Proteomic Characterisation of Lupin (Lupinus angustifolius) Milk as Influenced by Extraction Techniques, Seed Coat and Cultivars
Abstract
1. Introduction
2. Results and Discussion
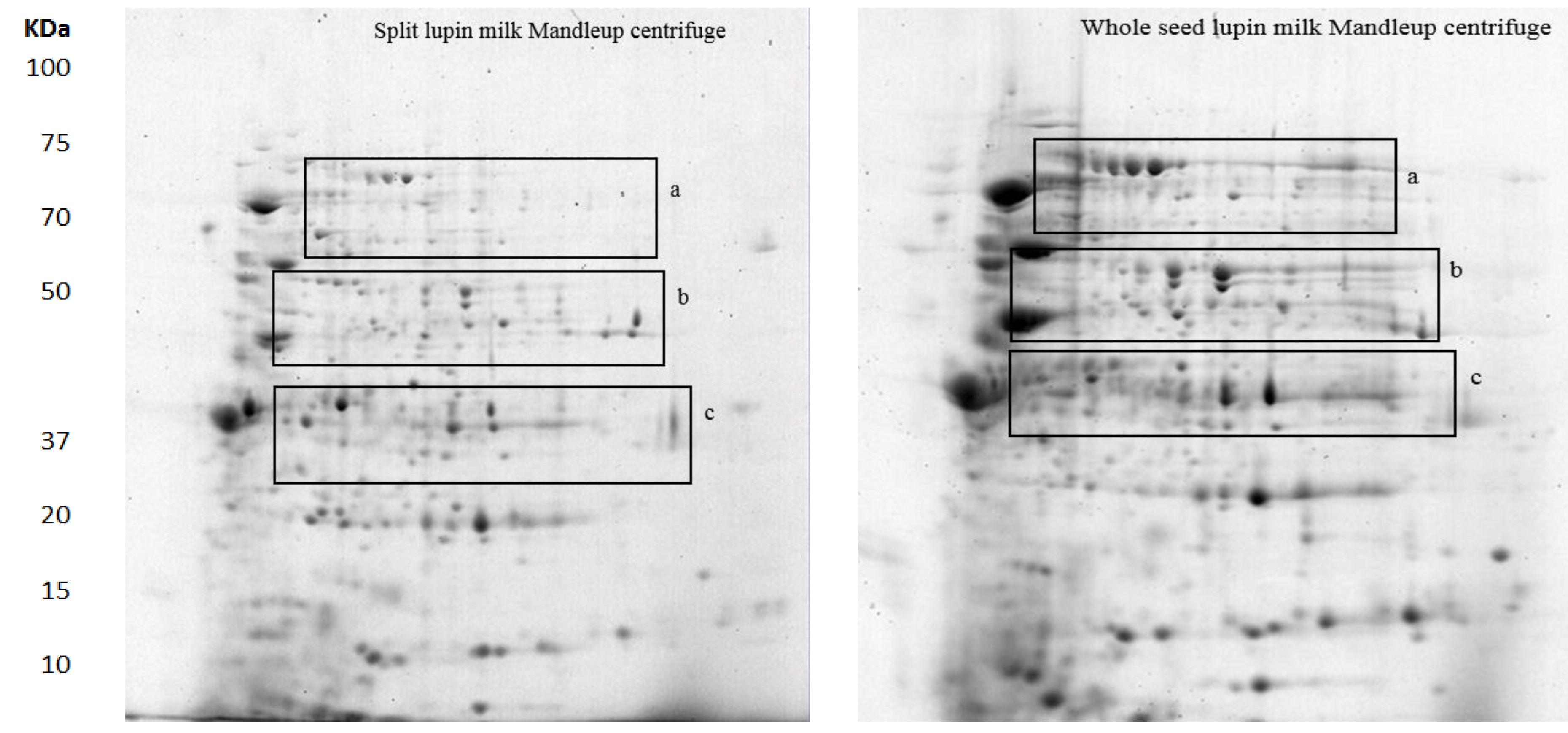
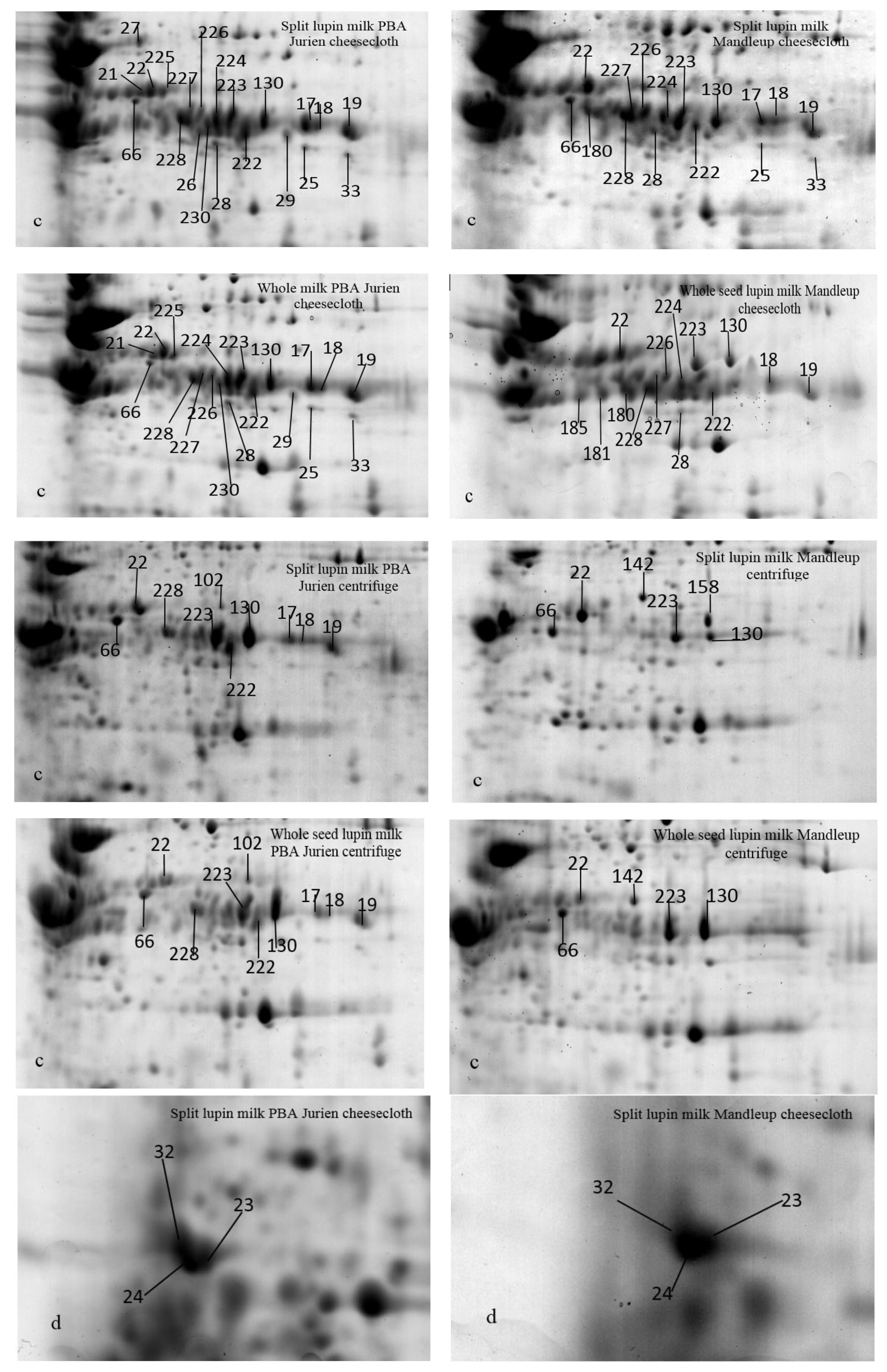
2.1. The Protein Profile of Lupin Milk
2.2. Effect of the Seed Types, Separation Method and Cultivars on Extractability of Proteins in the Lupin Milk
2.2.1. Seed Types
2.2.2. Separation Methods
2.2.3. Cultivars
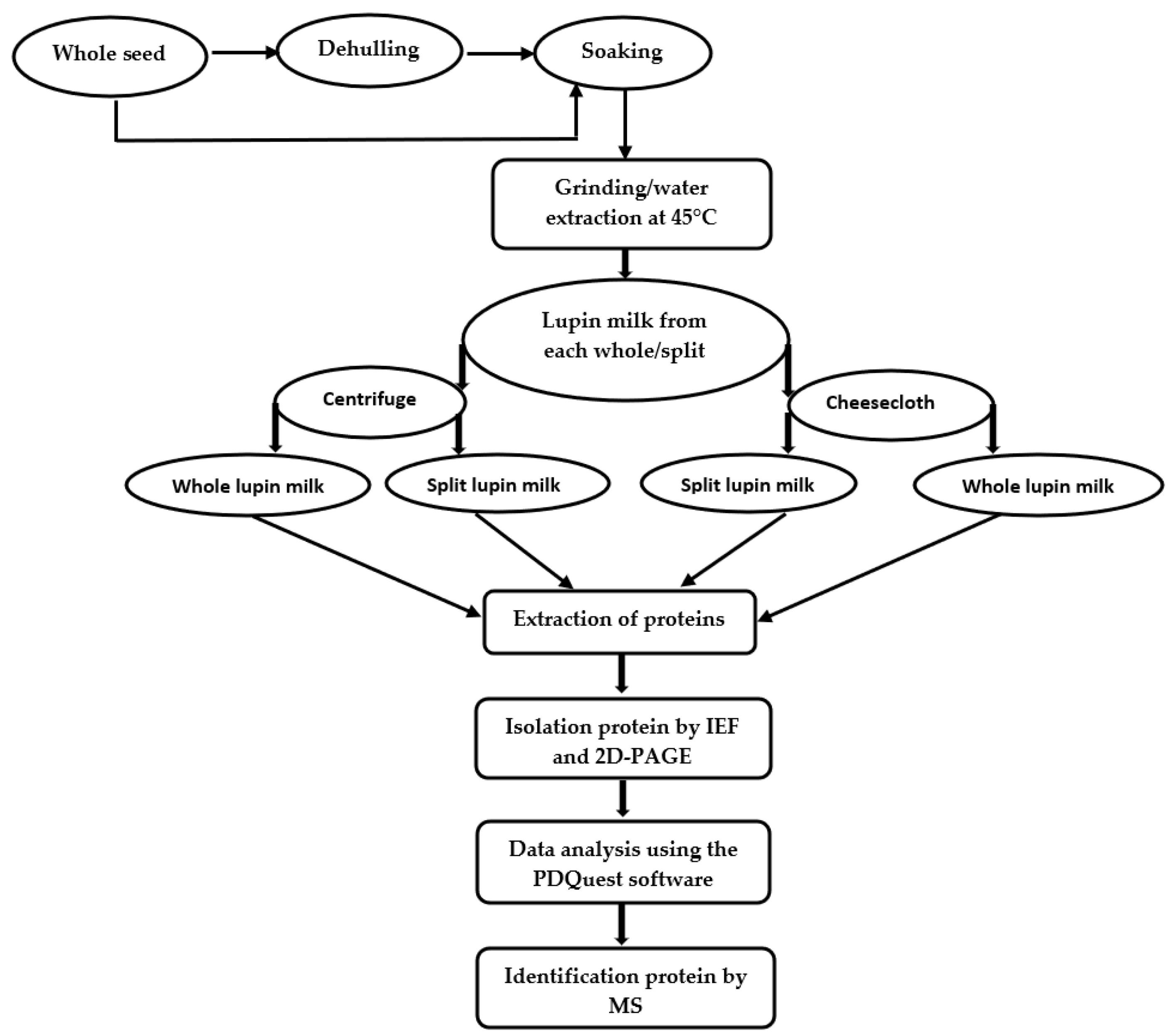
3. Materials and Methods
3.1. Chemicals
3.2. Preparation of Lupin Samples and Lupin Milk
3.3. Total Proteins
3.4. Extraction of Proteins
3.5. Separation of Proteins
3.6. Data Analysis
3.7. Identification of Protein
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chew, P.G.; Casey, A.J.; Johnson, S.K. Protein quality and physico-functionality of Australian sweet lupin (Lupinus angustifolius cv. Gungurru) protein concentrates prepared by isoelectric precipitation or ultrafiltration. Food Chem. 2003, 83, 575–583. [Google Scholar] [CrossRef]
- Johnson, S.K.; McQuillan, P.L.; Sin, J.H.; Ball, M.J. Sensory acceptability of white bread with added Australian sweet lupin (Lupinus angustifolius) kernel fibre and its glycaemic and insulinaemic responses when eaten as a breakfast. J. Sci. Food Agric. 2003, 83, 1366–1372. [Google Scholar] [CrossRef]
- Martínez-Villaluenga, C.; Zieliński, H.; Frias, J.; Piskuła, M.K.; Kozłowska, H.; Vidal-Valverde, C. Antioxidant capacity and polyphenolic content of high-protein lupin products. Food Chem. 2009, 112, 84–88. [Google Scholar] [CrossRef]
- Jayasena, V.; Chih, H.J.; Nasar-Abbas, S. Functional properties of sweet lupin protein isolated and tested at various pH levels. Res. J. Agric. Biol. Sci. 2010, 6, pp. 130–137. Available online: http://hdl.handle.net/20.500.11937/19417 (accessed on 19 March 2020).
- Kiosseoglou, A.; Doxastakis, G.; Alevisopoulos, S.; Kasapis, S. Physical characterization of thermally induced networks of lupin protein isolates prepared by isoelectric precipitation and dialysis. Int. J. Food Sci. Technol. 1999, 34, 253–263. [Google Scholar] [CrossRef]
- Martínez-Villaluenga, C.; Sironi, E.; Vidal-Valverde, C.; Duranti, M. Effects of oligosaccharide removing procedure on the protein profiles of lupin seeds. Eur. Food Res. Technol. 2006, 223, 691–696. [Google Scholar] [CrossRef]
- Siddique, K.; Walton, G.; Seymour, M. A comparison of seed yields of winter grain legumes in Western Australia. Aust. J. Exp. Agric. 1993, 33, 915–922. [Google Scholar] [CrossRef]
- Archer, B.J.; Johnson, S.K.; Devereux, H.M.; Baxter, A.L. Effect of fat replacement by inulin or lupin-kernel fibre on sausage patty acceptability, post-meal perceptions of satiety and food intake in men. Br. J. Nutr. 2004, 91, 591–599. [Google Scholar] [CrossRef]
- Foley, R.C.; Gao, L.-L.; Spriggs, A.; Soo, L.Y.; Goggin, D.E.; Smith, P.M.; Atkins, C.A.; Singh, K.B. Identification and characterisation of seed storage protein transcripts from Lupinus angustifolius. BMC Plant Biol. 2011, 11, 59. [Google Scholar] [CrossRef]
- Restani, P.; Duranti, M.; Cerletti, P.; Simonetti, P. Subunit composition of the seed globulins of Lupinus albus. Phytochemistry 1981, 20, 2077–2083. [Google Scholar] [CrossRef]
- Melo, T.S.; Ferreira, R.B.; Teixeira, A.N. The seed storage proteins from Lupinus albus. Phytochemistry 1994, 37, 641–648. [Google Scholar] [CrossRef]
- Salmanowicz, B. Comparative study of seed albumins in the Old-WorldLupinus species (Fabaceae) by reversed-phase HPLC. Plant Syst. Evol. 1995, 195, 77–86. [Google Scholar] [CrossRef]
- Duranti, M.; Restani, P.; Poniatowska, M.; Cerletti, P. The seed globulins of Lupinus albus. Phytochemistry 1981, 20, 2071–2075. [Google Scholar] [CrossRef]
- Goggin, D.E.; Mir, G.; Smith, W.B.; Stuckey, M.; Smith, P.M. Proteomic analysis of lupin seed proteins to identify conglutin β as an allergen, Lup an 1. J. Agric. Food Chem. 2008, 56, 6370–6377. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Martínez, C.; Hernández-Sánchez, H.; Dávila-Ortiz, G. Production of a yogurt-like product from Lupinus campestris seeds. J. Sci. Food Agric. 2003, 83, 515–522. [Google Scholar] [CrossRef]
- Jayasena, V.; Chih, H.J.; Nasar-Abbas, S. Efficient isolation of lupin protein. Food Aust. 2011, 63, pp. 306–309. Available online: http://hdl.handle.net/20.500.11937/22645 (accessed on 19 March 2020).
- Muranyi, I.S.; Otto, C.; Pickardt, C.; Koehler, P.; Schweiggert-Weisz, U. Microscopic characterisation and composition of proteins from lupin seed (Lupinus angustifolius L.) as affected by the isolation procedure. Food Res. Int. 2013, 54, 1419–1429. [Google Scholar] [CrossRef]
- Berghout, J.; Boom, R.; Van der Goot, A. The potential of aqueous fractionation of lupin seeds for high-protein foods. Food Chem. 2014, 159, 64–70. [Google Scholar] [CrossRef]
- Sirtori, E.; Resta, D.; Brambilla, F.; Zacherl, C.; Arnoldi, A. The effects of various processing conditions on a protein isolate from Lupinus angustifolius. Food Chem. 2010, 120, 496–504. [Google Scholar] [CrossRef]
- Chapleau, N.; De Lamballerie-Anton, M. Improvement of emulsifying properties of lupin proteins by high pressure induced aggregation. Food Hydrocoll. 2003, 17, 273–280. [Google Scholar] [CrossRef]
- Bader, S.; Bez, J.; Eisner, P. Can protein functionalities be enhanced by high-pressure homogenization?–A study on functional properties of lupin proteins. Procedia Food Sci. 2011, 1, 1359–1366. [Google Scholar] [CrossRef]
- Natarajan, S.S.; Krishnan, H.B.; Khan, F.; Chen, X.; Garrett, W.M.; Lakshman, D. Analysis of soybean embryonic axis proteins by two-dimensional gel electrophoresis and mass spectrometry. J. Basic Appl. Sci. 2013, 9, 302–308. [Google Scholar] [CrossRef]
- Magni, C.; Scarafoni, A.; Herndl, A.; Sessa, F.; Prinsi, B.; Espen, L.; Duranti, M. Combined 2D electrophoretic approaches for the study of white lupin mature seed storage proteome. Phytochemistry 2007, 68, 997–1007. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Fang, Z.; Wahlqvist, M.L.; Wu, G.; Hodgson, J.M.; Johnson, S.K. Seed coats of pulses as a food ingredient: Characterization, processing, and applications. Trends Food Sci. Technol. 2018, 80, 35–42. [Google Scholar] [CrossRef]
- Lampart-Szczapa, E.; Korczak, J.; Nogala-Kalucka, M.; Zawirska-Wojtasiak, R. Antioxidant properties of lupin seed products. Food Chem. 2003, 83, 279–285. [Google Scholar] [CrossRef]
- Hove, E.L. Composition and protein quality of sweet lupin seed. J. Sci. Food Agric. 1974, 25, 851–859. [Google Scholar] [CrossRef]
- Martínez-Maqueda, D.; Hernández-Ledesma, B.; Amigo, L.; Miralles, B.; Gómez-Ruiz, J.Á. Extraction/fractionation techniques for proteins and peptides and protein digestion. In Proteomics in Foods; Springer: Boston, MA, USA, 2013; pp. 21–50. [Google Scholar]
- Islam, S.; Yan, G.; Appels, R.; Ma, W. Comparative proteome analysis of seed storage and allergenic proteins among four narrow-leafed lupin cultivars. Food Chem. 2012, 135, 1230–1238. [Google Scholar] [CrossRef]
- Duranti, M.; Consonni, A.; Magni, C.; Sessa, F.; Scarafoni, A. The major proteins of lupin seed: Characterisation and molecular properties for use as functional and nutraceutical ingredients. Trends Food Sci. Technol. 2008, 19, 624–633. [Google Scholar] [CrossRef]
- Kamphuis, L.G.; Hane, J.K.; Nelson, M.N.; Gao, L.; Atkins, C.A.; Singh, K.B. Transcriptome sequencing of different narrow-leafed lupin tissue types provides a comprehensive uni-gene assembly and extensive gene-based molecular markers. Plant Biotechnol. J. 2015, 13, 14–25. [Google Scholar] [CrossRef]
- Chemists, A.O.A.C. Official Methods of Analysis: Changes in Official Methods of Analysis Made at the Annual Meeting. Supplement; Association of Official Analytical Chemists: Arlington, VA, USA, 1991. [Google Scholar]
- Bringans, S.; Eriksen, S.; Kendrick, T.; Gopalakrishnakone, P.; Livk, A.; Lock, R.; Lipscombe, R. Proteomic analysis of the venom of Heterometrus longimanus (Asian black scorpion). Proteomics 2008, 8, 1081–1096. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the authors. |


| Cultivars | Separation Method | Type of Lupin Seeds | Total Protein (g/L) Mean ± SD (n = 3) | Spots Numbers Mean ± SD (n = 3) |
|---|---|---|---|---|
| PBA Jurien | cheesecloth | Split | 27.53 ± 1.84 | 231.33 ± 1.15 |
| Whole | 20.98 ± 1.27 | 201 ± 1 | ||
| centrifuge | Split | 20.49 ± 1.51 | 196.33 ± 1.52 | |
| Whole | 14.49 ± 1.16 | 158.33 ± 0.57 | ||
| Mandelup | cheesecloth | Split | 27 ± 2.03 | 204 ± 1 |
| Whole | 20.38 ± 1.92 | 190.67 ± 0.57 | ||
| centrifuge | Split | 19.85 ± 1.17 | 189.33 ± 0.57 | |
| Whole | 14.94 ± 1.55 | 180 ± 1 |
| Split Lupin Milk PBA Jurien | Whole Lupin Milk PBA Jurien | Split Lupin Milk Mandelup | Whole Lupin Milk Mandelup | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Cheesecloth | Centrifuge | Cheesecloth | Centrifuge | Cheesecloth | Centrifuge | Cheesecloth | Centrifuge | ||
| Spot No | SSP | Mean ± SD (n = 3) | Mean ± SD (n = 3) | Mean ± SD (n = 3) | Mean ± SD (n = 3) | Mean ± SD (n = 3) | Mean ± SD (n = 3) | Mean ± SD (n = 3) | Mean ± SD (n = 3) |
| 1 | 4802 | 51 ± 1.41 | 29.53 ± 2.71 | 62.14 ± 1.16 | 20.75 ± 0.87 | 41.50 ± 2.67 | ND | ND | ND |
| 2 | 4803 | 56.69 ± 2.66 | 39.49 ± 1.17 | 75.29 ± 4.20 | 30.69 ± 1.12 | 43.89 ± 3.10 | ND | 38.19 ± 2.03 | 35.44 ± 1.44 |
| 3 | 5802 | 61.59 ± 3.60 | 27.95 ± 1.26 | 78.22 ± 1.40 | 22.03 ± 2.35 | 47.47 ± 2.06 | 33.60 ± 1.32 | 35.17 ± 0.57 | 30.61 ± 0.74 |
| 4 | 5804 | 92.63 ± 1.91 | 33.39 ± 1.10 | 104.57 ± 3.69 | 31.55 ± 2.15 | 67.07 ± 1.8 | 41.66 ± 1.23 | 52.95 ± 4.74 | 44.95 ± 0.95 |
| 5 | 5806 | 78.74 ± 1.22 | 47.74 ± 2.51 | 84.34 ± 2.95 | ND | 61.30 ± 0.61 | 50.95 ± 0.26 | 58.15 ± 0.58 | 55.38 ± 0.40 |
| 6 | 5808 | 79.40 ± 1.34 | ND | 81.45 ± 0.25 | ND | 71.63 ± 2.59 | ND | 53.62 ± 2.83 | 20.72 ± 1.18 |
| 7 | 2802 | 98.30 ± 0.84 | 60.37 ± 1.44 | 108.13 ± 4.50 | 44.48 ± 0.99 | ND | ND | ND | ND |
| 8 | 2803 | 113.25 ± 4.23 | 64.72 ± 1.13 | 160.5 ± 2.80 | 55.63 ± 0.75 | ND | ND | ND | ND |
| 9 | 3801 | 114.49 ± 5.20 | 37.48 ± 1.77 | 163.79 ± 3.75 | 26.1 ± 2.19 | 44.88 ± 2.99 | 21.06 ± 0.82 | ND | ND |
| 10 | 3802 | 147.59 ± 2.82 | 66.33 ± 1.52 | 155.21 ± 2.24 | 49.15 ± 2.73 | 83.81 ± 1.05 | 37.16 ± 0.76 | 52.77 ± 1.83 | 43.68 ± 2.36 |
| 11 | 3804 | 85.27 ± 1.48 | 74.63 ± 2.90 | 260.20 ± 3.63 | 73.24 ± 1.73 | 59.43 ± 2.95 | 23.86 ± 3.5 | 33.56 ± 0.58 | 30.53 ± 0.93 |
| 12 | 5801 | 46.01 ± 3.36 | ND | 30.57 ± 1.12 | ND | 30.59 ± 2.28 | ND | 21.25 ± 0.65 | ND |
| 13 | 5804 | 54.53 ± 1.60 | ND | 33.19 ± 2.34 | ND | 36.06 ± 0.55 | ND | 30 ± 1.12 | ND |
| 14 | 5807 | 61.05 ± 1.59 | ND | 38.47 ± 1.43 | ND | 41.77 ± 0.43 | ND | 33.11 ± 1.11 | ND |
| 15 | 6506 | 166.15 ± 5.16 | 251.43 ± 1.68 | 240.37 ± 3.75 | 266.77 ± 3.16 | 194.52 ± 3.84 | 206.84 ± 1.90 | 227.61 ± 4.49 | 238.54 ± 5.21 |
| 16 | 6505 | 130.97 ± 6.40 | 221.05 ± 1.79 | 238.83 ± 2.63 | 298.11 ± 1.78 | 191.46 ± 2.87 | 214.00 ± 1.55 | 215.04 ± 2.03 | 247.86 ± 5.04 |
| 17 | 6306 | 501.68 ± 5.11 | 54.85 ± 0.89 | 254.56 ± 2.98 | 32.66 ± 1.22 | 232.86 ± 3.69 | ND | ND | ND |
| 18 | 7304 | 393.13 ± 4.70 | 33.71 ± 2.83 | 311.65 ± 2.43 | 23.66 ± 1.79 | 160.93 ± 3.79 | ND | 76.18 ± 2.46 | ND |
| 19 | 8301 | 407.24 ± 3.58 | 40.51 ± 0.78 | 365.69 ± 0.90 | 20.76 ± 1.24 | 195.39 ± 3.92 | ND | 79.51 ± 0.53 | ND |
| 21 | 3405 | 567.09 ± 3.26 | ND | 105.35 ± 2.07 | ND | ND | ND | ND | ND |
| 22 | 4401 | 218.94 ± 4.51 | 135.26 ± 2.64 | 204.80 ± 3.82 | 46.16 ± 5.01 | 196.97 ± 2.20 | 102.37 ± 2.44 | 178.05 ± 0.15 | 25.53 ± 1.06 |
| 23 | 1208 | 417.46 ± 2.86 | ND | ND | ND | 548.56 ± 5.12 | ND | ND | ND |
| 24 | 1210 | 314.10 ± 1.80 | ND | ND | ND | 415.90 ± 6.44 | ND | ND | ND |
| 25 | 7204 | 78.68 ± 3.2 | ND | 49.85 ± 3.2 | ND | 40.72 ± 2.27 | ND | ND | ND |
| 26 | 5307 | 73.65 ± 0.68 | ND | ND | ND | ND | ND | ND | ND |
| 27 | 3404 | 178.12 ± 5.24 | ND | 61.05 ± 1.58 | ND | ND | ND | ND | ND |
| 28 | 5202 | 107.12 ± 4.67 | ND | 95.07 ± 4.79 | ND | 64.06 ± 3.56 | ND | 44.42 ± 1.42 | ND |
| 29 | 7201 | 145.77 ± 4.39 | ND | 126.81 ± 5.21 | ND | ND | ND | ND | ND |
| 32 | 1205 | 381.31 ± 3.20 | ND | ND | ND | 459.90 ± 5.21 | ND | ND | ND |
| 33 | 8201 | 92.23 ± 1.51 | ND | 71.89 ± 4.06 | ND | 30.04 ± 0.39 | ND | ND | ND |
| 46 | 6502 | 44.85 ± 0.70 | 183.11 ± 5.18 | 142.16 ± 3.93 | 190.45 ± 1.31 | 77.36 ± 1.04 | 95.33 ± 3.50 | 110.68 ± 2.30 | 132.66 ± 4.02 |
| 47 | 5502 | 21.88 ± 1.65 | 206.73 ± 3.82 | 178.29 ± 1.28 | 290.27 ± 0.81 | 80.54 ± 2.35 | 86.15 ± 1.81 | 98.30 ± 1.41 | 121.00 ± 1.00 |
| 66 | 3301 | 121.76 ± 0.92 | 76.06 ± 1.42 | 97.43 ± 4.20 | 62.81 ± 1.13 | 79.88 ± 0.58 | 74.25 ± 3.54 | ND | 65.79 ± 2.87 |
| 68 | 8501 | 50.81 ± 2.04 | 70.65 ± 3.52 | 76.39 ± 2.18 | 88.83 ± 1.35 | 81.82 ± 5.14 | 91.38 ± 3.85 | 30.55 ± 0.58 | 45.81 ± 1.57 |
| 69 | 5607 | ND | 21.59 ± 1.16 | ND | ND | ND | 18.09 ± 0.58 | ND | ND |
| 70 | 5507 | ND | 31.08 ± 1.13 | ND | ND | ND | 14.13 ± 0.80 | ND | ND |
| 71 | 8401 | 42.92 ± 2.68 | 78.65 ± 0.68 | 80.15 ± 2.60 | 89.99 ± 1.59 | 63.51 ± 3.84 | 82.74 ± 2.42 | 40.54 ± 0.49 | 68.21 ± 0.80 |
| 82 | 5801 | ND | ND | ND | 44.03 ± 0.71 | ND | ND | ND | ND |
| 88 | 2802 | ND | 22.39 ± 0.93 | 54.77 ± 1.39 | 33.63 ± 0.75 | ND | ND | ND | ND |
| 93 | 5401 | ND | 89.53 ± 1.05 | 136.34 ± 1.05 | 115.99 ± 0.73 | ND | 104.13 ± 1.88 | 34.51 ± 0.58 | 126.40 ± 2.12 |
| 102 | 6404 | ND | 21.97 ± 0.29 | ND | 32.94 ± 0.65 | ND | ND | ND | ND |
| 130 | 6306 | 549.00 ± 5.08 | 236.34 ± 1.05 | 265.33 ± 1.57 | 188.33 ± 2.54 | 355.63 ± 3.82 | 198.38 ± 1.51 | 167.82 ± 3.14 | 132.37 ± 3.20 |
| 142 | 7404 | ND | ND | ND | ND | ND | 81.11 ± 0.90 | ND | 74.04 ± 0.30 |
| 143 | 4505 | ND | ND | ND | ND | ND | 16.91 ± 0.77 | ND | ND |
| 144 | 4612 | ND | ND | ND | ND | ND | 18.58 ± 0.59 | ND | ND |
| 152 | 7401 | 190.41 ± 1.85 | ND | 179.77 ± 2.78 | ND | 98.466 ± 2.56 | ND | 50.31 ± 0.69 | ND |
| 153 | 6401 | 106.60 ± 3.01 | ND | 97.89 ± 1.75 | ND | 87.30 ± 2.99 | ND | 68.88 ± 0.39 | ND |
| 180 | 4303 | ND | ND | ND | ND | 85.41 ± 0.74 | ND | 72.82 ± 0.58 | ND |
| 181 | 3302 | ND | ND | ND | ND | ND | ND | 87.63 ± 1.19 | ND |
| 185 | 3406 | ND | ND | ND | ND | ND | ND | 90.88 ± 0.28 | ND |
| 222 | 6303 | 672.66 ± 2.64 | 230.70 ± 1.33 | 569.39 ± 5.81 | 189.54 ± 2.42 | 433.7 ± 2.31 | ND | 75.86 ± 0.81 | ND |
| 223 | 6301 | 431.56 ± 2.73 | 209.31 ± 3.40 | 388.92 ± 5.48 | 198.01 ± 1.95 | 324.37 ± 4.14 | 178.60 ± 1.09 | 235.81 ± 1.19 | 162.45 ± 0.64 |
| 224 | 5305 | 470.67 ± 1.28 | ND | 311.70 ± 4.91 | ND | 281.78 ± 5.33 | ND | 182.23 ± 2.16 | ND |
| 225 | 4402 | 201.33 ± 3.50 | ND | 150.53 ± 4.40 | ND | ND | ND | ND | ND |
| 226 | 5303 | 191.00 ± 4.16 | ND | 83.65 ± 5.11 | ND | 75.71 ± 2.47 | ND | 64.54 ± 0.82 | ND |
| 227 | 5302 | 146.33 ± 1.52 | ND | 104.45 ± 3.39 | ND | 69.81 ± 2.68 | ND | 35.188 ± 2.94 | ND |
| 228 | 5301 | 206.23 ± 1.66 | 42.07 ± 1.86 | 196.74 ± 5.00 | 34.49 ± 0.76 | 157.35 ± 2.93 | ND | 86.32 ± 3.96 | ND |
| 230 | 5304 | 581.00 ± 5.73 | ND | 354.70 ± 2.26 | ND | ND | ND | ND | ND |
| Cultivars | Separation Method | Type of Lupin Seeds to Make Milk | Protein Spots Present | Higher Level of Abundance |
|---|---|---|---|---|
| PBA Jurien | cheesecloth | Split | 23, 26 [β-conglutins] 24, 32 [α-conglutins] | 12–14, 17–19, 21, 22, 25, 27–29, 33 130, 153, 222–228, 230 [β-conglutins] 152, 66 [α-conglutins] |
| Whole | 93 [α-conglutins] 88 [β-conglutins] | 1–11, 46, 47, 71 [β-conglutins] 15, 16 [α-conglutins] 68 [hypothetical protein Tanjilg] | ||
| PBA Jurien | centrifuge | Split | 69, 70 [α-conglutins] 5 [β-conglutins] | 1–4, 7–11, 17–19, 22, 130, 222, 223, 228 [β-conglutins] 66 [α-conglutins] |
| Whole | 82 [Lupan Putative TAG factor protein] | 15, 16, 93 [α-conglutins] 46, 47, 71, 88, 102 [β-conglutins] 68 [hypothetical protein Tanjilg] | ||
| Mandelup | cheesecloth | Split | 1, 9, 23, 17, 25, 33, 66 [β-conglutins] 24, 32 [α-conglutins] | 2–6, 10–14, 18, 19, 22, 28, 71, 222–224, 226–228 130, 153, 180 [β-conglutins] 152 [α-conglutins] 68 [hypothetical protein Tanjilg] |
| Whole | 185, 93 [α-conglutins] 181 [β-conglutins] | 15, 16 [α-conglutins] 46, 47 [β-conglutins] | ||
| Mandelup | centrifuge | Split | 69, 70, 143, 144 [α-conglutins] 9 [β-conglutins] 158 [Lupan Putative TAG factor protein] | 66, 142 [α-conglutins] 22, 71, 130, 223 [β-conglutins] 68 [hypothetical protein Tanjilg] |
| Whole | 2, 6 [β-conglutins] | 15, 16, 93 [α-conglutins] 3–5, 10, 11, 46, 47 [β-conglutins] |
| Lupin Milk | Separation Method | Protein Spots Present | Higher Level of Abundance |
|---|---|---|---|
| Split seed lupin milk PBA Jurien | Cheesecloth | 6, 12–14, 21, 23, 25–29, 33, 153, 224–227, 230 [β-conglutins] 24, 32, 152 [α-conglutins] | 1–5, 7–11, 17–19, 22, 130, 222, 223, 228 [β-conglutins] 66 [α-conglutins] |
| Centrifuge | 88, 93, 102 [β-congutins] 69, 70 [α-conglutins] | 15, 16 [α-congutins] 46, 47, 71 [β-congutins] 68 [hypothetical protein Tanjilg] | |
| Whole seed lupin milk PBA Jurien | Cheesecloth | 5, 6, 12–14, 21, 25, 28, 29, 33, 153, 224–227, 230 [β-congutins] 152 [α-conglutins] | 1–4,7–11, 17–19, 22, 88, 130, 222, 223, 228 [β-congutins] 66, 93 [α-conglutins] |
| Centrifuge | 102 [β-conglutins] 82 [Lupan Putative TAG factor protein] | 15, 16 [α-conglutins] 46, 47, 71 [β-conglutins] 68 [hypothetical protein Tanjilg] | |
| Split seed lupin milk Mandelup | Cheesecloth | 1, 2, 6, 12–14, 17–19, 23, 25, 28, 33, 153, 180, 222, 224, 226–228 [β-conglutins] 24, 32, 152 [α-conglutins] | 3-5, 9–11, 22, 130, 223 [β-conglutins] 66 [α-conglutins] |
| Centrifuge | 69, 70, 93, 142–144 [α-conglutins] 158 [Lupan Putative TAG factor protein] | 46, 47, 71 [β-congutins] 16, 15 [α-conglutins] 68 [hypothetical protein Tanjilg] | |
| Whole seed lupin milk Mandelup | Cheesecloth | 12–14,18, 19, 28, 153, 180, 181, 222, 224, 226–228 [β-conglutins] 152, 185 [α-conglutins] | 2–6, 10, 11, 22,130, 223 [β-conglutins] |
| Centrifuge | 142, 66 [α-conglutins] | 46, 47, 71 [β-conglutins] 15, 16, 93 [α-conglutins] 68 [hypothetical protein Tanjilg] |
| Lupin Milk | Cultivars | Proteins Spots Present | Higher Level of Abundance |
|---|---|---|---|
| Split seed lupin milk cheesecloth separation | PBA Jurien | 7, 8, 21, 26, 29, 225, 230 [β-conglutins] | 1–6, 9–14, 17–19, 22, 25, 28, 33, 130, 153, 222–224, 226–228 [β-conglutins] 152 [α-conglutins] |
| Mandelup | 180 [β-conglutins] | 15, 16, 24, 32 [α-conglutins] 23, 46, 47, 71 [β-conglutins] 68 [hypothetical protein Tanjilg] | |
| Whole seed lupin milk cheesecloth separation | PBA Jurien | 1, 7–9, 17, 21, 25, 29, 33, 88, 225, 230 [β-conglutins] 66 [α-conglutins] | 15, 16, 152, 93 [α-conglutins] 2–6, 10–14, 18, 19, 22, 28, 46, 47, 130,153, 222–224, 226–228 [β-conglutins] |
| Mandelup | 180, 181 [β-conglutins] 185 [α-conglutins] | NPS | |
| Split seed lupin milk centrifuge separation | PBA Jurien | 1, 2, 7, 8, 17-19, 88, 102, 222, 228 [β-conglutins] | 15, 16, 70, 66, 69 [α-conglutins] 9–11, 22, 46, 47, 130, 223 [β-conglutins] |
| Mandelup | 143, 144, 142 [α-conglutins] 158 [Lupan Putative TAG factor protein] | 3–5, 71 [β-conglutins] 93 [α-conglutins] 68 [hypothetical protein Tanjilg] | |
| Whole seed lupin milk centrifuge separation | PBA Jurien | 1, 7–9, 17–19, 88, 102, 228 [β-conglutins] 82 [Lupan Putative TAG factor protein] | 15, 16 [α-conglutins] 10, 11, 22, 46, 47, 71, 130, 223 [β-conglutins] 68 [hypothetical protein Tanjilg] |
| Mandelup | 142 [α-conglutins] 5, 6 [β-conglutins] | 2–4 [β-conglutins] 93, 66 [α-conglutins] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Saedi, N.; Agarwal, M.; Ma, W.; Islam, S.; Ren, Y. Proteomic Characterisation of Lupin (Lupinus angustifolius) Milk as Influenced by Extraction Techniques, Seed Coat and Cultivars. Molecules 2020, 25, 1782. https://doi.org/10.3390/molecules25081782
Al-Saedi N, Agarwal M, Ma W, Islam S, Ren Y. Proteomic Characterisation of Lupin (Lupinus angustifolius) Milk as Influenced by Extraction Techniques, Seed Coat and Cultivars. Molecules. 2020; 25(8):1782. https://doi.org/10.3390/molecules25081782
Chicago/Turabian StyleAl-Saedi, Nadia, Manjree Agarwal, Wujun Ma, Shahidul Islam, and Yonglin Ren. 2020. "Proteomic Characterisation of Lupin (Lupinus angustifolius) Milk as Influenced by Extraction Techniques, Seed Coat and Cultivars" Molecules 25, no. 8: 1782. https://doi.org/10.3390/molecules25081782
APA StyleAl-Saedi, N., Agarwal, M., Ma, W., Islam, S., & Ren, Y. (2020). Proteomic Characterisation of Lupin (Lupinus angustifolius) Milk as Influenced by Extraction Techniques, Seed Coat and Cultivars. Molecules, 25(8), 1782. https://doi.org/10.3390/molecules25081782







