Abstract
Five 8,17-epoxybriaranes, including three new compounds—briarenols I–K (1–3), along with two known analogues, briaexcavatolide P (4) and briaexcavatin P (5), were isolated from the octocoral Briareum excavatum. The structures of briaranes 1–3 were elucidated by spectroscopic methods, including 1D and 2D NMR studies and (+)-HRESIMS. Briarane 4 exerted inhibition effects on inducible nitric oxide synthase (iNOS) and cyclooxygenase-2 (COX-2) release from RAW 264.7.
1. Introduction
Octocorals of the genus Briareum (family Briareidae) [1,2,3,4] are proven to be the most important source to produce briarane-type diterpenoids [5]. The compounds of this type are only found in marine invertebrates, particularly in octocorals and demonstrated a wide spectrum of bioactivities, such as anti-inflammatory activity [6] and cytotoxicity [7]. In our continuing research into the chemical constituents of an octocoral B. excavatum (Nutting 1911), which was distributed extensively in the coral reefs of Taiwan, have resulted in isolation of three previously unreported 8,17-epoxybriaranes–briarenols I–K (1–3) along with two known analogues, briaexcavatolide P (4) [8] and briaexcavatin P (5) [9], (Figure 1). In the current study, the comprehensive workflow of isolation, structure determination, and anti-inflammatory activity evaluation, was implemented on briaranes 1–5.
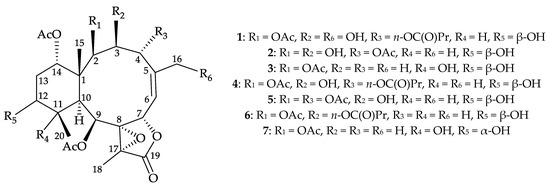
Figure 1.
Structures of briarenols I–K (1–3), briaexcavatolide P (4), briaexcavatin P (5), excavatolide B (6), and briareolide B (7).
2. Results and Discussion
Briarenol I (1) was isolated as an amorphous powder and displayed a sodiated adduct ion at m/z 649.24677 in the (+)-HRESIMS, which indicated its molecular formula was C30H42O14 (calculated for C30H42O14 + Na, 649.24668; unsaturation degrees = 10). The IR spectrum revealed absorptions for hydroxy (νmax 3524 cm–1), γ-lactone (νmax 1783 cm–1), and ester carbonyl (νmax 1736 cm–1) moieties. Resonances in the 13C NMR of 1 at δC 172.9, 172.3, 170.5, 170.0, and 170.0 (5 × C) supported the presence of a γ-lactone and four esters (Table 1). Three of the esters were identified as acetates by the presence of three methyl singlet resonances in the 1H NMR spectrum at δH 2.34, 2.15, and 2.08 (Table 2) and the remaining ester was found to be an n-butyroxy group based on 1H NMR studies, including a correlation spectroscopy (COSY) experiment, which revealed seven contiguous protons (δH 2.30, 2H, t, J = 7.2 Hz; 1.63, 2H, tq, J = 7.2 Hz; 0.95, 3H, t, J = 7.2 Hz). From the COSY spectrum (Figure 2), the proton sequences from H-6/H-7, H-9/H-10/H-11/H-12/H2-13/H-14, and H-11/H3-20 were established. The hydroxy proton signals at δH 4.30 (1H, d, J = 12.0 Hz), 1.49 (1H, d, J = 4.0 Hz), and 3.49 (1H, dd, J = 9.6, 4.4 Hz) were found to correlate with H-3 (δH 4.59, d, J = 12.0 Hz), H-12 (δH 4.10, m), and H2-16 (δH 4.35, 1H, dd, J = 13.6, 4.4 Hz; 4.04, 1H, dd, J = 13.6, 9.6 Hz), respectively. Thus, the hydroxy groups should be positioned at C-3, C-12, and C-16, respectively. Olefinic resonances in the 13C NMR at δC 125.5 (CH-6) and 142.0 (C-5) indicated the presence of a trisubstituted carbon–carbon double bond. On the basis of these data and the heteronuclear multiple bond correlation (HMBC) experiment (Figure 2), the connectivity from C-1 to C-14 was established. A hydroxymethyl group at C-5 was revealed by the HMBC between C-16 oxymethylene protons to C-4, C-5, and C-6. The C-15 methyl group at C-1 was confirmed by the HMBC between H3-15/C-1, C-2, C-10, C-14, and H-10/C-15. The n-butyrate positioned at C-4 was confirmed from the connectivity between H-4 (δH 6.14) and the carbonyl carbon of n-butyrate group (δC 172.3). HMBC from the oxymethine protons at δH 4.53 (H-2), 5.32 (H-9), and 4.88 (H-14) to the acetate carbonyls at δC 172.9, 170.0, and 170.0, placed the acetoxy groups on C-2, C-9, and C-14, respectively. Thirteen of the fourteen oxygen atoms in the molecular formula of 1 could be accounted for from the presence of a γ-lactone, four esters, and three hydroxy groups. The remaining oxygen atom had to be placed between C-8 and C-17 to form a tetrasubstituted epoxide based on the 13C NMR evidences at δC 70.8 (C-8) and 62.5 (C-17) and the 1H NMR chemical shift of a tertiary methyl at δH 1.66 (3H, s, H3-18).

Table 1.
13C NMR (δC 100 MHz, CDCl3) data for briaranes 1–3.

Table 2.
1H NMR (δH, 400 MHz in CDCl3) data (J in Hz) for briaranes 1–3.
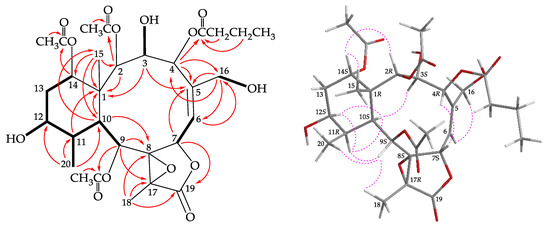
Figure 2.
The COSY ( ) correlations, selective HMBC (
) correlations, selective HMBC ( ), and protons with key NOESY correlations (
), and protons with key NOESY correlations ( ) of 1.
) of 1.
 ) correlations, selective HMBC (
) correlations, selective HMBC ( ), and protons with key NOESY correlations (
), and protons with key NOESY correlations ( ) of 1.
) of 1.
The stereochemistry of 1 was deduced from an NOESY experiment (Figure 2) and biogenetic considerations. The NOE correlations of H-10/H-11, H-10/H-12, and H-11/H-12 indicated that these protons were situated on the same face of the structure and were assigned as the α protons since the C-15 methyl is the β-substituent at C-1. The NOE correlation between H3-15 and H-14 implied that H-14 had a β-orientation. H-3 exhibited a correlation with H-10, and, as well as a lack of coupling constants were detected between H-2/H-3 and H-3/H-4, indicating the dihedral angles between H-2/H-3 and H-3/H-4 were approximately 90° and the 2-acetoxy, 3-hydroxy, and 4-n-butyroxy groups were β-, β-, and α-oriented, respectively. A correlation from H-4 to H-7, suggested that H-7 was β-oriented. The Z-configuration of C-5/6 double bond was confirmed based on the fact that the C-6 olefinic proton (δH 5.53) correlated to one of the C-16 hydroxymethyl protons (δH 4.04). H-9 was found to correlate with H-11, H3-18, and H3-20. From a consideration of molecular model, H-9 was found to be reasonably close to H-11, H3-18, and H3-20, thus, H-9 should be placed on the α face, and Me-18 was β-oriented in the γ-lactone moiety, and the 8,17-epoxy group should be α-oriented. It was found that the NMR signals of 1 were similar to those of a known briarane, briaexcavatolide P (4) (Figure 1) [8], except that the signals corresponding to the Me-16 vinyl methyl in 4 were replaced by signals for a hydroxymethyl group in 1. Additionally, as briaranes 1–5 were isolated along with a known briarane, excavatolide B (6) [6,10,11] from the same target organism, B. excavatum, and the absolute configuration of 6 was determined by a single-crystal X-ray diffraction analysis [6,11]. Therefore, on biogenetic grounds to assume that briaranes 1–5 had the same absolute stereochemistry as that of 6, tentatively, and the configurations of stereogenic carbons of 1 were determined as 1R,2R,3S,4R,7S,8S, 9S,10S,11R,12S,14S, and 17R (Supplementary Materials, Figures S1–S10).
Briarenol J (2) had a molecular formula C26H36O12 by its (+)-HRESIMS at m/z 563.21007 (calculated for C26H36O12 + Na, 563.20990). The IR spectrum showed bands at 3483, 1779, and 1727 cm−1, consistent with the presence of hydroxy, γ-lactone, and ester groups, respectively, in 2. From the 13C and DEPT data (Table 2), one trisubstituted double bond was deduced from the signals of two carbons at δC 139.3 (C-5) and 124.3 (CH-6). A methyl-containing tetrasubstituted epoxy group was confirmed from the signals of two oxygenated quaternary carbons at δC 69.9 (C-8) and 61.8 (C-17), and from the chemical shift of a tertiary methyl (δH 1.66, 3H, s; δC 10.3, CH3-18; Table 1 and Table 2). Four carbonyl resonances at δC 170.9, 170.0, 169.5, and 169.2 in the 13C spectrum confirmed the presence of a γ-lactone and three esters. All the esters were identified as acetates by the presence of three methyl singlet resonances in the 1H NMR spectrum at δH 2.32, 2.16, and 2.14, respectively.
Coupling constants information in the COSY spectrum of 2 enabled identification of H-6/H-7, H-9/H-10/H-11/H-12/H2-13/H-14, H-11/H3-20, and H-6/H3-16 (by allylic coupling; Figure 3), these data, together with the HMBC experiment (Figure 3), the molecular framework of 2 could be established. The HMBC also indicated that the acetoxy groups should be attached at C-4, C-9, and C-14, respectively. Thus, the remaining hydroxy groups have to be positioned at C-2, C-3, and C-12, as indicated by the COSY correlations between H-2/OH-2, H-3/OH-3, and H-12/OH-12.
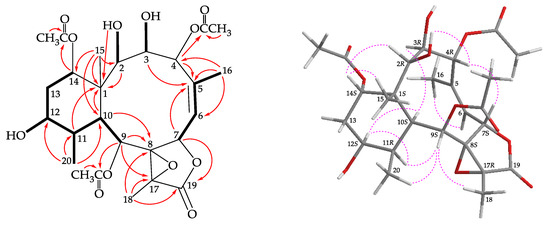
Figure 3.
The COSY ( ) correlations, selective HMBC (
) correlations, selective HMBC ( ), and protons with key NOESY correlations (
), and protons with key NOESY correlations ( ) of 2.
) of 2.
 ) correlations, selective HMBC (
) correlations, selective HMBC ( ), and protons with key NOESY correlations (
), and protons with key NOESY correlations ( ) of 2.
) of 2.
The stereochemistry of 2 was elucidated from the NOE interactions observed in a NOESY experiment (Figure 3) and by the vicinal 1H−1H coupling constant analysis. In the NOESY spectrum, correlations were observed between H-10 with H-3 and H-12; and H-12 correlated with H-11, indicating that these protons should be α-oriented. H-14 gave a correlation with H3-15, confirming the β-orientation for this proton. H-2 showed a correlation with H-14, and a lack of coupling constant was detected between H-2/H-3, indicating the dihedral angle between H-2/H-3 is approximately 90° and the 2-hydroxy group was β-oriented. H-4 exhibited correlations with H-7 and 2-hydroxy proton, confirming the β-orientations for H-4 and H-7. H-9 was found to show correlations with H-11, H3-18, and H3-20, and from molecular models, H-9 and H3-18 should be placed on the α- and β-face, respectively. The Z-configuration of C-5/C-6 double bond was elucidated by a correlation between H-6 and H3-16. The NMR data of 2 were found to be similar to those of a known briarane, briaexcavatin P (5) [9]. It was found that the 2-acetoxy substituent in 5 was replaced by a hydroxy group in 2. By comparison of the proton and carbon chemical shifts, coupling constants, NOESY correlations, and rotation value of 2 with those of 5, the stereochemistry of 2 was confirmed to be the same as that of 5, and the configurations of the stereogenic centers of 2 were assigned as 1S,2R,3R,4R, 7S,8S,9S,10S,11R,12S,14S, and 17R (Supplementary Materials, Figures S11–S20).
Briarane 3 (briarenol K) was found to have a molecular formula of C26H36O11 based on its (+)-HRESIMS peak at m/z 547.21514 (calculated for C26H36O11 + Na, 547.21498). Its absorption peaks in the IR spectrum showed ester carbonyl, γ-lactone, and broad OH stretching at 1739, 1780, and 3468 cm−1, respectively. The 13C NMR spectrum indicated that three esters and a γ-lactone were present, as carbonyl resonances were observed at δC 168.1, 170.2, 170.4, and 170.4, respectively (Table 1). The 1H NMR data also indicated that presence of three acetate methyls at δH 2.22, 2.03, and 2.00 (each 3H × s; Table 2). It was found that the spectroscopic data of 3 were similar to those of a known briarane, briareolide B (7) [12]; however, by comparison of the 1H and 13C NMR chemical shifts of CH-12 oxymethine (δH 3.72, 1H, dd, J = 12.4, 4.8 Hz; δC 73.4), CH2-13 sp3 methylene (δH 1.67, 1H, m; 2.04, 1H, m; δC 30.2), C-11 oxygenated quaternary carbon (δC 78.2), and Me-20 tertiary methyl (δH 1.15, 3H, s; δC 16.9) of 3 with those of 7 (δH 3.56, 1H, m; δC 73.9, CH-12; δH 2.03, 1H, m; 2.12, 1H, m; δC 27.6, CH2-13; δC 74.7, C-11; δH 1.16, 3H, s; δC 22.5, Me-20) [12] showed that the hydroxy group at C-12 in 3 was β-oriented. The locations of the functional groups were further confirmed by other HMBC and COSY correlations (Figure 4), and hence briarenol K was assigned the structure of 3. The NOESY spectrum exhibited a correlation from H-10 to H-12, further supporting that H-12 was α-oriented and the stereogenic centers of 3 were assigned as 1S,2S,7S,8S,9S,10S,11S,12S,14S, and 17R, by the correlations observed in a NOESY spectrum (Figure 4) and this compound was found to be the 12-epimer of briareolide B (7) [12] (Supplementary Materials, Figures S21–S30).
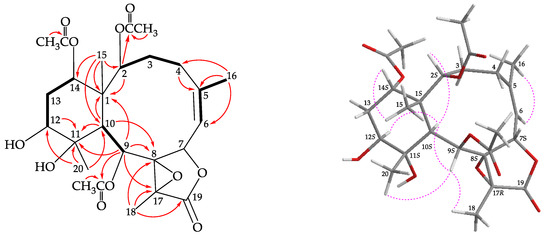
Figure 4.
The COSY ( ) correlations, selective HMBC (
) correlations, selective HMBC ( ), and protons with key NOESY correlations (
), and protons with key NOESY correlations ( ) of 3.
) of 3.
 ) correlations, selective HMBC (
) correlations, selective HMBC ( ), and protons with key NOESY correlations (
), and protons with key NOESY correlations ( ) of 3.
) of 3.
The inhibition effects of briaranes 1–5 on the release of inducible nitric oxide synthase (iNOS) and cyclooxygenase-2 (COX-2) protein from lipopolysaccharides (LPS)-stimulated RAW 264.7 were assessed. The results showed that briarane 4 reduced the release of iNOS and COX-2 to 35.37% and 54.61% at a concentration of 10 µM, respectively (Figure 5 and Table 3). Briarane 1 was found to be weaker than those of 4 in term of reducing the expression of iNOS and COX-2, indicating that the hydroxy group at C-16 in 1 reduced the activity.
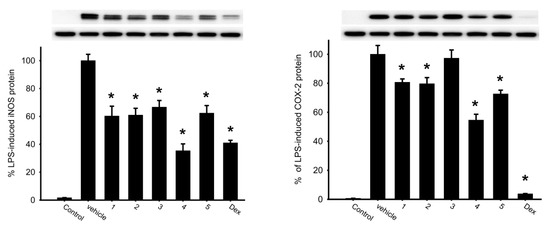
Figure 5.
Western blotting showed that briarane 4 downregulated the expression of iNOS and COX-2. Data were normalized to the cells treated with LPS only, and cells treated with dexamethasone (Dex; 10 µM) were used as a positive control. Data are expressed as the mean ± SEM (n = 2~4). * Significantly different from cells treated with LPS (p < 0.05).

Table 3.
Effects of briaranes 1–5 on LPS-induced pro-inflammatory iNOS and COX-2 protein expression in macrophages.
3. Materials and Methods
3.1. General Experimental Procedures
Optical rotation values were measured using a Jasco P-1010 digital polarimeter (Japan Spectroscopic, Tokyo, Japan). IR spectra were measured on a Thermo Scientific Nicolet iS5 FT-IR spectrophotometer (Waltham, MA, USA). NMR spectra were taken on a Jeol Resonance ECZ 400 S NMR spectrometer (Tokyo, Japan), using the residual CHCl3 signal (δH 7.26 ppm) and CDCl3 (δC 77.1 ppm) as the internal standard for 1H and 13C NMR, respectively; coupling constants (J) are presented in Hz. ESIMS and HRESIMS were recorded using a Bruker 7 Tesla solariX FTMS system. Column chromatography was carried out with silica gel (230–400 mesh, Merck, Darmstadt, Germany). TLC was performed on plates precoated with Kieselgel 60 F254 (0.25-mm-thick, Merck, Darmstadt, Germany), then sprayed with 10% H2SO4 solution followed by heating to visualize the spots. Normal-phase HPLC (NP-HPLC) was performed using a system comprised of a Hitachi L-7100 pump (Tokyo, Japan) and a Rheodyne 7725i injection port (Rohnert Park, CA, USA). Reverse-phase HPLC (RP-HPLC) was performed using a system comprised of a Hitachi L-2130 pump (Tokyo, Japan), a Hitachi L-2455 photodiode array detector (Tokyo, Japan), and a Rheodyne 7725i injection port (Rohnert Park, CA, USA). A semipreparative normal-phase column (YMC-Pack SIL, S-5 µm, 250 mm × 20 mm, Sigma-Aldrich, St. Louis, MO, USA) was used for NP-HPLC. A semipreparative reverse-phase column (Luna, 5 µm, C18(2) 100 Å, AXIA Packed, 250 mm × 21.2 mm; Phenomenex, Torrance, CA, USA) was used for RP-HPLC.
3.2. Animal Material
Specimens of B. excavatum were collected in June 2017 by hand with self-contained underwater breathing apparatus (SCUBA) divers off the coast of Lanyu Island (Orchid Island), Taiwan. The samples were then stored in a –20 °C freezer until extraction. A voucher specimen was deposited in the National Museum of Marine Biology and Aquarium, Taiwan (NMMBA-TW-SC-2017-418). Identification of the species of this organism was performed by comparison as described in previous publications [1,2,3,4].
3.3. Extraction and Isolation
The freeze-dried and sliced bodies (wet/dry weight = 1344/568 g) of the specimen were extracted with supercritical CO2 to give 58.9 g of extract. Partial extract (36.4 g) was then applied on silica gel column and eluted with gradients of n-hexane/EtOAc to furnish fractions A−K. Fraction F was purified by NP-HPLC using a mixture of n-hexane/acetone (4:1) to yield fractions F1−F13. Fraction F6 was repurified by RP-HPLC, using a mixture of MeOH/H2O (60:40; at a flow rate = 4 mL/min) to afford 4 (6.7 mg). Fraction G was separated by NP-HPLC, using a mixture of n-hexane/acetone (3:1) to yield fractions G1−G12. Fractions G6 and G7 were repurified by RP-HPLC using a mixture of MeOH/H2O (60:40; at a flow rate = 4.0 mL/min) to afford 5 (1.3 mg) and 3 (1.0 mg), respectively. Fraction H was separated by NP-HPLC using a mixture of n-hexane and acetone (3:1) to yield fractions H1−H18. Fractions H12 and H15 were repurified by RP-HPLC, using a mixture of MeOH/ H2O (60:40; at a flow rate = 4.0 mL/min) to afford 2 (2.1 mg) and 1 (0.6 mg), respectively.
Briarenol I (1): Amorphous powder; + 207 (c 0.03, CHCl3), IR (ATR) νmax 3524, 1783, 1736, 1222, 891 cm−1; 13C (100 MHz, CDCl3) and 1H (400 MHz, CDCl3) NMR data (see Table 1 and Table 2); ESIMS: m/z 649 [M + Na]+; HRESIMS m/z 649.24677 (calculated for C30H42O14 + Na, 649.24668).
Briarenol J (2): Amorphous powder; + 140 (c 0.08, CHCl3), IR (ATR) νmax 3483, 1779, 1727, 1220, 890 cm−1; 13C (100 MHz, CDCl3) and 1H (400 MHz, CDCl3) NMR data (see Table 1 and Table 2); ESIMS: m/z 563 [M + Na]+; HRESIMS m/z 563.21007 (calculated for C26H36O12 + Na, 563.20990).
Briarenol K (3): Amorphous powder; + 37 (c 0.06, CHCl3), IR (ATR) νmax 3468, 1780, 1739, 1255, 892 cm−1; 13C (100 MHz, CDCl3) and 1H (400 MHz, CDCl3) NMR data (see Table 1 and Table 2); ESIMS: m/z 547 [M + Na]+; HRESIMS m/z 547.21514 (calculated for C26H36O11 + Na, 547.21498).
Briaexcavatolide P (4): Amorphous powder; + 182 (c 0.3, CHCl3) (ref. [8], [α]27D + 167 (c 1.0, CHCl3)), IR (ATR) νmax 3513, 1783, 1731, 1218, 889 cm−1; 1H and 13C NMR data were found to be in agreement with previous study [8]; ESIMS: m/z 633 [M + Na]+.
Briaexcavatin P (5): Amorphous powder; + 134 (c 0.05, CHCl3) (ref. [9], + 198 (c 0.08, CHCl3)), IR (ATR) νmax 3503, 1785, 1735, 1240, 889 cm−1; 1H and 13C NMR data were found to be in agreement with previous study [9]; ESIMS: m/z 605 [M + Na]+.
3.4. In Vitro Anti-inflammatory Assay
The proinflammatory suppression assay was employed to assess the activities of the isolated compounds 1–5 against the release of iNOS and COX-2 from macrophage cells as the literature reported [13,14,15].
4. Conclusions
B. excavatum was demonstrated to have a wide structural diversity of briarane-type diterpenoids that possessed various pharmacological properties, especially in anti-inflammatory activity. In our continued study on B. excavatum, three previously unreported briaranes, briarenols I–K (1–3), along with the known analogues, briaexcavatolide P (4) and briaexcavatin P (5), were isolated. In the present study, the anti-inflammatory activity of 1–5 was assessed using inhibition of pro-inflammatory iNOS and COX-2 release from macrophages. The results indicated that briaexcavatolide P (4) showed the most potent suppressive effect on iNOS release.
Supplementary Materials
The Supplementary Materials are available online. ESIMS, HRESIMS, IR, 1D and 2D NMR spectra of new compounds 1–3.
Author Contributions
Conceptualization, L.-S.F., Y.-J.W., Z.-H.W. and P.-J.S; investigation, T.-H.H., Y.-H.C., B.-R.P., Y.-Y.C., L.-G.Z. and J.-J.C.; writing—original draft preparation, T.-H.H. and P.-J.S.; writing—review and editing, T.-C.L. and P.-J.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by grants from the National Museum of Marine Biology and Aquarium; the National Dong Hwa University; and the Ministry of Science and Technology, Taiwan (Grant Nos: MOST 106-2320-B-291-001-MY3 and 107-2320-B-291-001-MY3) awarded to Ping-Jyun Sung.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Bayer, F.M. Key to the genera of octocorallia exclusive of Pennatulacea (Coelenterata: Anthozoa), with diagnoses of new taxa. Proc. Biol. Soc. Wash. 1981, 94, 902–947. [Google Scholar]
- Benayahu, Y.; Jeng, M.-S.; Perkol-Finkel, S.; Dai, C.-F. Soft corals (Octocorallia: Alcyonacea) from Southern Taiwan. II. Species diversity and distributional patterns. Zool. Stud. 2004, 43, 548–560. [Google Scholar]
- Miyazaki, Y.; Reimer, J.D. Morphological and genetic diversity of Briareum (Anthozoa: Octocorallia) from the Ryukyu Archipelago, Japan. Zool. Sci. 2014, 31, 692–702. [Google Scholar] [CrossRef] [PubMed]
- Samimi-Namin, K.; van Ofwegen, L.P. Overview of the genus Briareum (Cnidaria, Octocorallia, Briareidae) in the Indo-Pacific, with the description of a new species. Zookeys 2016, 557, 1–44. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.-D.; Su, J.-H.; Hwang, T.-L.; Wen, Z.-H.; Sheu, J.-H.; Wu, Y.-C.; Sung, P.-J. Briarane diterpenoids isolated from octocorals between 2014 and 2016. Mar. Drugs 2017, 15, 44. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.-C.; Lin, N.-C.; Peng, B.-R.; Chen, Y.-Y.; Wen, Z.-H.; Chang, Y.-C.; Lee, G.-H.; Wu, Y.-C.; Sung, P.-J. New trihydroxybriarane diterpenoids from an octocoral Briareum sp. Phytochem. Lett. 2020, 35, 23–27. [Google Scholar] [CrossRef]
- Molina, S.L.; Forero, A.M.; Ayala, F.I.; Puyana, M.; Zea, S.; Castellanos, L.; Muñoz, D.; Arboleda, G.; Sandoval-Hernández, A.G.; Ramos, F.A. Metabolic profiling of the soft coral Erythropodium caribaeorum (Alcyonacea: Anthothelidae) from the Colombian Caribbean reveals different chemotypes. Mar. Drugs 2020, 18, 4. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.-L.; Sung, P.-J.; Chiang, M.Y.; Wu, J.-Y.; Sheu, J.-H. New polyoxygenated briarane diterpenoids, briaexcavatolides O–R, from the gorgonian Briareum excavatum. J. Nat. Prod. 2001, 64, 1415–1420. [Google Scholar] [CrossRef] [PubMed]
- Sung, P.-J.; Lin, M.-R.; Hwang, T.-L.; Fan, T.-Y.; Su, W.-C.; Ho, C.-C.; Fang, L.-S.; Wang, W.-H. Briaexcavatins M–P, four new briarane-related diterpenoids from cultured octocoral Briareum excavatum (Briareidae). Chem. Pharm. Bull. 2008, 56, 930–935. [Google Scholar] [CrossRef] [PubMed]
- Sheu, J.-H.; Sung, P.-J.; Cheng, M.-C.; Liu, H.-Y.; Fang, L.-S.; Duh, C.-Y.; Chiang, M.Y. Novel cytotoxic diterpenes, excavatolides A–E, isolated from the Formosan gorgonian Briareum excavatum. J. Nat. Prod. 1998, 61, 602–608. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.-W.; Chi, W.-C.; Lee, G.-H.; Su, J.-H.; Hwang, T.-L.; Wu, Y.-J.; Su, T.-R.; Sheu, J.-H.; Sung, P.-J. 2-Acetoxybriaranes from Briareum violaceum. Tetrahedron 2019, 75, 3751–3757. [Google Scholar] [CrossRef]
- Pordesimo, E.O.; Schmitz, F.J.; Ciereszko, L.S.; Hossain, M.B.; van der Helm, D. New briarein diterpenes from the Caribbean gorgonians Erythropodium caribaeorum and Briareum sp. J. Org. Chem. 1991, 56, 2344–2357. [Google Scholar] [CrossRef]
- Jean, Y.-H.; Chen, W.-F.; Sung, C.-S.; Duh, C.-Y.; Huang, S.-Y.; Lin, C.-S.; Tai, M.-H.; Tzeng, S.-F.; Wen, Z.-H. Capnellene, a natural marine compound derived from soft coral, attenuates chronic constriction injury-induced neuropathic in rats. Br. J. Pharmacol. 2009, 158, 713–725. [Google Scholar] [CrossRef] [PubMed]
- Jean, Y.-H.; Chen, W.-F.; Duh, C.-Y.; Huang, S.-Y.; Hsu, C.-H.; Lin, C.-S.; Sung, C.-S.; Chen, I.-M.; Wen, Z.-H. Inducible nitric oxide synthase and cyclooxygenase-2 participate in anti-inflammatory and analgesic effects of the natural marine compound lemnalol from Formosan soft coral Lemnalia cervicorni. Eur. J. Pharmacol. 2008, 578, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-C.; Lin, Y.-Y.; Jean, Y.-H.; Lu, Y.; Chen, W.-F.; Yang, S.-N.; Wang, H.-M.D.; Jang, I.-Y.; Chen, I.-M.; Su, J.-H.; et al. Anti-inflammatory and analgesic effects of the marine-derived compound comaparvin isolated from the crinoid Comanthus bennetti. Molecules 2014, 19, 14667–14686. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).