Constructed Wetland Revealed Efficient Sulfamethoxazole Removal but Enhanced the Spread of Antibiotic Resistance Genes
Abstract
:1. Introduction
2. Results and Discussion
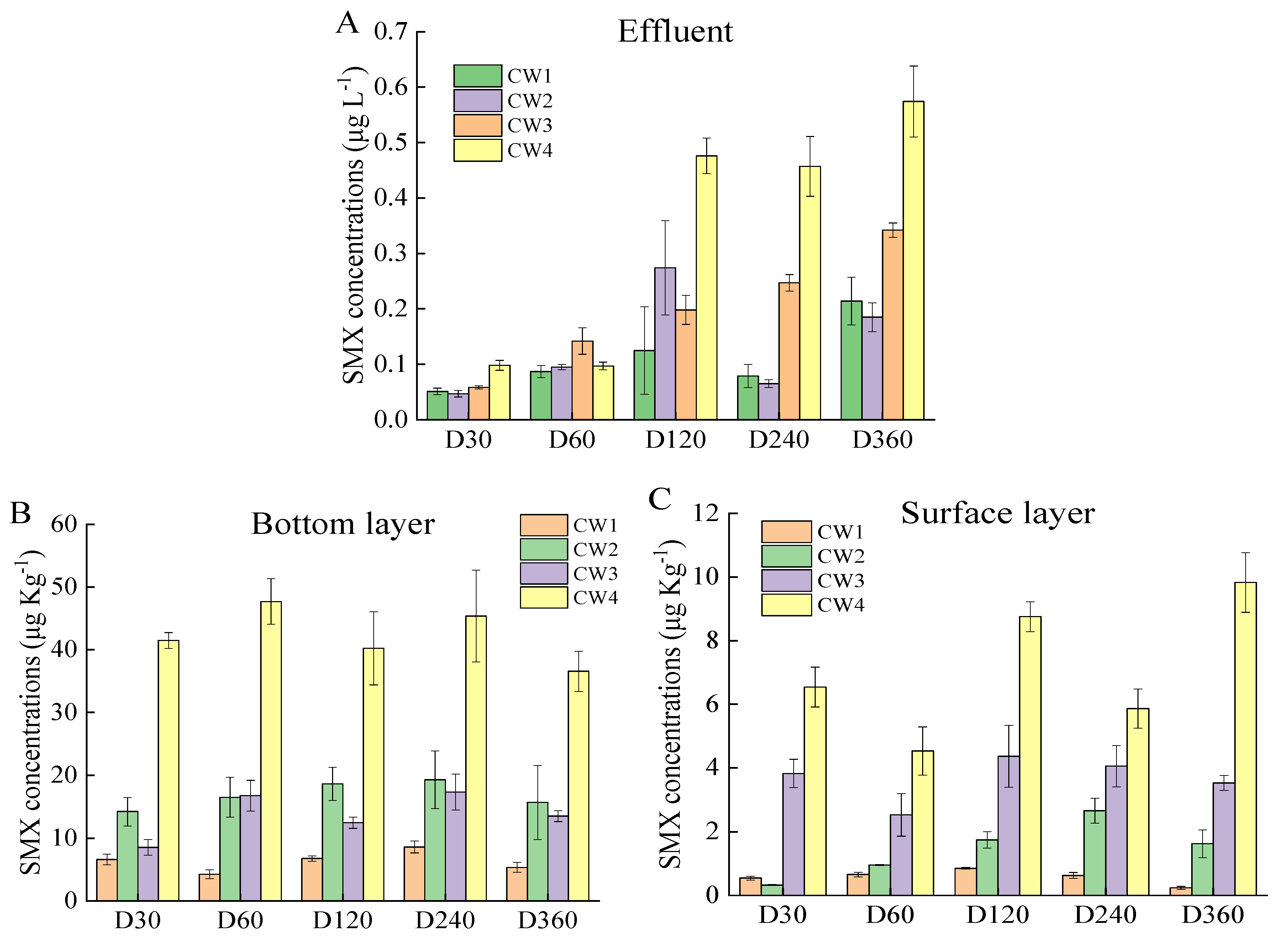
2.1. SMX Removal Efficiency
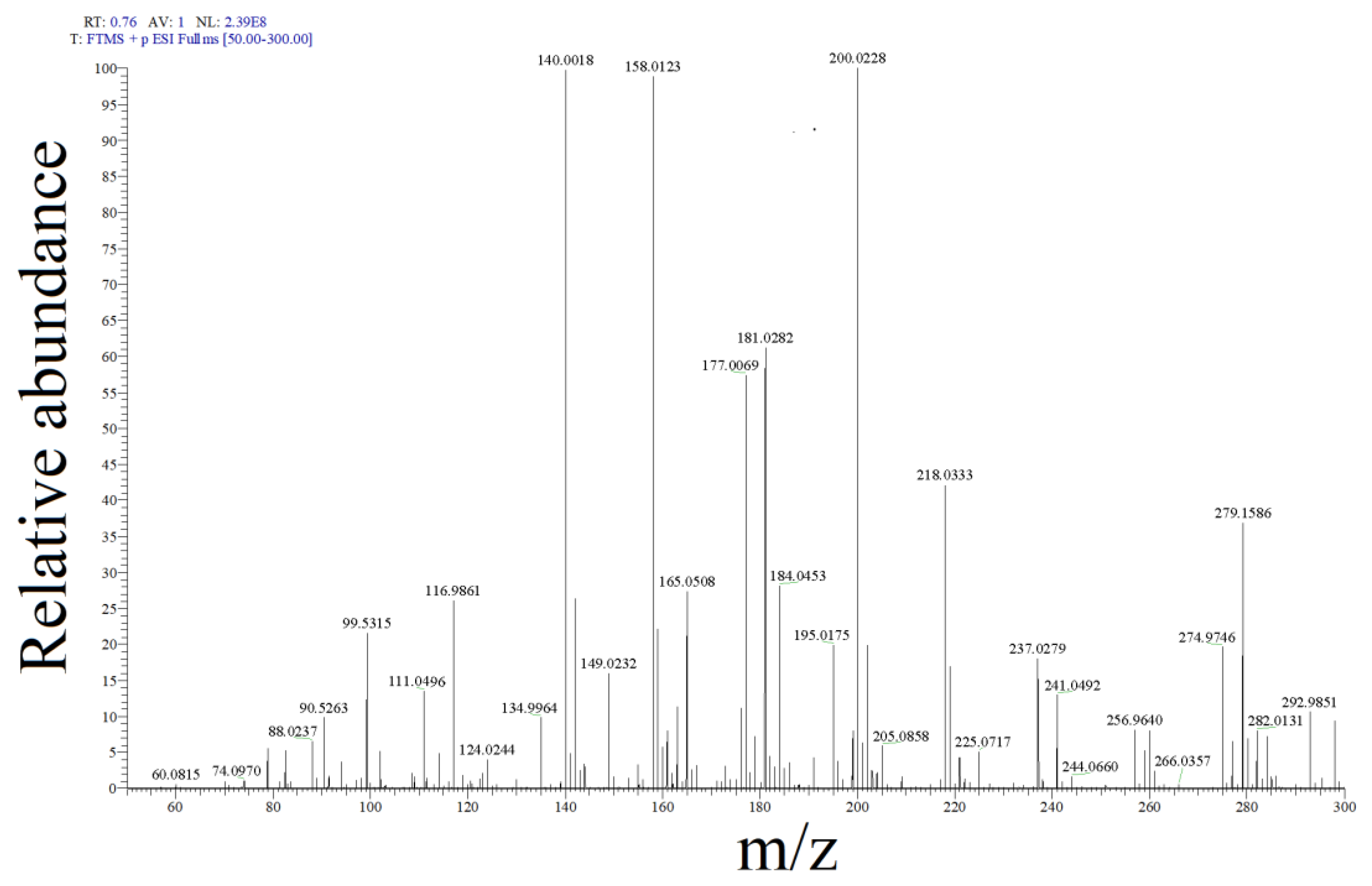
2.2. SMX Degradation Products
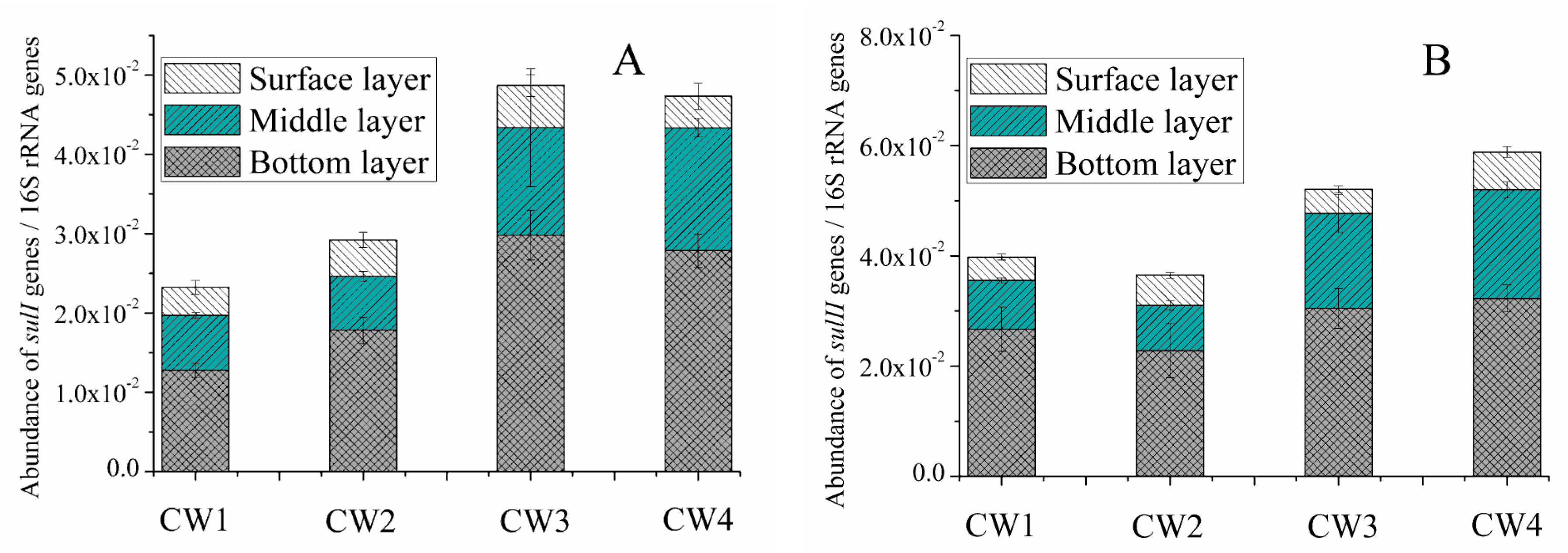
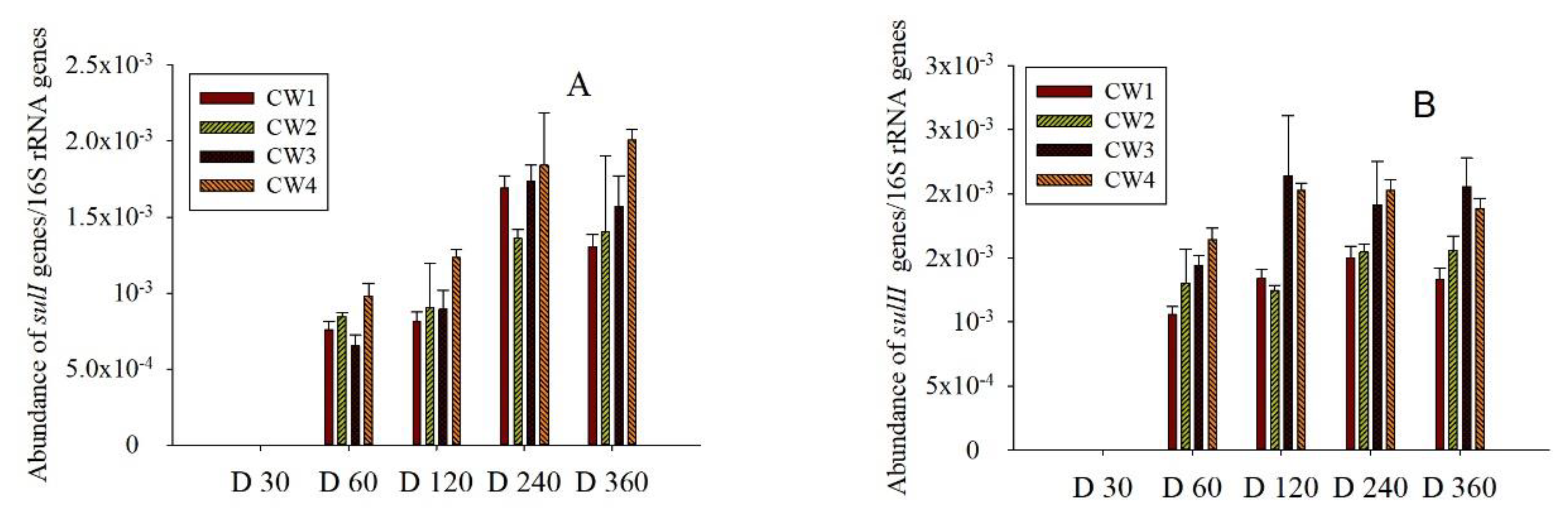
2.3. Sul Genes in the Effluent and Media
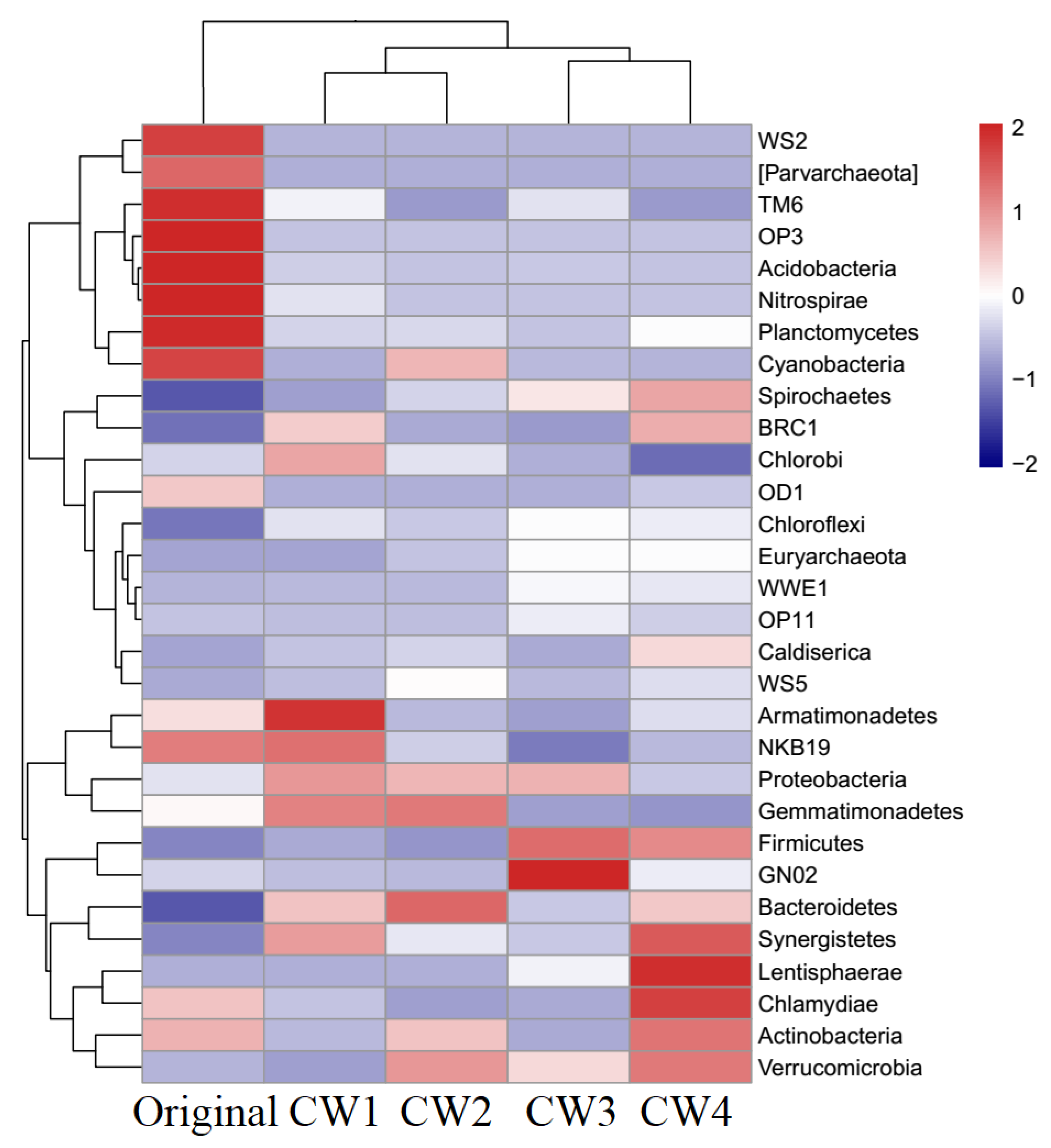
2.4. Composition of Bacterial Communities
3. Materials and Methods
3.1. Reactor Configuration
3.2. SMX Detection
3.3. DNA Extraction and ARG Analysis
3.4. High-Throughput Sequencing
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hou, J.; Wan, W.N.; Mao, D.Q.; Wang, C.; Mu, Q.H.; Qin, S.Y.; Luo, Y. Occurrence and distribution of sulfonamides, tetracyclines, quinolones, macrolides, and nitrofurans in livestock manure and amended soils of Northern China. Environ. Sci. Pollut. Res. 2015, 22, 4545–4554. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.J.; Ying, G.G.; Zhang, R.Q.; Liu, S.; Lai, H.J.; Chen, Z.F.; Yang, B.; Zhao, J.L. Use patterns, excretion masses and contamination profiles of antibiotics in a typical swine farm, south China. Environ. Sci. Proc. Imp. 2013, 15, 802–813. [Google Scholar] [CrossRef] [PubMed]
- Adegoke, A.A.; Faleye, C.A.; Singh, G.; Stenström, A.T. Antibiotic Resistant Superbugs: Assessment of the Interrelationship of Occurrence in Clinical Settings and Environmental Niches. Molecules 2017, 22, 29. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.D.; Vu, T.N.; Nguyen, H.L.; Le, P.H.P.; Hoang, T.S. Adsorptive removal of antibiotic ciprofloxacin from aqueous solution using protein-modified nanosilica. Polymers 2020, 12, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules 2018, 23, 795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKinney, C.W.; Loftin, K.A.; Meyer, M.T.; Davis, J.G.; Pruden, A. Tet and sul antibiotic resistance genes in livestock lagoons of various operation type, configuration, and antibiotic occurrence. Environ. Sci. Technol. 2010, 44, 6102–6109. [Google Scholar] [CrossRef] [PubMed]
- Storteboom, H.; Arabi, M.; Davis, J.G.; Crimi, B.; Pruden, A. Identification of antibiotic-resistance-gene molecular signatures suitable as tracers of pristine river, urban, and agricultural sources. Environ. Sci. Technol. 2010, 44, 1947–1953. [Google Scholar] [CrossRef]
- Ogawara, H. Comparison of Antibiotic Resistance Mechanisms in Antibiotic-Producing and Pathogenic Bacteria. Molecules 2019, 24, 3430. [Google Scholar] [CrossRef] [Green Version]
- Yuan, H.; Miller, J.H.; Abu-Reesh, I.M.; Pruden, A.; He, Z. Effects of electron acceptors on removal of antibiotic resistant Escherichia coli, resistance genes and class 1 integrons under anaerobic conditions. Sci. Total Environ. 2016, 569, 1587–1594. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Yun, Z.; Ha, U.H.; Lee, S.; Park, H.; Kwon, E.E.; Cho, Y.; Choung, S.; Oh, J.; Medriano, C.A.; et al. Transfer of antibiotic resistance plasmids in pure and activated sludge cultures in the presence of environmentally representative micro-contaminant concentrations. Sci. Total Environ. 2014, 468–469, 813–820. [Google Scholar] [CrossRef]
- Guo, J.H.; Li, J.; Chen, H.; Bond, P.L.; Yuan, Z.G. Metagenomic analysis reveals wastewater treatment plants as hotspots of antibiotic resistance genes and mobile genetic elements. Water Res. 2017, 123, 468–478. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Ying, G.G.; Wei, X.D.; Liu, Y.S.; Liu, S.S.; Hu, L.X.; He, L.Y.; Chen, Z.F.; Chen, F.R.; Yang, Y.Q. Removal of antibiotics and antibiotic resistance genes from domestic sewage by constructed wetlands: Effect of flow configuration and plant species. Sci. Total Environ. 2016, 571, 974–982. [Google Scholar] [CrossRef] [PubMed]
- Beekmann, S.E.; Heilmann, K.P.; Richter, S.S.; García-de-Lomas, J.; Doern, G.V.; Group, G.S. Antimicrobial resistance in Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis and group A β-haemolytic streptococci in 2002–2003: Results of the multinational GRASP Surveillance Program. Int. J. Antimicrob. Agents 2005, 25, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Penesyan, A.; Gillings, M.; Paulsen, I. Antibiotic Discovery: Combatting Bacterial Resistance in Cells and in Biofilm Communities. Molecules 2015, 20, 5286–5298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, P.H.; Juhrend, B.; Olson, T.M.; Marrs, C.F.; Wigginton, K.R. Degradation of extracellular antibiotic resistance genes with UV254 treatment. Environ. Sci. Technol. 2017, 51, 6185–6192. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Liu, C.; Li, K.; Su, J.; Zhu, G.; Liu, L. Performance of vertical up-flow constructed wetlands on swine wastewater containing tetracyclines and tet genes. Water Res. 2015, 70, 109–117. [Google Scholar] [CrossRef]
- Naquin, A.; Shrestha, A.; Sherpa, M.; Nathaniel, R.; Boopathy, R. Presence of antibiotic resistance genes in a sewage treatment plant in Thibodaux, Louisiana, USA. Bioresour. Technol. 2015, 188, 79–83. [Google Scholar] [CrossRef]
- Gao, P.; Munir, M.; Xagoraraki, I. Correlation of tetracycline and sulfonamide antibiotics with corresponding resistance genes and resistant bacteria in a conventional municipal wastewater treatment plant. Sci. Total Environ. 2012, 421–422, 173–183. [Google Scholar] [CrossRef]
- Zhang, S.; Song, H.-L.; Yang, X.-L.; Yang, Y.-L.; Yang, K.-Y.; Wang, X.-Y. Fate of tetracycline and sulfamethoxazole and their corresponding resistance genes in microbial fuel cell coupled constructed wetlands. RSC Adv. 2016, 6, 95999–96005. [Google Scholar] [CrossRef]
- Rosendahl, I.; Siemens, J.; Groeneweg, J.; Linzbach, E.; Laabs, V.; Herrmann, C.; Vereecken, H.; Amelung, W. Dissipation and sequestration of the veterinary antibiotic sulfadiazine and its metabolites under field conditions. Environ. Sci. Technol. 2011, 45, 5216–5222. [Google Scholar] [CrossRef]
- Liu, L.; Liu, C.; Zheng, J.; Huang, X.; Wang, Z.; Liu, Y.; Zhu, G. Elimination of veterinary antibiotics and antibiotic resistance genes from swine wastewater in the vertical flow constructed wetlands. Chemosphere 2013, 91, 1088–1093. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.A.; Prasher, S.O.; Patel, R.M. Removal of ionophoric antibiotics in free water surface constructed wetlands. Ecol. Eng. 2012, 41, 13–21. [Google Scholar] [CrossRef]
- Almeida, C.M.R.; Santos, F.; Ferreira, A.C.F.; Lourinha, I.; Basto, M.C.P.; Mucha, A.P. Can veterinary antibiotics affect constructed wetlands performance during treatment of livestock wastewater? Ecol. Eng. 2017, 102, 583–588. [Google Scholar] [CrossRef]
- Huang, X.; Zheng, J.; Liu, C.; Liu, L.; Liu, Y.; Fan, H. Removal of antibiotics and resistance genes from swine wastewater using vertical flow constructed wetlands: Effect of hydraulic flow direction and substrate type. Chem. Eng. J. 2017, 308, 692–699. [Google Scholar] [CrossRef]
- Chen, J.; Liu, Y.S.; Su, H.C.; Ying, G.G.; Liu, F.; Liu, S.S.; He, L.Y.; Chen, Z.F.; Yang, Y.Q.; Chen, F.R. Removal of antibiotics and antibiotic resistance genes in rural wastewater by an integrated constructed wetland. Environ. Sci. Pollut. Res. 2015, 22, 1794–1803. [Google Scholar] [CrossRef]
- Fang, H.S.; Zhang, Q.; Nie, X.P.; Chen, B.W.; Xiao, Y.D.; Zhou, Q.B.; Liao, W.; Liang, X.M. Occurrence and elimination of antibiotic resistance genes in a long-term operation integrated surface flow constructed wetland. Chemosphere 2017, 173, 99–106. [Google Scholar] [CrossRef]
- Almeida, C.M.R.; Santos, F.; Ferreira, A.C.F.; Gomes, C.R.; Basto, M.C.P.; Mucha, A.P. Constructed wetlands for the removal of metals from livestock wastewater—Can the presence of veterinary antibiotics affect removals? Ecotox. Environ. Saf. 2017, 137, 143–148. [Google Scholar] [CrossRef]
- Liu, L.; Liu, Y.H.; Wang, Z.; Liu, C.X.; Huang, X.; Zhu, G.F. Behavior of tetracycline and sulfamethazine with corresponding resistance genes from swine wastewater in pilot-scale constructed wetlands. J. Hazard. Mater. 2014, 278, 304–310. [Google Scholar] [CrossRef]
- Cetecioglu, Z.; Ince, B.; Orhon, D.; Ince, O. Anaerobic sulfamethoxazole degradation is driven by homoacetogenesis coupled with hydrogenotrophic methanogenesis. Water Res. 2016, 90, 79–89. [Google Scholar] [CrossRef] [Green Version]
- Soares, S.F.; Fernandes, T.; Trindade, T.; Daniel-da-Silva, A.L. Trimethyl Chitosan/Siloxane-Hybrid Coated Fe3O4 Nanoparticles for the Uptake of Sulfamethoxazole from Water. Molecules 2019, 24, 1958. [Google Scholar] [CrossRef] [Green Version]
- Barkovskii, A.L.; Bridges, C. Persistence and Profiles of Tetracycline Resistance Genes in Swine Farms and Impact of Operational Practices on Their Occurrence in Farms’ Vicinities. Water Air Soil Pollut. 2011, 223, 49–62. [Google Scholar] [CrossRef]
- Dan, A.; Yang, Y.; Dai, Y.N.; Chen, C.X.; Wang, S.Y.; Tao, R. Removal and factors influencing removal of sulfonamides and trimethoprim from domestic sewage in constructed wetlands. Bioresour. Technol. 2013, 146, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.Z.; Tran, N.H.; Yin, T.R.; He, Y.L.; Gin, K.Y.H. Removal of selected PPCPs, EDCs, and antibiotic resistance genes in landfill leachate by a full-scale constructed wetlands system. Water Res. 2017, 121, 46–60. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Xu, Y.; Wang, H.; Guo, C.; Qiu, H.; He, Y.; Zhang, Y.; Li, X.; Meng, W. Occurrence of antibiotics and antibiotic resistance genes in a sewage treatment plant and its effluent-receiving river. Chemosphere 2015, 119, 1379–1385. [Google Scholar] [CrossRef]
- Dordio, A.V.; Carvalho, A.J. Organic xenobiotics removal in constructed wetlands, with emphasis on the importance of the support matrix. J. Hazard. Mater. 2013, 252–253, 272–292. [Google Scholar] [CrossRef] [Green Version]
- Hijosa-Valsero, M.; Fink, G.; Schlusener, M.P.; Sidrach-Cardona, R.; Martin-Villacorta, J.; Ternes, T.; Becares, E. Removal of antibiotics from urban wastewater by constructed wetland optimization. Chemosphere 2011, 83, 713–719. [Google Scholar] [CrossRef]
- Gong, W.; Liu, X.; He, H.; Wang, L.; Dai, G. Quantitatively modeling soil-water distribution coefficients of three antibiotics using soil physicochemical properties. Chemosphere 2012, 89, 825–831. [Google Scholar] [CrossRef]
- Müller, E.; Schüssler, W.; Horn, H.; Lemmer, H. Aerobic biodegradation of the sulfonamide antibiotic sulfamethoxazole by activated sludge applied as co-substrate and sole carbon and nitrogen source. Chemosphere 2013, 92, 969–978. [Google Scholar] [CrossRef]
- Wang, L.; Liu, Y.; Ma, J.; Zhao, F. Rapid degradation of sulphamethoxazole and the further transformation of 3-amino-5-methylisoxazole in a microbial fuel cell. Water Res. 2016, 88, 322–328. [Google Scholar] [CrossRef]
- Wang, L.; Wu, Y.C.; Zheng, Y.; Liu, L.D.; Zhao, F. Efficient degradation of sulfamethoxazole and the response of microbial communities in microbial fuel cells. RSC Adv. 2015, 5, 56430–56437. [Google Scholar] [CrossRef]
- Antunes, P.; Machado, J.; Sousa, J.C.; Peixe, L. Dissemination of sulfonamide resistance genes (sul1, sul2, and sul3) in Portuguese Salmonella enterica strains and relation with integrons. Antimicrob. Agents Chemother. 2005, 49, 836–839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nolvak, H.; Truu, M.; Tiirik, K.; Oopkaup, K.; Sildvee, T.; Kaasik, A.; Mander, U.; Truu, J. Dynamics of antibiotic resistance genes and their relationships with system treatment efficiency in a horizontal subsurface flow constructed wetland. Sci. Total Environ. 2013, 461–462, 636–644. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.P.; Almeida, C.M.R.; Pereira, A.C.; Ribeiro, I.L.; Reis, I.; Carvalho, P.; Basto, M.C.P.; Mucha, A.P. Microbial community dynamics associated with veterinary antibiotics removal in constructed wetlands microcosms. Bioresour. Technol. 2015, 182, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Wang, H.; Zhang, S.; Nishimura, O.; Li, X. Azo dye degradation pathway and bacterial community structure in biofilm electrode reactors. Chemosphere 2018, 208, 219–225. [Google Scholar] [CrossRef]
- Huang, X.; Zheng, J.L.; Liu, C.X.; Liu, L.; Liu, Y.H.; Fan, H.Y.; Zhang, T.F. Performance and bacterial community dynamics of vertical flow constructed wetlands during the treatment of antibiotics-enriched swine wastewater. Chem. Eng. J. 2017, 316, 727–735. [Google Scholar] [CrossRef]
- Arroyo, P.; Saenz de Miera, L.E.; Ansola, G. Influence of environmental variables on the structure and composition of soil bacterial communities in natural and constructed wetlands. Sci. Total Environ. 2015, 506–507, 380–390. [Google Scholar] [CrossRef]
- Srinivasan, P.; Sarmah, A.K. Dissipation of sulfamethoxazole in pasture soils as affected by soil and environmental factors. Sci. Total Environ. 2014, 479, 284–291. [Google Scholar] [CrossRef]
- Wu, D.; Huang, Z.T.; Yang, K.; Graham, D.; Xie, B. Relationships between Antibiotics and Antibiotic Resistance Gene Levels in Municipal Solid Waste Leachates in Shanghai, China. Environ. Sci. Technol. 2015, 49, 4122–4128. [Google Scholar] [CrossRef]
- Rodriguez-Mozaz, S.; Chamorro, S.; Marti, E.; Huerta, B.; Gros, M.; Sanchez-Melsio, A.; Borrego, C.M.; Barcelo, D.; Balcazar, J.L. Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river. Water Res. 2015, 69, 234–242. [Google Scholar] [CrossRef]
- Pruden, A.; Pei, R.; Storteboom, H.; Carlson, K.H. Antibiotic Resistance Genes as Emerging Contaminants: Studies in Northern Colorado†. Environ. Sci. Technol. 2006, 40, 7445–7450. [Google Scholar] [CrossRef]
- Wang, W.; Cao, J.; Yang, F.; Wang, X.L.; Zheng, S.S.; Sharshov, K.; Li, L.X. High-throughput sequencing reveals the core gut microbiome of Bar-headed goose (Anser indicus) in different wintering areas in Tibet. MicrobiologyOpen 2016, 5, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Zhu, H.; Szewzyk, U.; Geissen, S.U. Enhanced removal of sulfamethoxazole with manganese-adapted aerobic biomass. Int. Biodeterior. Biodegrad. 2017, 116, 171–174. [Google Scholar] [CrossRef]
- Mikkelson, K.M.; Lozupone, C.A.; Sharp, J.O. Altered edaphic parameters couple to shifts in terrestrial bacterial community structure associated with insect-induced tree mortality. Soil Biol. Biochem. 2016, 95, 19–29. [Google Scholar] [CrossRef] [Green Version]
Sample Availability: Not available. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Lu, Y.-X.; Zhang, J.-J.; Liu, S.; Song, H.-L.; Yang, X.-L. Constructed Wetland Revealed Efficient Sulfamethoxazole Removal but Enhanced the Spread of Antibiotic Resistance Genes. Molecules 2020, 25, 834. https://doi.org/10.3390/molecules25040834
Zhang S, Lu Y-X, Zhang J-J, Liu S, Song H-L, Yang X-L. Constructed Wetland Revealed Efficient Sulfamethoxazole Removal but Enhanced the Spread of Antibiotic Resistance Genes. Molecules. 2020; 25(4):834. https://doi.org/10.3390/molecules25040834
Chicago/Turabian StyleZhang, Shuai, Yu-Xiang Lu, Jia-Jie Zhang, Shuai Liu, Hai-Liang Song, and Xiao-Li Yang. 2020. "Constructed Wetland Revealed Efficient Sulfamethoxazole Removal but Enhanced the Spread of Antibiotic Resistance Genes" Molecules 25, no. 4: 834. https://doi.org/10.3390/molecules25040834
APA StyleZhang, S., Lu, Y.-X., Zhang, J.-J., Liu, S., Song, H.-L., & Yang, X.-L. (2020). Constructed Wetland Revealed Efficient Sulfamethoxazole Removal but Enhanced the Spread of Antibiotic Resistance Genes. Molecules, 25(4), 834. https://doi.org/10.3390/molecules25040834






