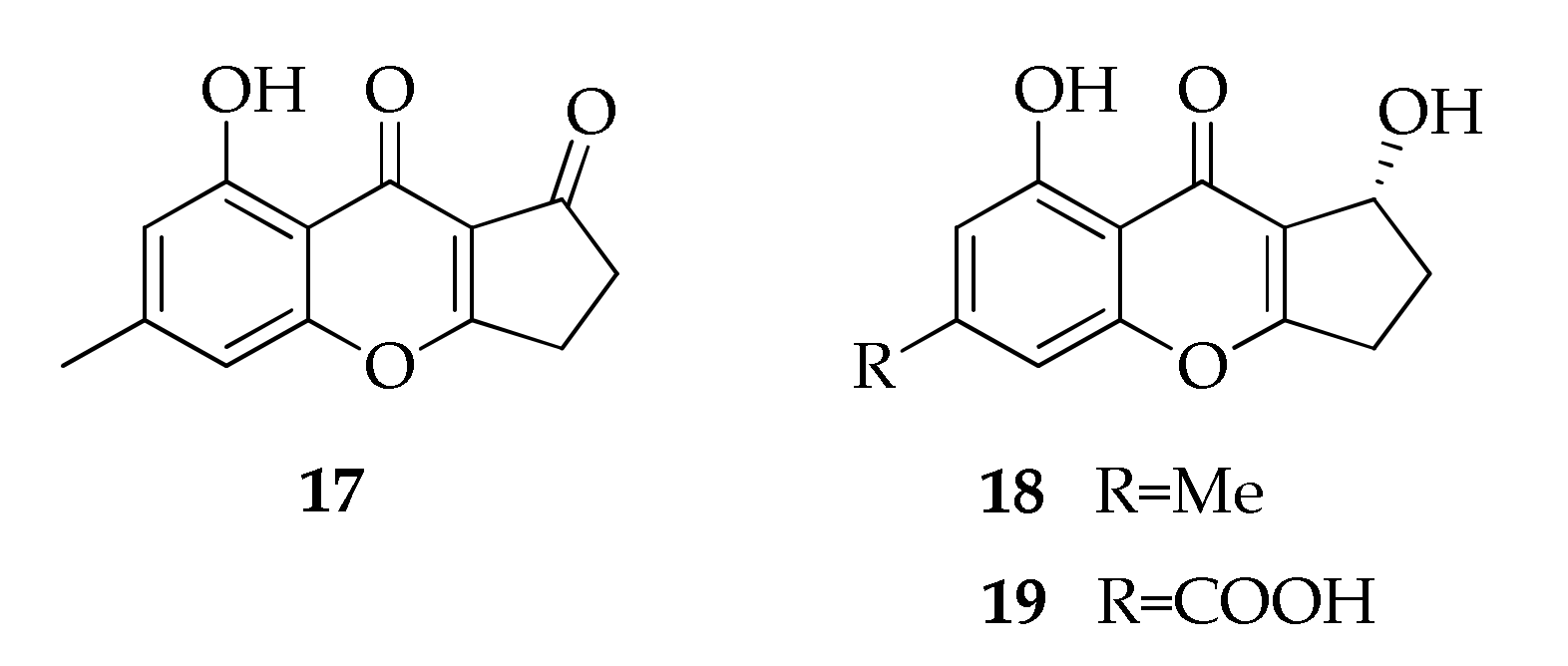
2.4. Benzopyrones
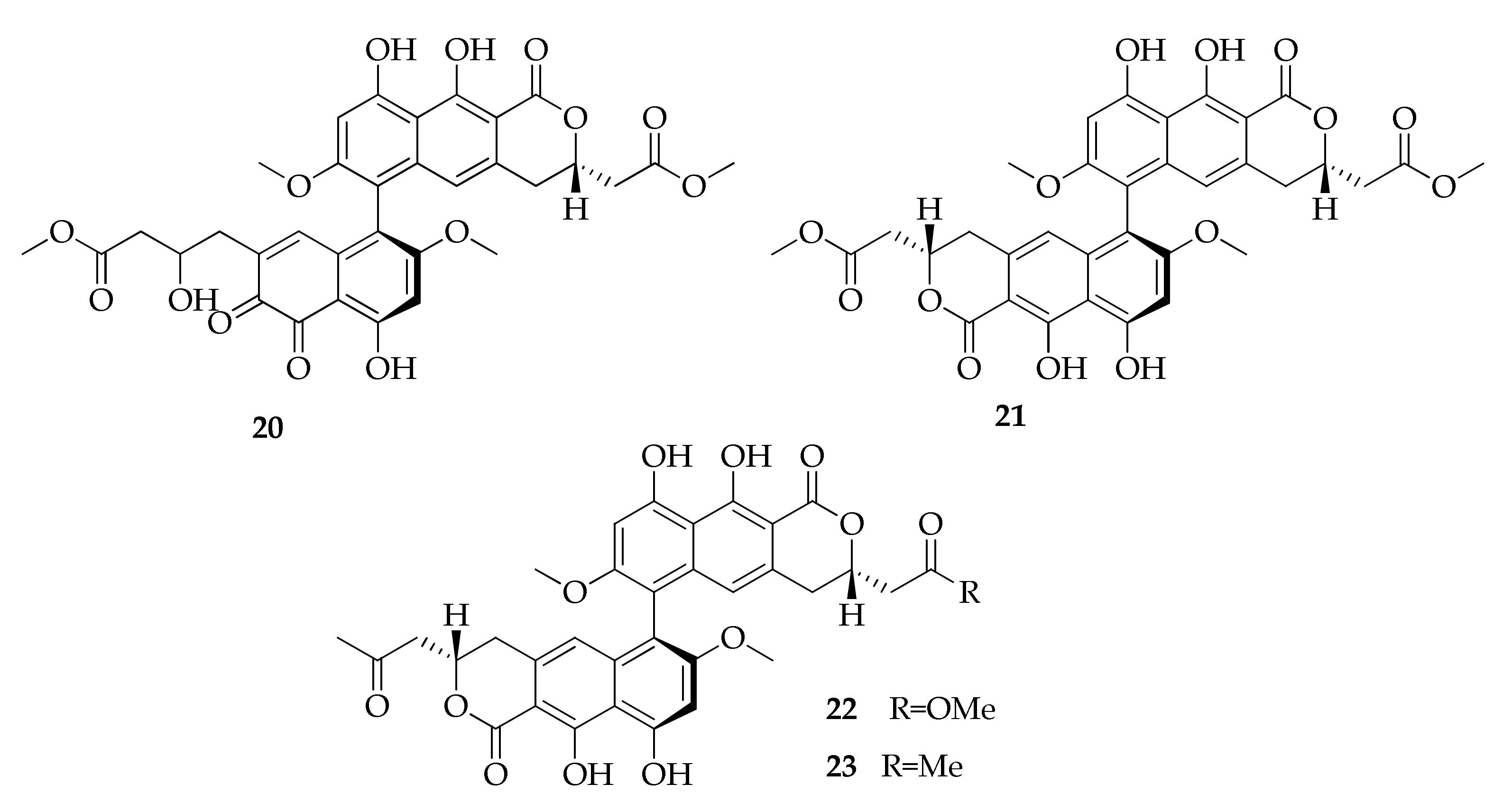
2.5. Binaphtopyrones
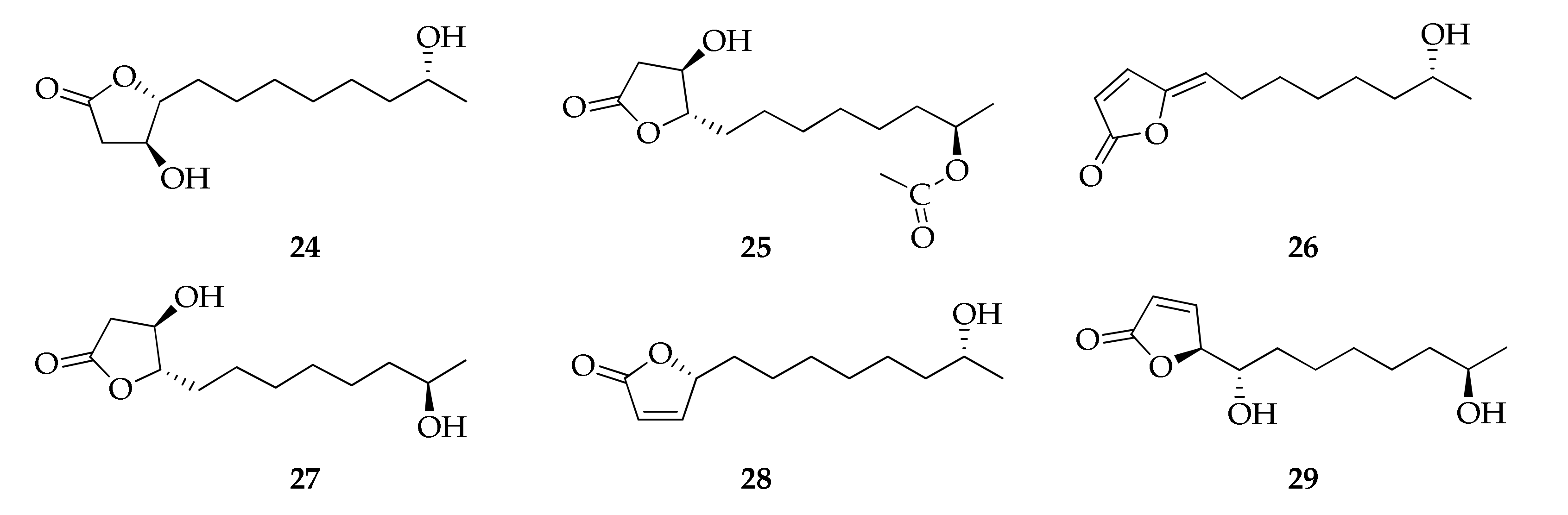
2.6. Butanolides and Butenolides
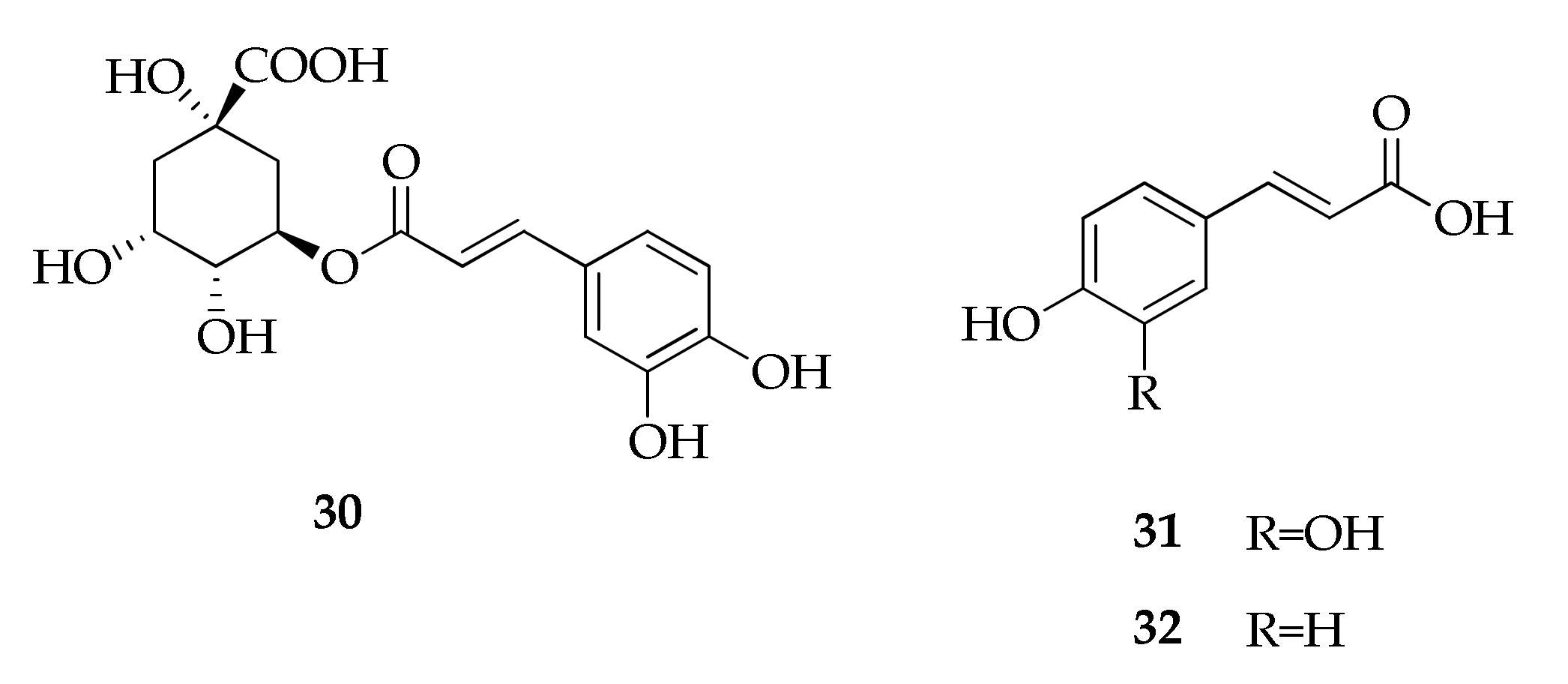
2.7. Cinnamic acid Derivatives
2.8. Citrinin Derivatives
2.9. Coumarins and Isocoumarins
2.10. Cyclohexene Derivatives
2.11. Depsides
2.12. Diketopiperazines
2.13. Flavonoids
2.14. Gibberellins
2.15. Fusicoccane Diterpene Glycosides
2.16. Lactones
2.17. Macrolides
2.18. Naphthalene Derivatives
2.19. Naphthalenones
2.20. Naphtoquinones and Anthraquinones
2.21. Perylenquinones
2.22. Pyrones
2.23. Seco Acids
2.24. Sterols
2.25. Tetramic Acids
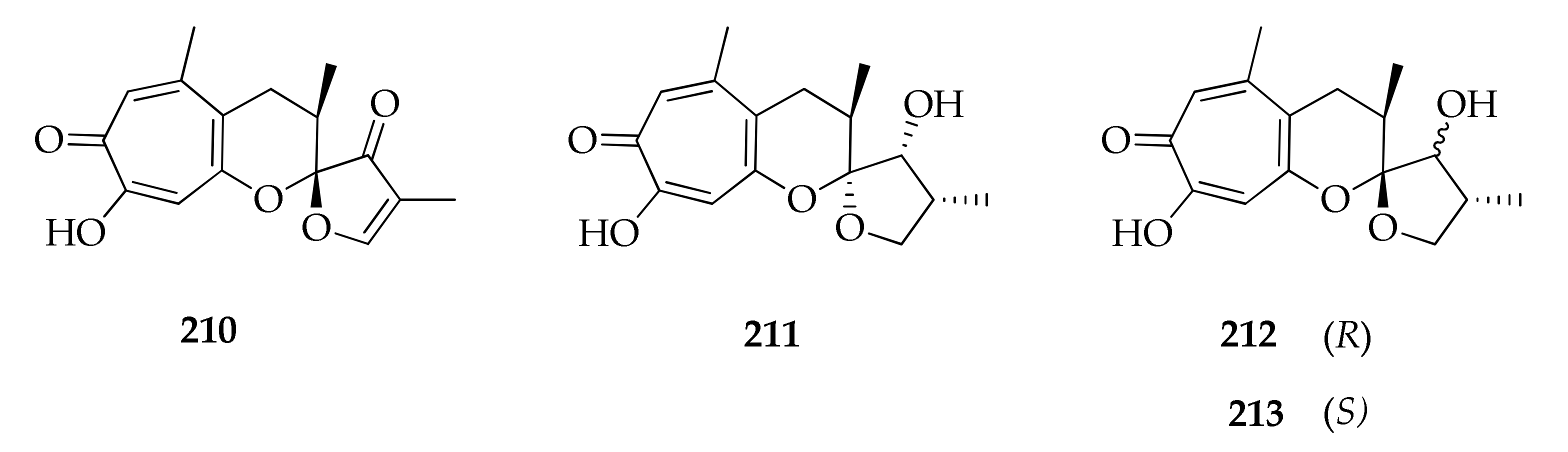
2.26. Tropolones
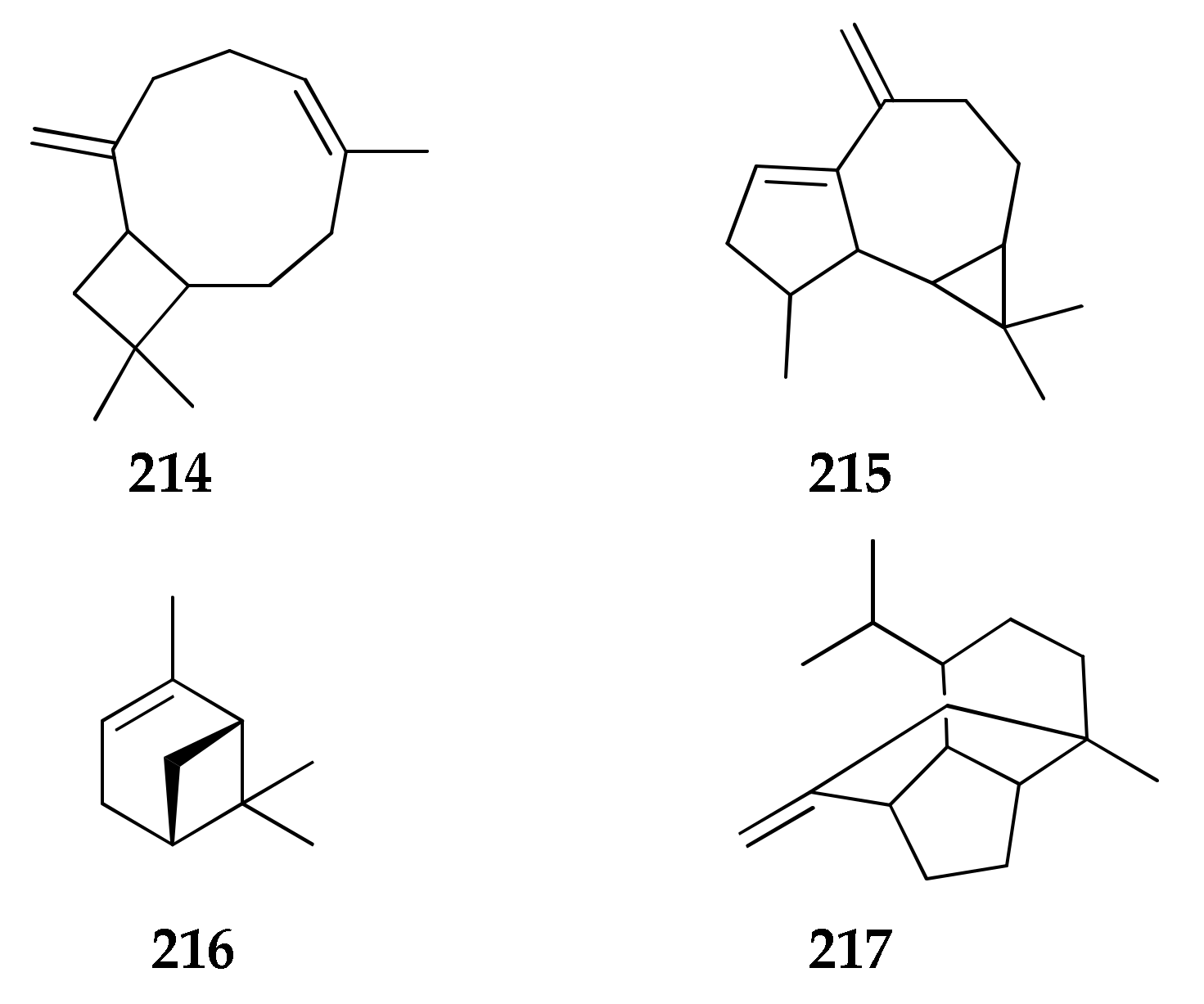
2.27. Volatile Terpenes
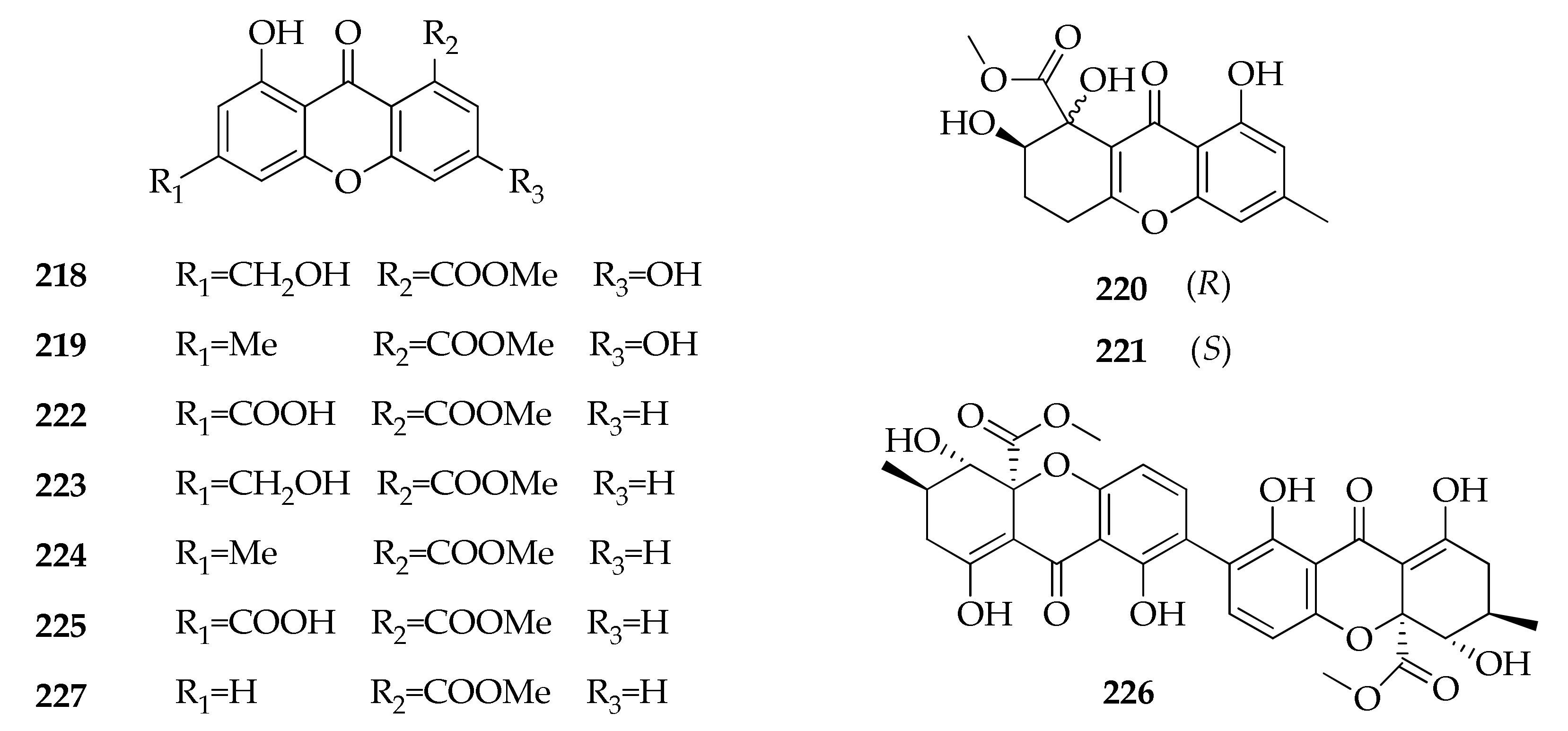
2.28. Xanthones
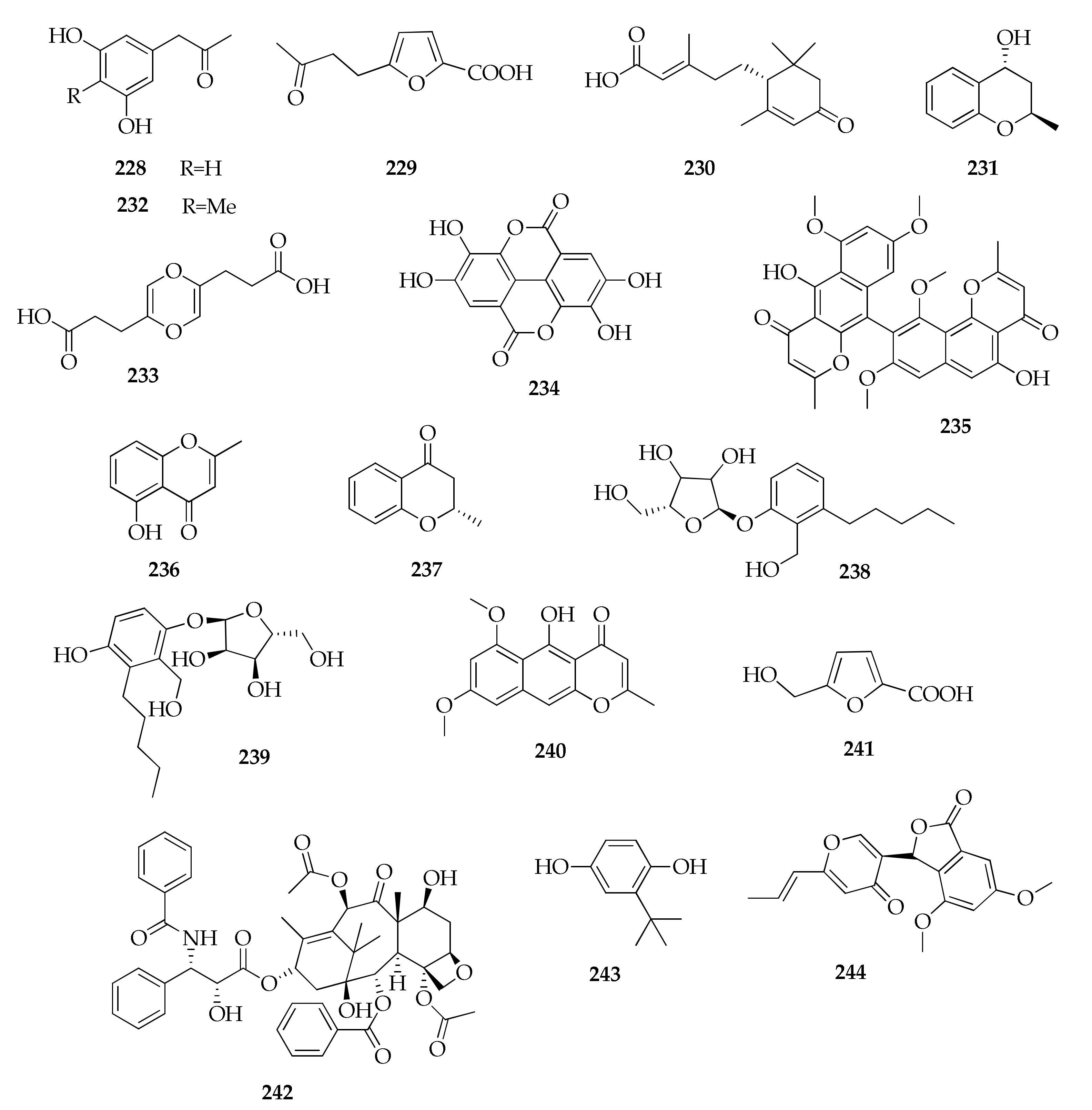
2.29. Miscellaneous
3. Biological Activities of Secondary Metabolites
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bensch, K.; Groenewald, J.Z.; Braun, U.; Dijksterhuis, J.; de Jesús Yáñez-Morales, M.; Crous, P.W. Common but different: The expanding realm of Cladosporium. Stud. Mycol. 2015, 82, 23–74. [Google Scholar] [CrossRef] [PubMed]
- Segers, F.J.J.; Meijer, M.; Houbraken, J.; Samson, R.A.; Wösten, H.A.B.; Dijksterhuis, J. Xerotolerant Cladosporium sphaerospermum are predominant on indoor surfaces compared to other Cladosporium species. PLoS ONE 2015, 10, e0145415. [Google Scholar] [CrossRef]
- Bensch, K.; Groenewald, J.Z.; Meijer, M.; Dijksterhuis, J.; Jurjević, Ž.; Andersen, B.; Houbraken, J.; Crous, P.W.; Samson, R.A. Cladosporium species in indoor environments. Stud. Mycol. 2018, 89, 177–301. [Google Scholar] [CrossRef] [PubMed]
- Yew, S.M.; Chan, C.L.; Ngeow, Y.F.; Toh, Y.F.; Na, S.L.; Lee, K.W.; Hoh, C.C.; Yee, W.Y.; Ng, K.P.; Kuan, C.S. Insight into different environmental niches adaptation and allergenicity from the Cladosporium sphaerospermum genome, a common human allergy-eliciting Dothideomycetes. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef]
- Cheng, S.C.H.; Lin, Y.Y.; Kuo, C.N.; Lai, L.J. Cladosporium keratitis—A case report and literature review. BMC Ophthalmol. 2015, 15, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Sandoval-Denis, M.; Gené, J.; Sutton, D.A.; Wiederhold, N.P.; Cano-Lira, J.F.; Guarro, J. New species of Cladosporium associated with human and animal infections. Persoonia Mol. Phylogeny Evol. Fungi 2016, 36, 281–298. [Google Scholar] [CrossRef] [PubMed]
- Manos, P.S.; Zhou, Z.K.; Cannon, C.H. Systematics of Fagaceae: Phylogenetic tests of reproductive trait evolution. Int. J. Plant Sci. 2016, 162, 1361–1379. [Google Scholar] [CrossRef]
- Batra, N.; Kaur, H.; Mohindra, S.; Singh, S.; Shamanth, A.S.; Rudramurthy, S.M. Cladosporium sphaerospermum causing brain abscess, a saprophyte turning pathogen: Case and review of published reports. J. Mycol. Med. 2019, 29, 180–184. [Google Scholar] [CrossRef]
- Rosado, A.W.C.; Custódio, F.A.; Pinho, D.B.; Ferreira, A.P.S.; Pereira, O.L. Cladosporium species associated with disease symptoms on Passiflora edulis and other crops in Brazil, with descriptions of two new species. Phytotaxa 2019, 409, 239–260. [Google Scholar] [CrossRef]
- Iturrieta-González, I.; García, D.; Gené, J. Novel species of Cladosporium from environmental sources in Spain. MycoKeys 2021, 77, 1–25. [Google Scholar] [CrossRef]
- Zimowska, B.; Becchimanzi, A.; Krol, E.D.; Furmanczyk, A.; Bensch, K.; Nicoletti, R. New Cladosporium species from normal and galled flowers of Lamiaceae. Pathogens 2021, 10, 369. [Google Scholar] [CrossRef]
- Scott, P.M.; Van Walbeek, W.; MacLean, W.M. Cladosporin, a new antifungal metabolite from Cladosporium cladosporioides. J. Antibiot. 1971, 24, 747–755. [Google Scholar] [CrossRef]
- Xiong, H.; Qi, S.; Xu, Y.; Miao, L.; Qian, P.Y. Antibiotic and antifouling compound production by the marine-derived fungus Cladosporium sp. F14. J. Hydro-Environ. Res. 2009, 2, 264–270. [Google Scholar] [CrossRef]
- Setyaningrum, A.F.; Pratiwi, R.; Nasution, N.E.; Indrayanto, G. Identification of 4-4’-(1-methylethylidene)-bisphenol from the endophytic fungus Cladosporium oxysporum derived from Aglaia odorata. Alchemy J. Penelit. Kim. 2018, 14, 193–201. [Google Scholar] [CrossRef][Green Version]
- Ding, L.; Qin, S.; Li, F.; Chi, X.; Laatsch, H. Isolation, antimicrobial activity, and metabolites of fungus Cladosporium sp. associated with red alga Porphyra yezoensis. Curr. Microbiol. 2008, 56, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Waqas, M.; Khan, A.L.; Ali, L.; Kang, S.M.; Kim, Y.H.; Lee, I.J. Seed germination-influencing bioactive secondary metabolites secreted by the endophyte Cladosporium cladosporioides LWL5. Molecules 2013, 18, 15519–15530. [Google Scholar] [CrossRef]
- Shaker, N.; Ahmed, G.; El-Sawy, M.; Ibrahim, H.; Ismail, H. Isolation, characterization and insecticidal activity of methylene chloride extract of Cladosporium cladosporioides secondary metabolites against Aphis gossypii (Glov.). J. Plant Prot. Pathol. 2019, 10, 115–119. [Google Scholar] [CrossRef]
- Shaker, N.O.; Ahmed, G.M.M.; Ibrahim, H.Y.E.-S.; El-Sawy, M.M.; Mostafa, M.E.-H.; Ismail, H.N.A.E.-R. Secondary metabolites of the entomopathogenic fungus, Cladosporium cladosporioides and its relation to toxicity of cotton aphid. Int. J. Entomol. Nematol. 2019, 5, 115–120. [Google Scholar]
- Gallo, M.L.; Seldes, A.M.; Cabrera, G.M. Antibiotic long-chain and α,β-unsaturated aldehydes from the culture of the marine fungus Cladosporium sp. Biochem. Syst. Ecol. 2004, 32, 545–551. [Google Scholar] [CrossRef]
- Sunesson, A.L.; Vaes, W.H.J.; Nilsson, C.A.; Blomquist, G.; Andersson, B.; Carlson, R. Identification of volatile metabolites from five fungal species cultivated on two media. Appl. Environ. Microbiol. 1995, 61, 2911–2918. [Google Scholar] [CrossRef]
- Hulikere, M.M.; Joshi, C.G.; Ananda, D.; Poyya, J.; Nivya, T. Antiangiogenic, wound healing and antioxidant activity of Cladosporium cladosporioides (endophytic fungus) isolated from seaweed (Sargassum wightii). Mycology 2016, 7, 203–211. [Google Scholar] [CrossRef]
- Liu, Y.; Kurt’n, T.; Wang, C.Y.; Han Lin, W.; Orfali, R.; Müller, W.E.G.; Daletos, G.; Proksch, P. Cladosporinone, a new viriditoxin derivative from the hypersaline lake derived fungus Cladosporium cladosporioides. J. Antibiot. 2016, 69, 702–706. [Google Scholar] [CrossRef]
- San-Martin, A.; Painemal, K.; Díaz, Y.; Martinez, C.; Rovirosa, J. Metabolites from the marine fungus Cladosporium cladosporioides. An. Asoc. Quim. Argent. 2005, 93, 247–251. [Google Scholar]
- Yehia, R.S.; Osman, G.H.; Assaggaf, H.; Salem, R.; Mohamed, M.S.M. Isolation of potential antimicrobial metabolites from endophytic fungus Cladosporium cladosporioides from endemic plant Zygophyllum mandavillei. S. Afr. J. Bot. 2020, 134, 296–302. [Google Scholar] [CrossRef]
- Wang, X.; Radwan, M.M.; Taráwneh, A.H.; Gao, J.; Wedge, D.E.; Rosa, L.H.; Cutler, H.G.; Cutler, S.J. Antifungal activity against plant pathogens of metabolites from the endophytic fungus Cladosporium cladosporioides. J. Agric. Food Chem. 2013, 61, 4551–4555. [Google Scholar] [CrossRef]
- Sakagami, Y.; Sano, A.; Hara, O.; Mikawa, T.; Marumo, S. Cladosporol, β-1, 3-glucan biosynthesis inhibitor, isolated from fungus, Cladosporium cladosporioides. Tetrahedron Lett. 1995, 36, 1469–1472. [Google Scholar] [CrossRef]
- Wrigley, S.K.; Ainsworth, A.M.; Kau, D.A.; Martin, S.M.; Bahl, S.; Tang, J.S.; Hardick, D.J.; Rawlins, P.; Sadheghia, R.; Moorea, M. Novel reduced benzo [j] fluoranthen-3-ones from Cladosporium cf cladosporioides properties with cytokine production and tyrosine kinase Inhibitory. J. Antibiot. 2001, 54, 479–488. [Google Scholar]
- Assante, G.; Bava, A.; Nasini, G. Enhancement of a pentacyclic tyrosine kinase inhibitor production in Cladosporium cf. cladosporioides by cladosporol. Appl. Microbiol. Biotechnol. 2006, 69, 718–721. [Google Scholar] [CrossRef]
- Kobayashi, E.; Ando, K.; Nakano, H.; Tamaoki, T. UCN-1028A, a novel and specific inhibitor of protein kinase C, from Cladosporium. J. Antibiot. 1989, 42, 153–155. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, E.; Ando, K.; Nakano, H.; Iida, T.; Tamaoki, T.; Ohno, H.; Morimoto, M. Calphostins (ucn-1028), novel and specific inhibitors of protein kinase C. I. Fermentation, isolation, physico-chemical properties and biological activities. J. Antibiot. 1989, 42, 1470–1474. [Google Scholar] [CrossRef] [PubMed]
- Arnone, A.; Assante, G.; Merlini, L.; Nasini, G. Structure and stereochemistry of cladochrome D and E, novel perylenequinone pigments from Cladosporium cladosporioides. Gazz. Chim. Ital. 1990, 21, 557–559. [Google Scholar] [CrossRef]
- Zhang, F.; Li, X.; Li, X.; Yang, S.; Meng, L. Polyketides from the mangrove-derived endophytic fungus Cladosporium cladosporioides. Mar. Drugs 2019, 17, 296. [Google Scholar] [CrossRef]
- Zhang, F.Z.; Li, X.M.; Yang, S.Q.; Meng, L.H.; Wang, B.G. Thiocladospolides A-D, 12-membered macrolides from the mangrove-derived endophytic fungus Cladosporium cladosporioides MA-299 and structure revision of pandangolide 3. J. Nat. Prod. 2019, 82, 1535–1541. [Google Scholar] [CrossRef]
- Zhang, F.Z.; Li, X.M.; Meng, L.H.; Wang, B.G. Cladocladosin A, an unusual macrolide with bicyclo 5/9 ring system, and two thiomacrolides from the marine mangrove-derived endophytic fungus, Cladosporium cladosporioides MA-299. Bioorg. Chem. 2020, 101, 103950. [Google Scholar] [CrossRef]
- He, Z.H.; Zhang, G.; Yan, Q.X.; Zou, Z.B.; Xiao, H.X.; Xie, C.L.; Tang, X.X.; Luo, L.Z.; Yang, X.W. Cladosporactone A, a unique polyketide with 7-methylisochromen-3-one skeleton from the deep-sea-derived fungus Cladosporium cladosporioides. Chem. Biodivers. 2020, 17, e2000158. [Google Scholar] [CrossRef]
- Reese, P.B.; Rawlings, B.J.; Ramer, S.E.; Vederas, J.C. Comparison of stereochemistry of fatty acid and cladosporin biosynthesis in Cladosporium cladosporioides using deuterium-decoupled proton, carbon-13 NMR shift correlation. J. Am. Chem. Soc. 1988, 110, 316–318. [Google Scholar] [CrossRef]
- Jacyno, J.M.; Harwood, J.S.; Cutler, H.G.; Lee, M.K. Isocladosporin, a biologically active isomer of cladosporin from Cladosporium cladosporioides. J. Nat. Prod. 1993, 56, 1397–1401. [Google Scholar] [CrossRef]
- Paul, D.; Park, K.S. Identification of volatiles produced by Cladosporium cladosporioides CL-1, a fungal biocontrol agent that promotes plant growth. Sensors 2013, 13, 13969–13977. [Google Scholar] [CrossRef] [PubMed]
- Li, H.L.; Li, X.M.; Mandi, A.; Antus, S.; Li, X.; Zhang, P.; Liu, Y.; Kurtan, T.; Wang, B.G. Characterization of cladosporols from the marine algal-derived endophytic fungus Cladosporium cladosporioides EN-399 and configurational revision of the previously reported cladosporol derivatives. J. Org. Chem. 2017, 82, 9946–9954. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; He, X.; Liu, C.; Che, Q.; Zhu, T.; Gu, Q.; Li, D. Clindanones A and B and cladosporols F and G, polyketides from the deep-sea derived fungus: Cladosporium cladosporioides HDN14-342. RSC Adv. 2016, 6, 76498–76504. [Google Scholar] [CrossRef]
- Liu, H.X.; Tan, H.B.; Li, S.N.; Chen, Y.C.; Li, H.H.; Qiu, S.X.; Zhang, W.M. Two new 12-membered macrolides from the endophytic fungal strain Cladosprium colocasiae A801 of Callistemon viminalis. J. Asian Nat. Prod. Res. 2019, 21, 696–701. [Google Scholar] [CrossRef]
- Venkateswarulu, N.; Shameer, S.; Bramhachari, P.V.; Basha, S.K.T.; Nagaraju, C.; Vijaya, T. Isolation and characterization of plumbagin (5- hydroxyl- 2- methylnaptalene-1,4-dione) producing endophytic fungi Cladosporium delicatulum from endemic medicinal plants: Isolation and characterization of plumbagin producing endophytic fungi from endemic. Biotechnol. Rep. 2018, 20, e00282. [Google Scholar] [CrossRef]
- Wang, C.N.; Lu, H.M.; Gao, C.H.; Guo, L.; Zhan, Z.Y.; Wang, J.J.; Liu, Y.H.; Xiang, S.T.; Wang, J.; Luo, X.W. Cytotoxic benzopyranone and xanthone derivatives from a coral symbiotic fungus Cladosporium halotolerans GXIMD 02502. Nat. Prod. Res. 2020, 1–8. [Google Scholar] [CrossRef]
- Jadulco, R.; Proksch, P.; Wray, V.; Sudarsono; Berg, A.; Gräfe, U. New macrolides and furan carboxylic acid derivative from the sponge-derived fungus Cladosporium herbarum. J. Nat. Prod. 2001, 64, 527–530. [Google Scholar] [CrossRef] [PubMed]
- Jadulco, R.; Brauers, G.; Edrada, R.A.; Ebel, R.; Wray, V.; Sudarsono; Proksch, P. New metabolites from sponge-derived fungi Curvularia lunata and Cladosporium herbarum. J. Nat. Prod. 2002, 65, 730–733. [Google Scholar] [CrossRef]
- Robeson, D.J.; Jalal, M.A. Formation of Ent-isophleichrome by Cladosporium herbarum isolated from sugar beet. Chem. Pharm. Bull. 1992, 56, 949–952. [Google Scholar]
- Ye, Y.H.; Zhu, H.L.; Song, Y.C.; Liu, J.Y.; Tan, R.X. Structural revision of aspernigrin A, reisolated from Cladosporium herbarum IFB-E002. J. Nat. Prod. 2005, 68, 1106–1108. [Google Scholar] [CrossRef]
- Hartanti, D.; Purwanti, D.I.; Putro, H.S.; Rateb, M.E.; Wongso, S.; Sugijanto, N.E.; Ebel, R.; Indrayanto, G. Isolation of pandangolide 1 from Cladosporium oxysporum, an endophyte of the terrestrial plant Alyxia reinwardtii. Makara J. Sci. 2015, 18, 123–126. [Google Scholar] [CrossRef]
- Lu, Y.H.; Li, S.; Shao, M.W.; Xiao, X.H.; Kong, L.C.; Jiang, D.H.; Zhang, Y.L. Isolation, identification, derivatization and phytotoxic activity of secondary metabolites produced by Cladosporium oxysporum DH14, a locust-associated fungus. J. Integr. Agric. 2016, 15, 832–839. [Google Scholar] [CrossRef]
- Wang, W.; Feng, H.; Sun, C.; Che, Q.; Zhang, G.; Zhu, T.; Li, D. Thiocladospolides F-J, antibacterial sulfur containing 12-membered macrolides from the mangrove endophytic fungus Cladosporium oxysporum HDN13-314. Phytochemistry 2020, 178, 112462. [Google Scholar] [CrossRef]
- Gokul Raj, K.; Manikandan, R.; Arulvasu, C.; Pandi, M. Anti-proliferative effect of fungal taxol extracted from Cladosporium oxysporum against human pathogenic bacteria and human colon cancer cell line HCT 15. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 138, 667–674. [Google Scholar] [CrossRef]
- Fan, Z.; Sun, Z.H.; Liu, H.X.; Chen, Y.C.; Li, H.H.; Zhang, W.M. Perangustols A and B, a pair of new azaphilone epimers from a marine sediment-derived fungus Cladosporium perangustm FS62. J. Asian Nat. Prod. Res. 2016, 18, 1024–1029. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, T.; Shimanuki, T.; Araki, T.; Sakamura, S. Phleichrome; a new phytotoxic compound by Cladosporium phlei. Agric. Biol. Chem. 1975, 39, 1683–1684. [Google Scholar] [CrossRef]
- Arnone, A.; Camarda, L.; Nasini, G.; Merlini, L. Secondary mould metabolites. Part 13. Fungal perylenequinones: Phleichrome, isophleichrome, and their endoperoxides. J. Chem. Soc. Perkin Trans. 1 1985, 1387–1392. [Google Scholar] [CrossRef]
- Wu, G.; Sun, X.; Yu, G.; Wang, W.; Zhu, T.; Gu, Q.; Li, D. Cladosins A-E, hybrid polyketides from a deep-sea-derived fungus, Cladosporium sphaerospermum. J. Nat. Prod. 2014, 77, 270–275. [Google Scholar] [CrossRef]
- Yu, G.H.; Wu, G.W.; Zhu, T.J.; Gu, Q.Q.; Li, D.H. Cladosins F and G, two new hybrid polyketides from the deep-sea-derived Cladosporium sphaerospermum 2005-01-E3. J. Asian Nat. Prod. Res. 2015, 17, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Hamayun, M.; Afzal Khan, S.; Ahmad, N.; Tang, D.S.; Kang, S.M.; Na, C.I.; Sohn, E.Y.; Hwang, Y.H.; Shin, D.H.; Lee, B.H.; et al. Cladosporium sphaerospermum as a new plant growth-promoting endophyte from the roots of Glycine max (L.) Merr. World J. Microbiol. Biotechnol. 2009, 25, 627–632. [Google Scholar] [CrossRef]
- Liang, X.; Huang, Z.H.; Ma, X.; Qi, S.H. Unstable tetramic acid derivatives from the deep-sea-derived fungus Cladosporium sphaerospermum EIODSF 008. Mar. Drugs 2018, 16, 448. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.-h.; Nong, X.-h.; Liang, X.; Qi, S.-h. New tetramic acid derivatives from the deep-sea-derived fungus Cladosporium sp. SCSIO z0025. Tetrahedron 2018, 74, 2620–2626. [Google Scholar] [CrossRef]
- Lee, S.R.; Lee, D.; Eom, H.J.; Rischer, M.; Ko, Y.J.; Kang, K.S.; Kim, C.S.; Beemelmanns, C.; Kim, K.H. Hybrid polyketides from a hydractinia-associated Cladosporium sphaerospermum SW67 and their putative biosynthetic origin. Mar. Drugs 2019, 17, 606. [Google Scholar] [CrossRef]
- Rischer, M.; Lee, S.R.; Eom, H.J.; Park, H.B.; Vollmers, J.; Kaster, A.K.; Shin, Y.H.; Oh, D.C.; Kim, K.H.; Beemelmanns, C. Spirocyclic cladosporicin A and cladosporiumins I and J from a: Hydractinia-associated Cladosporium sphaerospermum SW67. Org. Chem. Front. 2019, 6, 1084–1093. [Google Scholar] [CrossRef]
- Lee, S.R.; Kang, H.; Yoo, M.J.; Yi, S.A.; Beemelmanns, C.; Lee, J.; Kim, K.H. Anti-adipogenic pregnane steroid from a Hydractinia-associated fungus, Cladosporium sphaerospermum sw67. Nat. Prod. Sci. 2020, 26, 230–235. [Google Scholar]
- Pan, F.; El-Kashef, D.H.; Kalscheuer, R.; Müller, W.E.G.; Lee, J.; Feldbrügge, M.; Mándi, A.; Kurtán, T.; Liu, Z.; Wu, W.; et al. Cladosins L-O, new hybrid polyketides from the endophytic fungus Cladosporium sphaerospermum WBS017. Eur. J. Med. Chem. 2020, 191, 112159. [Google Scholar] [CrossRef]
- Fujii, Y.; Fukuda, A.; Hamasaki, T.; Ichimoto, I.; Nakajima, H. Twelve-membered lactones produced by Cladosporium tenuissimum and the plant growth retardant activity of cladospolide B. Phytochemistry 1995, 40, 1443–1446. [Google Scholar] [CrossRef]
- Fadhillah; Elfita; Muharni; Yohandini, H.; Widjajanti, H. Chemical compound isolated from antioxidant active extract of endophytic fungus Cladosporium tenuissimum in Swietenia mahagoni leaf stalks. Biodiversitas 2019, 20, 2645–2650. [Google Scholar]
- Nasini, G.; Arnone, A.; Assante, G.; Bava, A.; Moricca, S.; Ragazzi, A. Secondary mould metabolites of Cladosporium tenuissimum, a hyperparasite of rust fungi. Phytochemistry 2004, 65, 2107–2111. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.Q.; Kang, Q.J.; Li, G.H.; Shen, Y.M. Three new polyketide metabolites from the endophytic fungal strain Cladosporium tenuissimum LR463 of Maytenus hookeri. Helv. Chim. Acta 2006, 89, 527–531. [Google Scholar] [CrossRef]
- Naseer, S.; Bhat, K.A.; Qadri, M.; Riyaz-Ul-Hassan, S.; Malik, F.A.; Khuroo, M.A. Bioactivity-guided isolation, antimicrobial and cytotoxic evaluation of secondary metabolites from Cladosporium tenuissimum associated with Pinus wallichiana. ChemistrySelect 2017, 2, 1311–1314. [Google Scholar] [CrossRef]
- De Medeiros, L.S.; Murgu, M.; de Souza, A.Q.L.; Rodrigues-Fo, F. Antimicrobial depsides produced by Cladosporium uredinicola, an endophytic fungus isolated from Psidium guajava fruits. Helv. Chim. Acta 2011, 94, 1077. [Google Scholar] [CrossRef]
- Singh, B.; Sharma, P.; Kumar, A.; Chadha, P.; Kaur, R.; Kaur, A. Antioxidant and in vivo genoprotective effects of phenolic compounds identified from an endophytic Cladosporium velox and their relationship with its host plant Tinospora cordifolia. J. Ethnopharmacol. 2016, 194, 450–456. [Google Scholar] [CrossRef]
- Gesner, S.; Cohen, N.; Ilan, M.; Yarden, O.; Carmeli, S. Pandangolide 1a, a metabolite of the sponge-associated fungus Cladosporium sp., and the absolute stereochemistry of pandangolide 1 and iso-cladospolide B. J. Nat. Prod. 2005, 68, 1350–1353. [Google Scholar] [CrossRef]
- Sassa, T.; Ooi, T.; Nukina, M.; Ikeda, M.; Katct, N. Structural confirmation of Cotylenin A, a novel Fusicoccane-diterpene glycoside with potent plant growth-regulating activity from Cladosporium fungus sp. 501-7W. Biosci. Biotechnol. Biochem. 1998, 62, 1815–1818. [Google Scholar] [CrossRef]
- Sassa, T.; Togashi, M.; Kitaguchi, T. The structures of cotylenins A, B, C, D and E. Agric. Biol. Chem. 1975, 39, 1735–1744. [Google Scholar] [CrossRef]
- Sassa, T. Cotylenins, leaf growth substances produced by a fungus part I. Isolation and characterization of cotylenins A and B. Agric. Biol. Chem. 1971, 35, 1415–1418. [Google Scholar]
- Qi, S.H.; Xu, Y.; Xiong, H.R.; Qian, P.Y.; Zhang, S. Antifouling and antibacterial compounds from a marine fungus Cladosporium sp. F14. World J. Microbiol. Biotechnol. 2009, 25, 399–406. [Google Scholar] [CrossRef]
- Fan, C.; Zhou, G.; Wang, W.; Zhang, G.; Zhu, T.; Che, Q.; Li, D. Tetralone derivatives from a deep-sea-derived fungus Cladosporium sp. HDN17-58. Nat. Prod. Commun. 2021, 16, 1–4. [Google Scholar]
- Wang, F.W.; Jiao, R.H.; Cheng, A.B.; Tan, S.H.; Song, Y.C. Antimicrobial potentials of endophytic fungi residing in Quercus variabilis and brefeldin A obtained from Cladosporium sp. World J. Microbiol. Biotechnol. 2007, 23, 79–83. [Google Scholar] [CrossRef]
- Wuringe; Guo, Z.K.; Wei, W.; Jiao, R.H.; Yan, T.; Zang, L.Y.; Jiang, R.; Tan, R.X.; Ge, H.M. Polyketides from the plant endophytic fungus Cladosporium sp. IFB3lp-2. J. Asian Nat. Prod. Res. 2013, 15, 928–933. [Google Scholar] [CrossRef]
- Hosoe, T.; Okada, H.; Itabashi, T.; Nozawa, K.; Okada, K.; de Campos Takaki, G.M.; Fukushima, K.; Miyaji, M.; Kawai, K. A new pentanorlanostane derivative, cladosporide A, as a characteristic antifungal agent against Aspergillus fumigatus, isolated from Cladosporium sp. Chem. Pharm. Bull. 2000, 48, 1422–1426. [Google Scholar] [CrossRef] [PubMed]
- Hosoe, T.; Okamoto, S.; Nozawa, K.; Kawai, K.I.; Okada, K.; De Campos Takaki, G.M. New pentanorlanostane derivatives, cladosporide B-D, as characteristic antifungal agents against Aspergillus fumigatus, isolated from Cladosporium sp. J. Antibiot. 2001, 54, 747–750. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.T.; Zheng, C.J.; Zhang, B.; Zhou, X.M.; Zhou, Q.; Chen, G.Y.; Zeng, Z.E.; Xie, J.L.; Han, C.R.; Lyu, J.X. Two new secondary metabolites from a mangrove-derived fungus Cladosporium sp. JJM22. Nat. Prod. Res. 2019, 33, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wu, J.T.; Zheng, C.J.; Zhou, X.M.; Yu, Z.X.; Li, W.S.; Chen, G.Y.; Zhu, G.Y. Bioactive cyclohexene derivatives from a mangrove-derived fungus Cladosporium sp. JJM22. Fitoterapia 2021, 149, 104823. [Google Scholar] [CrossRef]
- Ma, R.Z.; Zheng, C.J.; Zhang, B.; Yang, J.Y.; Zhou, X.M.; Song, X.M. Two new naphthalene-chroman coupled derivatives from the mangrove-derived fungus Cladosporium sp. JJM22. Phytochem. Lett. 2021, 43, 114–116. [Google Scholar] [CrossRef]
- Han, X.; Bao, X.-F.; Wang, C.-X.; Xie, J.; Song, X.-J.; Dai, P.; Chen, G.-D.; Hu, D.; Yao, X.-S.; Gao, H. Cladosporine A, a new indole diterpenoid alkaloid with antimicrobial activities from Cladosporium sp. Nat. Prod. Res. 2019, 35, 1–7. [Google Scholar] [CrossRef]
- Bai, M.; Zheng, C.J.; Tang, D.Q.; Zhang, F.; Wang, H.Y.; Chen, G.Y. Two new secondary metabolites from a mangrove-derived fungus Cladosporium sp. JS1-2. J. Antibiot. 2019, 72, 779–782. [Google Scholar] [CrossRef]
- Ai, W.; Lin, X.; Wang, Z.; Lu, X.; Mangaladoss, F.; Yang, X.; Zhou, X.; Tu, Z.; Liu, Y. Cladosporone A, a new dimeric tetralone from fungus Cladosporium sp. KcFL6′ derived of mangrove plant Kandelia candel. J. Antibiot. 2015, 68, 213–215. [Google Scholar] [CrossRef]
- Zhang, F.; Zhou, L.; Kong, F.; Ma, Q.; Xie, Q.; Li, J.; Dai, H.; Guo, L.; Zhao, Y. Altertoxins with quorum sensing inhibitory activities from the marine-derived fungus Cladosporium sp. KFD33. Mar. Drugs 2020, 18, 67. [Google Scholar] [CrossRef]
- Shigemori, H.; Kasai, Y.; Komatsu, K.; Tsuda, M.; Mikami, Y.; Kobayashi, J. Sporiolides A and B, new cytotoxic twelve-membered macrolides from a marine-derived fungus Cladosporium species. Mar. Drugs 2004, 2, 164–169. [Google Scholar] [CrossRef]
- Zhang, Z.; He, X.; Wu, G.; Liu, C.; Lu, C.; Gu, Q.; Che, Q.; Zhu, T.; Zhang, G.; Li, D. Aniline-tetramic acids from the deep-sea-derived fungus Cladosporium sphaerospermum L3P3 cultured with the HDAC inhibitor SAHA. J. Nat. Prod. 2018, 81, 1651–1657. [Google Scholar] [CrossRef]
- Khan, M.I.H.; Sohrab, M.H.; Rony, S.R.; Tareq, F.S.; Hasan, C.M.; Mazid, M.A. Cytotoxic and antibacterial naphthoquinones from an endophytic fungus, Cladosporium sp. Toxicol. Rep. 2016, 3, 861–865. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Chen, T.; Yan, Z.; Guo, H.; Hou, X.; Jiang, L.; Long, Y. Thiocladospolide E and cladospamide A, novel 12-membered macrolide and macrolide lactam from mangrove endophytic fungus Cladosporium sp. SCNU-F0001. Fitoterapia 2019, 137, 104246. [Google Scholar] [CrossRef]
- Amin, M.; Zhang, X.Y.; Xu, X.Y.; Qi, S.H. New citrinin derivatives from the deep-sea-derived fungus Cladosporium sp. SCSIO z015. Nat. Prod. Res. 2020, 34, 1219–1226. [Google Scholar] [CrossRef]
- Rotinsulu, H.; Yamazaki, H.; Sugai, S.; Iwakura, N.; Wewengkang, D.S.; Sumilat, D.A.; Namikoshi, M. Cladosporamide A, a new protein tyrosine phosphatase 1B inhibitor, produced by an Indonesian marine sponge-derived Cladosporium sp. J. Nat. Med. 2018, 72, 779–783. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Gao, H.; Wu, C.; Zhu, T.; Che, Q.; Gu, Q.; Guo, P.; Li, D. Lipid-lowering polyketides from a soft coral-derived fungus Cladosporium sp. TZP29. Bioorg. Med. Chem. Lett. 2015, 25, 3606–3609. [Google Scholar] [CrossRef]
- Silber, J.; Ohlendorf, B.; Labes, A.; Wenzel-Storjohann, A.; Näther, C.; Imhoff, J.F. Malettinin E, an antibacterial and antifungal tropolone produced by a marine Cladosporium strain. Front. Mar. Sci. 2014, 1, 35. [Google Scholar] [CrossRef]
- Zhu, G.; Kong, F.; Wang, Y.; Fu, P.; Zhu, W. Cladodionen, a cytotoxic hybrid polyketide from the marine-derived Cladosporium sp. OUCMDZ-1635. Mar. Drugs 2018, 16, 71. [Google Scholar] [CrossRef]
- Grove, J.F. New metabolic products of Aspergiullus flavus. Part I. Asperentin, its methyl ethers, and 5′-hydroxyasperentin. J. Chem. Soc. Perkin Trans. 1 1972, 2400–2406. [Google Scholar] [CrossRef]
- De Medeiros, L.S.; Murgu, M.; de Souza, A.Q.L.; Rodrigues-Fo, E. Erratum: Antimicrobial depsides produced by Cladosporium uredinicola, an endophytic fungus isolated from Psidium guajava fruits. Helv. Chim. Acta 2011, 94, 1077–1084, Erratum in 2013, 96, 1406–1407. [Google Scholar] [CrossRef]
- Mino, Y.; Idonuma, T.; Sakai, R. Effect of phleichrome by the Timothy leaf spot fungus, Cladosporium phlei on the invertases from the host leaves. Ann. Phytopath. Soc. Jpn. 1979, 45, 463–467. [Google Scholar] [CrossRef]
- Hiort, J.; Maksimenka, K.; Reichert, M.; Perović-Ottstadt, S.; Lin, W.H.; Wray, V.; Steube, K.; Schaumann, K.; Weber, H.; Proksch, P.; et al. New natural products from the sponge-derived fungus Aspergillus niger. J. Nat. Prod. 2004, 67, 1532–1543. [Google Scholar] [CrossRef]
- Gao, J.; Yang, S.; Qin, J. Azaphilones: Chemistry and Biology. Chem. Rev. 2013, 113, 4755–4811. [Google Scholar] [CrossRef]
- Nicoletti, R.; Salvatore, M.M.; Andolfi, A. Secondary metabolites of mangrove-associated strains of Talaromyces. Mar. Drugs 2018, 16, 12. [Google Scholar] [CrossRef]
- Bao, J.; Zhang, X.Y.; Dong, J.J.; Xu, X.Y.; Nong, X.H.; Qi, S.H. Cyclopentane-condensed chromones from marine-derived fungus Penicillium oxalicum. Chem. Lett. 2014, 43, 837–839. [Google Scholar] [CrossRef]
- Zhang, X.; Li, S.J.; Li, J.J.; Liang, Z.Z.; Zhao, C.Q. Novel natural products from extremophilic fungi. Mar. Drugs 2018, 16, 194. [Google Scholar] [CrossRef] [PubMed]
- Dewick, P.M. Medicinal Natural Products: A Biosyntheic Approach, 3rd ed.; John Wiley & Sons: Hoboken, NJ, USA, 2016; ISBN 978-0-470-74168-9. [Google Scholar]
- Flajs, D.; Peraica, M. Toxicological properties of citrinin. Arh. Hig. Rada Toksikol. 2009, 60, 457–464. [Google Scholar] [CrossRef]
- Saeed, A. Isocoumarins, miraculous natural products blessed with diverse pharmacological activities. Eur. J. Med. Chem. 2016, 116, 290–317. [Google Scholar] [CrossRef]
- Reveglia, P.; Masi, M.; Evidente, A. Melleins—Intriguing natural compounds. Biomolecules 2020, 10, 772. [Google Scholar] [CrossRef] [PubMed]
- Ellestad, G.A.; Mirando, P.; Kunstmann, M.P. Structure of the metabolite LL-S490 beta from an unidentified Aspergillus species. J. Org. Chem. 1973, 38, 24–25. [Google Scholar] [CrossRef] [PubMed]
- Cutler, H.G.; Crumley, F.G.; Cox, R.H.; Hernandez, O.; Cole, R.J.; Dorner, J.W. Orlandin: A nontoxic fungal metabolite with plant growth inhibiting properties. J. Agric. Food Chem. 1979, 27, 592–595. [Google Scholar] [CrossRef]
- Büchi, G.; Luk, K.C.; Kobbe, B.; Townsend, J.M. Four new mycotoxins of Aspergillus clavatus related to tryquivaline. J. Org. Chem. 1977, 42, 244–246. [Google Scholar] [CrossRef] [PubMed]
- Hedden, P.; Sponsel, V. A century of gibberellin research. J. Plant Growth Regul. 2015, 34, 740–760. [Google Scholar] [CrossRef]
- Smith, C.J.; Abbanat, D.; Bernan, V.S.; Maiese, W.M.; Greenstein, M.; Jompa, J.; Tahir, A.; Ireland, C.M. Novel polyketide metabolites from a species of marine fungi. J. Nat. Prod. 2000, 63, 142–145. [Google Scholar] [CrossRef]
- Hirota, A.; Sakai, H.; Isogai, A. New plant growth regulators, cladospolide A and B, macrolides produced by Cladosporium cladosporioides. Agric. Biol. Chem. 1985, 49, 731–735. [Google Scholar] [CrossRef]
- Hirota, A.; Sakai, H.; Isogai, A.; Kitano, Y.; Ashida, T.; Hirota, H.; Takahashi, T. Absolute stereochemistry of cladospolide a, a phytotoxic macrolide from Cladosporium cladospor hides. Agric. Biol. Chem. 1985, 49, 903–904. [Google Scholar]
- Hirota, H.; Hirota, A.; Sakai, H.; Isogai, A.; Takahashi, T. Absolute stereostructure determination of cladospolide A using MTPA ester method. Bull. Chem. Soc. Jpn. 1985, 58, 2147–2148. [Google Scholar] [CrossRef]
- Félix, C.; Salvatore, M.M.; Dellagreca, M.; Meneses, R.; Duarte, A.S.; Salvatore, F.; Naviglio, D.; Gallo, M.; Jorrín-Novo, J.V.; Alves, A.; et al. Production of toxic metabolites by two strains of Lasiodiplodia theobromae, isolated from a coconut tree and a human patient. Mycologia 2018, 110, 642–653. [Google Scholar] [CrossRef] [PubMed]
- Salvatore, M.M.; Alves, A.; Andolfi, A. Secondary metabolites of Lasiodiplodia theobromae: Distribution, chemical diversity, bioactivity, and implications of their occurrence. Toxins 2020, 12, 457. [Google Scholar] [CrossRef]
- Qiu, H.Y.; Wang, P.F.; Lin, H.Y.; Tang, C.Y.; Zhu, H.L.; Yang, Y.H. Naphthoquinones: A continuing source for discovery of therapeutic antineoplastic agents. Chem. Biol. Drug Des. 2018, 91, 681–690. [Google Scholar] [CrossRef]
- Salvatore, M.M.; Alves, A.; Andolfi, A. Secondary metabolites produced by Neofusicoccum species associated with plants: A review. Agriculture 2021, 11, 149. [Google Scholar] [CrossRef]
- Iida, T.; Kobayashi, E.; Yoshida, M. Calphostins, novel and specific inhibitors of protein kinase C. J. Antibiot. 1989, 42, 1475–1481. [Google Scholar] [CrossRef] [PubMed]
- Stack, M.E.; Prival, M.J. Mutagenicity of the Alternaria metabolites altertoxins I, II, and III. Appl. Environ. Microbiol. 1986, 52, 718–722. [Google Scholar] [CrossRef] [PubMed]
- Bhat, Z.S.; Rather, M.A.; Maqbool, M.; Lah, H.U.; Yousuf, S.K.; Ahmad, Z. α-Pyrones: Small molecules with versatile structural diversity reflected in multiple pharmacological activities-an update. Biomed. Pharmacother. 2017, 91, 265–277. [Google Scholar] [CrossRef] [PubMed]
- McGlacken, G.P.; Fairlamb, I.J.S. 2-Pyrone natural products and mimetics: Isolation, characterisation and biological activity. Nat. Prod. Rep. 2005, 22, 369–385. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Nakano, M.; Biju Kumar, G.; Kishihara, K. Efficient conditions for conversion of 2-substituted furans into 4-oxygenated 2-enoic acids and its application to synthesis of (+)-aspicilin, (+)-patulolide a, and (−)-pyrenophorin. J. Org. Chem. 1998, 63, 7505–7515. [Google Scholar] [CrossRef]
- Dos Santos Dias, A.C.; Couzinet-Mossion, A.; Ruiz, N.; Lakhdar, F.; Etahiri, S.; Bertrand, S.; Ory, L.; Roussakis, C.; Pouchus, Y.F.; Nazih, E.H.; et al. Steroids from marine-derived fungi: Evaluation of antiproliferative and antimicrobial activities of eburicol. Mar. Drugs 2019, 17, 372. [Google Scholar] [CrossRef]
- Rodrigues, M.L. The multifunctional fungal ergosterol. MBio 2018, 9, e01755. [Google Scholar] [CrossRef]
- De Tommaso, G.; Salvatore, M.M.; Nicoletti, R.; DellaGreca, M.; Vinale, F.; Bottiglieri, A.; Staropoli, A.; Salvatore, F.; Lorito, M.; Iuliano, M.; et al. Bivalent metal-chelating properties of harzianic acid produced by Trichoderma pleuroticola associated to the gastropod Melarhaphe neritoides. Molecules 2020, 25, 2147. [Google Scholar] [CrossRef] [PubMed]
- De Tommaso, G.; Salvatore, M.M.; Nicoletti, R.; Dellagreca, M.; Vinale, F.; Staropoli, A.; Salvatore, F.; Lorito, M.; Iuliano, M.; Andolfi, A. Coordination properties of the fungal metabolite harzianic acid toward toxic heavy metals. Toxics 2021, 9, 19. [Google Scholar] [CrossRef]
- Nicoletti, R.; Fiorentino, A. Plant bioactive metabolites and drugs produced by endophytic fungi of Spermatophyta. Agriculture 2015, 5, 918–970. [Google Scholar] [CrossRef]
- Negi, J.S.; Bisht, V.K.; Singh, P.; Rawat, M.S.M.; Joshi, G.P. Naturally occurring xanthones: Chemistry and biology. J. Appl. Chem. 2013, 3, 1–11. [Google Scholar] [CrossRef]
- Nicoletti, R.; Manzo, E.; Ciavatta, M.L. Occurence and bioactivities of funicone-related compounds. Int. J. Mol. Sci. 2009, 10, 1430–1444. [Google Scholar] [CrossRef]
- Houbraken, J.; Kocsubé, S.; Visagie, C.M.; Yilmaz, N.; Wang, X.C.; Meijer, M.; Kraak, B.; Hubka, V.; Bensch, K.; Samson, R.A.; et al. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Stud. Mycol. 2020, 95, 5–169. [Google Scholar] [CrossRef] [PubMed]
- Zwickel, T.; Kahl, S.M.; Rychlik, M.; Müller, M.E.H. Chemotaxonomy of mycotoxigenic small-spored Alternaria fungi—Do multitoxin mixtures act as an indicator for species differentiation? Front. Microbiol. 2018, 9, 1368. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.; Kim, J.; Choi, J.N.; Liu, K.H.; Lee, C.H. Chemotaxonomy of Trichoderma spp. using mass spectrometry-based metabolite profiling. J. Microbiol. Biotechnol. 2011, 21, 5–13. [Google Scholar] [CrossRef] [PubMed]
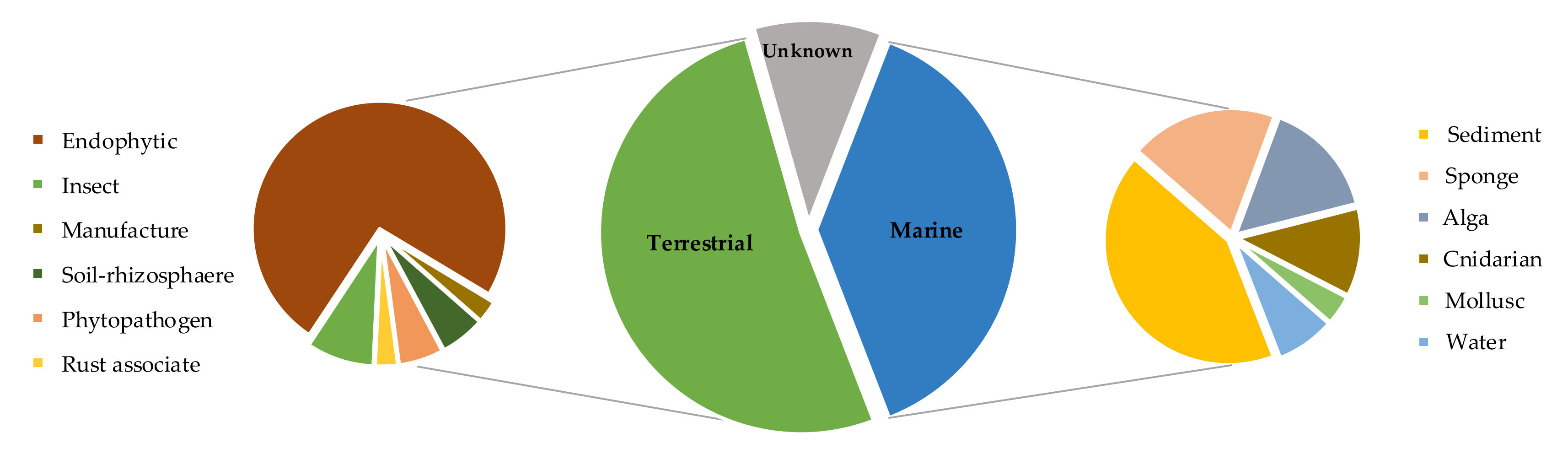

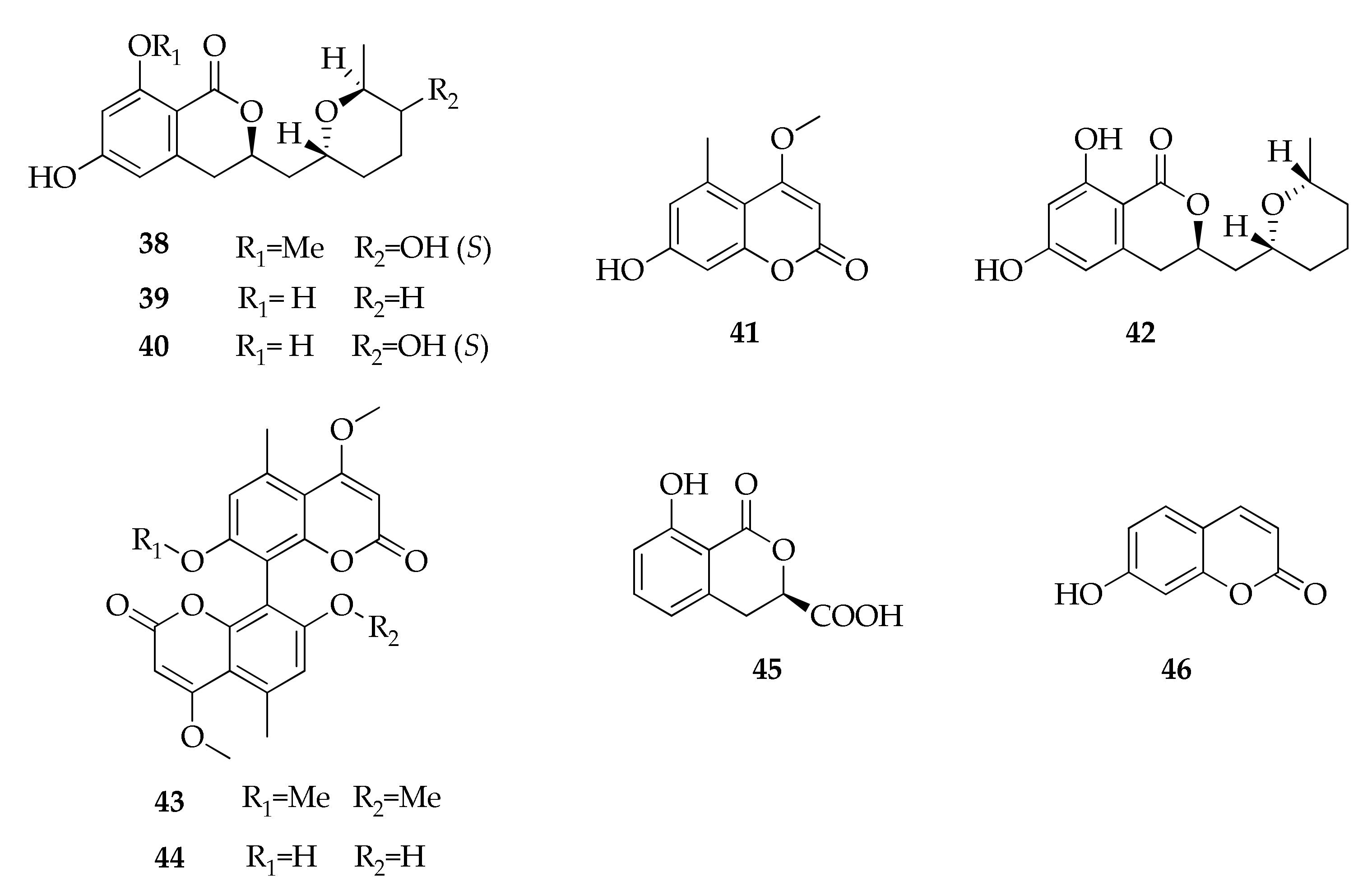

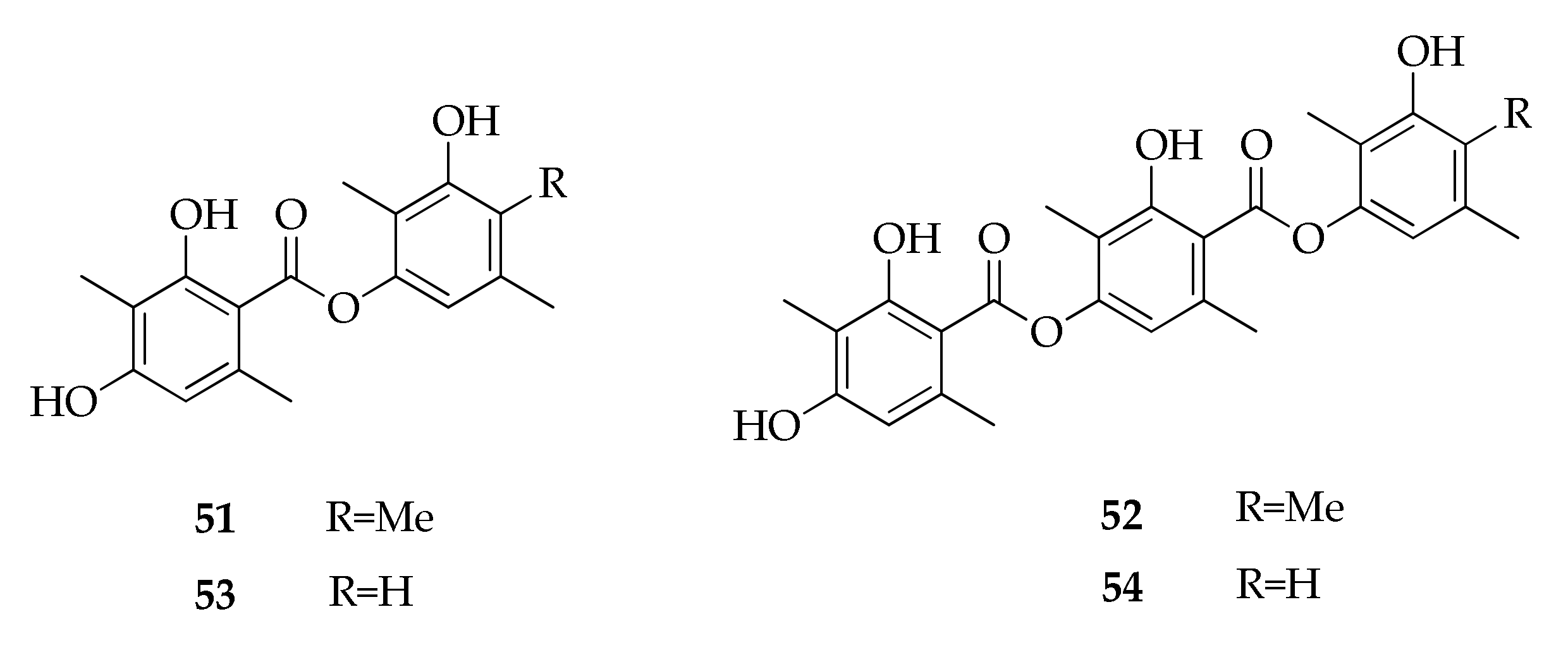

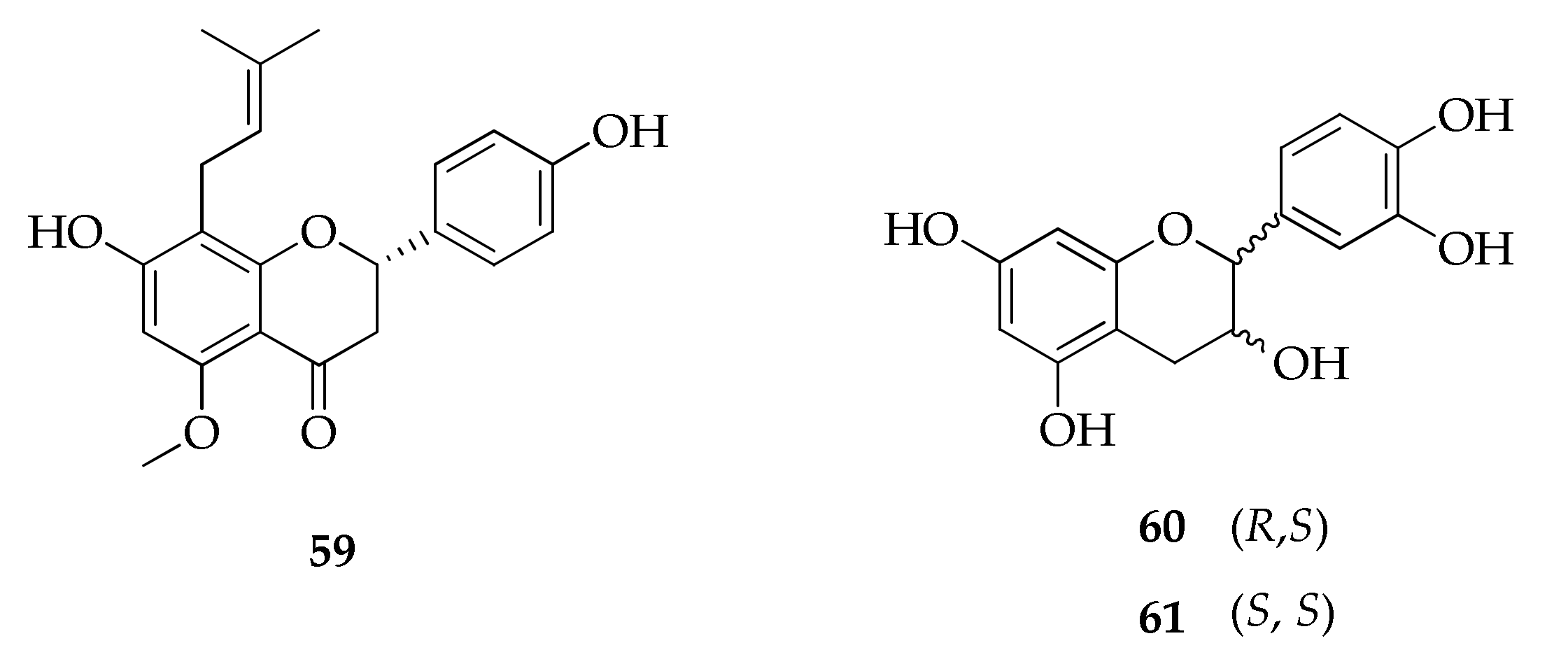
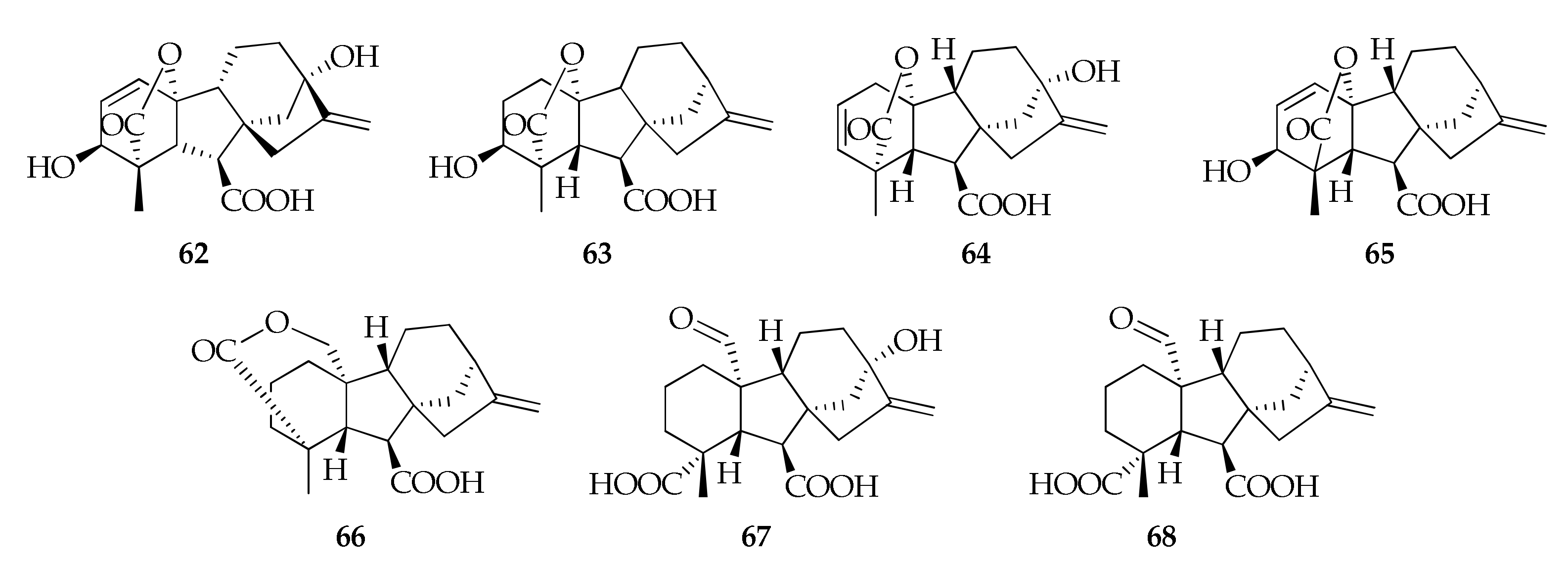





| Species/Strain | Substrate | Location | Refs. |
|---|---|---|---|
| C. cladosporioides | - | - | [12] |
| C. cladosporioides | sediment of hypersaline lake | El Hamra, Egypt | [22] |
| C. cladosporioides | sponge (Cliona sp.) | Los Molles, Chile | [23] |
| C. cladosporioides | aphid (Aphis craccivora) | Egypt | [17,18] |
| C. cladosporioides | endophytic in Zygophyllum mandavillei | Al-Ahsa, Saudi Arabia | [24] |
| C. cladosporioides | endophytic in unspecified plant | Tifton, United States | [25] |
| C. cladosporioides | - | Japan | [26] |
| C. cladosporioides/CBUK20700 | dead insect | Thailand | [27,28] |
| C. cladosporioides/FERMBP-1285 | block fence | Osaka, Japan | [29,30] |
| C. cladosporioides/IPV-F167 | - | Italy | [31] |
| C. cladosporioides/LWL5 | endophytic in Helianthus annuus | Daegu, South Korea | [16] |
| C. cladosporioides/MA-299 | endophytic in Bruguiera gymnorrhiza | Hainan, China | [32,33,34] |
| C. cladosporioides/MCCC3A00182 | deep sea sediment | Pacific Ocean | [35] |
| C. cladosporioides/MUPGBIOCHEM-CC-07-2015 | brown alga (Sargassum wightii) | Tamil Nadu, India | [21] |
| C. cladosporioides/NRRL5507 | - | - | [36,37] |
| C. cladosporioides/CL-1 | rhizosphere of red pepper | South Korea | [38] |
| C. cladosporioides/EN-399 | red alga (Laurencia okamurai) | Qingdao, China | [39] |
| C. cladosporioides/HDN14-342 | deep sea sediment | Indian Ocean | [40] |
| C. colocasiae/A801 | endophytic in Callistemon viminalis | Guangzhou, China | [41] |
| C. delicatulum/EF33 | endophytic in Terminalia pallida | Andhra Pradesh, India | [42] |
| C. halotolerans/GXIMD 02502 | coral (Porites lutea) | Weizhou islands, China | [43] |
| C. herbarum | sponge (Callyspongia aerizusa) | Bali, Indonesia | [44,45] |
| C. herbarum/FC27P | endophytic in Beta vulgaris | Dublin, United States | [46] |
| C. herbarum/IFB-E002 | endophytic in Cynodon dactylon | Yancheng reserve, China | [47] |
| C. oxysporum | endophytic in Aglaia odorata | Java, Indonesia | [14] |
| C. oxysporum | endophytic in Alyxia reinwardtii | Java, Indonesia | [48] |
| C. oxysporum/DH14 | locust (Oxya chinensis) | Jinhua, China | [49] |
| C. oxysporum/HDN13-314 | endophytic in Avicennia marina | Hainan, China | [50] |
| C. oxysporum/RM1 | endophytic in Moringa oleifera | Tamil Nadu, India | [51] |
| C. perangustum/FS62 | deep sea sediment | South China Sea | [52] |
| C. phlei/C-273 w | pathogenic on Phleum pratense | Hokkaido, Japan | [53] |
| C. phlei/CBS 358.69 | pathogenic on Phleum pratense | Germany | [54] |
| C. sphaerospermum/2005-01-E3 | deep sea sludge | Pacific Ocean | [55,56] |
| C. sphaerospermum/DK-1-1 | endophytic in Glycine max | South Korea | [57] |
| C. sphaerospermum/EIODSF 008 | deep sea sediment | Indian Ocean | [58] |
| C. sphaerospermum/L3P3 | deep sea sediment | Mariana Trench | [59] |
| C. sphaerospermum/SW67 | hydroid (Hydractinia echinata) | South Korea | [60,61,62] |
| C. sphaerospermum/WBS017 | endophytic in Fritillaria unibracteata var. wabuensis | China | [63] |
| C. tenuissimum | soil | Karo-cho, Japan | [64] |
| C. tenuissimum/DMG 3 | endophytic in Swietenia mahagoni | Sumatra, Indonesia | [65] |
| C. tenuissimum/ITT21 | pine rust (Cronartium flaccidum) | Tuscany, Italy | [66] |
| C. tenuissimum/LR463 | endophytic in Maytenus hookeri | Yunnan, China | [67] |
| C. tenuissimum/P1S11 | endophytic in Pinus wallichiana | Kashmir, India | [68] |
| C. uredinicola | endophytic in Psidium guajava | São Carlos, Brazil | [69] |
| C. velox/TN-9S | endophytic in Tinospora cordifolia | Amritsar, India | [70] |
| Cladosporium sp. | sponge (Niphates rowi) | Gulf of Aqaba, Israel | [71] |
| Cladosporium sp./486 | intertidal sediment | San Antonio Oeste, Argentina | [19] |
| Cladosporium sp./501-7w | - | Japan | [72,73,74] |
| Cladosporium sp./F14 | sea water | Sai Kung, China | [13,75] |
| Cladosporium sp./HDN17-58 | deep sea sediment | Pacific Ocean | [76] |
| Cladosporium sp./I(R)9-2 | endophytic in Quercus variabilis | Nanjing, China | [77] |
| Cladosporium sp./IFB3lp-2 | endophytic in Rhizophora stylosa | Hainan, China | [78] |
| Cladosporium sp./IFM 49189 | - | Japan | [79,80] |
| Cladosporium sp./JJM22 | endophytic in Ceriops tagal | Hainan, China | [81,82,83] |
| Cladosporium sp./JNU17DTH12-9-0 | unknown | China | [84] |
| Cladosporium sp./JS1-2 | endophytic in Ceriops tagal | Hainan, China | [85] |
| Cladosporium sp./KcFL6’ | endophytic in Kandelia candel | Daya Bay, China | [86] |
| Cladosporium sp./KFD33 | blood cockle | Hainan, China | [87] |
| Cladosporium sp./L037 | brown alga (Actinotrichia fragilis) | Okinawa, Japan | [88] |
| Cladosporium sp./N5 | red alga (Porphyra yezoensis) | Lianyungang, China | [15] |
| Cladosporium sp./OUCMDZ-1635 | sponge | Xisha Islands, China | [89] |
| Cladosporium sp./RSBE-3 | endophytic in Rauwolfia serpentina | Bangladesh | [90] |
| Cladosporium sp./SCNU-F0001 | endophytic in unspecified mangrove | Zhuhai, China | [91] |
| Cladosporium sp./SCSIO z01 | deep sea sediment | East China Sea | [92] |
| Cladosporium sp./TPU1507 | unidentified sponge | Manado, Indonesia | [93] |
| Cladosporium sp./TZP-29 | unidentified soft coral | Guangzhou, China | [94] |
| Cladosporium sp./KF501 | water sample | Wadden Sea, Germany | [95] |
| Cladosporium sp./SCSIO z0025 | deep sea sediment | Okinawa, Japan | [96] |
| Code | Name | Formula | Nominal Mass (U) | Refs. |
|---|---|---|---|---|
| Alkaloids | ||||
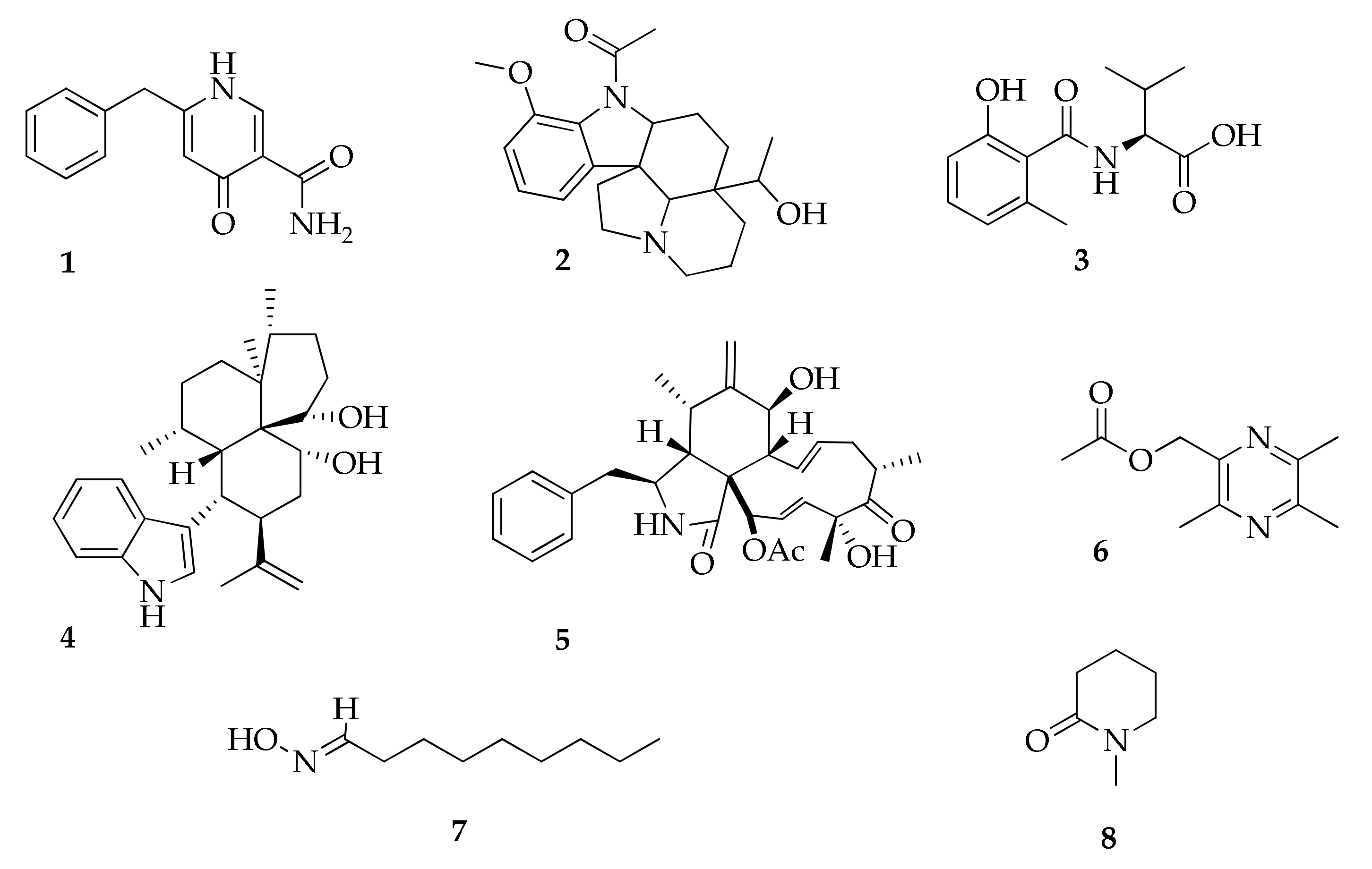
| 1 | Aspernigrin A | C13H12N2O2 | 228 | [47] |
| 2 | Aspidospermidin-20-ol, 1-acetyl-17-methoxy | C22H30N2O3 | 370 | [24] |
| 3 | Cladosin E | C13H17NO4 | 251 | [55] |
| 4 | Cladosporine A | C28H39NO2 | 421 | [84] |
| 5 | Cytochalasin D | C30H37NO6 | 507 | [85] |
| 6 | 2-Methylacetate-3,5,6-trimethylpyrazine | C10H14N2O2 | 194 | [85] |
| 7 | Nonanal oxime | C9H19NO | 157 | [17] |
| 8 | 2-Piperidinone methyl | C6H11NO | 113 | [17] |
| Azaphilones | ||||
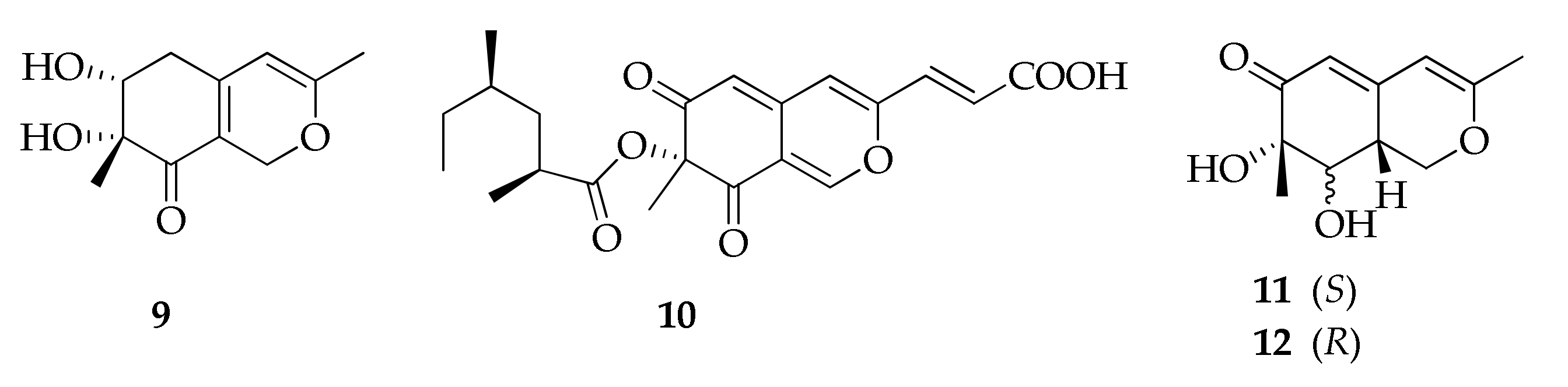
| 9 | Bicyclic diol | C11H14O4 | 210 | [52] |
| 10 | Lunatoic acid A | C21H24O7 | 388 | [49] |
| 11 | Perangustol A | C11H14O4 | 210 | [52] |
| 12 | Perangustol B | C11H14O4 | 210 | [52] |
| Benzofluoranthenones | ||||
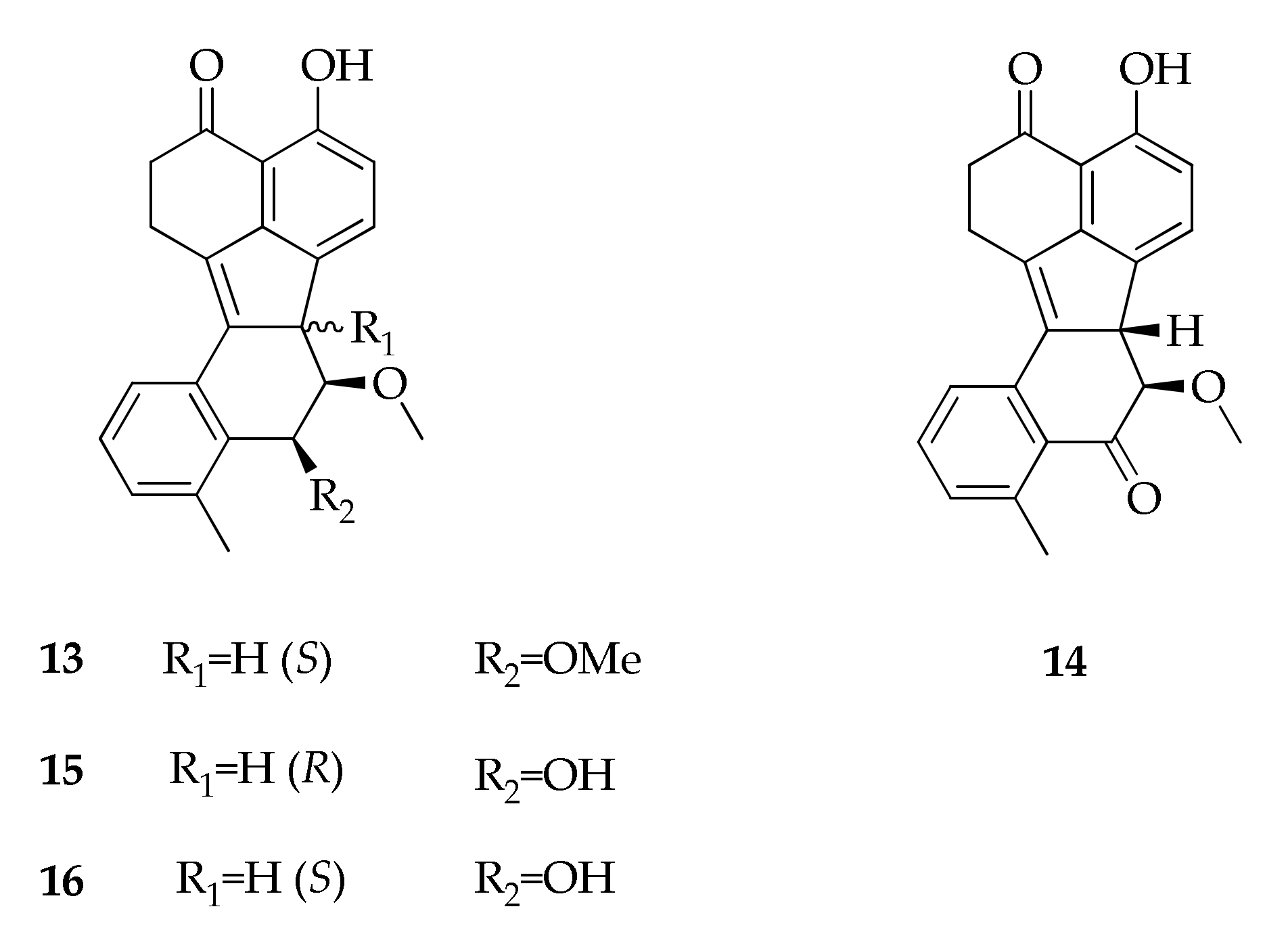
| 13 | (6bS,7R,8S)-4,9-Dihydroxy-7,8- dimethoxy-1,6b,7,8-tetra-hydro-2H- benzo[J]fluoranthen-3-one | C23H22O4 | 362 | [27] |
| 14 | (6bS,7R)-4,9-Dihydroxy-7- methoxy-1,2,6b,7-tetrahydrobenzo[J]fluoranthen-3,8-dione | C22H18O4 | 346 | [27] |
| 15 | (6bR,7R,8S)-7- Methoxy-4,8,9-trihydroxy- 1,6b,7,8-tetrahydro-2H- benzo[J]fluoranthen-3-one | C22H20O4 | 348 | [27] |
| 16 | (6bS,7R,8S)-7- Methoxy-4,8,9-trihydroxy- 1,6b,7,8-tetrahydro-2H- benzo[J]fluoranthen-3-one | C22H20O4 | 348 | [27,28] |
| Benzopyranones | ||||
| 17 | Coniochaetone A | C13H10O4 | 230 | [43] |
| 18 | Coniochaetone B | C13H12O4 | 232 | [43] |
| 19 | Coniochaetone K | C13H10O6 | 262 | [43] |
| Binaphthopyrones | ||||
| 20 | Cladosporinone | C33H30O14 | 650 | [22] |
| 21 | Viriditoxin | C34H30O14 | 662 | [22] |
| 22 | Viriditoxin SC-28763 | C34H30O13 | 646 | [22] |
| 23 | Viriditoxin SC-30532 | C34H30O12 | 630 | [22] |
| Butenolides and butanolides | ||||
| 24 | Cladospolide F | C12H22O4 | 230 | [94] |
| 25 | Cladospolide G | C14H24O5 | 272 | [32] |
| 26 | Cladospolide H | C12H18O3 | 210 | [32] |
| 27 | ent-Cladospolide F | C12H22O4 | 230 | [32] |
| 28 | 11-Hydroxy-γ-dodecalactone | C12H20O3 | 212 | [94] |
| 29 | iso-Cladospolide B | C12H20O4 | 228 | [32,44,50,67,71,94] |
| Cinnamic acid derivatives | ||||
| 30 | Chlorogenic acid | C16H18O9 | 354 | [70] |
| 31 | Caffeic acid | C9H8O4 | 180 | [70] |
| 32 | Coumaric acid | C9H8O3 | 164 | [70] |
| Citrinin derivatives | ||||
| 33 | Citrinin H1 | C25H30O7 | 442 | [85] |
| 34 | Cladosporin A | C13H15NO3 | 233 | [92] |
| 35 | Cladosporin B | C13H15NO3 | 233 | [92] |
| 36 | Cladosporin C | C14H16O4 | 248 | [92] |
| 37 | Cladosporin D | C12H16O4 | 224 | [92] |
| Coumarins and Isocoumarins | ||||
| 38 | Asperentin-8-methyl ether (= cladosporin-8-methyl ether) | C17H22O6 | 322 | [25] |
| 39 | Cladosporin (= asperentin) | C16H20O5 | 292 | [12,24,25,36,37] |
| 40 | 5′-Hydroxyasperentin | C16H20O6 | 308 | [24,25] |
| 41 | 7-Hydroxy-4-methoxy-5-methylcoumarin | C11H10O4 | 206 | [47] |
| 42 | Isocladosporin | C16H20O5 | 292 | [24,25,37] |
| 43 | Kotanin | C24H22O8 | 438 | [47] |
| 44 | Orlandin | C22H18O8 | 410 | [47] |
| 45 | Phomasatin | C10H8O5 | 208 | [35] |
| 46 | Umbelliferone | C9H6O3 | 162 | [70] |
| Cyclohexene derivatives | ||||
| 47 | Cladoscyclitol A | C12H20O5 | 244 | [82] |
| 48 | Cladoscyclitol B | C13H22O7 | 290 | [82] |
| 49 | Cladoscyclitol C | C12H22O4 | 230 | [82] |
| 50 | Cladoscyclitol D | C12H22O5 | 246 | [82] |
| Despsides | ||||
| 51 | 3-Hydroxy-2,4,5-trimethylphenyl 2,4-dihydroxy-3,6-dimethylbenzoate | C18H20O5 | 316 | [69,98] |
| 52 | 3-Hydroxy-2,4,5-trimethylphenyl 4-[(2,4-dihydroxy-3,6-dimethylbenzoyl)oxy]-2-hydroxy-3,6-dimethylbenzoate | C27H28O8 | 480 | [69,98] |
| 53 | 3-Hydroxy-2,5-dimethylphenyl 2,4-dihydroxy- 3,6-dimethylbenzoate | C17H18O5 | 302 | [69,98] |
| 54 | 3-Hydroxy-2,5-dimethylphenyl 4-[(2,4-dihydroxy-3,6-dimethylbenzoyl)oxy]-2-hydroxy-3,6-dimethylbenzoate | C26H26O8 | 466 | [69,98] |
| Diketopiperazines | ||||
| 55 | (3R,8aR)-Cyclo(leucylprolyl) | C11H18N2O2 | 210 | [17] |
| 56 | (3S,8aS)-Cyclo(leucylprolyl) | C11H18N2O2 | 210 | [17] |
| 57 | (3R,8aR)-Cyclo(phenylalanylprolyl) | C14H16N2O2 | 244 | [17] |
| 58 | (3S,8aS)-Cyclo(phenylalanylprolyl) | C14H16N2O2 | 244 | [17] |
| Flavonoids | ||||
| 59 | (2S)-7,4′-Dihydroxy-5-methoxy-8-(γ,γ-dimethylallyl)-flavanone | C21H22O5 | 354 | [93] |
| 60 | Catechin | C15H14O6 | 290 | [70] |
| 61 | Epicatechin | C15H14O6 | 290 | [70] |
| Gibberelines | ||||
| 62 | GA3 | C19H22O6 | 346 | [57] |
| 63 | GA4 | C19H24O5 | 332 | [57] |
| 64 | GA5 | C19H22O5 | 330 | [57] |
| 65 | GA7 | C19H22O5 | 330 | [57] |
| 66 | GA15 | C20H26O4 | 330 | [57] |
| 67 | GA19 | C20H26O6 | 362 | [57] |
| 68 | GA24 | C20H26O5 | 346 | [57] |
| Fusicoccane diterpene glycosides | ||||
| 69 | Cotylenin A | C33H50O11 | 622 | [72,73,74] |
| 70 | Cotylenin B | C33H51ClO11 | 659 | [73,74] |
| 71 | Cotylenin C | C33H52O11 | 624 | [73] |
| 72 | Cotylenin D | C33H52O12 | 640 | [73] |
| 73 | Cotylenin E | C28H46O9 | 526 | [73] |
| Lactones | ||||
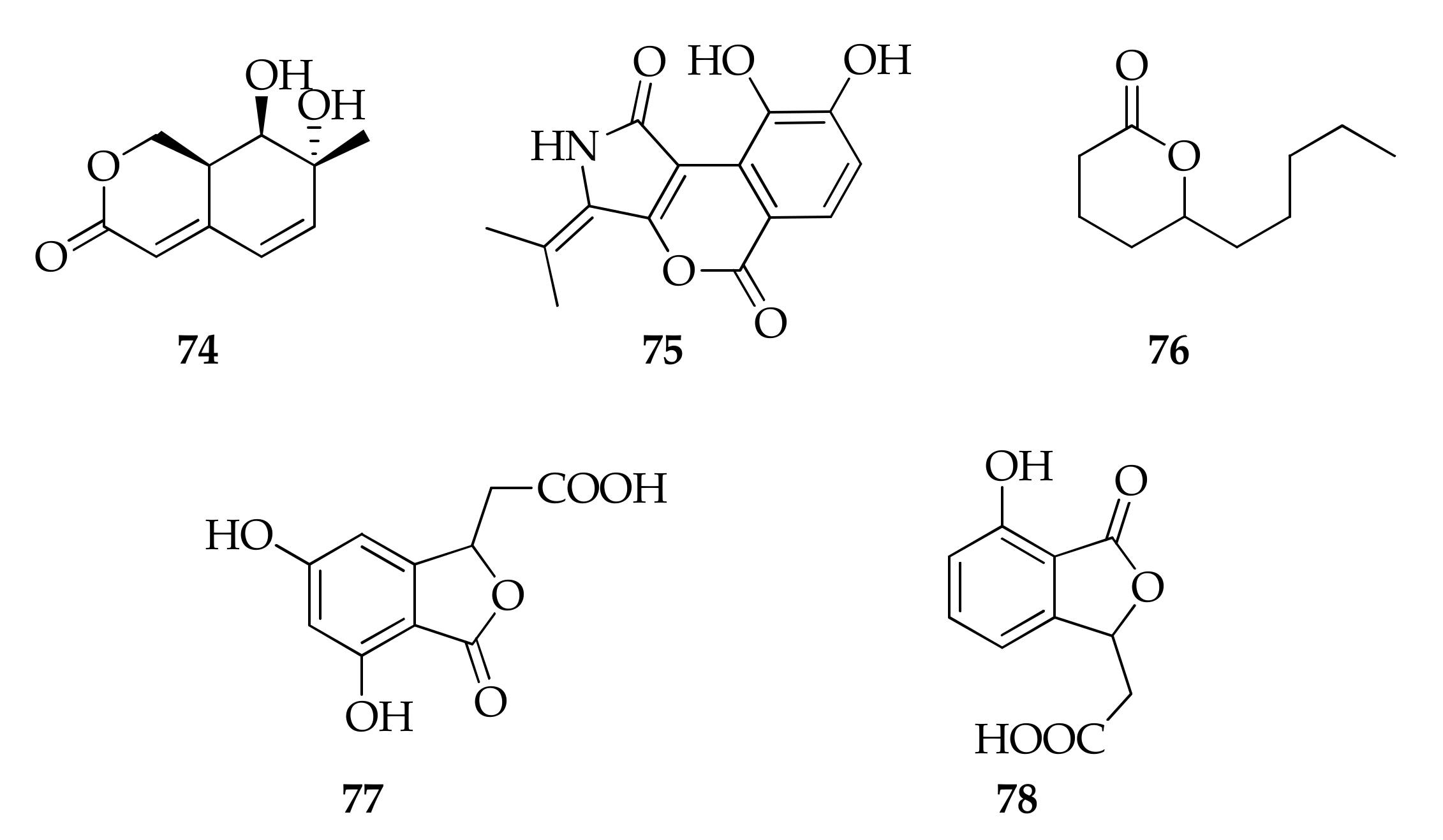
| 74 | Cladosporactone A | C10H12O4 | 196 | [35] |
| 75 | Cladosporamide A | C14H11NO5 | 273 | [93] |
| 76 | 5-Decanolide | C10H18O2 | 170 | [17] |
| 77 | Herbaric acid | C10H8O6 | 224 | [45] |
| 78 | Isochracinic acid | C10H8O5 | 208 | [35] |
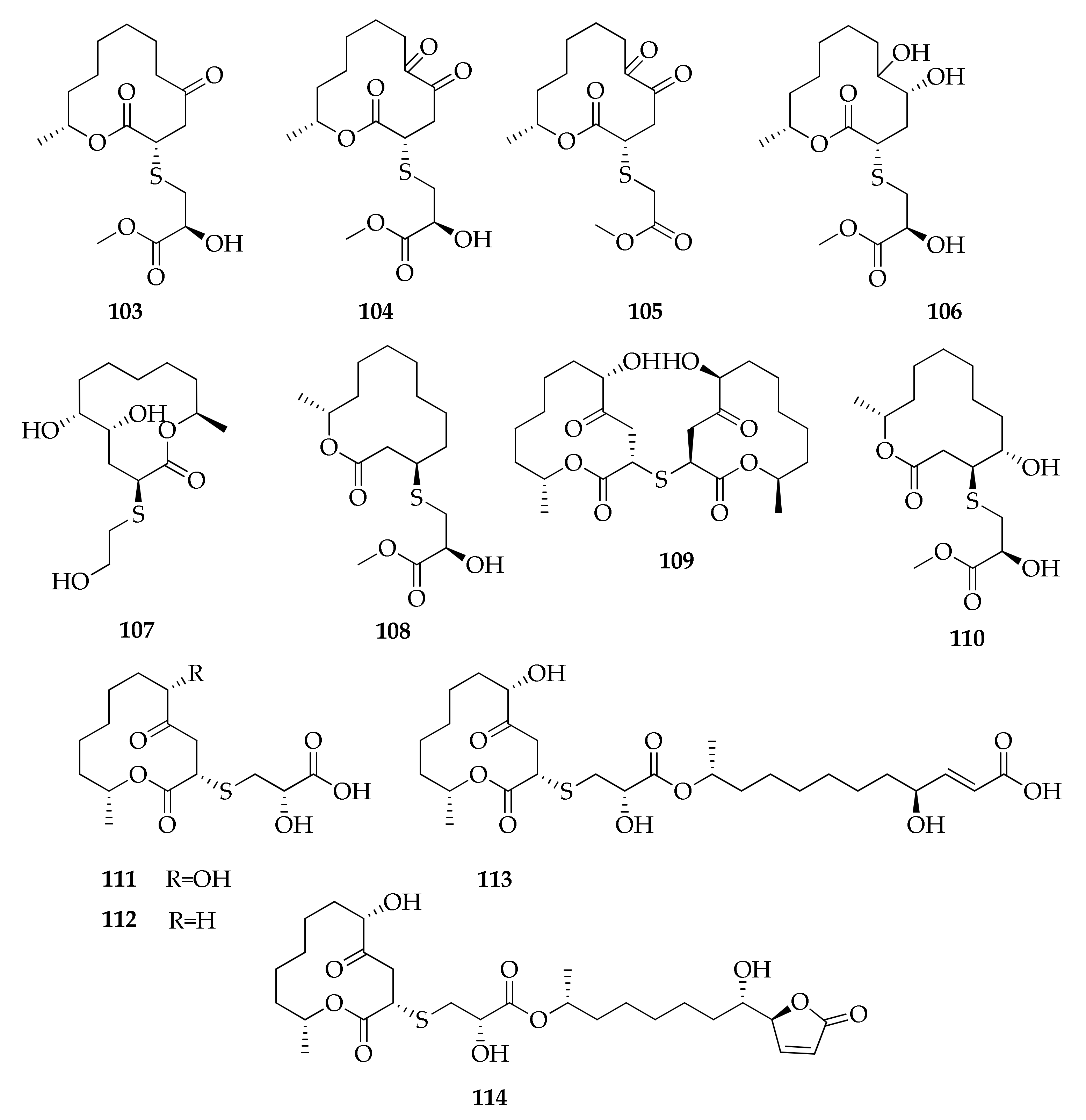
| Macrolides | ||||
| 79 | Brefeldin A | C16H24O4 | 280 | [77] |
| 80 | Cladocladosin A | C13H18O3 | 222 | [34] |
| 81 | Cladospamide A | C13H20N2O4 | 268 | [91] |
| 82 | Cladospolide A | C12H20O4 | 228 | [41,64,67,78,91] |
| 83 | Cladospolide B | C12H20O4 | 228 | [44,64,67,78] |
| 84 | Cladospolide C | C12H20O4 | 228 | [64] |
| 85 | 4,5-Dihydroxy-12-methyloxacyclododecan-2-one | C12H22O4 | 230 | [67] |
| 86 | (6R,12S)-6-Hydroxy-12-methyl- oxacyclodoecane-2,5-dione | C12H20O4 | 228 | [67] |
| 87 | (10S,12S)-10-Hydroxy-12-methyloxacyclododecane-2,5-dione | C12H20O4 | 228 | [67] |
| 88 | 4-Hydroxy-12-methyloxacyclododecane-2,5,6-trione | C12H18O5 | 242 | [41] |
| 89 | (E)-(3R,6S)-6-Hydroxy-12-methyl-2,5-dioxooxacyclododecan-3-yl 4,11-dihydroxydodec-2-enoate | C24H40O8 | 456 | [78] |
| 90 | 5R-Hydroxyrecifeiolide | C12H20O3 | 212 | [32] |
| 91 | 5S-Hydroxyrecifeiolide | C12H20O3 | 212 | [32] |
| 92 | 12-Methyloxacyclododecane-2,5,6-trione | C12H18O4 | 226 | [41] |
| 93 | Methyl 2-(((4R,6S,12R)-6-hydroxy-12-methyl-2,5-dioxooxacyclododecan-4-yl)thio)acetate | C15H24O6S | 332 | [78] |
| 94 | 5Z-7-Oxozeaenol | C19H22O7 | 362 | [49] |
| 95 | Pandangolide 1 | C12H20O5 | 244 | [32,41,48,71,78] |
| 96 | Pandangolide 1a | C12H20O5 | 244 | [71,78] |
| 97 | Pandangolide 2 | C14H22O6S | 318 | [44,78] |
| 98 | Pandangolide 3 | C16H26O7S | 362 | [33,44,50,78] |
| 99 | Pandangolide 4 | C24H38O8S | 486 | [44] |
| 100 | Patulolide B | C13H20O3 | 224 | [41] |
| 101 | Sporiolide A | C19H24O6 | 348 | [88] |
| 102 | Sporiolide B | C14H24O4 | 256 | [88] |
| 103 | Thiocladospolide A | C16H26O6S | 346 | [33,50] |
| 104 | Thiocladospolide B | C16H24O7S | 360 | [33] |
| 105 | Thiocladospolide C | C15H22O6S | 330 | [33] |
| 106 | Thiocladospolide D | C16H28O7S | 364 | [33] |
| 107 | Thiocladospolide E | C14H26O5S | 306 | [91] |
| 108 | Thiocladospolide F | C16H28O5S | 332 | [34] |
| 109 | Thiocladospolide F bis | C24H38O8S | 486 | [50] |
| 110 | Thiocladospolide G | C16H28O6S | 348 | [34] |
| 111 | Thiocladospolide G bis | C15H24O7S | 348 | [50] |
| 112 | Thiocladospolide H | C15H24O6S | 332 | [50] |
| 113 | Thiocladospolide I | C27H44O10S | 560 | [50] |
| 114 | Thiocladospolide J | C27H42O10S | 558 | [50] |
| 115 | Zeaenol | C19H24O7 | 364 | [49] |
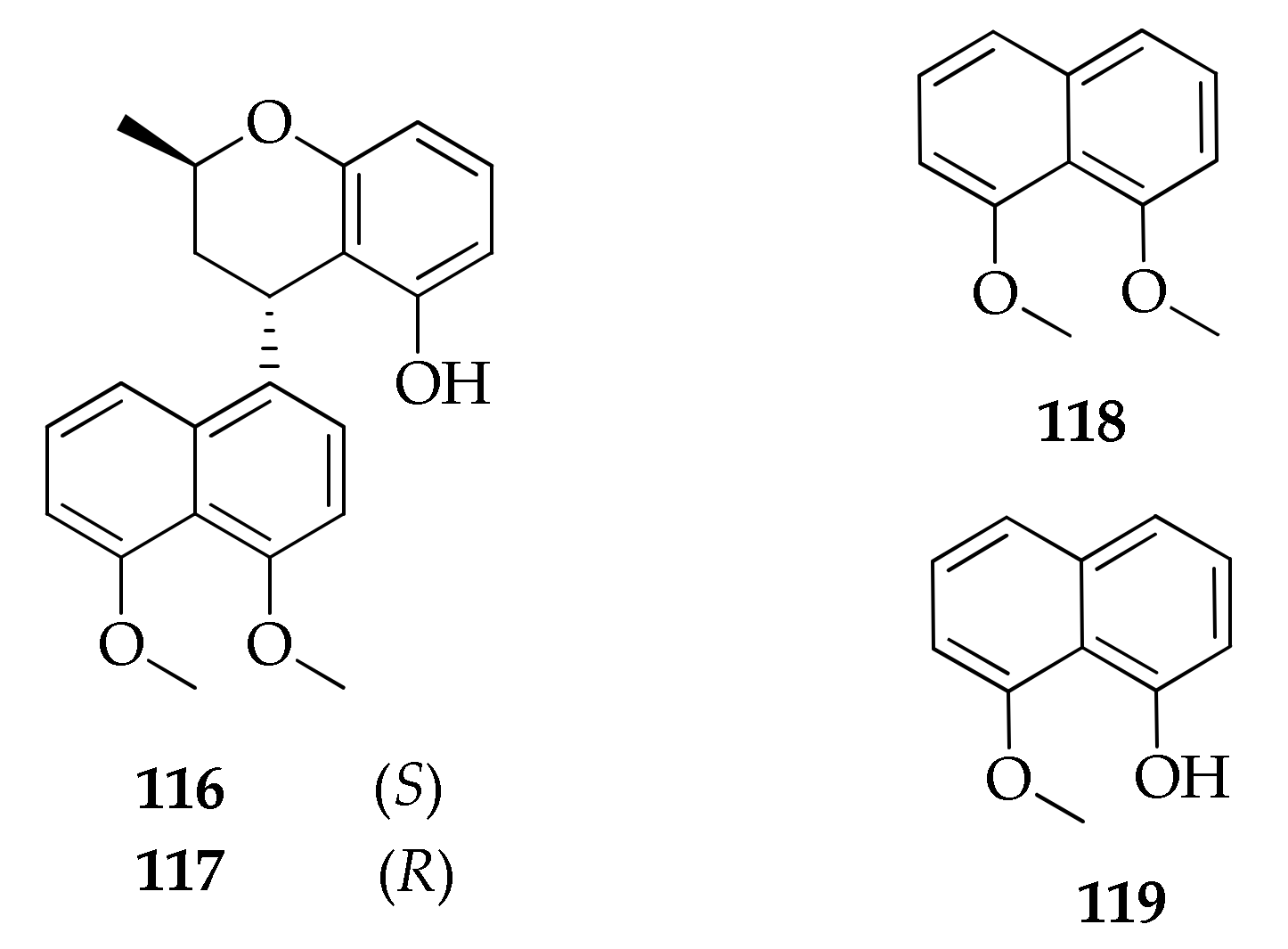
| Naphthalene derivatives | ||||
| 116 | Cladonaphchrom A | C22H22O4 | 350 | [83] |
| 117 | Cladonaphchrom B | C22H22O4 | 350 | [83] |
| 118 | 1,8-Dimethoxynaphthalene | C12H12O2 | 188 | [81,83] |
| 119 | 8-Methoxynaphthalen-1-ol | C11H10O2 | 174 | [83] |
| Naphtalenones | ||||
| 120 | Altertoxin XII | C20H18O4 | 322 | [87] |
| 121 | Cladosporol A | C20H16O6 | 352 | [26,66,86] |
| 122 | Cladosporol B | C20H14O6 | 350 | [66] |
| 123 | Cladosporol C | C20H18O5 | 338 | [35,39,40,66,85,86] |
| 124 | Cladosporol D | C20H18O6 | 354 | [66,86] |
| 125 | Cladosporol E | C20H18O7 | 370 | [40,66,85] |
| 126 | Cladosporol F | C21H20O5 | 352 | [39,40] |
| 127 | Cladosporol G | C20H17ClO6 | 388 | [40] |
| 128 | Cladosporol G bis | C21H20O5 | 352 | [39] |
| 129 | Cladosporol H | C20H16O5 | 336 | [39] |
| 130 | Cladosporol I | C20H18O5 | 338 | [39,92] |
| 131 | Cladosporol J | C20H18O5 | 338 | [39] |
| 132 | Cladosporone A | C20H16O6 | 352 | [86] |
| 133 | Clindanone A | C22H18O7 | 394 | [40] |
| 134 | Clindanone B | C22H18O7 | 394 | [40] |
| 135 | (3R,4R)-3,4-Dihydro-3,4,8-trihydroxy-1(2H)-napthalenone | C10H10O4 | 194 | [81] |
| 136 | 4,8-Dihydroxy-1-tetralone | C10H10O3 | 178 | [52] |
| 137 | (3S)-3,8-Dihydroxy-6,7-dimethyl-α-tetralone | C12H14O3 | 206 | [81] |
| 138 | Isosclerone | C10H10O3 | 178 | [40] |
| 139 | Scytalone | C10H10O4 | 194 | [68] |
| 140 | (−)-(4R)-Regiolone | C10H10O3 | 178 | [81] |
| 141 | (−)-trans-(3R,4R)-3,4,8-Trihydroxy-6,7-dimethyl-3,4- dihydronaphtha- len-1(2H)-one | C12H14O4 | 222 | [81] |
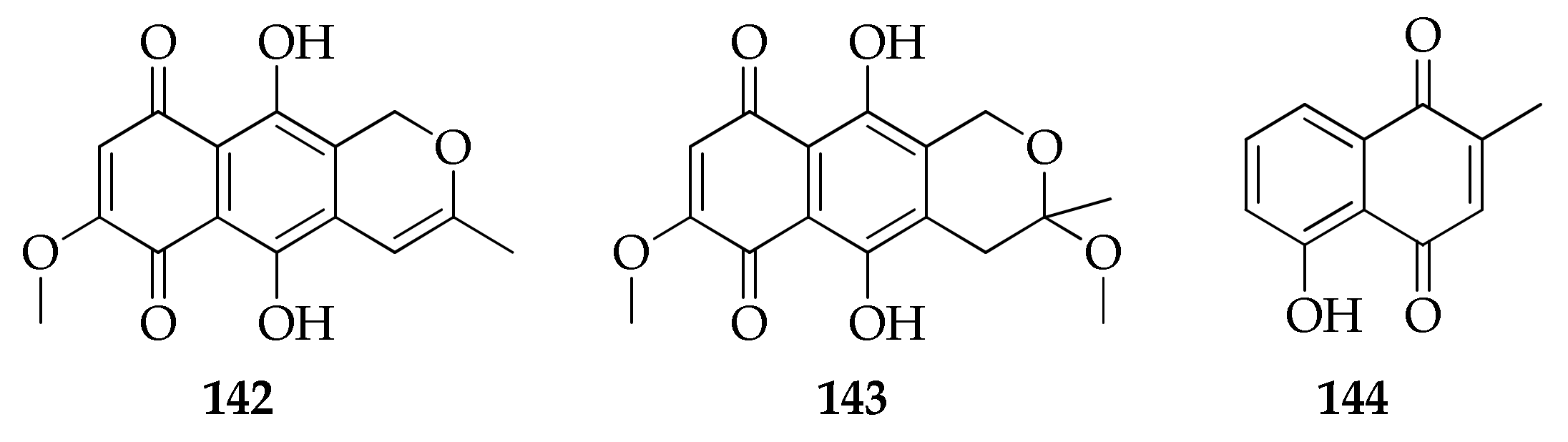
| Naphthoquinones and anthraquinones | ||||
| 142 | Anhydrofusarubin | C15H12O6 | 288 | [90] |
| 143 | Methyl ether of fusarubin | C16H16O7 | 320 | [90] |
| 144 | Plumbagin | C11H8O3 | 188 | [42] |
| Perylenequinones | ||||
| 145 | Altertoxin VIII | C20H16O3 | 304 | [87] |
| 146 | Altertoxin IX | C20H18O2 | 290 | [87] |
| 147 | Aterotoxin X | C20H18O2 | 290 | [87] |
| 148 | Altertoxin XI | C21H20O2 | 304 | [87] |
| 149 | Calphostin A (= UCN-1028A) | C44H38O12 | 758 | [29,30] |
| 150 | Calphostin B | C37H34O11 | 654 | [30] |
| 151 | Calphostin C (= Cladochrome E) | C44H38O14 | 790 | [30,31] |
| 152 | Calphostin D (= ent-isophleinchrome) | C30H30O10 | 550 | [30,46] |
| 153 | Calphostin I (= Cladochrome D) | C44H38O15 | 806 | [30,31] |
| 154 | Phleichrome | C30H30O10 | 550 | [53,54,99] |
| Pyrones | ||||
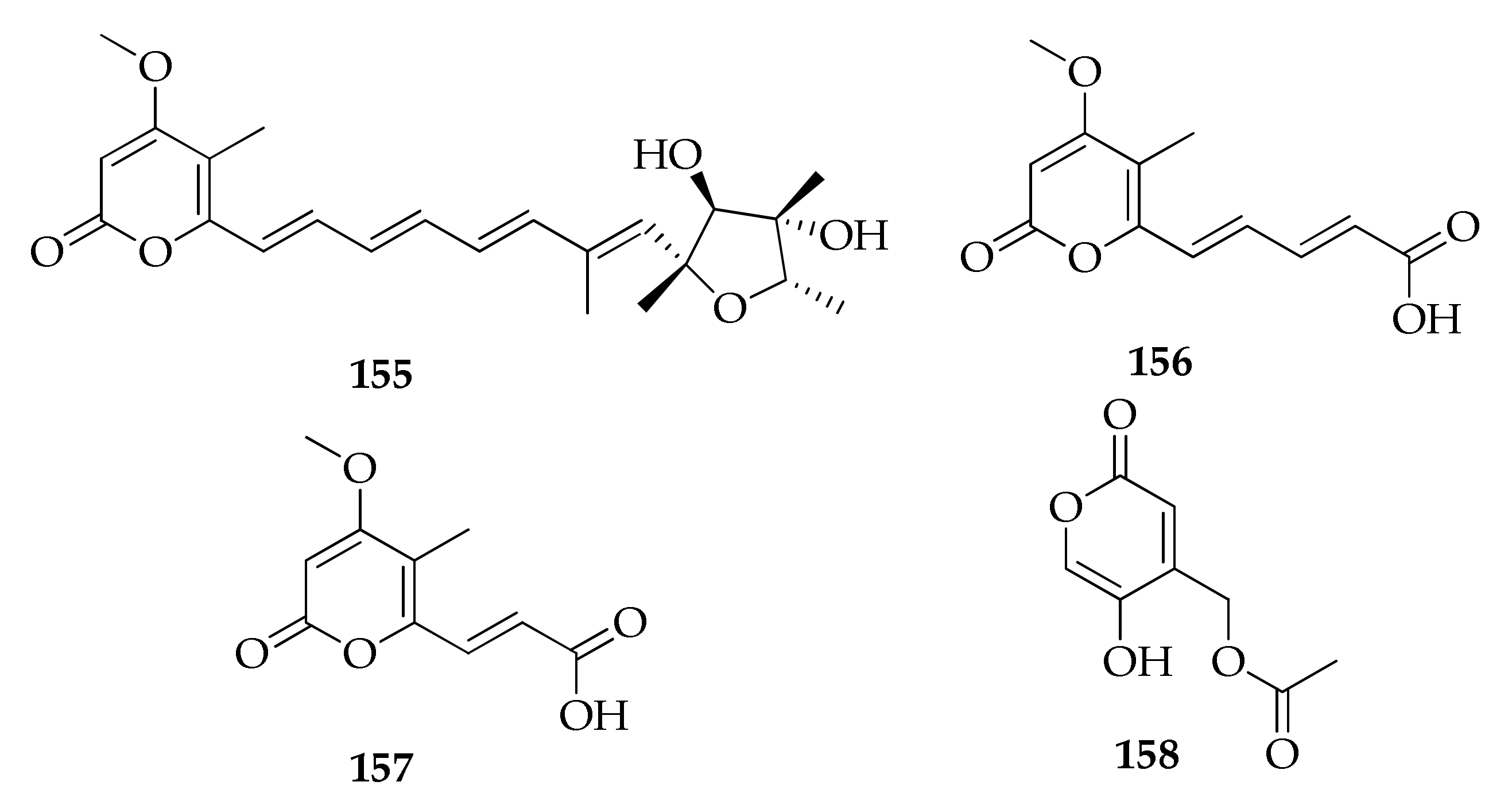
| 155 | Citreoviridin A | C23H30O6 | 402 | [45] |
| 156 | Herbarin A | C12H12O5 | 236 | [45] |
| 157 | Herbarin B | C10H10O5 | 210 | [45] |
| 158 | 5-Hydroxy-2-oxo-2H-piran-4-yl) methyl acetate | C8H8O5 | 184 | [65] |
| Seco acids | ||||
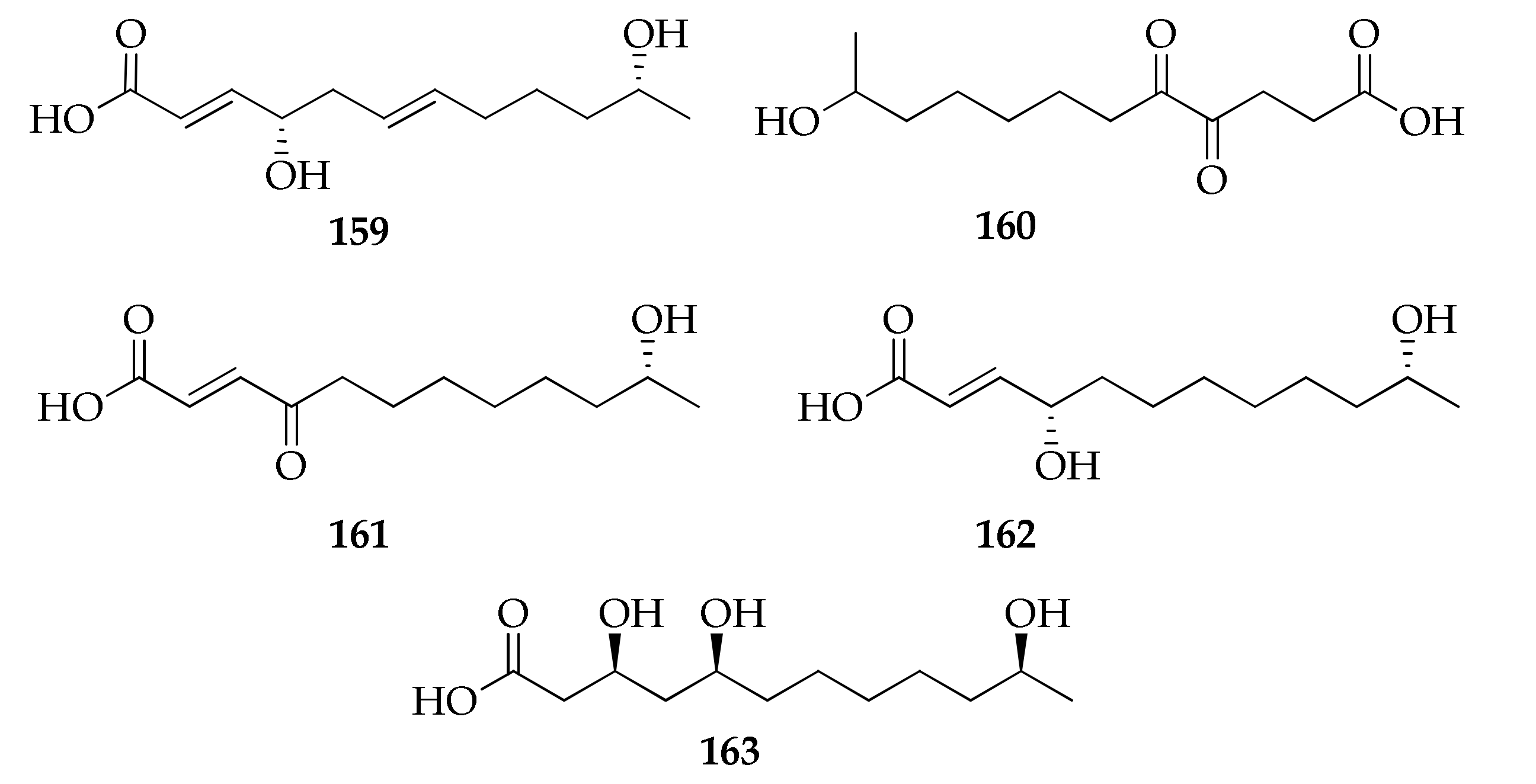
| 159 | Cladospolide E | C12H20O4 | 228 | [94] |
| 160 | 11-Hydroxy-4,5-dioxododecanoic acid | C12H20O5 | 244 | [78] |
| 161 | seco-Patulolide A | C12H20O4 | 228 | [94] |
| 162 | seco-Patulolide C | C12H22O4 | 230 | [33,41,50,94] |
| 163 | (3S,5S, 11S)-Trihydroxydodecanoic acid | C12H24O5 | 248 | [68] |
| Sterols | ||||
| 164 | Cladosporide A | C25H40O3 | 388 | [79] |
| 165 | Cladosporide B | C25H38O3 | 386 | [80] |
| 166 | Cladosporide C | C25H40O3 | 388 | [80] |
| 167 | Cladosporide D | C25H38O3 | 386 | [80] |
| 168 | Cladosporisteroid B | C21H30O3 | 330 | [35] |
| 169 | (22E,24R)-3β,5α-Dihydroxyergosta-7,22-dien-6-one | C28H44O3 | 428 | [35] |
| 170 | Ergosterol | C28H44O | 396 | [79] |
| 171 | 3α-Hydroxy-pregn-7-ene-6,20-dione | C21H30O3 | 330 | [62] |
| 172 | 23,24,25,26,27-Pentanorlanost-8-ene-3β,22-diol | C28H42O5 | 458 | [79] |
| 173 | Peroxyergosterol (= (22E)-5α,8α-epidioxyergosta-6,22-dien-3β-ol) | C28H44O3 | 428 | [23,35,79] |
| 174 | 3β,5α,6β-Trihydroxyergosta-7,22-diene | C29H48O3 | 444 | [47] |
| 175 | 3β,5α,9α-Trihydroxy- (22E,24R)-ergosta-6-one | C28H44O4 | 444 | [35] |
| Tetramic acids | ||||
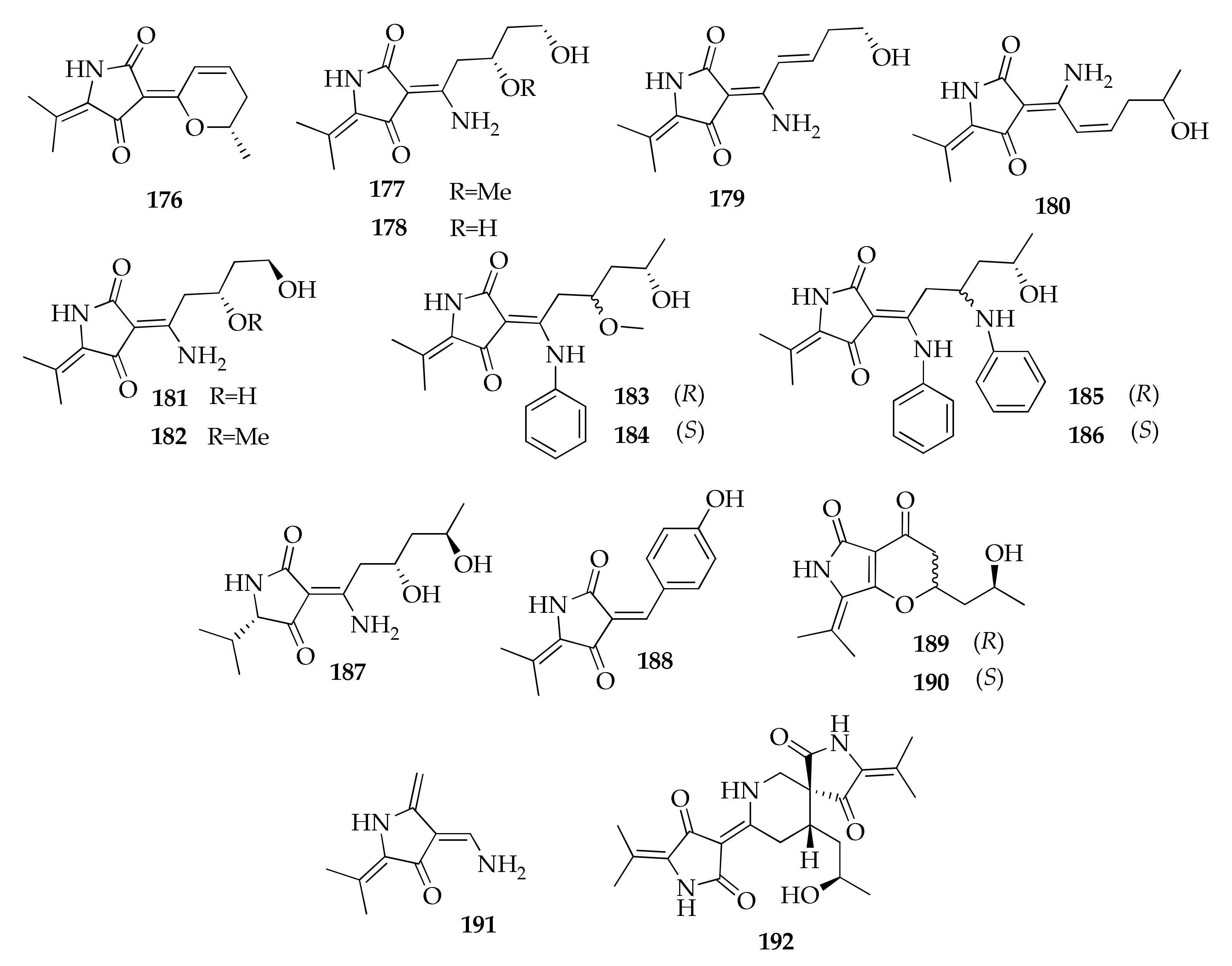
| 176 | Cladodionen | C13H15NO3 | 233 | [58,63,89,96] |
| 177 | Cladosin A | C13H20N2O4 | 268 | [55] |
| 178 | Cladosin B | C12H18N2O4 | 254 | [55,60,63] |
| 179 | Cladosin C | C12H16N2O3 | 236 | [55,60,63] |
| 180 | Cladosin D | C13H18N2O3 | 250 | [55] |
| 181 | Cladosin F | C12H18N2O4 | 254 | [56,60,63] |
| 182 | Cladosin G | C13H20N2O4 | 268 | [56] |
| 183 | Cladosin H | C20H26N2O4 | 358 | [89] |
| 184 | Cladosin I | C20H26N2O4 | 358 | [89] |
| 185 | Cladosin J | C25H29N3O3 | 419 | [89] |
| 186 | Cladosin K | C25H29N3O3 | 419 | [89] |
| 187 | Cladosin L | C13H22N2O4 | 270 | [60] |
| 188 | Cladosin L bis | C14H13NO3 | 243 | [63] |
| 189 | Cladosin M | C13H17NO4 | 251 | [63] |
| 190 | Cladosin N | C13H17NO4 | 251 | [63] |
| 191 | Cladosin O | C9H12N2O | 164 | [63] |
| 192 | Cladosporicin A | C21H27N3O5 | 401 | [61] |
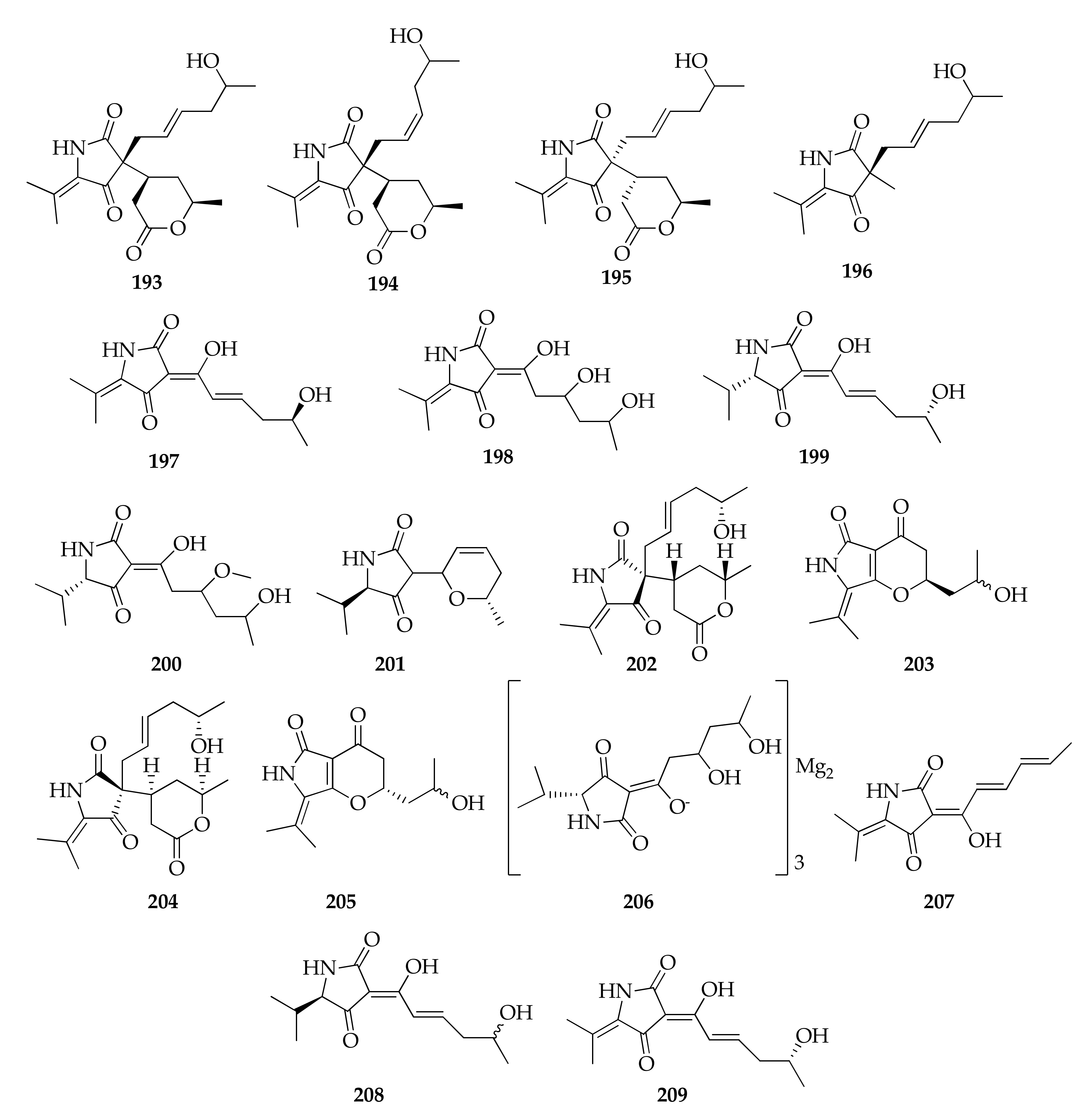
| 193 | Cladosporiumin A | C19H27NO5 | 349 | [59] |
| 194 | Cladosporiumin B | C19H27NO5 | 349 | [59] |
| 195 | Cladosporiumin C | C19H27NO5 | 349 | [59] |
| 196 | Cladosporiumin D | C14H21NO3 | 251 | [59] |
| 197 | Cladosporiumin E | C13H17NO4 | 251 | [58,59] |
| 198 | Cladosporiumin F | C13H19NO5 | 269 | [59] |
| 199 | Cladosporiumin G | C13H19NO4 | 253 | [58,59] |
| 200 | Cladosporiumin H | C14H23NO5 | 285 | [59] |
| 201 | Cladosporiumin I | C13H19NO3 | 237 | [58] |
| 202 | Cladosporiumin I bis | C19H27NO5 | 349 | [61] |
| 203 | Cladosporiumin J | C13H17NO4 | 251 | [58] |
| 204 | Cladosporiumin J bis | C19H27NO5 | 349 | [61] |
| 205 | Cladosporiumin K | C13H17NO4 | 251 | [58] |
| 206 | Cladosporiumin L | (C13H20NO5)3Mg2 | 889 | [58] |
| 207 | Cladosporiumin M | C13H15NO3 | 233 | [58] |
| 208 | Cladosporiumin N | C13H19NO4 | 253 | [58] |
| 209 | Cladosporiumin O | C13H17NO4 | 251 | [58] |
| Tropolones | ||||
| 210 | Malettinin A | C16H16O5 | 288 | [95] |
| 211 | Malettinin B | C16H20O5 | 292 | [95] |
| 212 | Malettinin C | C16H20O5 | 292 | [95] |
| 213 | Malettinin E | C16H20O5 | 292 | [95] |
| Volatile terpenes | ||||
| 214 | (−)-trans-Caryophyllene | C15H24 | 204 | [38] |
| 215 | Dehydro aromdendrene | C15H22 | 202 | [38] |
| 216 | α-Pinene | C10H16 | 136 | [38] |
| 217 | (+)-Sativene | C15H24 | 204 | [38] |
| Xanthones | ||||
| 218 | Conioxanthone A | C16H12O7 | 316 | [43] |
| 219 | 3,8-Dihydroxy-6-methyl-9-oxo-9H-xanthene-1-carboxylate | C16H12O6 | 300 | [43] |
| 220 | α-Diversonolic ester | C16H16O7 | 320 | [43] |
| 221 | β-Diversonolic ester | C16H16O7 | 320 | [43] |
| 222 | 8-Hydroxy-6-methylxanthone-1-carboxylic acid | C15H12O5 | 272 | [43] |
| 223 | Methyl 8-hydroxy-6-(hydroxymethyl)- 9-oxo-9H-xanthene-1-carboxylate | C16H12O6 | 300 | [43] |
| 224 | Methyl 8-hydroxy-6-methyl-9-oxo-9H-xanthene-1- carboxylate | C16H12O5 | 284 | [43] |
| 225 | 8-(Methoxycarbonyl)-1-hydroxy-9-oxo-9H-xanthene-3-carboxylic acid | C16H10O7 | 314 | [43] |
| 226 | Secalonic acid D | C32H30O14 | 638 | [85] |
| 227 | Vertixanthone | C15H10O5 | 270 | [43] |
| Miscellaneous | ||||
| 228 | α-Acetylorcinol | C9H10O3 | 166 | [52] |
| 229 | Acetyl Sumiki’s acid | C9H10O4 | 182 | [44] |
| 230 | Cladosacid | C15H22O3 | 250 | [96] |
| 231 | (2R*,4R*)-3,4-dihydro-5-methoxy-2-methyl-1(2H)-benzopyran-4-ol | C10H12O2 | 164 | [81] |
| 232 | 1-(3,5-Dihydroxy-4-methylphenyl)propan-2-one | C10H12O3 | 180 | [52] |
| 233 | 1,1′-Dioxine-2,2′-dipropionic acid | C10H12O6 | 228 | [85] |
| 234 | Ellagic acid | C14H6O8 | 302 | [70] |
| 235 | Fonsecinone A | C32H26O10 | 570 | [47] |
| 236 | 5-Hydroxy-2-methyl-4H-chromen-4-one | C10H8O3 | 176 | [83] |
| 237 | (2S)-5-Hydroxy-2-methyl-chroman-4-one | C10H10O2 | 162 | [81,83] |
| 238 | 4-O-α-d-Ribofuranose-2-pentyl-3-phemethylol | C17H26O6 | 326 | [82] |
| 239 | 4-O-α-d-Ribofuranose-3-hydroxymethyl-2-pentylphenol | C17H26O7 | 342 | [81] |
| 240 | Rubrofusarin B | C16H14O5 | 286 | [47] |
| 241 | Sumiki’s acid | C6H6O4 | 142 | [44] |
| 242 | Taxol | C47H51NO14 | 853 | [51] |
| 243 | tert-Butylhydroquinone | C10H14O2 | 166 | [70] |
| 244 | Vermistatin | C18H16O6 | 328 | [85] |
| Name (Code) | Biological Activity | Concentration | Results | Ref. |
|---|---|---|---|---|
| Alkaloids | ||||
| Aspidospermidin-20-ol, 1-acetyl-17-methoxy (2) | Antimicrobial | 125 µg mL−1; | Xanthomonas oryzae (MIC); | [24] |
| 62.50 µg mL−1; | Pseudomonas syringae (MIC); | |||
| 320.5 µg mL−1 | Aspergillus flavus (MIC) | |||
| Cladosporine A (4) | Antimicrobial | 4 μg mL−1; | Staphylococcus aureus (MIC); | [84] |
| 16 μg mL−1 | Candida albicans (MIC) | |||
| Cytochalasin D (5) | Antibacterial | 25 μg mL–1 | S. aureus (MIC) | [85] |
| 2-Methylacetate-3,5,6-trimethylpyrazine (6) | Antibacterial | 12.5 μg mL–1 | S. aureus (MIC) | [85] |
| Azaphilones | ||||
| Lunatoic acid A (10) | Phytotoxic | 100 μg mL–1 | Brassica rapa; Sorghum durra; Brassica campestris; Capsicum annuum; Raphanus sativus | [49] |
| Benzofluoranthenones | ||||
| (6bS,7R,8S)-4,9-Dihydroxy-7,8-dimethoxy-1,6b,7,8-tetra-hydro-2H-benzo[J]fluoranthen-3-one (13) | Inhibition of anti-CD28-induced IL2 | 2.4 µM | IC50 | [27] |
| (6bR,7R,8S)-7-Methoxy-4,8,9-trihydroxy-1,6b,7,8-tetrahydro-2H-benzo[J]fluoranthen-3-one (15) | Inhibition of anti-CD28-induced IL2 | 2.5 µM | IC50 | [27] |
| Abl tyrosine kinase | 0.76 µM | IC50 | ||
| (6bS,7R,8S)-7-Methoxy-4,8,9-trihydroxy-1,6b,7,8-tetrahydro-2H-benzo[J]fluoranthen-3-one (16) | Inhibition of anti-CD28-induced IL2 | 0.4 µM | IC50 | [27] |
| Abl tyrosine kinase | 0.06 µM | IC50 | ||
| Benzopyranones | ||||
| Coniochaetone A (17) | Cytotoxic | 10 µM | 22RV1 (67.4%), C4-2B (13.87%), RWPE-1 (17.3%) | [43] |
| Coniochaetone B (18) | Cytotoxic | 10 µM | 22RV1 (32.7%), C4-2B (2.9%), RWPE-1 (19.7%) | [43] |
| Coniochaetone K (19) | Cytotoxic | 10 µM | 22RV1 (64.6%), C4-2B (7.2%), RWPE-1 (11.7%) | [43] |
| Binaphthopyrones | ||||
| Cladosporinone (20) | Cytotoxic | 53.7 µM | L5178Y (IC50) | [22] |
| Antibacterial | 64 µg mL−1 | S. aureus (MIC) | ||
| Viriditoxin (21) | Cytotoxic | 0.1 µM | L5178Y (IC50) | [22] |
| Antibacterial | 0.015 µg mL−1 | S. aureus (MIC) | ||
| Viriditoxin SC-28763 (22) | Cytotoxic | 0.25 µM | L5178Y (IC50) | [22] |
| Antibacterial | 2 µg mL−1 | S. aureus (MIC) | ||
| Viriditoxin SC-30532 (23) | Antibacterial | 16 µg mL−1 | S. aureus (MIC) | [22] |
| Butenolides and butanolides | ||||
| Cladospolide F (24) | Lipid accumulation | 10 µM | Oleic acid | [94] |
| Cladospolide G (25) | Antimicrobial | 32 µg mL−1; | E. coli (MIC); | [32] |
| 1 µg mL−1; | Glomerella cingulata (MIC); | |||
| 32 µg mL−1; | Bipolaris sorokiniana (MIC); | |||
| 1 µg mL−1 | Fusarium oxysporum f. sp. cucumerinum (MIC) | |||
| ent-Cladospolide F (27) | Antibacterial | 8 µg mL−1; | S. aureus (MIC); | [32] |
| 16 µg mL−1; | Edwardsiella ictarda (MIC); | |||
| 64 µg mL−1 | P. aeruginosa (MIC) | |||
| Acetylcholinesterase | 40.46 µM | IC50 | ||
| 11-Hydroxy-γ-dodecalactone (28) | Lipid accumulation | 10 µM | Oleic acid | [94] |
| iso-Cladospolide B (29) | Antimicrobial | 32 µg mL−1; | E. coli (MIC); | [32] |
| 32 µg mL−1; | Edwardsiella tarda (MIC); | |||
| 16 µg mL−1; | E. ictarda (MIC); | |||
| 64 µg mL−1 | G. cingulata (MIC) | |||
| Antimicrobial | 16 µg mL−1; | E. tarda (MIC); | [50] | |
| 8 µg mL−1; | E. ictarda (MIC); | |||
| 8 µg mL−1; | Clematis mandshurica Miura (MIC); | |||
| 16 µg mL−1; | Colletotrichum gloeosporioides (MIC); | |||
| 32 µg mL−1; | B. sorokiniana (MIC); | |||
| 32 µg mL−1 | F. oxysporum f. sp. cucumerinum (MIC) | |||
| Citrinin dervatives | ||||
| Citrinin H1 (33) | Antibacterial | 6.25 μg mL−1; | S. aureus (MIC); | [85] |
| 12.5 μg mL−1; | E. coli (MIC); | |||
| 12.5 μg mL−1 | B. cereus (MIC) | |||
| Cladosporin A (34) | Toxic | 72.0 µM | brine shrine nauplii (IC50) | [92] |
| Cladosporin B (35) | Toxic | 81.7 µM | brine shrine nauplii (IC50) | [92] |
| Cladosporin C (36) | Toxic | 49.9 µM | brine shrine nauplii (IC50) | [92] |
| Cladosporin D (37) | Antioxidant | 16.4 µM | DPPH radicals (IC50) | [92] |
| Toxic | 81.4 µM | brine shrine nauplii (IC50) | ||
| Coumarins and isocoumarins | ||||
| Cladosporin (39) | Antimicrobial | 75 µg mL−1; | dermatophytes (100%); | [12] |
| 40 µg mL−1 | spore germination of Penicillium sp. (100%) and Aspergillus sp. (100%) | |||
| 30 µM | Colletotrichum acutatum (92.7%), Colletotrichum fragariae (90.1%), C. gloeosporioides (95.4%), Plasmopara viticola (79.9%) | [25] | ||
| 500 µg mL−1; | X. oryzae (MIC), A. flavus (MIC); | [24] | ||
| 62.50 µg mL−1 | Fusarium solani (MIC) | |||
| Phytotoxic | 10−3 M | etiolated wheat (81%) | [37] | |
| 5′-Hydroxyasperentin (40) | Antimicrobial | 15.62 µg mL−1; | X. oryzae (MIC); | [24] |
| 62.50 µg mL−1; | P. syringae (MIC); | |||
| 15.62 µg mL−1; | A. flavus (MIC); | |||
| 7.81 µg mL−1 | F. solani (MIC) | |||
| 30 µM | P. viticola (53.9%), Phomopsis obscurans (25.6%) | [25] | ||
| Isocladosporin (41) | Antimicrobial | 30 µM | C. fragariae (50.4%), C. gloeosporioides (60.2%), P. viticola (83.0%) | [25] |
| 15.62 µg mL−1; | X. oryzae (MIC), P. syringae (MIC); | [24] | ||
| 125 µg mL−1; | A. flavus (MIC); | |||
| 62.50 µg mL−1 | F. solani (MIC) | |||
| Phytotoxic | 10−3 M | etiolated wheat (100%) | [37] | |
| Cyclohexene derivatives | ||||
| Cladoscyclitol B (48) | Inhibition of α-glucosidase | 2.95 µM | IC50 | [82] |
| Depsides | ||||
| 3-Hydroxy-2,4,5-trimethylphenyl 4-[(2,4-dihydroxy-3,6-dimethylbenzoyl)oxy]-2-hydroxy-3,6-dimethylbenzoate (51) | Antimicrobial | 25 µg mL−1; | B. subtilis (bacteriostatic); | [69,98] |
| 25 µg mL−1; | P. aeruginosa (bacteriostatic); | |||
| 250 µg mL−1; | E. coli (bacteriostatic); | |||
| 250 µg mL−1 | S. aureus (bacteriostatic) | |||
| 3-Hydroxy-2,5-dimethylphenyl 2,4-dihydroxy-3,6-dimethylbenzoate (53) | Antimicrobial | 25 µg mL−1; | B. subtilis (MIC); | [69,98] |
| 25 µg mL−1; | P. aeruginosa (MIC); | |||
| 250 µg mL−1; | E. coli (MIC); | |||
| 250 µg mL−1; | S. aureus (MIC) | |||
| 3-Hydroxy-2,5-dimethylphenyl 4-[(2,4-dihydroxy-3,6-dimethylbenzoyl)oxy]-2-hydroxy-3,6-dimethylbenzoate (54) | Antimicrobial | 250 µg mL−1; | B. subtilis (bacteriostatic); | [69,98] |
| 250 µg mL−1; | P. aeruginosa (bacteriostatic); | |||
| 250 µg mL−1; | E. coli (bacteriostatic); | |||
| 250 µg mL−1 | S. aureus (bacteriostatic) | |||
| Flavonoids | ||||
| (2S)-7,4′-Dihydroxy-5-methoxy-8-(γ,γ-dimethylallyl)- flavanone (59) | Enzymatic inhibitory | 11 µM; | PTP1B (IC50); | [93] |
| 27 µM | TCPTP (IC50) | |||
| Lactones | ||||
| Cladosporamide A (75) | Enzymatic inhibitory | 48 µM; | PTP1B (IC50); | [93] |
| 54 µM | TCPTP (IC50) | |||
| Macrolides | ||||
| Cladocladosin A (80) | Antimicrobial | 16 µg mL−1; | E. coli (MIC); | [34] |
| 1 µg mL−1; | E. tarda (MIC); | |||
| 4 µg mL−1; | P. aeruginosa (MIC); | |||
| 2 µg mL−1; | Vibrio anguillarum (MIC); | |||
| 32 µg mL−1; | F. oxysporium f. sp. momordicae (MIC); | |||
| 32 µg mL−1; | Penicillium digitatum (MIC); | |||
| 8 µg mL−1 | Harpophora maydis (MIC) | |||
| Cladospolide B (83) | Phytotoxic | 1 µg plant−1 | Oryza sativa (37.8%) | [64] |
| 5R-Hydroxyrecifeiolide (90) | Antimicrobial | 32 µg mL−1; | E. ictarda (MIC); | [32] |
| 32 µg mL−1 | P. aeruginosa (MIC) | |||
| 5S-Hydroxyrecifeiolide (91) | Antimicrobial | 16 µg mL−1 | G. cingulata (MIC) | [32] |
| 5Z-7-Oxozeaenol (94) | Phytotoxic | 4.8 µg mL−1 | Amaranthus retroflexus (IC50) | [49] |
| Pandangolide 1 (95) | Antimicrobial | 32 µg mL−1; | S. aureus (MIC); | [32] |
| 4 µg mL−1; | E. ictarda (MIC); | |||
| 1 µg mL−1; | G. cingulata (MIC); | |||
| 32 µg mL−1 | P. aeruginosa (MIC) | |||
| Pandangolide 3 (98) | Antimicrobial | 2 µg mL−1; | C. gloeosporioides (MIC); | [33] |
| 8 µg mL−1 | B. sorokiniana (MIC) | |||
| 32 µg mL−1; | E. tarda (MIC); | [50] | ||
| 32 µg mL−1; | E. ictarda (MIC); | |||
| 32 µg mL−1; | C. gloeosporioides (MIC); | |||
| 16 µg mL−1 | F. oxysporum f. sp. cucumerinum (MIC) | |||
| Sporiolide A (101) | Antimicrobial | 16.7 µg mL−1; | Micrococcus luteus (MIC); | [88] |
| 16.7 µg mL−1; | C. albicans (MIC); | |||
| 8.4 µg mL−1; | Cryptococcus neoformans (MIC); | |||
| 16.7 µg mL−1; | Aspergillus niger (MIC); | |||
| 8.4 µg mL−1 | Neurospora crassa (MIC) | |||
| Cytotoxic | 0.13 µg mL−1 | L1210 (IC50) | ||
| Sporiolide B (102) | Antimicrobial | 16.7 µg mL−1 | M. luteus (MIC) | [88] |
| Cytotoxic | 0.81 µg mL−1 | L1210 (IC50) | ||
| Thiocladospolide A (103) | Antimicrobial | 1 µg mL−1; | E. tarda (MIC); | [33] |
| 8 µg mL−1; | E. ictarda (MIC); | |||
| 2 µg mL−1 | C. glecosporioides (MIC) | |||
| 32 µg mL−1; | E. tarda (MIC); | [50] | ||
| 32 µg mL−1; | E. ictarda (MIC); | |||
| 16 µg mL−1 | C. gloeosporioides (MIC) | |||
| Thiocladospolide B (104) | Antimicrobial | 2 µg mL−1; | C. gloeosporioides (MIC); | [33] |
| 32 µg mL−1; | Physalospora piricola (MIC); | |||
| 1 µg mL−1ù | F. oxysporum (MIC) | |||
| Thiocladospolide C (105) | Antimicrobial | 1 µg mL−1; | C. gloeosporioides (MIC); | [33] |
| 32 µg mL−1; | P. piricola (MIC); | |||
| 32 µg mL−1 | F. oxysporum (MIC) | |||
| Thiocladospolide D (106) | Antimicrobial | 1 µg mL−1; | E. ictarda (MIC); | [33] |
| 1 µg mL−1; | C. gloeosporioides (MIC); | |||
| 32 µg mL−1; | P. piricola (MIC); | |||
| 1 µg mL−1 | F. oxysporum (MIC) | |||
| Thiocladospolide F (108) | Antimicrobial | 16 µg mL−1; | E. coli (MIC); | [34] |
| 2 µg mL−1; | E. tarda (MIC); | |||
| 2 µg mL−1; | V. anguillarum (MIC); | |||
| 16 µg mL−1; | F. oxysporium f. sp. momordicae (MIC); | |||
| 16 µg mL−1; | P. digitatum (MIC); | |||
| 4 µg mL−1 | H. maydis (MIC) | |||
| Thiocladospolide F bis (109) | Antimicrobial | 32 µg mL−1; | E. tarda (MIC); | [50] |
| 16 µg mL−1; | E. ictarda (MIC); | |||
| 16 µg mL−1 | B. sorokiniana (MIC) | |||
| Thiocladospolide G (110) | Antimicrobial | 2 µg mL−1; | E. tarda (MIC); | [34] |
| 2 µg mL−1; | V. anguillarum (MIC); | |||
| 32 µg mL−1; | F. oxysporium f. sp. momordicae (MIC); | |||
| 32 µg mL−1; | P. digitatum (MIC); | |||
| 8 µg mL−1 | H. maydis (MIC) | |||
| Thiocladospolide G bis (111) | Antimicrobial | 4 µg mL−1; | E. tarda (MIC); | [50] |
| 32 µg mL−1; | E. ictarda (MIC); | |||
| 32 µg mL−1; | C. mandshurica Miura (MIC); | |||
| 16 µg mL−1; | C. gloeosporioides (MIC); | |||
| 32 µg mL−1 | F. oxysporum f. sp. cucumerinum (MIC) | |||
| Thiocladospolide H (112) | Antimicrobial | 16 µg mL−1; | E. tarda (MIC); | [50] |
| 8 µg mL−1; | E. ictarda (MIC); | |||
| 16 µg mL−1; | C. gloeosporioides (MIC); | |||
| 16 µg mL−1 | B. sorokiniana (MIC) | |||
| Thiocladospolide I (113) | Antimicrobial | 32 µg mL−1; | E. tarda (MIC); | [50] |
| 32 µg mL−1; | E. ictarda (MIC); | |||
| 16 µg mL−1 | F. oxysporum f. sp. cucumerinum (MIC) | |||
| Thiocladospolide J (114) | Antimicrobial | 16 µg mL−1; | E. tarda (MIC); | [50] |
| 16 µg mL−1; | E. ictarda (MIC); | |||
| 16 µg mL−1; | C. mandshurica Miura (MIC); | |||
| 16 µg mL−1; | C. gloeosporioides (MIC); | |||
| 32 µg mL−1; | B. sorokiniana (MIC); | |||
| 16 µg mL−1 | F. oxysporum f. sp. cucumerinum (MIC) | |||
| Zeaenol (115) | Phytotoxic | 8.16 µg mL−1 | A. retroflexus (IC50) | [49] |
| Naphthalene derivatives | ||||
| Cladonaphchrom A (116) | Antimicrobial | 1.25 µg mL−1; | Scaphirhynchus albus (MIC); | [83] |
| 2.5 µg mL−1; | E. coli (MIC); | |||
| 10 µg mL−1; | B. subtilis (MIC); | |||
| 5 µg mL−1; | Micrococcus tetragenus (MIC); | |||
| 10 µg mL−1; | M. luteus (MIC); | |||
| 50 µg mL−1; | Alternaria brassicicola (MIC); | |||
| 50 µg mL−1; | Phytophthora parasitica var. nicotianae (MIC); | |||
| 25 µg mL−1; | Colletotrichum capsici (MIC); | |||
| 100 µg mL−1; | B. oryzae (MIC); | |||
| 50 µg mL−1; | Diaporthe medusaea (MIC); | |||
| 50 µg mL−1 | Cyanophora paradoxa (MIC) | |||
| Cladonaphchrom B (117) | Antibacterial | 2.5 µg mL−1; | S. albus (MIC); | [83] |
| 2.5 µg mL−1; | E. coli (MIC); | |||
| 5 µg mL−1; | B. subtilis (MIC); | |||
| 5 µg mL−1; | M. tetragenus (MIC); | |||
| 10 µg mL−1; | M. luteus (MIC); | |||
| 25 µg mL−1; | A. brassicicola (MIC); | |||
| 50 µg mL−1; | P. parasitica var. nicotianae (MIC); | |||
| 25 µg mL−1; | C. capsici (MIC); | |||
| 100 µg mL−1; | D. medusaea (MIC); | |||
| 50 µg mL−1 | C. paradoxa (MIC) | |||
| Naphtalenones | ||||
| Altertoxin XII (120) | Quorum sensing inhibitory | 20 µg well−1 | Chromobacterium violaceum (MIC) | [87] |
| Cladosporol A (121) | Antifungal | 100 ppm | Uromyces appendiculatus (84.2%) | [66] |
| Β-1,3-glucan biosynthesis inhibitor | 10 µg ml | IC50 | [26] | |
| Cladosporol B (122) | Antifungal | 100 ppm | U. appendiculatus (100%) | [66] |
| Cladosporol C (123) | Antifungal | 100 ppm | U. appendiculatus (77.6%) | [66] |
| Antibacterial | 8 µg mL−1; | E. coli (MIC); | [39] | |
| 64 µg mL−1; | M. luteus (MIC); | |||
| 16 µg mL−1 | Vibrio harveyi (MIC) | |||
| Cytotoxic | 33.9 µM; | A549 (100%); | [86] | |
| 45.6 µM; | H1975 (100%); | |||
| 72.5 µM; | HL60 (100%); | |||
| 11.4 µM | MOLT-4 (100%) | |||
| 14 µM; | A549 (IC50); | [39] | ||
| 4 µM | H446 (IC50) | |||
| Cladosporol D (124) | Antifungal | 100 ppm | U. appendiculatus (69.4%) | [66] |
| Anti-COX-2 | 60.2 µM | IC50 | [86] | |
| Cladosporol E (125) | Antifungal | 100 ppm | U. appendiculatus (74.8%) | [66,85] |
| Cladosporol F (126) | Antibacterial | 32 µg mL−1; | E. coli (MIC); | [39] |
| 64 µg mL−1; | M. luteus (MIC); | |||
| 32 µg mL−1 | V. harveyi (MIC) | |||
| Cytotoxic | 15 µM; | A549 (IC50); | [39,40] | |
| 10 µM; | HeLa (IC50); | |||
| 23 µM; | K562 (IC50); | |||
| 23 µM | HCT-116 (IC50) | |||
| Cladosporol G (127) | Cytotoxic | 3.9 µM; | HeLa (IC50); | [40] |
| 8.8 µM; | K562 (IC50); | |||
| 19.5 µM | HCT-116 (IC50) | |||
| Cladosporol G bis (128) | Antibacterial | 64 µg mL−1; | E. coli (MIC); | [39] |
| 128 µg mL−1; | M. luteus (MIC); | |||
| 64 µg mL−1 | V. harveyi (MIC) | |||
| Cytotoxic | 13 µM; | A549 (IC50); | ||
| 11 µM; | HeLa (IC50); | |||
| 10 µM; | Huh7 (IC50); | |||
| 11 µM; | L02 (IC50); | |||
| 14 µM; | LM3 (IC50); | |||
| 15 µM | SW1990 (IC50) | |||
| Cladosporol H (129) | Antibacterial | 32 µg mL−1; | E. coli (MIC); | [39] |
| 64 µg mL−1; | M. luteus (MIC); | |||
| 4 µg mL−1 | V. harveyi (MIC) | |||
| Cytotoxic | 5 µM; | A549 (IC50); | ||
| 10 µM; | H446 (IC50); | |||
| 1 µM; | Huh7 (IC50); | |||
| 4.1 µM; | LM3 (IC50); | |||
| 10 µM; | MCF-7 (IC50); | |||
| 14 µM | SW1990 (IC50) | |||
| Cladosporol I (130) | Quorum sensing inhibitory | 30 µg well−1 | C. violaceum (MIC) | [87] |
| Antibacterial | 64 µg mL−1; | E. coli (MIC); | [39] | |
| 64 µg mL−1; | M. luteus (MIC); | |||
| 16 µg mL−1 | V. harveyi (MIC) | |||
| Cytotoxic | 10.8 µM | HeLa (IC50) | ||
| Cladosporol J (131) | Antibacterial | 16 µg mL−1; | E. coli (MIC); | [39] |
| 64 µg mL−1; | M. luteus (MIC); | |||
| 32 µg mL−1 | V. harveyi (MIC) | |||
| Cytotoxic | 15 µM; | A549 (IC50); | ||
| 4 µM; | H446 (IC50); | |||
| 4.9 µM; | HeLa (IC50); | |||
| 6.2 µM; | Huh7 (IC50); | |||
| 13 µM; | L02 (IC50); | |||
| 9.1 µM; | LM3 (IC50); | |||
| 1.8 µM; | MCF-7 (IC50); | |||
| 2.2 µM | SW1990 (IC50) | |||
| Cladosporone A (132) | Cytotoxic | 14.3 µM; | K562 (100%); | [86] |
| 15.7 µM; | A549 (100%); | |||
| 29.9 µM; | Huh-7 (100%); | |||
| 40.6 µM; | H1975 (100%); | |||
| 21.3 µM; | MCF-7 (100%): | |||
| 10.5 µM; | U937 (100%); | |||
| 17.0 µM; | BGC823 (100%); | |||
| 10.1 µM; | HL60 (100%); | |||
| 53.7 µM; | HeLa (100%) | |||
| 14.6 µM | MOLT-4 (100%) | |||
| Anti-COX-2 | 49.1 µM | IC50 | ||
| (3S)-3,8-Dihydroxy-6,7-dimethyl-α- tetralone (137) | Antibacterial | 20 µM | S. aureus, B. cereus, E. coli, Vibrio alginolyticus, Vibrio parahemolyticus, methicillin-resistant S. aureus | [81] |
| Scytalone (139) | Antibacterial | 63.6 µg mL−1; | Bacillus cereus (IC50); | [68] |
| 95.5 µg mL−1 | E. coli (IC50) | |||
| Naphtoquinones and anthraquinones | ||||
| Anhydrofusarubin (142) | Cytotoxic | 3.97 μg mL–1 | K-562 (IC50) | [90] |
| Methyl ether of fusarubin (143) | Cytotoxic | 3.58 μg mL–1 | K-562 (IC50) | [90] |
| Antibacterial | 40 μg disc–1 | S. aureus (27 mm), Bacillus megaterium (22 mm), E coli (25 mm), P. aeruginosa (24 mm) | ||
| Toxic | 81.4 µM | brine shrine naupalii (IC50) | ||
| Perylenequinones | ||||
| Altertoxin VIII (145) | Quorum sensing inhibitory | 30 µg well−1 | C. violaceum (MIC) | [87] |
| Altertoxin IX (146) | Quorum sensing inhibitory | 30 µg well−1 | C. violaceum (MIC) | [87] |
| Aterotoxin X (147) | Quorum sensing inhibitory | 20 µg well−1 | C. violaceum (MIC) | [87] |
| Altertoxin XI (148) | Quorum sensing inhibitory | 30 µg well−1 | C. violaceum (MIC) | [87] |
| Calphostin A (=UCN-1028A) (149) | PK inhibition | 0.19 µg mL−1; | PKC (IC50); | [29,30] |
| 40 µg mL−1 | PKA (IC50) | |||
| Cytotoxic | 0.29 µg mL−1; | HeLa S3 (IC50); | ||
| 0.21 µg mL−1 | MCF-7 (IC50) | |||
| Calphostin B (150) | PK inhibition | 1.04 µg mL−1; | PKC (IC50); | [7] |
| 22.9 µg mL−1 | PKA (IC50) | |||
| Cytotoxic | 2.56 µg mL−1; | HeLa S3 (IC50); | ||
| 1.61 µg mL−1 | MCF-7 (IC50) | |||
| Calphostin C (=cladochrome E) (151) | PK inhibition | 0.05 µg mL−1 | PKC (IC50) | [30] |
| Cytotoxic | 0.23 µg mL−1; | HeLa S3 (IC50); | ||
| 0.18 µg mL−1 | MCF-7 (IC50) | |||
| Calphostin D (= ent-isophleinchrome) (152) | PK inhibition | 6.36 µg mL−1; | PKC (IC50); | [30] |
| 12.7 µg mL−1 | PKA (IC50) | |||
| Cytotoxic | 8.45 µg mL−1; | HeLa S3 (IC50); | ||
| 2.69 µg mL−1 | MCF-7 (IC50) | |||
| Phytotoxic | 5 µg L−1 | Sugar beet cells (100% inhibition in the light, 37–64% inhibition in the dark) | [46] | |
| 33 µg L−1 | Necrosis on sugar beet leaves | |||
| Calphostin I (= Cladochrome D) (153) | PK inhibition | 6.36 µg mL−1; | PKC (IC50); | [30] |
| 12.7 µg mL−1 | PKA (IC50) | |||
| Cytotoxic | 0.24 µg mL−1; | HeLa S3 (IC50); | ||
| 0.16 µg mL−1 | MCF-7 (IC50) | |||
| Phleichrome (154) | Invertase I inhibition | 0.5 mM | 62% | [99] |
| Seco acids | ||||
| Cladospolide E (159) | Lipid accumulation | 10 µM | Oleic acid; Triglycerides (~170 µg mg−1 protein); Total cholesterol (~3 µg mg−1 protein) | [94] |
| Seco-patulolide A (161) | Lipid accumulation | 10 µM | Oleic acid; Triglycerides (~150 µg mg−1 protein); Total cholesterol (~3 µg mg−1 protein) | [94] |
| Seco-patulolide C (162) | Lipid accumulation | 10 µM | Oleic acid; Triglycerides (~150 µg mg−1 protein); Total cholesterol (~3 µg mg−1 protein) | [94] |
| Antimicrobial | 32 µg mL−1; | E. tarda (MIC); | [50] | |
| 32 µg mL−1; | E. ictarda (MIC); | |||
| 16 µg mL−1 | C. gloeosporioides (MIC) | |||
| (3S,5S,11S)-Trihydroxydodecanoic acid (163) | Antibacterial | 63.6 µg mL−1 | B. cereus (MIC) | [68] |
| Cytotoxic | 42 µM; | MCF-7; | ||
| 82 µM | T47D | |||
| Antibacterial | 40 μg disc–1 | S. aureus (27 mm), B. megaterium (22 mm), E coli (25 mm), P. aeruginosa (24 mm) | ||
| Toxic | 81.4 µM | brine shrine naupalii (IC50) | ||
| Sterols | ||||
| Cladosporide A (164) | Antifungal | 0.5 µg mL−1 | Aspergillus fumigatus (IC50) | [79] |
| Cladosporide B (165) | Antifungal | 3 µg disc−1 | A. fumigatus (11 mm) | [80] |
| Cladosporide C (166) | Antifungal | 1.5 µg disc−1 | A. fumigatus (11 mm) | [80] |
| 3α-Hydroxy-pregn-7-ene-6,20-dione (171) | Anti-adipogenic | 1.25 – 10 µM | 3T3-L1 | [62] |
| Tetramic acids | ||||
| Cladodionen (176) | Cytotoxic | 28.6 µM | HL-60 (IC50) | [58] |
| 4.5 µM; | K562 (IC50); | [89] | ||
| 6.6 µM; | HL-60 (IC50); | |||
| 12 µM; | HCT-116 (IC50); | |||
| 11 µM; | PC-3 (IC50); | |||
| 15 µM; | SH-SYSY (IC50); | |||
| 22 µM | MGC-803 (IC50) | |||
| 18.7 µM; | MCF-7 (IC50); | [96] | ||
| 19.1 µM; | HeLa (IC50); | |||
| 17.9 µM; | HCT-116 (IC50); | |||
| 9.1 µM | HL-60 (IC50) | |||
| 3.7 µM | L5178 (IC50) | [63] | ||
| Antifungal | 100 mg/plate; | Ustilago maydis (0.97 cm); | ||
| 100 mg/plate | Saccharomyces cerevisiae (3.27 cm) | |||
| Cladosin B (178) | Renoprotective effects against cisplatin-induced kidney cell damage | 25 µM; 50 µM; 100 µM | LLC-PK1 (dose-dependent) | [60] |
| Cladosin C (179) | Antiviral | 276 µM | H1N1 (IC50) | [55] |
| Cladosin F (181) | Renoprotective effects against cisplatin-induced kidney cell damage | 25 µM; 50 µM; 100 µM | LLC-PK1 (dose-dependent) | [60] |
| Cladosin I (184) | Cytotoxic | 4.1 µM; | K562 (IC50); | [89] |
| 2.8 µM; | HL-60 (IC50); | |||
| 11 µM; | HCT-116 (IC50); | |||
| 13 µM; | PC-3 (IC50); | |||
| 12 µM; | SH-SYSY (IC50); | |||
| 19 µM | MGC-803 (IC50) | |||
| Cladosin J (185) | Cytotoxic | 6.8 µM; | K562 (IC50); | [89] |
| 7.8 µM | HL-60 (IC50) | |||
| Cladosin K (186) | Cytotoxic | 5.9 µM; | K562 (IC50); | [89] |
| 7.5 µM; | HL-60 (IC50); | |||
| 14 µM; | HCT-116 (IC50); | |||
| 18 µM | PC-3 (IC50) | |||
| Cladosin L (187) | Renoprotective effects against cisplatin-induced kidney cell damage | 25 µM; 50 µM; 100 µM | LLC-PK1 (dose-indipendent) | [60] |
| Cladosin L bis (188) | Antibacterial | 25 µM; | S. aureus ATCC 700699 (IC50); | [63] |
| 50 µM | S. aureus ATCC 29213 (IC50) | |||
| Cladosporicin A (192) | Cytotoxic | 70.88 µM; | Bt549 (IC50); | [61] |
| 74.48 µM; | HCC70 (IC50); | |||
| 75.54 µM; | MDA-MB-231 (IC50); | |||
| 79.36 µM | MDA-MB-468 (IC50) | |||
| Cladosporiumin I bis (202) | Cytotoxic | 76.18 µM; | Bt549 (IC50); | [61] |
| 85.29 µM; | HCC70 (IC50); | |||
| 82.37 µM; | MDA-MB-231 (IC50); | |||
| 81.44 µM | MDA-MB-468 (IC50) | |||
| Cladosporiumin J bis (204) | Cytotoxic | 78.96 µM; | Bt549 (IC50); | [61] |
| 76.41 µM; | HCC70 (IC50); | |||
| 79.27 µM; | MDA-MB-231 (IC50); | |||
| 74.64 µM | MDA-MB-468 (IC50) | |||
| Tropolones | ||||
| Malettinin A (210) | Antimicrobial | 33.1 µM; | Trichophyton rubrum (IC50); | [95] |
| 100 µM | C. albicans (81%) | |||
| Malettinin B (211) | Antimicrobial | 28.3 µM; | Xanthomonas campestris (IC50); | [95] |
| 60.6 µM; | T. rubrum (IC50); | |||
| 100 µM; | Staphylococcus epidermidis (<80%); | |||
| 100 µM; | B. subtilis (<80%); | |||
| 100 µM | C. albicans (<80%) | |||
| Malettinin C (212) | Antimicrobial | 37.9 µM; | X. campestris (IC50); | [95] |
| 83.2 µM; | T. rubrum (IC50); | |||
| 100 µM; | S. epidermidis (<80%); | |||
| 100 µM; | B. subtilis (<80%); | |||
| 100 µM | C. albicans (<80%) | |||
| Malettinin E (213) | Antimicrobial | 28.7 µM; | X. campestris (IC50); | [95] |
| 30.7 µM; | T. rubrum (IC50) | |||
| Xanthones | ||||
| Conioxanthone A (218) | Cytotoxic | 10 µM | 22RV1 (36.8%), C4-2B (3.3%), RWPE-1 (20.3%) | [43] |
| 3,8-Dihydroxy-6-methyl-9-oxo-9H-xanthene-1-carboxylate (219) | Cytotoxic | 10 µM | 22RV1 (82.1%), C4-2B (77.7%), RWPE-1 (11.5%) | [43] |
| α-Diversonolic ester (220) | Cytotoxic | 10 µM | 22RV1 (28.8%), C4-2B (12.9%), RWPE-1 (24.3%) | [43] |
| β-Diversonolic ester (221) | Cytotoxic | 10 µM | 22RV1 (40.2%), C4-2B (2.8%), RWPE-1 (10.3%) | [43] |
| 8-Hydroxy-6-methylxanthone-1-carboxylic acid (222) | Cytotoxic | 10 µM | 22RV1 (71.3%), C4-2B (60.7%), RWPE-1 (19.7%) | [43] |
| Methyl 8-hydroxy-6-(hydroxymethyl)- 9-oxo-9H-xanthene-1-carboxylate (223) | Cytotoxic | 10 µM | 22RV1 (68.1%), C4-2B (20.2%), RWPE-1 (19.0%) | [43] |
| Methyl 8-hydroxy-6-methyl-9-oxo-9H-xanthene-1-carboxylate (224) | Cytotoxic | 10 µM | 22RV1 (55.8%), C4-2B (8.1%), RWPE-1 (5.3%) | [43] |
| 8-(Methoxycarbonyl)-1-hydroxy-9-oxo-9H-xanthene-3-carboxylic acid (225) | Cytotoxic | 10 µM | 22RV1 (63.9%), C4-2B (12.2%), RWPE-1 (27.0%) | [43] |
| Vertixanthone (227) | Cytotoxic | 10 µM | 22RV1 (27.1%), RWPE-1 (25.0%) | [43] |
| Miscellaneous | ||||
| Acetyl Sumiki’s acid (229) | Antibacterial | 5 μg disc−1 | B. subtilis (7 mm), S. aureus (7 mm) | [44] |
| 1,1′-Dioxine-2,2′-dipropionic acid (233) | Antibacterial | 25 μg mL−1; | S. aureus (MIC); | [85] |
| 25 μg mL−1; | E. coli (MIC); | |||
| 12.5 μg mL−1 | B. cereus (MIC) | |||
| 4-O-α-d-Ribofuranose-2-pentyl-3-phemethylol (238) | Inhibition of α-glucosidase | 2.50 µM | IC50 | [82] |
| Sumiki’s acid (241) | Antibacterial | 5 μg disc−1 | B. subtilis (7 mm), S. aureus (7 mm) | [44] |
| Taxol (242) | Cytotoxic | 3.5 µM | HCT 15 (IC50) | [51] |
| Antibacterial | 30 µL disc−1; | Pseudomonas aeruginosa (2 mm); | ||
| 20 µL disc−1; | Escherichia coli (3 mm); | |||
| 30 µL disc−1; | Klebsiella pneumoniae (2 mm); | |||
| 20 µL disc−1; | Acetobacter sp. (2 mm); | |||
| 40 µL disc−1 | Bacillus subtilis (1 mm) | |||
| Vermistatin (244) | Antibacterial | 25 μg mL−1; | S. aureus (MIC); | [85] |
| 25 μg mL−1 | B. cereus (MIC) | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salvatore, M.M.; Andolfi, A.; Nicoletti, R. The Genus Cladosporium: A Rich Source of Diverse and Bioactive Natural Compounds. Molecules 2021, 26, 3959. https://doi.org/10.3390/molecules26133959
Salvatore MM, Andolfi A, Nicoletti R. The Genus Cladosporium: A Rich Source of Diverse and Bioactive Natural Compounds. Molecules. 2021; 26(13):3959. https://doi.org/10.3390/molecules26133959
Chicago/Turabian StyleSalvatore, Maria Michela, Anna Andolfi, and Rosario Nicoletti. 2021. "The Genus Cladosporium: A Rich Source of Diverse and Bioactive Natural Compounds" Molecules 26, no. 13: 3959. https://doi.org/10.3390/molecules26133959
APA StyleSalvatore, M. M., Andolfi, A., & Nicoletti, R. (2021). The Genus Cladosporium: A Rich Source of Diverse and Bioactive Natural Compounds. Molecules, 26(13), 3959. https://doi.org/10.3390/molecules26133959








