Abstract
Nucleic acids are essential biomolecules in living systems and represent one of the main targets of chemists, biophysics, biologists, and nanotechnologists. New small molecules are continuously developed to target the duplex (ds) structure of DNA and, most recently, RNA to be used as therapeutics and/or biological tools. Stimuli-triggered systems can promote and hamper the interaction to biomolecules through external stimuli such as light and metal coordination. In this work, we report on the interaction with ds-DNA and ds-RNA of two aza-macrocycles able to coordinate Zn2+ metal ions and form binuclear complexes. The interaction of the aza-macrocycles and the Zn2+ metal complexes with duplex DNA and RNA was studied using UV thermal and fluorescence indicator displacement assays in combination with theoretical studies. Both ligands show a high affinity for ds-DNA/RNA and selectivity for ds-RNA. The ability to interact with these duplexes is blocked upon Zn2+ coordination, which was confirmed by the low variation in the melting temperature and poor displacement of the fluorescent dye from the ds-DNA/RNA. Cell viability assays show a decrease in the cytotoxicity of the metal complexes in comparison with the free ligands, which can be associated with the observed binding to the nucleic acids.
1. Introduction
Nucleic acids (DNA and RNA) play essential roles in storing genetic information and templating the formation of proteins in living systems. Modulation of the nucleic acid metabolism, including DNA replication, translation, and transcription, by synthetic or natural molecules has been the basis of current clinical chemotherapy. Initially, DNA became the most interesting biomolecule to target, being cis-diamminedichloridoplatinum(II) (cisplatin), the first anticancer molecule used in clinics. The action mechanism of cisplatin involves DNA binding, halting of replication, and induction of apoptosis [1]. In order to treat human diseases such as microorganism infections and genetic disorders, which rank among the most difficult diseases to tackle, the paradigm has recently shifted to RNA targeting with small molecules [2,3,4,5].
To overcome the limitations of nucleic acid agents such as poor solubility, side effects, dose-limiting toxicity, distribution in tissues, or acquired resistance, several approaches have emerged combining chemo- and radiotherapies with more effective drug delivery systems [6,7]. A promising strategy is the development of new stimuli-responsive nucleic acid binders in which a given stimulus promotes or hinders the nucleic acid binding to the molecule and, therefore, modulates the biological activity associated with the nucleic acid metabolism. Mascareñas, Vazquez, and co-workers have finely designed several stimuli-responsive systems based mainly on synthetic polypeptide helices that bind to DNA upon light irradiation and metal coordination and are compiled in a recent review [8]. Among the triggering stimuli, metal coordination is one of the most challenging strategies due to the abundance of metal ions in cells that can occupy the coordination sites of the synthetic systems and hamper the envisaged DNA interaction process. In this regard, Peacok’s team reported two short peptides connected by a bipyridine or a terpyridine unit that undergo conformational changes upon Zn2+ or Cu2+ coordination, prompting their interaction with double-stranded DNA (ds-DNA) [9]. An interesting system described by Learte-Aymami et al. is composed of a peptide moiety containing histidine units, which is unable to bind DNA because of the unstructured nature in solution, but Pd2+ coordination to the nitrogen atoms of the histidines promotes its folding, adopting a helical conformation with a concomitant high ds-DNA binding [10]. In contrast, metal coordination can also inhibit the binding to nucleic acids. For instance, Futaki and co-workers described as Co2+ coordination to some peptide dimeric systems containing iminodiacetic acid groups decreases their structural helical arrangement reducing thereby, their interaction with DNA [11]. In this line, our group is devoted to the development of polyamine-based systems because they present interesting structural features for modulating the binding of anionic species, in particular, (i) the significant changes in net charge within short pH ranges [12] and (ii) the ability to strongly coordinate metal ions [13,14].
Regarding nucleic acids, we have developed a series of scorpiand-like ligands containing a macrocyclic core and a pendant arm with an aromatic moiety, which display an open extended or a closed conformation depending on the pH and metal coordination [15]. While the open extended conformation of the scorpiand favors an intercalative binding to DNA of condensed aromatic groups such as anthracene or pyrene introduced in the pending arm, the closed conformation of the ligand achieved by metal binding or by an increase in pH blocks the intercalation [16,17]. In addition to metal coordination, we have explored the ability of polyprotonable ligands to modulate the binding to either DNA or RNA by changing the pH. A change in the pH from 7.4 to 5.0 increases the net charge of the ligand favoring the interaction with RNA over DNA by means of electrostatic interactions [18,19].
Herein we explored the ability of two ligands containing two different coordinating sites to bind double-stranded DNA and RNA (ds-DNA/RNA) differing in the conformation, B- and A-type, respectively, either as free ligands or coordinated to Zn2+ ions (see Chart 1). We studied the acid-base behavior and metal coordination by potentiometry to determine the net charge and characterize the Zn2+ complexes formed in solution. Then, we combined thermal stabilization experiments, fluorescent indicator displacement assays, and theoretical multiscale studies to investigate the binding of the ligands and their corresponding Zn2+ complexes to ds-DNA/RNA. Finally, the cell viability was assessed for the ligand and their corresponding metal complexes to determine the biological activity.
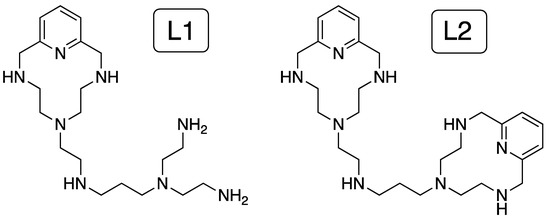
Chart 1.
Ligands (L1 and L2) studied in this work.
2. Results
2.1. Characterization of L1–L2 and Their Zn2+ Complexes
2.1.1. Speciation in Solution of L1–L2
The knowledge of the species prevailing in aqueous solution and their protonation state is key to understand the effect of electrostatic forces and hydrogen bonding in host-guest complexation events. However, these parameters are often difficult to determine in water due to solubility issues. In our study, the ligands L1 and L2 (described and characterized previously in reference [20]) are totally water soluble within the pH range, and thus, we studied the acid-base and metal coordination behavior of both L1 and L2 by pH-metric titrations. Stepwise protonation constants of the ligands determined in 0.15 M NaCl at 298.1 K, as well as their distribution diagrams, are collected in Supplementary Materials (Table S1 and Figure S1) in ESI. As discussed in reference [20], L1 and L2 present five protonation steps in the pH range of study, being their net charge at physiological pH 7.4, calculated from the distribution diagrams, +4.4 and +4.3, respectively (Figure 1).
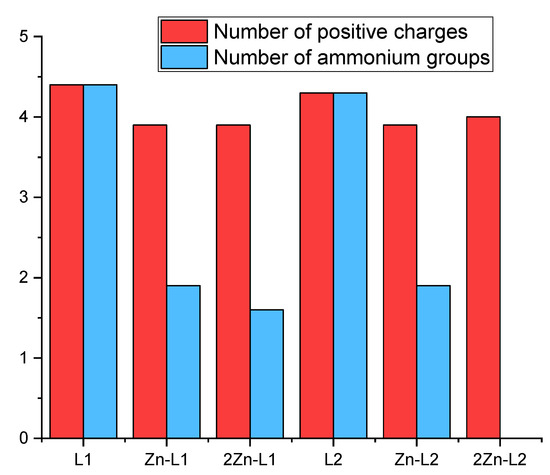
Figure 1.
Average number of positive charges and number of ammonium groups at pH 7.4 (L = 1 × 10−5 M, Zn2+ = 1 × 10−5 M and 2 × 10−5 M for Zn-L and 2Zn-L complexes, respectively) derived from the values of stability and protonation constants previously obtained by potentiometric titrations in aqueous solution at 298.1 K using NaCl 0.15 M as supporting electrolyte.
Then, the formation of Zn2+ complexes with L1 and L2 was studied in solution by potentiometric measurements [20]. The stability constants provided by the pH-metric titrations (Table S2) allowed us to establish the distribution diagrams gathered in Figures S2 and S3. Such representations show the formation of the binuclear species Zn2L+, Zn2L(OH)3+ and Zn2L(OH)22+ for both ligands in a 2:1 Zn2+:L molar ratio (Figure S3). The net charges of the complexes at pH = 7.4, calculated for 1:1 and 2:1 Zn2+:L mole ratios from the distribution diagrams, are just a little bit lower than those of the free ligands. (see Figure 1). However, the coordination of the Zn2+ ions, particularly in the case of the binuclear complexes, drastically reduces the number of ammonium groups present at pH 7.4, altering, therefore, the possible pattern of hydrogen bonds of the ligand with nucleic acids (Figure 1). The stability constant values for the formation of the mononuclear species of both ligands are within the same range, suggesting that the entry of the metal is occurring in the common structural core, which is the macrocycle. The lower value of the stability constant obtained for the entry of the second metal for L1 (7.89) in comparison with L2 (11.70) suggests a stronger interaction of the metal ion with the nitrogen atoms of the macrocycle than the nitrogen atoms of the acyclic part in L1.
2.1.2. QM Modeling of the Zn2+ Complexes
We initially optimized the geometry of the Zn2+ binuclear complexes of both ligands using DFT to afford the minimum energy conformers shown in Figure 2 (structural coordinates are collected in Tables S3 and S4). The metal ions in Zn2L14+ show different coordination modes depending on the coordination site. The Zn2+ ion at the macrocyclic site is coordinated by the four nitrogen atoms of the macrocyclic moiety and by two water molecules in a distorted octahedral geometry, while the second metal ion is coordinated by the four nitrogen atoms of the acyclic side chain and a water molecule with a trigonal bipyramid geometry (Figure 2A). In contrast, in the binuclear Zn2L24+ complex, both metal centers present an octahedral coordination geometry in which each metal ion is coordinated by the four nitrogen atoms of each macrocyclic moiety. However, the coordination sphere of each Zn2+ is completed differently. In the metallic center separated from the amine group of the linker by an ethylenic chain, besides the nitrogens of the macrocycle, the metal ion is coordinated by a water molecule and the amino group of the linker. On the other hand, the second metal ion completes its coordination with just one water molecule (Figure 2B).
Figure 2.
Visualization of the minimum energy structure optimized by DFT for the complex Zn2L14+ (a) and Zn2L24+ (b).
2.2. Interaction Studies of Ligands to Double-Stranded Nucleic Acids
2.2.1. Thermal Denaturation Experiments
We investigated the interaction of the ligand with ds-DNA/RNA by means of thermal stabilization assays. Double-stranded oligomers are held together by hydrogen bonds, which can be dissociated upon heating to provide single-stranded oligomers. During the thermal denaturation process, a melting temperature or transition temperature (Tm) can be determined as the temperature in which half of the DNA strands are in the random coil or single-stranded state. High CG contents or non-covalent interactions with small molecules usually stabilize the double-stranded helical structure. The variation between the melting temperature of the free ds-DNA/RNA and of the adducts formed with small molecules (ΔTm) is a qualitative parameter for the interaction. The ΔTm values for the interaction of L1 and L2 with a ds-DNA model of B-type conformation (calf thymus DNA/ctDNA) and a ds-RNA model (poly A-poly U) with an A-form duplex are gathered in Table S5 for different molar ratios (r = [ligand]/[polynucleotide]).
Interaction of L1/L2 with ds-DNA and ds-RNA leads to thermal stabilization of both double helixes (see Figure 3). The ΔTm values obtained for ctDNA ranged between 0.1 and 3.2 °C, while for poly A-poly U were higher than 15 °C, indicating a stronger and selective interaction with the RNA duplex model. The strong interaction of these polyamine-based ligands with poly A-poly U is probably due to the A-type conformation of this ds-RNA, which exposes the negative charges of the nucleic acid outwards the helix and allows a more efficient interaction with the positively charged ligands than in the case of the B-type helix adopted by ds-DNA. The higher ΔTm values achieved for L1 and L2 at high r = [ligand]/[polynucleotide] ratios (r = 0.2 and 0.5) are among the largest ones observed in the literature for these duplex models [21].
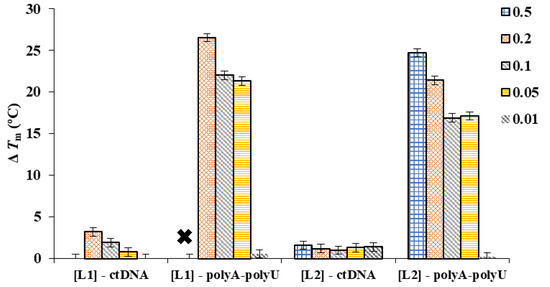
Figure 3.
ΔTm values determined for the interaction of L1 and L2 toward ctDNA and poly A-poly U at different r = [ligand]/[polynucleotide]. Each bar stands for an r value, as is described in the legend. The “×” indicates a ΔTm value too high to be determined.
2.2.2. Fluorescent Indicator Displacement Assays
Once established the strong interaction of L1 and L2 with duplexes and, in particular, with ds-RNA, we used fluorescence indicator displacement assays to estimate the binding affinity. Indirect measurements were employed due to the non-emissive character of both ligands, as well as to the fact that their absorption bands overlap with those of the nucleic acids, which hampers to perform direct fluorimetric and UV-Vis titrations. In addition, we used a well-established intercalative dye (ethidium bromide, EB, Chart 2) and a minor groove binder (DAPI) to study the binding mode. Both dyes are poorly emissive in solution but become highly emissive when bound to duplex DNA/RNA.
Chart 2.
Fluorescent dyes used for displacement assays, ethidium bromide (EB) and DAPI.
EB Displacement Assays
The addition of the ligands to a solution of duplex DNA/RNA slightly saturated with the intercalator EB induced a decrease in the fluorescence emission of the EB dye (Figure 4) because EB is displaced from the hydrophobic nucleic acid matrix to the solution. As shown in Figure 4, the addition of the ligands yields a larger decrease in emission intensity when interacting with poly A-poly U than with ctDNA. Since a decrease in emission is directly associated with the extent of binding, we can conclude that L1 and L2 interact stronger with ds-RNA than with ds-DNA.
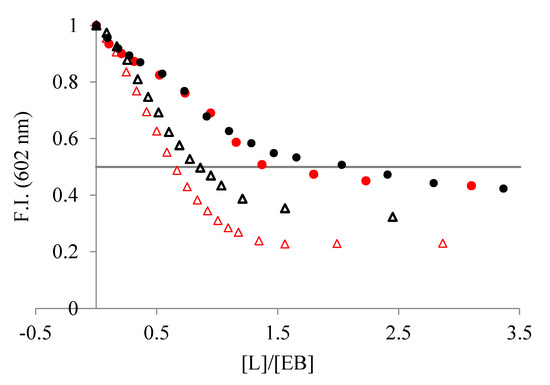
Figure 4.
Changes in fluorescent emission obtained from EB displacement assays of (●) ctDNA and (Δ) poly A-poly U, 3–4 × 10−5 M, when titrated with (red) L1 and (black) L2 (NaCac 50 mM, pH 7.4). The line represents the emission decrease by 50% with respect to the initial value.
From the displacement assay isotherms, we calculated the IC50 values to estimate the binding affinities, in which the IC50 is the [ligand]/[DNA] ratio corresponding to the emission decrease by 50% with respect to the initial value (Figure 4). Both ligands show lower IC50 values for poly A-poly U than for ctDNA, indicating a higher affinity for the ds-RNA (see Table 1). Moreover, L1 binds to both duplexes stronger than L2, suggesting that it has a structure better suited to interact with the double-stranded nucleic acid structure.

Table 1.
IC50 values determined for the displacement of EB and DAPI from ctDNA and poly A-poly U by addition of L1 and L2 (NaCac 50 mM, pH 7.4).
The values of the affinity constants (Kapp) can be calculated by taking into account such IC50 values and the affinity constants for the interaction of EB with ctDNA and poly A-poly U under the same experimental conditions (pH 7.0, log Kapp = 106) [22]. The affinity constants range between Kapp ~ 105–106 M−1 (see Table 1).
DAPI Displacement Assays
In order to find out whether the ligands displayed an intercalative or a minor groove binding mode, displacement measurements were performed using DAPI as a fluorescent dye (Figure 5). A decrease in DAPI emission was only observed for ds-RNA, suggesting that both ligands bind to the minor groove of poly A-poly U and are not able to significantly interact with ds-DNA. The values of IC50 are collected in Table 1 together with the values obtained from the EB assay. The values obtained from the DAPI displacement assay for ds-RNA are within the same range as those obtained from the EB assay (see Table 1).
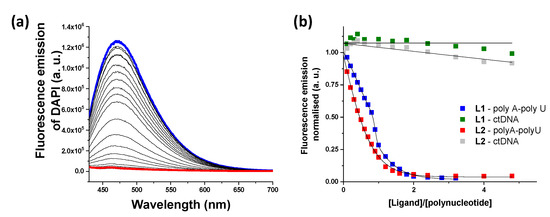
Figure 5.
(a) Changes in fluorescent emission obtained from DAPI displacement assays with poly A-poly U and L1 (blue line, initial spectrum and red line, final spectrum). (b) Plot of the fluorescence emission vs. ratio in the DAPI displacement assays.
2.2.3. Molecular Modeling
Once we assessed the interaction of the ligands with duplexes by means of UV melting and fluorescent indicator displacement assays, multiscale QM/MM-MD studies were conducted to obtain insights into the binding mode. The structures shown in Figure 6 (Supplementary Materials Figures S4 and S5) show the minimum energy conformers determined for the interaction of the ligands with both the DNA and RNA duplexes through the minor groove. Strikingly, both ligands bind externally to the duplex DNA by means of electrostatic interactions with the phosphate backbone while they are inserted into the minor groove of RNA duplex model forming a larger number of electrostatic and hydrogen bond contacts. The ability to bind in the minor groove of RNA can be ascribed to the ligand shape that fits better in the shallow and wide minor groove of RNA ((3.70 Å deep and 10.36 Å wide) than in the narrower and deeper one of DNA (5.96 Å deep and 5.37 Å wide).
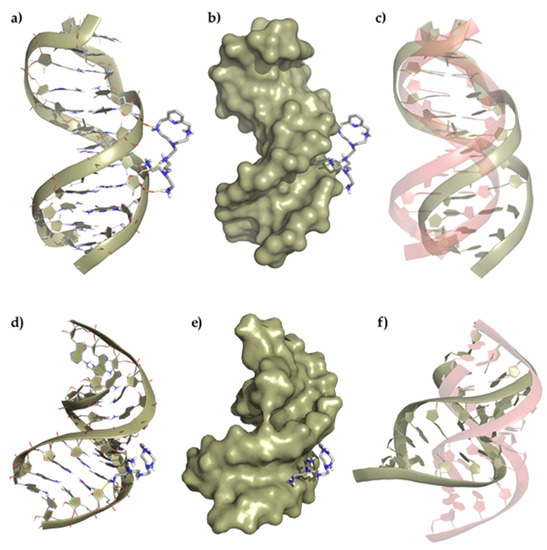
Figure 6.
Top panel, representation of (a) the interaction of L1 with the ds-DNA, (b) the polynucleotide surface, and (c) deviation of the duplex DNA structure after interaction regarding its original conformation (represented in red). Bottom panel, representation of (d) the interaction of L1 with the duplex RNA, (e) the polynucleotide surface, and (f) deviation of the ds-RNA structure after interaction regarding its original conformation (represented in red).
A detailed analysis of the minimum energy conformers of L1 with ds-DNA shows the formation of three salt bridges between the oxygen atoms of the phosphate groups and some of the protonated amines of the ligand (dO···H–N = 1.64–1.76 Å), which slightly distorts the helical structure. On the other hand, the adduct formed between L1 and ds-RNA gives rise to minimum energy conformers stabilized by the formation of a larger number of salt bridges between the oxygens of the phosphate groups and some of the protonated amines of the ligand (dO···H–N = 2.08 Å), which has a greater impact on the RNA structure. The insertion of L1 into the minor groove duplex bends the structure, forming an even wider and deeper minor groove (amplitude widens from 10.36 to 17.61 Å, and the depth decreases from 3.70 to 2.73 Å). A similar interaction is observed for the ligand L2 and the DNA/RNA duplexes (Figures S4 and S5).
2.3. Metal-Induced Modulation of RNA Binding
Once determined the strong interaction and high-affinity constants of L1–L2 toward ds-DNA/RNA, we explored the binding of the metal complexes with ds-RNA. Both ligands form mono- and binuclear metal complexes with Zn2+ at pH 7.4 depending on the molar ratio M:L (see Figures S2 and S3). We envisaged that the metal complexes of the ligands would induce a structural arrangement and/or will disable the ammonium groups for electrostatic contacts with ds-RNA because of their participation in the metal coordination.
2.3.1. Thermal Denaturation Experiments of Metal Complexes
We studied the interaction of the mono- and binuclear Zn2+ complexes of L1–L2 toward poly A-poly U by UV thermal denaturation experiments, and the ΔTm values are collected in Table 2. In a previous work [20], we showed the formation of the mononuclear species ZnH2L4+ at pH 7.4 for both ligands at 1:1 molar ratio (M:L) while at 2:1 molar ratios, L2 forms the species Zn2L4+ and L1 presents a mixture of species, being predominant the species Zn2L4+ (~ 80% Zn2L4+, 15% Zn2HL5+ and free Zn2+) [20]. Therefore, we assume that the species interacting with ds-RNA will be ZnH2L4+ and Zn2L4+ for a 1:1 and 2:1 molar ratio, respectively. Interestingly, the coordination of the first Zn2+ ion is accompanied by a large reduction in the ΔTm values, while the coordination of the second Zn2+ ion either induces a small stabilization or even a slight destabilization.

Table 2.
ΔTm values determined for the metal complexes of L1–L2 with ctDNA (r = 0.05, NaCac 50 mM, pH 7.4).
2.3.2. Fluorescence Displacement Studies of Metal Complexes
Once determined the ds-RNA stability with the L1–L2 Zn2+ metal complexes, we evaluated the affinity of the binuclear species (Zn2L4+) toward ds-RNA by using the fluorescent indicator displacement assays with EB and DAPI. The displacement of the binuclear species depended on the fluorescence dye used (see Figure 7). The addition of the binuclear Zn2+ complexes released only 20% of the EB bound to poly A-poly U whereas they displaced 50% and 80% of the DAPI, respectively, indicating that the Zn2L4+ species interacts mostly through the minor groove of poly A-poly U. Nevertheless, both ligands displace completely both indicators from ds-RNA, indicating the larger affinity of the ligands in comparison with their metal complexes.
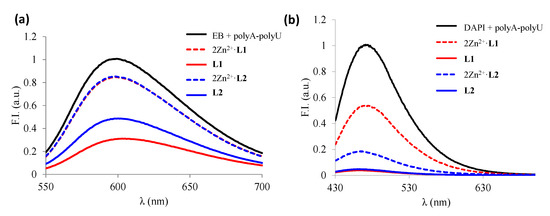
Figure 7.
Changes in fluorescence spectra obtained from EB (a) and DAPI (b) displacement assays under the addition of free ligands and the corresponding 2:1 Zn2+:L complexes.
2.3.3. Molecular Modeling
Multiscale QM/MM-MD methods were employed to generate a model for the interaction of the Zn2+ binuclear complexes with the DNA/RNA duplexes (Figure 8 and Supplementary Materials Figures S6–S8). The binuclear Zn2L4+ complexes of both ligands show a similar binding mode toward the ds-DNA and RNA models characterized by the interaction of the complexes through the major groove of the nucleic acid models. Interestingly, the low flexibility of the metal complexes, together with their limited capacity to form hydrogen bonds and to interact electrostatically, compared to the free ligands, leading to the formation of adducts with a smaller number of contacts and located more externally in the groove than the ligands alone. Therefore, the interaction of the Zn2+ complexes with the duplexes is weaker than that of the free ligands, as supported by the denaturalization and displacement studies.
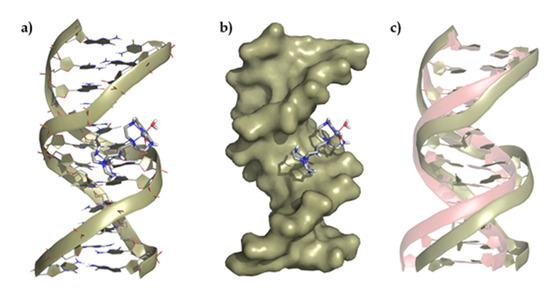
Figure 8.
Representation of (a) the interaction of Zn2L14+ with the ds-DNA model, (b) the polynucleotide surface, and (c) deviation of the ds-DNA structure after interaction regarding its original conformation (represented in red).
2.4. Viability Assays of the Ligands and the Metal Complexes
Before determining the interaction of the ligands and their Zn2+ complexes with DNA and RNA, we investigated the effects of L1, L2, and their Zn complexes in cell viability. Two bladder cancer cell lines (T24 and 253J) with different mutations in the tumor suppressor genes p53 and RB1 and a human fibroblast cell line (VERO) were treated with different concentrations of ligands and complexes (Figure 9). While, in all cell lines, L1 or L2 produce a diminution of cell viability, which was maximal at higher doses, L1 or L2 metal complexes did not affect the cellular viability at 10 μM and always less than the uncoordinated ligands at higher doses (Figure 9). This result, in agreement with the biophysical studies, suggests that metal coordination strongly influences the cytotoxic activity of these ligands.
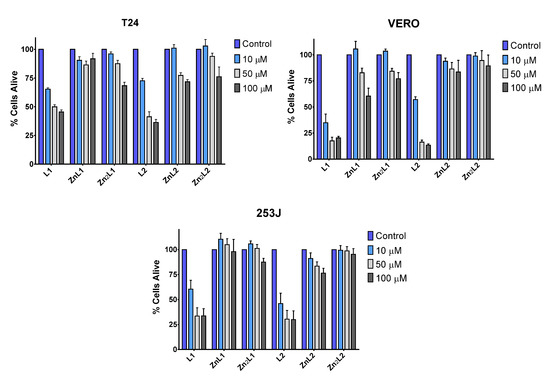
Figure 9.
Low toxicity of L1 and L2 metal complexes. Toxicity of L1, L2 and their metal complexes in mammalian cell lines. T24, VERO, and 253J cell lines were treated with 10, 50, and 100 μM of the compounds for 48 h to assess their toxicity based on the cell viability of the cell lines. The data are two independent experiments performed in triplicate.
3. Discussion
The coordination with Zn2+ and protonation degree of two polyamine-based ligands were investigated by potentiometry. L1 contains only one macrocyclic core with a pendant polyamine acyclic chain, while L2 contains two macrocyclic cores connected by an ethylenaminopropyl linker. Both ligands form stable binuclear complexes with Zn2+ along all the pH range studied. Such Zn2+ binding leads to a reduction in the net charge of the systems due to the participation of the nitrogen atoms in the coordination of the Zn2+ ions. Theoretical studies afforded the minimum energy conformers of the metal complexes. L2 presents octahedral geometry for both metal centers in the binuclear Zn2+ complex ([Zn2L2]4+ whereas L1 shows one metal center with an octahedral geometry corresponding to the macrocyclic core and a trigonal bipyramid geometry for the acyclic core. Both ligands show solvent molecules completing the metal ion coordination sphere, which can potentially coordinate the bases and phosphate of DNA/RNA.
Then, the binding ability of both ligands was assessed by UV melting experiments and fluorescent displacement assays, which show a large stabilization effect and high affinity to RNA duplexes for both ligands. The higher ΔTm values and lower IC50 values from the FID of L1 can be associated with its higher flexibility in comparison with the more rigid structure of L2 with two macrocyclic cores. The higher flexibility of L1 facilitates adopting a conformation in a solution that maximizes the interaction contacts with the nucleic acids. Interestingly, FID assays using the minor groove binder DAPI show a displacement only in ds-RNA, suggesting that both ligands interact through this binding mode with RNA. These studies are supported by molecular modeling, which highlights the more external interaction of both ligands to the B-type conformation of ds-DNA in comparison with the minor groove binding to the A-type conformation of the ds-RNA model.
When forming the binuclear Zn2+ complex, the thermal denaturation and FID studies show a negligible stabilization and lower indicator displacement, indicating the modulation of the interaction to ds-RNA upon Zn2+ coordination. Moreover, theoretical calculations show a lower number of contacts with duplexes because the nitrogen atoms of the ligands are involved in the metal coordination.
To obtain some insights into the biological activity, the cell viability was assessed by means of MTT assay with three cell lines and show the same cytotoxic modulation as the biophysical studies. The free ligands show high cytotoxicity toward all the cell lines while the metal coordination increases the viability of cells. This effect can be attributed to the DNA/RNA binding shown by our study.
4. Materials and Methods
4.1. Reagents
ctDNA and poly A-poly U were purchased from Sigma-Aldrich (Merck, Europe) and used without further purification. The stock solution was prepared by dissolved a small portion of the lyophilized polynucleotide in 200 μL of NaCac 50 mM buffer and stored at 5 °C overnight. The concentration of the stock solution was checked by measuring the absorption of four accumulative additions of the stock solution to a 1 mL of NaCac 50 mM. Concentration was then calculated by using the molar extinction coefficient given by the supplier (6600 M−1 cm−1 for ctDNA and 6000 M−1 cm−1 for poly A-poly U). Stock solutions were maintained at −10 °C and defrost before use. Ethidium bromide (EB) (3,8-diamino-5-ethyl-6-phenylphenanthridinium bromide) and DAPI (2-(4-amidinophenyl)-6-indolecarbamidine dihydrochloride, 4′,6-diamidino-2-phenylindole dihydrochloride) were purchased from commercial sources and used without further purification.
4.2. Synthesis of Ligands
Synthesis of the ligands 6-[7-(diaminoethyl)-3,7-diazaheptyl]-3,6,9-triaza-1-(2,6-pyridina)cyclodecaphane (L1) and 6-[6′-[3,6,9-triaza-1-(2,6-pyridina)cyclodecaphan-6-yl]-3-azahexyl]-3,6,9-triaza-1-(2,6-pyridina)cyclodecaphane (L2) was carried out following the general procedures described in the literature [20].
4.3. EMF Measurements
The potentiometric titrations were carried out at 298.1 ± 0.1 K using NaCl 0.15 M as supporting electrolyte (Merck, Europe). The acquisition of the emf data was performed with the computer program PASAT. The reference electrode was an Ag/AgCl electrode in saturated KCl solution. The glass electrode was calibrated as a hydrogen-ion concentration probe by titration of previously standardized amounts of HCl with CO2-free NaOH solutions and the equivalent point determined by the Gran’s method, which gives the standard potential, E°’, and the ionic product of water (pKw = 13.73(1)) [23,24].
The computer program HYPERQUAD was used to calculate the protonation and stability constants [25]. The pH range investigated was 2.0–11.0, and the concentration of the metal ions and the ligands ranged from 1 × 10−3 to 5 × 10−3 M with M:L molar ratios varying from 2:1 to 1:2. The different titration curves for each system (at least two) were treated either as a single set or as separated curves without significant variations in the values of the stability constants. Finally, the sets of data were merged together and treated simultaneously to provide the final stability constants.
4.4. Thermal Melting Experiments
Thermal melting curves were measured on an Agilent 8453 spectrometer equipped with a Peltier temperature controller system (±0.1 °C). The ratio of nucleic acid to compound used ranges from 20:1 to 2:1. Thermal melting curves were determined by following the absorption change at 260 nm in 50 mM cacodylate buffer (pH 7.4) as a function of temperature in the absence or presence of polyamine derivatives. The absorbance of the ligand was subtracted from every curve, and the absorbance scale was normalized. The Tm values were taken as the midpoints of the transition curves as determined from the maximum of the first derivative. ΔTm values were calculated by subtracting Tm for the free nucleic acid from Tm for the complex [22]. Every ΔTm value reported here was the average of at least two measurements; the error in ΔTm was ±0.5 °C.
4.5. Fluorescent Indicator Displacement Assays
Displacement experiments were recorded on a PTI spectrofluorimeter in the 540−680 nm range for EB and 430–700 nm range for DAPI with an excitation wavelength of 520 nm and 400 nm, respectively. The concentration of polynucleotide ranges from 3 to 4 × 10−5 M for EB displacement assays and 1 to 2 × 10−5 M for DAPI displacement assays. The fluorescence was normalized by the maximum fluorescence signal when the marker was bound to the nucleic acid in the absence of competition for binding and was corrected for background fluorescence of the free marker in solution.
4.6. Theoretical Studies
The generation of the computational models for the proposed complexes was performed using the density functional theory computational method as well as the Becke three-parameter Lee-Yang-Parr hybrid functional (B3LYP) [26,27,28]. All the gas-phase optimizations were carried out using the Ahlrichs’ basis set def2-SV(P) [29] for all atoms except for Zn2+ for which it was employed the MDF10 Stuttgart-Dresden effective core potential [30]. The influence of the dispersion was also taken into account by means of the Grimme’s dispersion (IOp(3/124 = 30)) correction [31]. Computations were carried out using the program Gaussian09 C.01. [32], gMolden [33], and PyMOL [34] were used for visual inspection and to create the molecular graphics. Finally, in order to use the obtained complexes for the MD and QM/MM-MD modeling, additional parameters were calculated using the MCPB parameter builder [35]. Briefly, the force field parameters (bond, angle, dihedral, electrostatic, van der Waals terms) were obtained at the B3LYP/def2-SV(P) level of theory, and the ESP punctual charges were derived by fitting the electrostatic potential according to the MDF10 Stuttgart-Dresden scheme.
The interaction studies of the Zn2+ complexes with the nucleic acids were performed through a QM/MM-MD approach. For the building of the double helix of calf thymus DNA systems, a representative duplex B-type DNA was taken from the Protein Data Bank (PDB, ID 1BNA) as a starting structure [36,37]; while the RNA one was build using the 3D-NuS program due to the absence of crystalline structures [38]. Once the duplex DNA/RNA systems were built, the ligand/complex systems were approximated, and, finally, the resulting system was solvated with the addition of 4258 water residues using TIP3PBOX and neutralized. Then, each system was energetically minimized and, after an equilibration stage at 300 K, a total of 20 ps molecular dynamics (MD) was performed. All the studies have been performed using AMBER16 [39] (Assisted Model Building with Energy Refinement) software. The organic compounds have been modeled using the gaff [40] force field, while the ff12SB force field was used for the nuclear bases. After the MD modeling, a final QM/MM-MD study was carried out at 300 K for 3 ns. The external Gaussian package was used for the quantum mechanical calculations. The QM region encompassed the complex, whereas the MM region included all the remaining solvent, counterions, and poly-nucleic strands. Therefore, no covalent bond had been cut when defining the QM/MM-MD boundary. The inner QM region was treated at the B3LYP-D3/def2-SV(P) level of theory. A cutoff of 10 Å was employed for the evaluation of the electrostatic interactions within the MM-MD region and of 10 Å for the QM region. The MD and QM/MM-MD simulation trajectories were analyzed using the cpptraj [41] module within AmberTools17 and PyMOL was used for visual inspection and to create the molecular graphics.
4.7. MTT Assay
T24 and 253J transitional-cell human bladder carcinoma cells and VERO green monkey kidney cells were cultured at 37 °C in a 5% CO2 in air atmosphere. VERO cells were grown in Eagle’s minimum essential medium and T24 and 253J in McCoy’s 5A media supplemented with 10% FBS, 2 mM L-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin. Cells were seeded at 2.5 × 104 cells/mL in 96-well plates and treated with L1, L2, and their metal complexes at different concentrations (10, 50, and 100 μM) for 48 h.
The MTT assay is based on the ability of mitochondrial dehydrogenases of viable cells to reduce MTT to a purple formazan product (insoluble in water), which can be quantified spectrophotometrically (after solubilization in DMSO). For each well, the culture medium was aspirated, and 100 µL of MTT solution (5 mg/mL in PBS) was added to the cells, which were incubated for 4 h at 37 °C. After this period, the formazan crystals formed were dissolved upon the addition of 100 µL of DMSO, and the absorbance of these solutions was measured at 570 nm (against a blank containing MTT and DMSO in a 1:1 ratio). The results were expressed as the percentage of MTT reduction, assuming the absorbance of control cells as 100%.
5. Conclusions
In summary, the interaction of two polyamine-based ligands and their Zn2+ metal complexes with duplex DNA and RNA was evaluated using UV thermal and fluorescence indicator displacement assays in combination with theoretical studies. Both ligands show a high affinity for ds-DNA/RNA and selectivity for ds-RNA. The ability to interact with these duplexes was blocked upon Zn2+ coordination, which was confirmed by the low variation in the melting temperature and poor displacement of the fluorescent dye from the ds-DNA/RNA. All the experimental findings were supported using molecular modeling, which indicates that a large number of electrostatic contacts for the free ligands are hampered upon metal ion coordination to the nitrogen atoms of the ligands. Finally, cell viability assays show a decrease in the cytotoxicity of the metal complexes in comparison with free ligand, which can be associated with the observed binding to ds-DNA/RNA.
Supplementary Materials
The following are available online. Figures S1–S8, Tables S1–S5. Tables S1 and S2 gather the logarithms of the stepwise protonation constants of L1 and L2 determined and equilibrium constants for the interaction of Zn2+ with L1 and L2. Tables S3 and S4 collect the structural data for complex Zn2L1 and Zn2L2 and Table S5 the ΔTm values determined for the interaction of L1 and L2 toward ds-DNA and ds-RNA. Figures S1–S3 collect the distribution diagrams for L1 and L2 and the metal complex systems Figures S4–S6 gather the representation of the interaction of ligands and metal complexes with ds-DNA and ds-RNA.
Author Contributions
Conceptualization, J.G.-G., B.V., and E.G.-E.; performed the experiments, L.G., Á.M.-C., J.U.C. and M.I.; methodology and statistical analysis, L.G., Á.M.-C., J.U.C., A.G.-E., J.G.-G., and E.G.-E., writing—original draft preparation, J.G.-G., B.V., and E.G.-E.; writing—review and editing, J.G.-G., B.V., and E.G.-E.; funding acquisition, J.G.-G., A.G.-E., and E.G.-E. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Spanish MICINN and MEC and FEDER funds from the European Union (grants PID2019-110751RB-I00, PID2019-108643GA-I00, CTQ2017-90852-REDC, RED2018-102331-T, Unidad de Excelencia María de Maeztu CEX2019-000919 and FIS PI16/00504), Conselleria de Innovación, Universidades, Ciencia y Sociedad Digital of the Generalitat Valenciana (CIDEGENT/2018/015). A.M.-C wants to thank Conselleria de Innovación, Universidades, Ciencia y Sociedad Digital of the Generalitat Valenciana for his postdoctoral grant.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets generated during the current study are available from the corresponding author on reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compounds are available from the authors on reasonable request and depending on the stock.
References
- Johnstone, T.C.; Suntharalingam, K.; Lippard, S.J. The Next Generation of Platinum Drugs: Targeted Pt(II) Agents, Nanoparticle Delivery, and Pt(IV) Prodrugs. Chem. Rev. 2016, 116, 3436–3486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costales, M.G.; Childs-Disney, J.L.; Haniff, H.S.; Disney, M.D. How we think about targeting RNA with small molecules. J. Med. Chem. 2020, 63, 8880–8900. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.-M.; Choi, Y.H.; Tu, M.-J. RNA Drugs and RNA Targets for Small Molecules: Principles, Progress, and Challenges. Pharmac. Rev. 2020, 72, 862–898. [Google Scholar] [CrossRef]
- Meyer, S.M.; Williams, C.C.; Akahori, Y.; Tanaka, T.; Aikawa, H.; Tong, Y.; Childs-Disney, J.L.; Disney, M.D. Small molecule recognition of disease-relevant RNA structures. Chem. Soc. Rev. 2020, 49, 7167–7199. [Google Scholar] [CrossRef]
- Giorgio, A.D.; Duca, M. Synthetic small-molecule RNA ligands: Future prospects as therapeutic agents. Med. Chem. Commun. 2019, 10, 1242–1255. [Google Scholar] [CrossRef]
- Anselmo, A.C.; Mitragotri, S. An overview of clinical and commercial impact of drug delivery systems. J. Control Release 2014, 190, 15–28. [Google Scholar] [CrossRef] [Green Version]
- Patra, J.K.; Das, G.; Fraceto, L.F.; Ramos Campos, E.V.; Rodriguez-Torres, M.P.; Acosta-Torres, L.S.; Diaz-Torres, L.A.; Grillo, R.; Swamy, M.K.; Sharma, S.; et al. Nano based drug delivery systems: Recent developments and future prospects. J. Nanobiotechnol. 2018, 16, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez, J.; Mosquera, J.; Learte-Aymamí, S.; Vázquez, M.E.; Mascareñas, J.L. Stimuli-Responsive DNA Binding by Synthetic Systems. Acc. Chem. Res. 2020, 53, 2286–2298. [Google Scholar] [CrossRef] [PubMed]
- Oheix, E.; Peacock, A.F.A. Metal-ion-regulated miniature DNA-binding proteins based on GCN4 and non-native regulation sites. Chem. Eur. J. 2014, 20, 2829–2839. [Google Scholar] [CrossRef]
- Learte-Aymamí, S.; Curado, N.; Rodríguez, J.; Vázquez, M.E.; Mascareñas, J.L. Metal-dependent DNA recognition and cell internalization of designed, basic peptides. J. Am. Chem. Soc. 2017, 139, 16188–16193. [Google Scholar] [CrossRef] [PubMed]
- Azuma, Y.; Imanishi, M.; Yoshimura, T.; Kawabata, T.; Futaki, S. Cobalt(II)-responsive DNA binding of a GCN4-bZIP protein containing cysteine residues functionalized with iminodiacetic acid. Angew. Chem. Int. Ed. 2009, 48, 6853–6856. [Google Scholar] [CrossRef]
- Gonzalez, J.; Llinares, J.M.; Belda, R.; Pitarch, J.; Soriano, C.; Tejero, R.; Verdejo, B.; Garcia-España, E. Tritopic phenanthroline and pyridine tail-tied aza-scorpiands. Org. Biomol. Chem. 2010, 8, 2367–2376. [Google Scholar] [CrossRef]
- Galiana-Rosello, C.; Aceves-Luquero, C.; Gonzalez, J.; Martinez-Camarena, A.; Villalonga, R.; de Mattos, S.F.; Soriano, C.; Llinares, J.; Garcia-España, E.; Villalonga, P.; et al. Toward a Rational Design of Polyamine-Based Zinc-Chelating Agents for Cancer Therapies. J. Med. Chem. 2020, 63, 1199–1215. [Google Scholar] [CrossRef] [PubMed]
- Miranda, C.; Escarti, F.; Lamarque, L.; Garcia-España, E.; Navarro, P.; Latorre, J.; Lloret, F.; Jimenez, H.R.; Yunta, M.J.R. CuII and ZnII Coordination Chemistry of Pyrazole-Containing Polyamine Receptors—Influence of the Hydrocarbon Side Chain Length on the Metal Coordination. Eur. J. Inorg. Chem. 2005, 1, 189–208. [Google Scholar] [CrossRef]
- Verdejo, B.; Ferrer, A.; Blasco, S.; Castillo, C.E.; Gonzalez, J.; Latorre, J.; Mañez, M.A.; Basallote, M.G.; Soriano, C.; Garcia-España, E. Hydrogen and Copper Ion-Induced Molecular Reorganizations in Scorpionand-like Ligands. A Potentiometric, Mechanistic, and Solid-State Study. Inorg. Chem. 2007, 46, 5707–5719. [Google Scholar] [CrossRef] [PubMed]
- Inclán, M.; Albelda, M.T.; Frías, J.C.; Blasco, S.; Verdejo, B.; Serena, C.; Salat-Canela, C.; Díaz, M.L.; García-España, A.; García-España, E. Modulation of DNA binding by reversible metal-controlled molecular reorganizations of scorpiand-like ligands. J. Am. Chem. Soc. 2012, 134, 9644–9656. [Google Scholar] [CrossRef]
- Inclán, M.; Guijarro, L.; Pont, I.; Frías, J.C.; Rotger, C.; Orvay, F.; Costa, A.; García-España, E.; Albelda, M.T. Binding Mode and Selectivity of a Scorpiand-Like Polyamine Ligand to Single- and Double-Stranded DNA and RNA: Metal- and pH-Driven Modulation. Chem. Eur. J. 2017, 23, 15966–15973. [Google Scholar] [CrossRef] [PubMed]
- González-García, J.; Uzelac, L.; Kralj, M.; Llinares, J.M.; García-España, E.; Piantanida, I. The size of the aryl linker between two polyaza-cyclophane moieties controls the binding selectivity to ds-RNA vs. ds-DNA. Org. Biomol. Chem. 2013, 11, 2154–2161. [Google Scholar] [CrossRef]
- Sornosa-Ten, A.; Albelda, M.T.; Frias, J.C.; Garcia-España, E.; Llinares, J.M.; Budimir, A.; Piantanida, I. Acid–base properties of functionalised tripodal polyamines and their interaction with nucleotides and nucleic acids. Org. Biomol. Chem. 2010, 8, 2567–2574. [Google Scholar] [CrossRef] [Green Version]
- Guijarro, L.; Inclan, M.; Pitarch-Jarque, J.; Domenech-Carbo, A.; Chicote, J.U.; Trefler, S.; García-España, E.; García-España, A.; Verdejo, B. Homo- and Heterobinuclear Cu2+ and Zn2+ Complexes of Ditopic Aza Scorpiand Ligands as Superoxide Dismutase Mimics. Inorg. Chem. 2017, 56, 13748–13758. [Google Scholar] [CrossRef] [PubMed]
- Malojcic, G.; Piantanida, I.; Marinic, M.; Zinic, M.; Marjanovic, M.; Kralj, M.; Pavelic, K.; Schneider, H.-J. A novel bis-phenanthridine triamine with pH controlled binding to nucleotides and nucleic acids. Org. Biomol. Chem. 2005, 3, 4373–4381. [Google Scholar]
- Piantanida, I.; Palm, B.S.; Žinić, M.; Schneider, H.-J. A new 4,9-diazapyrenium intercalator for single- and double-stranded nucleic acids: Distinct differences from related diazapyrenium compounds and ethidium bromide. J. Chem. Soc. Perkin Trans. 2001, 2, 1808–1816. [Google Scholar] [CrossRef]
- Gran, G. Determination of the equivalence point in potentiometric titrations. Part II. Analyst 1952, 77, 661–671. [Google Scholar] [CrossRef]
- Rossotti, F.J.; Rossotti, H. Potentiometric titrations using Gran plots: A textbook omission. J. Chem. Educ. 1965, 42, 375–378. [Google Scholar] [CrossRef]
- Gans, P.; Sabatini, A.; Vacca, A. Investigation of equilibria in solution. Determination of equilibrium constants with the HYPERQUAD suite of programs. Talanta 1996, 43, 1739–1753. [Google Scholar] [CrossRef]
- Perdew, J.P. Density-functional approximation for the correlation energy of the inhomogeneous electron gas. Phys. Rev. B 1986, 33, 8822–8824. [Google Scholar] [CrossRef]
- Becke, A.D. Density-functional exchange-energy approximation with correct asymptotic behavior. Phys. Rev. A 1988, 38, 3098–3100. [Google Scholar] [CrossRef]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 1988, 37, 785–789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weigend, F.; Ahlrichs, R. Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: Design and assessment of accuracy. Phys. Chem. Chem. Phys. 2005, 7, 3297–3305. [Google Scholar] [CrossRef] [PubMed]
- Andrae, D.; Häußermann, U.; Dolg, M.; Stoll, H.; Preuß, H. Energy-adjustedab initio pseudopotentials for the second and third row transition elements. Theor. Chim. Acta 1990, 77, 123–141. [Google Scholar] [CrossRef]
- Grimme, S.; Antony, J.; Ehrlich, S.; Krieg, H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 2010, 132, 154104. [Google Scholar] [CrossRef] [Green Version]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 16, Revision C.01; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Schaftenaar, G.; Noordik, J.H. Molden: A pre- and post-processing program for molecular and electronic structures. J. Comput. Mol. Des. 2000, 14, 123–134. [Google Scholar] [CrossRef] [PubMed]
- The PyMOL Molecular Graphics System; Version 1.5.0.4.; Schrödinger, LLC: New York, NY, USA, 2015.
- Li, P.; Merz, K.M. MCPB.py: A Python Based Metal Center Parameter Builder. J. Chem. Inf. Model. 2016, 56, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Drew, H.R.; Wing, R.M.; Takano, T.; Broka, C.; Tanaka, S.; Itakura, K.; Dickerson, R.E. Structure of a B-DNA dodecamer: Conformation and dynamics. Proc. Natl. Acad. Sci. USA 1981, 78, 2179–2183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chai, J.; Wang, J.; Xu, Q.; Hao, F.; Liu, R. Multi-spectroscopic methods combined with molecular modeling dissect the interaction mechanisms of ractopamine and calf thymus DNA. Mol. Biosyst. 2012, 8, 1902–1907. [Google Scholar] [CrossRef]
- Patro, L.P.P.; Kumar, A.; Kolimi, N.; Rathinavelan, T. 3D-NuS: A Web Server for Automated Modeling and Visualization of Non-Canonical 3-Dimensional Nucleic Acid Structures. J. Mol. Biol. 2017, 429, 2438–2448. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Betz, R.M.; Cerutti, D.S.; Cheatham, T.E., III; Darden, T.A.; Duke, R.E.; Giese, T.J.; Gohlke, H.; Goetz, A.W.; Homeyer, N.; et al. AMBER 2016; University of California: San Francisco, CA, USA, 2016. [Google Scholar]
- Wang, J.M.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E., III. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Com. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).