Interaction with Soil Bacteria Affects the Growth and Amino Acid Content of Piriformospora indica
Abstract
1. Introduction
2. Results
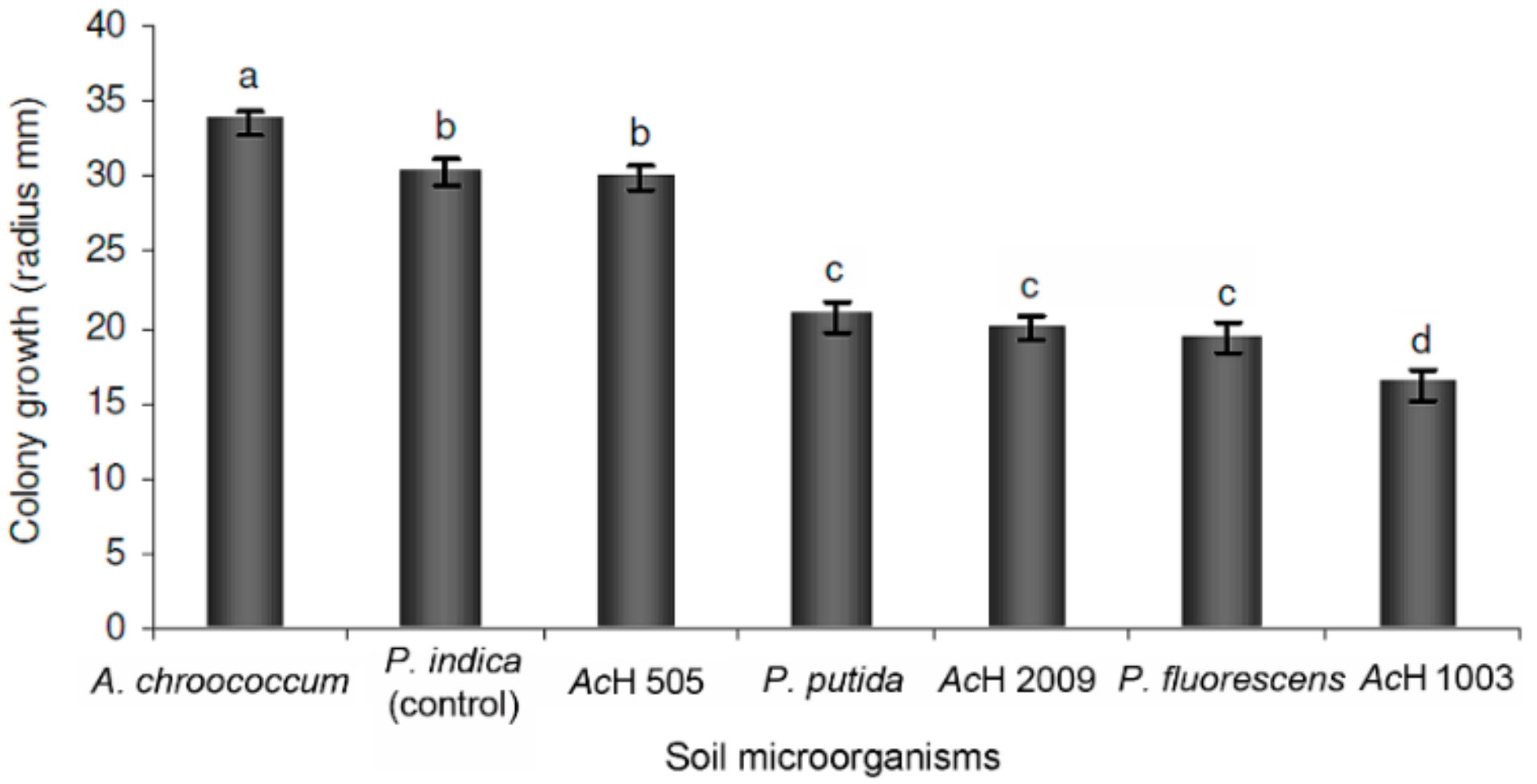
2.1. Dual Culture Assay of P. indica with Soil Microorganisms
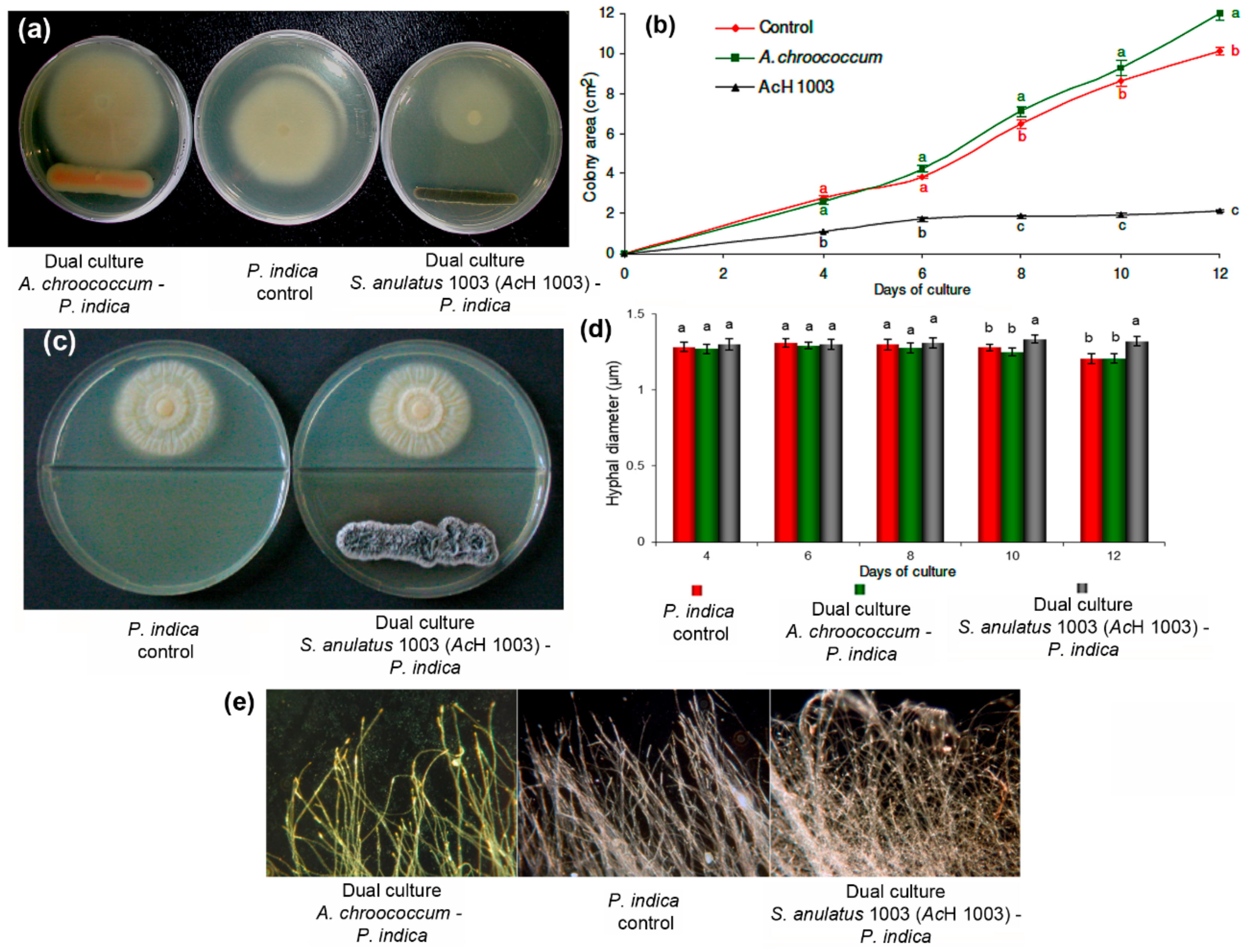
2.2. P. indica in Dual Culture with A. chroococcum and S. anulatus 1003
Morphology of P. indica Hyphae during Dual Culture with Soil Bacteria
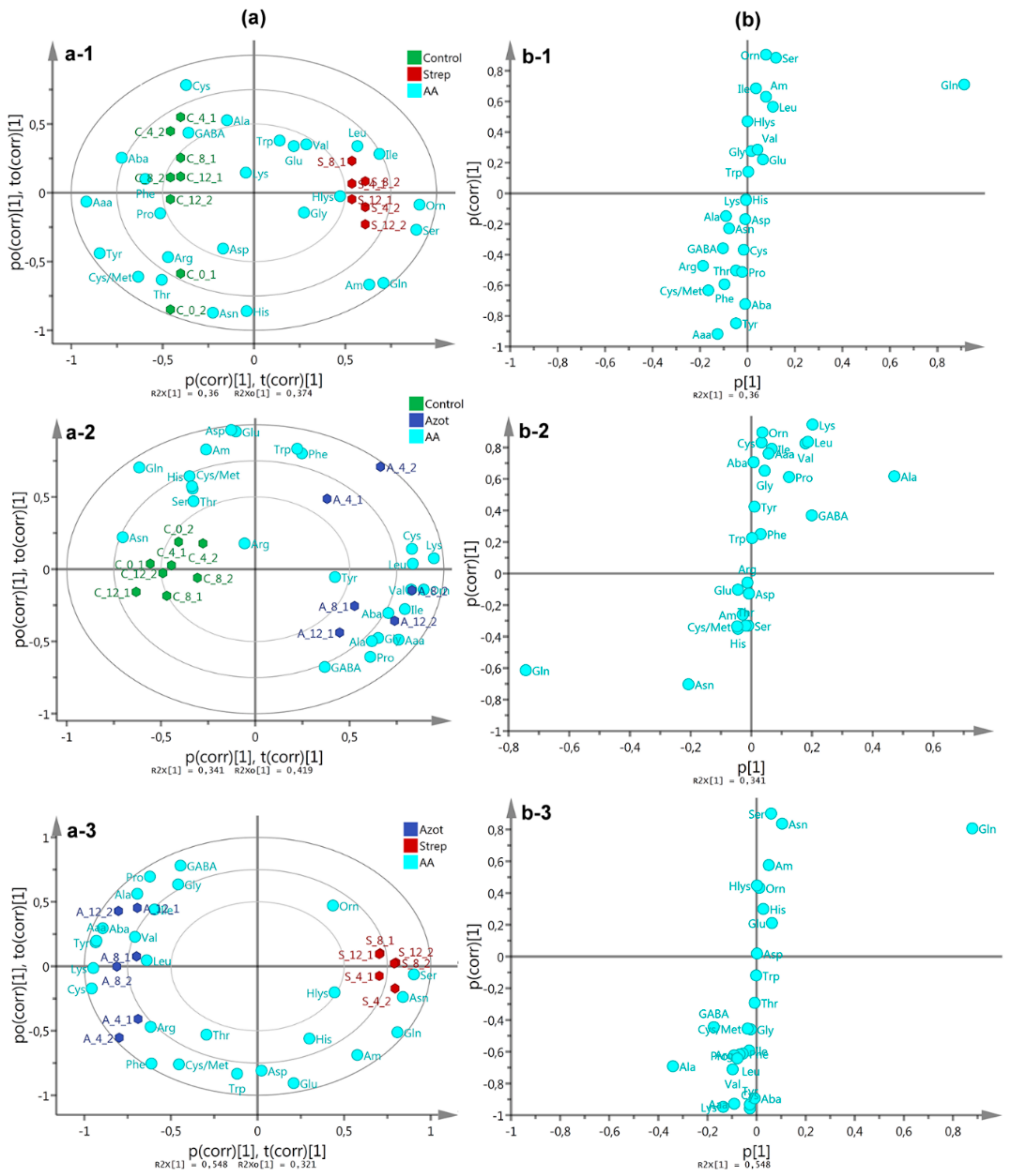
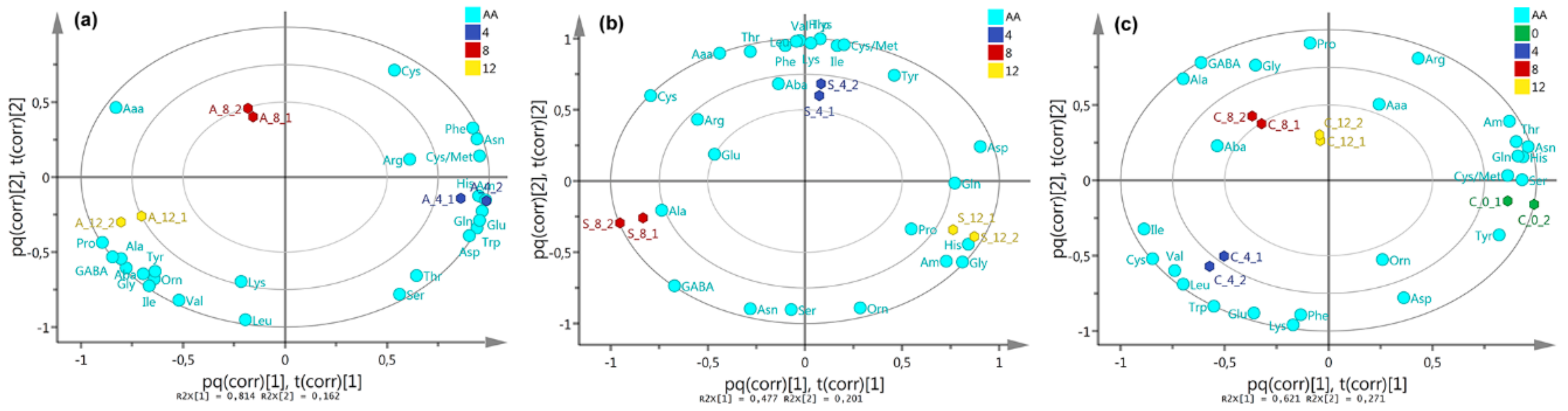
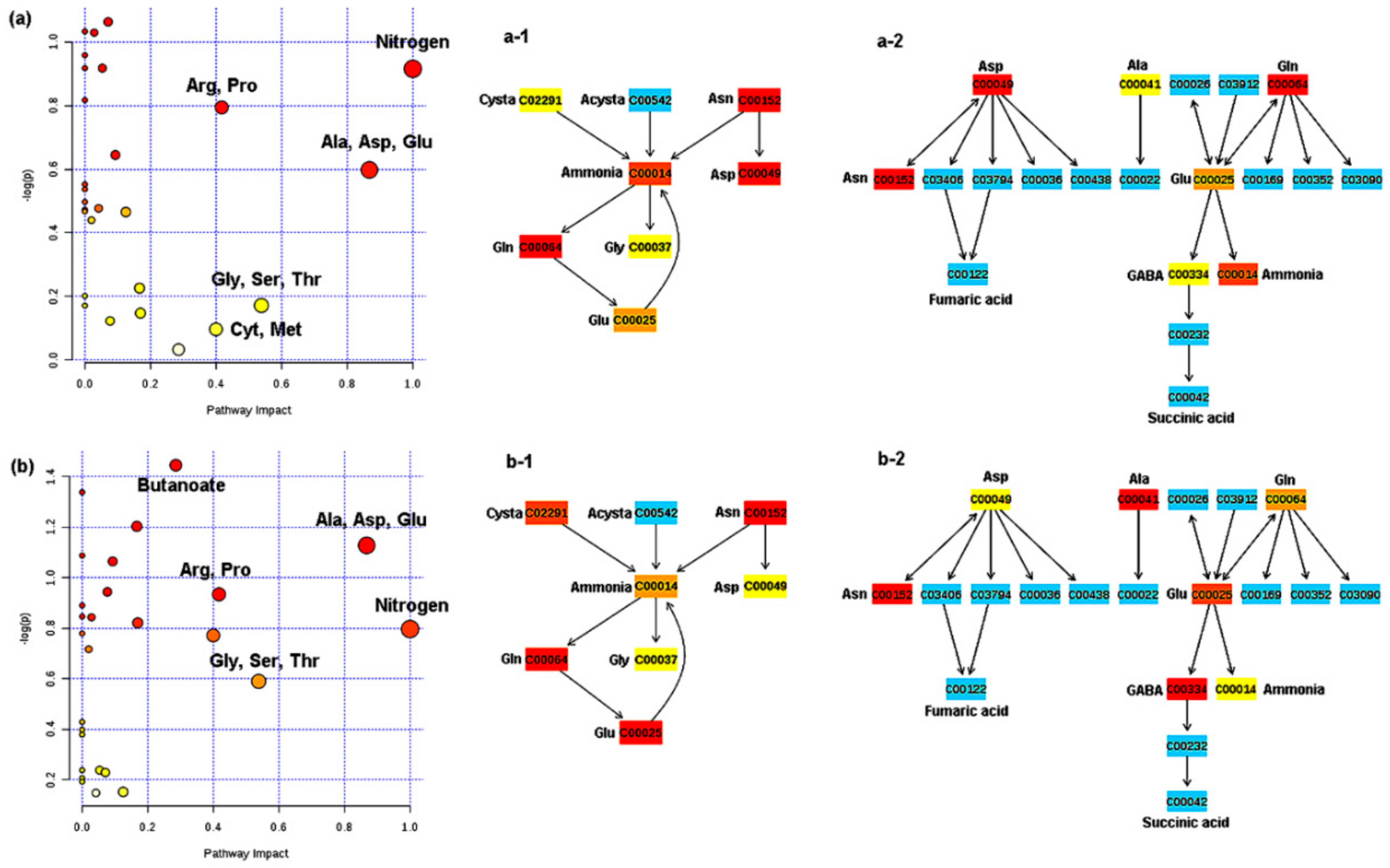
2.3. Free Amino Acid Content in P. indica
3. Discussion
3.1. Effect of the Soil Bacteria on the Growth of P. indica
3.2. P. indica Amino Acid Content During DC with A. chroococcum and AcH 1003
4. Materials and Methods
4.1. Dual Culture of Piriformospora indica with Soil Microorganisms and Sample Collection
4.2. Influence of Soil Bacteria on Fungal Growth
4.3. Morphology of Piriformospora indica Hyphae
4.4. Soil Bacteria Culture
4.5. Determination of Amino Acid Content
4.6. Data Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| No | Name | Abbreviation | No | Name | Abbreviation |
|---|---|---|---|---|---|
| 1 | glycine | Gly | 14 | hydroxylysine | Hlys |
| 2 | serine | Ser | 15 | glutamic acid | Glu |
| 3 | cysteine | Cys | 16 | glutamine | Gln |
| 4 | ornithine | Orn | 17 | alanine | Ala |
| 5 | histidine | His | 18 | arginine | Arg |
| 6 | tyrosine | Tyr | 19 | asparagine | Asn |
| 7 | isoleucine | Ile | 20 | proline | Pro |
| 8 | threonine | Thr | 21 | valine | Val |
| 9 | α-aminobutyric acid | Aba | 22 | leucine | Leu |
| 10 | cystathionine/methionine a | Cys/Met | 23 | lysine | Lys |
| 11 | Phenylalanine | Phe | 24 | ammonia | Am |
| 12 | aminoapidic acid | Aaa | 25 | aspartic acid | Asp |
| 13 | Tryptophan | Trp | 26 | γ-aminobutyric acid | Gaba |
References
- Deveau, A.; Bonito, G.; Uehling, J.; Paoletti, M.; Becker, M.; Bindschedler, S.; Hacquard, S.; Hervé, V.; Labbé, J.; Lastovetsky, O.A.; et al. Bacterial-fungal interactions: Ecology, mechanisms and challenges. FEMS Microbiol. Rev. 2018, 42, 335–352. [Google Scholar] [CrossRef]
- Churchland, C.; Grayston, S.J. Specificity of plant-microbe interactions in the tree mycorrhizosphere biome and consequences for soil cycling. Front. Microbiol. 2014, 5, 1–20. [Google Scholar] [CrossRef]
- Bonfante, P.; Anca, I.-A. Plants, mycorrhizal fungi, and bacteria: a network of interactions. Annu. Rev. Microbiol. 2009, 63, 363–383. [Google Scholar] [CrossRef]
- Kim, M.; Yoon, H.; Kim, Y.E.; Kim, Y.J.; Kong, W.S.; Kim, J.G. Comparative analysis of bacterial diversity and communities inhabiting the fairy ring of Tricholoma matsutake by barcoded pyrosequencing. J. Appl. Microbiol. 2014, 117, 699–710. [Google Scholar] [CrossRef]
- Schrey, S.D.; Tarkka, M.T. Friends and foes: Streptomycetes as modulators of plant disease and symbiosis. Antonie van Leeuwenhoek, Int. J. Gen. Mol. Microbiol. 2008, 94, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Xing, R.; Yan, H.Y.; Gao, Q.B.; Zhang, F.Q.; Wang, J.L.; Chen, S.L. Microbial communities inhabiting the fairy ring of Floccularia luteovirens and isolation of potential mycorrhiza helper bacteria. J. Basic Microbiol. 2018, 58, 554–563. [Google Scholar] [CrossRef] [PubMed]
- Garbaye, J. Helper bacteria: A new dimension to the mycorrhizal symbiosis. New Phytol. 1994, 128, 197–210. [Google Scholar] [CrossRef]
- Takács, T.; Cseresnyés, I.; Kovács, R.; Parádi, I.; Kelemen, B.; Szili-Kovács, T.; Füzy, A. Symbiotic effectivity of dual and tripartite associations on soybean (Glycine max L. Merr.) cultivars inoculated with Bradyrhizobium japonicum and AM Fungi. Front. Plant Sci. 2018, 9, 1–14. [Google Scholar] [CrossRef]
- Turrini, A.; Avio, L.; Giovannetti, M.; Agnolucci, M. Functional complementarity of arbuscular mycorrhizal fungi and associated microbiota: The challenge of translational research. Front. Plant Sci. 2018, 9, 10–13. [Google Scholar] [CrossRef]
- Maier, A.; Riedlinger, J.; Fiedler, H.-P.; Hampp, R. Actinomycetales bacteria from a spruce stand: Characterization and effects on growth of root symbiotic and plant parasitic soil fungi in dual culture. Mycol. Prog. 2004, 3, 129–136. [Google Scholar] [CrossRef]
- Riedlinger, J.; Schrey, S.D.; Tarkka, M.T.; Hampp, R.; Kapur, M.; Fiedler, H.-P. Auxofuran, a novel metabolite that stimulates the growth of fly agaric, is produced by the mycorrhiza helper bacterium Streptomyces strain AcH 505. Appl. Environ. Microbiol. 2006, 72, 3550–3557. [Google Scholar] [CrossRef] [PubMed]
- Keilhofer, N.; Nachtigall, J.; Kulik, A.; Ecke, M.; Hampp, R.; Süssmuth, R.D.; Fiedler, H.P.; Schrey, S.D. Streptomyces AcH 505 triggers production of a salicylic acid analogue in the fungal pathogen Heterobasidion abietinum that enhances infection of Norway spruce seedlings. Antonie van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2018, 111, 691–704. [Google Scholar] [CrossRef] [PubMed]
- Varma, A.; Verma, S.; Sudha; Sahay, N.; Bütehorn, B.; Franken, P. Piriformospora indica, a cultivable plant-growth-promoting root endophyte. Appl. Environ. Microbiol. 1999, 65, 2741–2744. [Google Scholar] [CrossRef]
- Franken, P. The plant strengthening root endophyte Piriformospora indica: Potential application and the biology behind. Appl. Microbiol. Biotechnol. 2012, 96, 1455–1464. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Gill, R.; Trivedi, D.K.; Anjum, N.A.; Sharma, K.K.; Ansari, M.W.; Ansari, A.A.; Johri, A.K.; Prasad, R.; Pereira, E.; et al. Piriformospora indica: Potential and significance in plant stress tolerance. Front. Microbiol. 2016, 7, 1–20. [Google Scholar] [CrossRef]
- Pham, G.H.; Singh, A.; Malla, R.; Kumari, R.; Prasad, R.; Sachdev, M.; Rexer, K.H.; Kost, G.; Luis, P.; Kaldorf, M.; et al. Interaction of Piriformospora indica with diverse microorganisms and plants. In Plant Surface Microbiology; Springer: Berlin/Heidelberg, Germany, 2008; pp. 237–265. ISBN 9783540740506. [Google Scholar]
- Bonfante, P.; Genre, A. Mechanisms underlying beneficial plant–fungus interactions in mycorrhizal symbiosis. Nat. Commun. 2010, 1, 48. [Google Scholar] [CrossRef]
- Wang, W.; Shi, J.; Xie, Q.; Jiang, Y.; Yu, N.; Wang, E. Nutrient exchange and regulation in arbuscular mycorrhizal symbiosis. Mol. Plant 2017, 10, 1147–1158. [Google Scholar] [CrossRef]
- Johansson, J.F.; Paul, L.R.; Finlay, R.D. Microbial interactions in the mycorrhizosphere and their significance for sustainable agriculture. FEMS Microbiol. Ecol. 2004, 48, 1–13. [Google Scholar] [CrossRef]
- Kumar Bhuyan, S.; Bandyopadhyay, P.; Kumar, P.; Kumar Mishra, D.; Prasad, R.; Kumari, A.; Chandra Upadhyaya, K.; Varma, A.; Kumar Yadava, P. Interaction of Piriformospora indica with Azotobacter chroococcum. Sci. Rep. 2015, 5, 1–13. [Google Scholar] [CrossRef]
- Bhattacharyya, P.N.; Jha, D.K. Plant growth-promoting rhizobacteria (PGPR): Emergence in agriculture. World J. Microbiol. Biotechnol. 2012, 28, 1327–1350. [Google Scholar] [CrossRef]
- Shishehbor, M.; Madani, H.; Ardakani, M.R. Effect of vermicompost and biofertilizers on yield and yield components of common millet (Panicum miliaceum). Ann. Biol. Res. 2013, 4, 174–180. [Google Scholar]
- Vafadar, F.; Amooaghaie, R.; Otroshy, M. Effects of plant-growth-promoting rhizobacteria and arbuscular mycorrhizal fungus on plant growth, stevioside, NPK, and chlorophyll content of Stevia rebaudiana. J. Plant Interact. 2014, 9, 128–136. [Google Scholar] [CrossRef]
- Sharma, S.D.; Sharma, N.C.; Sharma, C.L.; Kumar, P.; Chandel, A. Glomus-Azotobacter symbiosis in apple under reduced inorganic nutrient fertilization for sustainable and economic orcharding enterprise. Sci. Hortic. (Amsterdam) 2012, 146, 175–181. [Google Scholar] [CrossRef]
- Arora, M.; Saxena, P.; Choudhary, D.K.; Abdin, M.Z.; Varma, A. Dual symbiosis between Piriformospora indica and Azotobacter chroococcum enhances the artemisinin content in Artemisia annua L. World J. Microbiol. Biotechnol. 2016, 32, 19. [Google Scholar] [CrossRef]
- Maheshwari, D.K.; Dubey, R.C.; Aeron, A.; Kumar, B.; Kumar, S.; Tewari, S.; Arora, N.K. Integrated approach for disease management and growth enhancement of Sesamum indicum L. utilizing Azotobacter chroococcum TRA2 and chemical fertilizer. World J. Microbiol. Biotechnol. 2012, 28, 3015–3024. [Google Scholar] [CrossRef]
- Schrey, S.D.; Salo, V.; Raudaskoski, M.; Hampp, R.; Nehls, U.; Tarkka, M.T. Interaction with mycorrhiza helper bacterium Streptomyces sp. AcH 505 modifies organisation of actin cytoskeleton in the ectomycorrhizal fungus Amanita muscaria (fly agaric). Curr. Genet. 2007, 52, 77–85. [Google Scholar] [CrossRef]
- Walther, A.; Wendland, J. Apical localization of actin patches and vacuolar dynamics in Ashbya gossypii depend on the WASP homolog Wal1p. J. Cell Sci. 2004, 117, 4947–4958. [Google Scholar] [CrossRef]
- Jones, S.E.; Elliot, M.A. Streptomyces Exploration: Competition, volatile communication and new bacterial behaviours. Trends Microbiol. 2017, 25, 522–531. [Google Scholar] [CrossRef]
- Tyc, O.; Song, C.; Dickschat, J.S.; Vos, M.; Garbeva, P. The ecological role of volatile and soluble secondary metabolites produced by soil bacteria. Trends Microbiol. 2017, 25, 280–292. [Google Scholar] [CrossRef]
- Adams, A.S.; Currie, C.R.; Cardoza, Y.; Klepzig, K.D.; Raffa, K.F. Effects of symbiotic bacteria and tree chemistry on the growth and reproduction of bark beetle fungal symbionts. Can. J. For. Res. 2009, 39, 1133–1147. [Google Scholar] [CrossRef]
- Rainey, P.B. Effect of Pseudomonas putida on hyphal growth of Agaricus bisporus. Mycol. Res. 1991, 95, 699–704. [Google Scholar] [CrossRef]
- Schrey, S.D.; Schellhammer, M.; Ecke, M.; Hampp, R.; Tarkka, M.T. Mycorrhiza helper bacterium Streptomyces AcH 505 induces differential gene expression in the ectomycorrhizal fungus Amanita muscaria. New Phytol. 2005, 168, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Becker, D.M.; Bagley, S.T.; Podila, G.K. Effects of mycorrhizal-associated Streptomycetes on growth of Laccaria bicolor, Cenococcum geophilum, and Armillaria species and on gene expression in Laccaria bicolor. Mycologia 1999, 91, 33. [Google Scholar] [CrossRef]
- Martin, F.; Ramstedt, M.; Söderhäll, K.; Canet, D. Carbohydrate and amino acid metabolism in the ectomycorrhizal ascomycete Sphaerosporella brunnea during glucose utilization: A 13C NMR Study. Plant Physiol. 1988, 86, 935–940. [Google Scholar] [CrossRef]
- Breuninger, M.; Trujillo, C.G.; Serrano, E.; Fischer, R.; Requena, N. Different nitrogen sources modulate activity but not expression of glutamine synthetase in arbuscular mycorrhizal fungi. Fungal Genet. Biol. 2004, 41, 542–552. [Google Scholar] [CrossRef]
- Márquez, A.J.; Betti, M.; Ottonello, S.; Bonfante, P.; Montanini, B.; Balestrini, R. Distinctive properties and expression profiles of glutamine synthetase from a plant symbiotic fungus. Biochem. J. 2003, 373, 357–368. [Google Scholar]
- Botton, B.; Chalot, M.; Garnier, A.; Martin, F. L’assimilation de l’azote minéral chez les ectomycorhizes. Acta Bot. Gall. 1994, 141, 469–481. [Google Scholar] [CrossRef]
- Martin, F.; Canet, D. Biosynthesis of amino acids during [13C]glucose utilization by the ectomycorrhizal ascomycete Cenococciiin geophiluin monitored by 13C nuclear magnetic resonance. Physiol Vég 1986, 24, 209–218. [Google Scholar]
- Ceccaroli, P.; Saltarelli, R.; Cesari, P.; Pierleoni, R.; Sacconi, C.; Vallorani, L.; Rubini, P.; Stocchi, V.; Martin, F. Carbohydrate and amino acid metabolism in Tuber borchii mycelium during glucose utilization: A 13C NMR study. Fungal Genet. Biol. 2003, 39, 168–175. [Google Scholar] [CrossRef]
- Kim, J.K.; Mulrooney, S.B.; Hausinger, R.P. The UreEF fusion protein provides a soluble and functional form of the UreF urease accessory protein. J. Bacteriol. 2006, 188, 8413–8420. [Google Scholar] [CrossRef]
- Fichman, Y.; Gerdes, S.Y.; Kovács, H.; Szabados, L.; Zilberstein, A.; Csonka, L.N. Evolution of proline biosynthesis: Enzymology, bioinformatics, genetics, and transcriptional regulation. Biol. Rev. 2015, 90, 1065–1099. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, I.; Carleton, T.; Malloch, D.; Hellebust, J. Nitrogen metabolism in the ectomycorrhizal fungus Laccaria bicolor (R. Mre.) Orton. New Phytol. 1990, 116, 431–441. [Google Scholar] [CrossRef]
- Pati, B.R.; Sengupta, S.; Chandra, A.K. Studies on the amino acids released by phyllosphere diazotrophic bacteria. Microbiol. Res. 1994, 149, 287–290. [Google Scholar] [CrossRef]
- Thuler, D.S.; Floh, E.I.S.; Handro, W.; Barbosa, H.R. Beijerinckia derxii releases plant growth regulators and amino acids in synthetic media independent of nitrogenase activity. J. Appl. Microbiol. 2003, 95, 799–806. [Google Scholar] [CrossRef]
- Pócsi, I.; Prade, R.A.; Penninckx, M.J. Glutathione, altruistic metabolite in fungi. In Advances in Microbial Physiology; Academic Press: Cambridge, MA, USA, 2004; Volume 49, pp. 1–76. ISBN 0120277492. [Google Scholar]
- Ono, B.; Shirahige, Y.; Nanjoh, A.; Andou, N.; Ohue, H.; Ishino-Arao, Y. Cysteine biosynthesis in Saccharomyces cerevisiae: Mutation that confers cystathionine beta-synthase deficiency. J. Bacteriol. 1988, 170, 5883–5889. [Google Scholar] [CrossRef]
- Savin, M.A.; Flavin, M. Cystationine synthesis in yeast: An alternative pathway for homocysteine biosynthesis. J. Bacteriol. 1972, 112, 299–303. [Google Scholar] [CrossRef]
- Riedlinger, J. Die stofflichen Grundlagen der Modulation des Myzelwachstums von symbiotischen und pathogenen Pilzen durch Streptomyceten; Eberhard-Karls University of Tübingen: Tübingen, Germany, 2006. [Google Scholar]
- Ogita, A.; Matsumoto, K.; Fujita, K.; Usuki, Y.; Hatanaka, Y.; Tanaka, T. Synergistic fungicidal activities of amphotericin B and N-methyl-N”-dodecylguanidine: A constituent of polyol macrolide antibiotic niphimycin. J. Antibiot. (Tokyo) 2007, 60, 27–35. [Google Scholar] [CrossRef][Green Version]
- Käfer, E. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 1977, 19, 33–131. [Google Scholar]
- Pilot, G.; Stransky, H.; Bushey, D.F.; Pratelli, R.; Ludewig, U.; Wingate, V.P.; Frommer, W.B. Overexpression of GLUTAMINE DUMPER1 leads to hypersecretion of glutamine from hydathodes of Arabidopsis leaves. Plant Cell 2004, 16, 1827–1840. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the authors. |

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leyva-Rojas, J.A.; Coy-Barrera, E.; Hampp, R. Interaction with Soil Bacteria Affects the Growth and Amino Acid Content of Piriformospora indica. Molecules 2020, 25, 572. https://doi.org/10.3390/molecules25030572
Leyva-Rojas JA, Coy-Barrera E, Hampp R. Interaction with Soil Bacteria Affects the Growth and Amino Acid Content of Piriformospora indica. Molecules. 2020; 25(3):572. https://doi.org/10.3390/molecules25030572
Chicago/Turabian StyleLeyva-Rojas, Jorge A., Ericsson Coy-Barrera, and Rüdiger Hampp. 2020. "Interaction with Soil Bacteria Affects the Growth and Amino Acid Content of Piriformospora indica" Molecules 25, no. 3: 572. https://doi.org/10.3390/molecules25030572
APA StyleLeyva-Rojas, J. A., Coy-Barrera, E., & Hampp, R. (2020). Interaction with Soil Bacteria Affects the Growth and Amino Acid Content of Piriformospora indica. Molecules, 25(3), 572. https://doi.org/10.3390/molecules25030572