Abstract
Aspergillus pachycristatus is an industrially important fungus for the production of the antifungal echinocandin B and is closely related to model organism A. nidulans. Its secondary metabolism is largely unknown except for the production of echinocandin B and sterigmatocystin. We constructed mutants for three genes that regulate secondary metabolism in A. pachycristatus NRRL 11440, and evaluated the secondary metabolites produced by wild type and mutants strains. The secondary metabolism was explored by metabolic networking of UPLC-HRMS/MS data. The genes and metabolites of A. pachycristatus were compared to those of A. nidulans FGSC A4 as a reference to identify compounds and link them to their encoding genes. Major differences in chromatographic profiles were observable among the mutants. At least 28 molecules were identified in crude extracts that corresponded to nine characterized gene clusters. Moreover, metabolic networking revealed the presence of a yet unexplored array of secondary metabolites, including several undescribed fellutamides derivatives. Comparative reference to its sister species, A. nidulans, was an efficient way to dereplicate known compounds, whereas metabolic networking provided information that allowed prioritization of unknown compounds for further metabolic exploration. The mutation of global regulator genes proved to be a useful tool for expanding the expression of metabolic diversity in A. pachycristatus.
1. Introduction
Secondary metabolites from fungi are a plentiful and valuable source of molecules that exhibit a wide range of chemical structures and biological activities. Manipulation of culture media composition and genetic mutations can influence the metabolic outcome of culturing a fungus and expand overall numbers of detectable metabolites. However, recognition of effective combinations of parameters that will stimulate the biosynthesis of novel or cryptic metabolites often requires growing a strain in question in many different experiments, leading to cumbersome workflows and repeated encounters of the same compounds [1]. Development and application of efficient methods for molecular dereplication in complex samples with minimal sample work-up are needed to guide the decision-making process. One such method employs untargeted liquid chromatography coupled to tandem high-resolution mass spectrometry (LC-HRMS/MS) and takes advantage of both high-resolution monoisotopic mass measurements and MS/MS fragmentation profiles to generate data which can be used to detect and classify molecules within a single run [2,3]. The significant amounts of data generated are interrogated against databases to identify known substances without the need for purification of each individual component. These spectra can further be used as input for generation of molecular networks, clusters of known and unknown molecules linked based on similarities found in their MS and MS/MS spectra, thus allowing for the classification of a large number of molecules, including previously unidentified substances [4].
Aspergillus pachycristatus NRRL 11440 (= ATCC 58397) and its mutants, also previously referred to as A. nidulans var. roseus and Emericella rugulosa [5,6], have been used for industrial-scale production of echinocandin B, a lipohexapeptide used as starting material for the semisynthetic antifungal drug anidulafungin [7,8,9]. A draft genome sequence of A. pachycristatus NRRL 11440 revealed a remarkable similarity between A. pachycristatus and A. nidulans, a model organism that has had its secondary metabolism studied extensively during the past decade. The collection of studies aimed at understanding A. nidulans secondary metabolome has led to the discovery of at least 44 gene clusters responsible for the biosynthesis of over 100 secondary metabolites [10,11]. However, only a few secondary metabolites have been described for A. pachycristatus, including echinocandins and sterigmatocystin [5,7]. Recently we have determined that the LaeA (Apc.laeA) and VeA (Apc.veA) global regulatory system was operative in A. pachycristatus NRRL 11440 [12]. Disruption of genes encoding these key proteins negatively affected the production of echinocandin B and nearly abolished sterigmatocystin production. We also observed that Apc.laeA and Apc.veA were essential for normal conidiation and formation of ascomata in A. pachycristatus. These experiments focused exclusively on the echinocandin and sterigmatocystin pathways, but we recognized that these mutations also profoundly affected multiple secondary pathways in these mutant strains in a predictable and reproducible manner [12].
We hypothesized that by comparisons to a closely related reference species, viz. A. nidulans, as a genetic and metabolic database, we could quickly characterize a portion of the metabolome of A. pachycristatus while gaining a measurement of the extent of novelty available for further chemical exploration. Therefore, we aimed to interrogate the secondary metabolism of A. pachycristatus NRRL 11440 with previous data from the A. nidulans FGSC A4 metabolome and genome as a proxy to characterize its metabolites and tentatively attribute them to their corresponding gene clusters. These analyses provided new insights regarding the degree of divergence in secondary metabolism expected between sibling species of Aspergillus. We analyzed extracts from two modified wild-type strains and three mutant strains of NRRL 11440 with disruptions in three global regulator genes known to remodel Aspergillus secondary metabolism. These disrupted genes included laeA and veA, two global regulators of secondary metabolism in Aspergillus species [13,14,15,16,17], and mcrA, a recently discovered master transcription regulator of secondary metabolism [18].
2. Results and Discussion
2.1. Metabolic Diversity in Aspergillus pachycristatus
Five strains of A. pachycristatus, designated wt1, △Apc.laeA, △Apc.veA, wt3, and △Apc.mcrA (Figure S1, Table S1) were grown in the same set of culture conditions, and the crude extracts were analyzed by UPLC-HRMS/MS. The resulting chromatograms (Figure 1) were plotted on the same scale corresponding to a maximum peak height to highlight their differences.
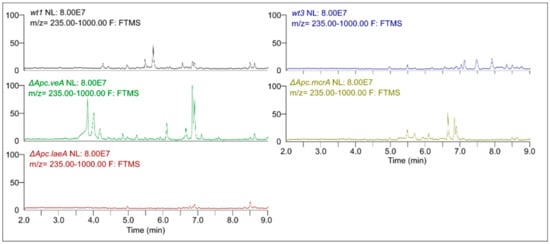
Figure 1.
Representative UPLC-HRMS chromatograms obtained for extracts from wt1, △Apc.laeA, △Apc.veA, wt3, and △Apc.mcrA strains of A. pachycristatus NRRL 11440 grown in SMY medium.
Remarkable qualitative differences were evident among them indicating changes in secondary metabolism. It was possible to observe the appearance of several peaks in △Apc.veA and △Apc.mcrA when compared to their respective parental strains, wt1 and wt3, helping to expand the array of detectable molecules. On the other hand, as we had observed in our previous experiments [12], the △Apc.laeA mutant showed remarkable absence of peaks, with almost no detectable signals.
The raw MS data were submitted to the GNPS platform (https://gnps.ucsd.edu; ID=5b9e80c94f054e0d91f194be81594019) in order to build a molecular network containing the detected metabolites of A. pachycristatus. A total of 2183 different MS2 spectra collected from among the five samples were clustered in 920 nodes according to its MS2 fragmentation. The databases in GNPS displayed positive matches for 11 of those nodes. Verification based on identity, HRMS monoisotopic mass deviation and MS2 spectra led to the confirmation of three of those secondary metabolites as 4, 14, and 21. The MS1 exact masses were compared with known A. nidulans secondary metabolites, which allowed the putative identification of 19 additional molecules (1–3, 5–13, 15–20) (Table 1). MS2 spectra were used to validate identified molecules when possible. Echinocandins were not detected by UPLC-HRMS/MS because the mass cutoff was set at m/z = 1000, although HPLC-MS found echinocandins in extracts of all strains (Figure S2).

Table 1.
Secondary metabolites identified from Aspergillus pachycristatus NRRL 11440.
From the 920 nodes, the GNPS generated 38 network clusters containing two or more nodes. Of the 136 nodes used, 11 corresponded to molecules identified by GNPS or HRMS (Figure 2).
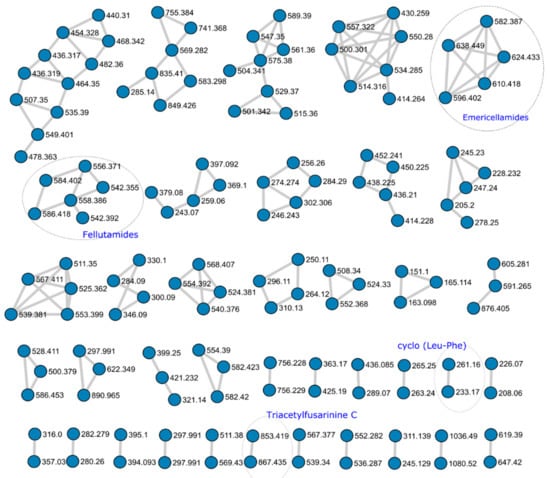
Figure 2.
Metabolic Network cluster obtained from the UPLC-HRMS analysis of Aspergillus pachycristatus mutants grown in SMY medium showing network clusters with two or more nodes. Numbers represent the HRMS m/z observed for each node. Circled clusters indicate molecules positively identified.
Two network clusters were further investigated to identify putative unknown nodes based on MS/MS fragment evaluation.
The GNPS database identified the molecule N,N’,N’’-triacetylfusarinine (TAFC) based on MS1 and MS2 spectra (Figure S3). TAFC is a cyclic molecule composed of three N5-cis-anhydromevalonyl-N5-hydroxy-N2-acetyl-L-ornithine residues. Aspergillus nidulans, A. fumigatus and other Aspergillus species produce it as a major extracellular iron siderophore [19,20]. The TAFC network node was clustered with an unknown nodule showing remarkable similarity in MS2 fragmentation (Figure 2). The major difference arose from an increment of m/z = 14.0157 Da in MS1 adducts, and MS2 evaluation indicated the presence of one extra methylene in one of the repeating units of the structure (22; Figure S4) The absence of an ion with m/z = 741.36 suggested the presence of a N6-cis-anhydromevalonyl-N6-hydroxy-N2-acetyl-L-lysine Although there are a few examples of bacterial siderophores containing -N6-hydroxy-N2-acetyl-L-lysine [21,22,23], fungal siderophores have not been reported to incorporate this unit [24,25,26,27]. Further MS1 evidence suggested the presence of di- and tri- substituted units, as observed by the MS1 spectra of ions m/z = 881 (23) and m/z = 895 (24) showing the same clustered adduct ions in MS1, but they were not obtained with enough intensity to produce observable MS2 spectra (Figure S5). It is possible to infer that the substitution might be observed in any number of the three N5-cis-anhydromevalonyl-N5-hydroxy-L-ornithine residues, resulting in derivatives of N,N’,N’’-triacetylfusarinine. To the best of our knowledge, derivatives bearing extra methylene units for this class of siderophore have not been reported.
Close inspection of MS1 and MS2 spectra and comparison with databases and literature enabled the identification of four fellutamide analogs in a constellation of six nodes, antibiotic 1656G (10; m/z = 584.402) antibiotic 3127 (11; m/z = 542.355), fellutamide B (12; m/z = 556.371) and fellutamide C (13; m/z = 558.386). Fellutamides are tripeptides aldehydes obtained from Penicillium, Aspergillus, and Metulocladosporiella species that inhibit the eukaryotic proteasome [28,29,30,31]. The fellutamide C identified here is the same as identified by Lee and co-workers [28], and not the one characterized from a Metulocladosporiella species [31] that was later renamed fellutamide E [32].
Analysis of MS2 spectra of the four fellutamides showed a common fragment associated with peptides fragmentation mechanisms (Figure 3) that allowed us to propose putative structures for the two other unidentified nodes (25 and 26). Furthermore, manual curation of chromatograms permitted the identification of two other nodes with similar MS/MS fragmentation patterns (27 and 28), even though they were not grouped into the molecular network due to the settings used (Table 2). Their MS1 and MS2 spectra are shown in Figure S6.
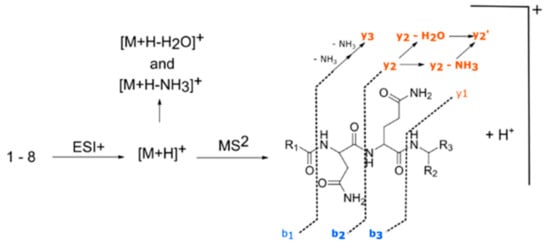
Figure 3.
Description of molecular diversity for the different fellutamides observed.

Table 2.
Fellutamide derivatives identified from Aspergillus pachycristatus NRRL 11440.
When comparing spectra of fellutamides B and C (12 and 13), there was a mass increment of m/z = 2 in [M + H]+ MS1 ion, as well as in MS2 ions [M + H − NH3]+, [M + H − H2O]+ and “y” series of fragments, while the MS2 “b” series of fragments shows the same nominal masses (Figure S5c,d). This feature confirmed the same structure throughout the alkyl side chain, asparagine and glutamine residues, with a modification at the terminal leucinal residue that was reduced to a leucinol. Comparison of spectra from compounds 10 and 25 (Figure S5a,e) showed the exact same characteristics as observed for the pair of spectra 12 and 13, from which we deduced the structure of 25 as being the leucinol derivative of 10, thus identifying it as fellutamide derivative 1.
Compound 26 has the same nominal mass from 11 (m/z = 542), although both have different retention times and monoisotopic masses for [M + H]+ (11, m/z = 542.3557; 26, m/z = 542.3914). HRMS monoisotopic mass indicated that the molecular formula of 26 differed from 11 by having one oxygen atom replaced by incorporation of one carbon and four hydrogens. Evaluation of MS2 spectra from 11 and 26 (Figure S5b,f) showed that there was a consistent pattern between them, with a decrease of m/z = 16 in the “b” series on the MS2 and an increase of m/z = 16 in “y” series of 26. This difference indicated changes with the same overall m/z occurring in the alkyl side chain and terminal amino acid residues, with loss of oxygen in the former and acquisition of carbon and hydrogens in the latter. The MS2 “y” series ions in compound 26 matched the ones observed in compound 13, indicating the same terminal leucinol residue. HRMS m/z from “b” series MS2 ions in 26 indicated the absence of hydroxyl in the alkyl side chain. As such, we identified compound 26 as a novel saturated fatty acid fellutamide derivative 2.
Compound 27 had the same m/z values for MS2 “y3” fragment as in compounds 10 and 12 indicating the same asn-gln-leucinal residue (Figure S5g). Differences were observed in the MS ion [M + H]+, as well as MS2 ions [M + H − NH3]+, [M + H-H2O]+ and “b” series fragments, with m/z values for 27 showing an increase of m/z = 14 when compared to 12, and a decrease of m/z = 14 when matched to compound 10. As the difference between 10 and 12 relies on an extra C2H4 on the alkyl side chain, it is possible to infer that compound 27 possesses an intermediate alkyl side chain between them. The pair of MS2 spectra from 27 and 28 (Figure S5g,h) showed the same patterns observed for 12/13 and 10/25, indicating that 28 was the leucinol analog of 27. We, therefore, identified 27 as fellutamide derivative 3, and 28 as fellutamide derivative 4.
An intense [M + H + H2O]+ peak in MS2 spectra of compounds 13, 25, 26, and 28 was observed and was indicative of a molecular structure that facilitates water loss during gas phase fragmentation, which further corroborates the presence of the reduced leucinol residue. To the best of our knowledge, molecules with structures 25–28 have not yet been described from natural sources. Moreover, we note that the MS2 spectra alone cannot determine with absolute certainty if alkane portions of these new fellutamides are linear, iso- or anteiso- alkanes.
In total, 28 molecules could be putatively identified by combining dereplication strategies using GNPS molecular network, database searches, literature comparisons of HRMS data from A. nidulans metabolites, molecular network evaluation, and manual curation of UPLC-HRMS/MS data (Figure 4).
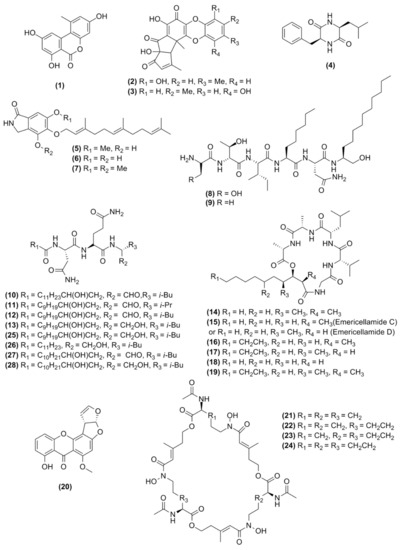
Figure 4.
Chemical structure of the molecules observed in this study.
The overall patterns of secondary metabolite expressed by the different A. pachycristatus strains were remarkably different, as evidenced by their chromatograms, but the patterns of metabolite production for the wt1, ΔApc.laeA and ΔApc.veA strains were remarkably similar to those observed previously [12].
We observed various qualitative differences in metabolite expression in the mutants relative to their parent strain controls grown in the same conditions (Figure 1, Table S2). The laeA deletion mutant exhibited overall repression of secondary metabolite production that was consistent with our observations for NRRL 11440 [12] and for other Aspergillus species [14,16,17,33]. The veA mutant showed a more complex metabolite pattern, with repression of several metabolites such as aspercryptins. However, it also showed an increased production of other metabolites such emericellamide, and also the observation of metabolites not detected in wild types such as F9775-A and F-9775-B, a result consistent with previous observations in A. nidulans FGSC A4 [15]. For the mcrA mutant, a general increase of secondary metabolite production was observed, which parallels previous experiments with the same mutation in A. nidulans [18].
In addition to the compounds identified above, several compounds observed in A. pachycristatus extracts could not be identified by comparison with the A. nidulans metabolome. Neither could their structures be deduced solely from HRMS and MS/MS experiments. It was evident from the molecular networking (Figure 2) that several of those metabolites were grouped in clusters indicating the production of families of related analogs that could be prioritized for further metabolic studies. These results clearly showed that even though A. pachycristatus shares many secondary metabolites with A. nidulans, this species produces many more that remain unidentified, of which some are not likely to be produced by A. nidulans. In addition, the results from the molecular networking highlight its utility as a dereplication strategy to prioritize unknown metabolites for further evaluation.
2.2. Confirmation of Homologous Gene Clusters in A. pachycristatus for the Identified Metabolites
To understand how species-level phylogenetic divergence is reflected in divergence in secondary metabolism, we also compared the overall correspondence of secondary metabolic biosynthetic gene clusters (BGCs) between the strains of A. pachycristatus and A. nidulans (Figure 5, Table S4). At least 49 core biosynthetic genes showed high amino acid similarities (>63%) between these two strains, of which 22 were PKS, 21 NRPS, 4 DMATS, 1 FAS, and 1 NRPS-like biosynthetic pathways. Furthermore, bioinformatic analysis indicated a capacity for each strain to encode BGCs unique to that strain. Aspergillus pachycristatus NRRL 11440 had 17 BGCs exclusive to its genome, while A. nidulans had 22. Thus, the two sibling species share approximately two-thirds of their respective secondary metabolite BGCs (Figure 5, Table S4). This degree of similarity was higher than between the sibling species pair of A. fumigatus and A. fishcheri where 10/33 of A. fumigatus BGCs are conserved in A. fischeri and only 13/48 A. fischeri BGCs are conserved in A. fumigatus [34].
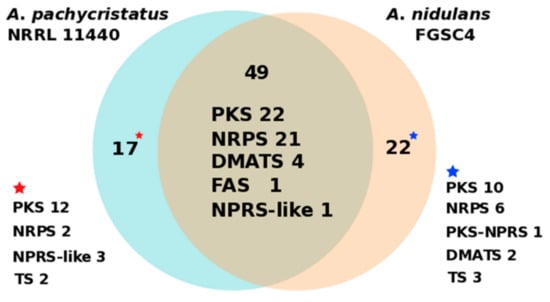
Figure 5.
Numbers of distinct and shared secondary metabolite core catalytic genes between A. nidulans FGSC4 and A. pachycristatus NRRL 11440. PKS (polyketide synthase), NRPS (nonribosomal peptide synthetase), DMATS (dimethylallyltryptophan synthase), FAS (fatty acid synthase), and TS (terpene synthase).
Biosynthetic gene clusters in the genome of A. nidulans FGSC4 responsible for encoding the enzymes involved in the biosynthesis of targeted molecules were used as templates for searching homologous gene clusters in A. pachycristatus. Table 3 shows that for each identified secondary metabolite or metabolite family, the corresponding homologous gene cluster could be found in A. pachycristatus. The nonribosomal peptide synthetases (NRPSs) and polyketide synthases (PKSs) from each BGC homolog pair showed remarkably high amino acid similarity (>79%) between these two species (Table 3). Thus, the genetic data confirmed that A. pachycristatus genome encodes the biosynthetic capacity for all the identified secondary metabolites.

Table 3.
Core biosynthetic genes from inferred biosynthetic clusters for metabolites identified in Aspergillus pachycristatus and their homologs in A. nidulans.
Microsynteny of emericellamide, fellutamide, aspercryptin, aspernidine A, orsellinic acid/9775, and sterigmatocystin BGCs was analyzed between A. pachycristatus and A. nidulans (Figure S7). As expected, all the evaluated BGCs in A. pachycristatus consisted of close homologs of the genes encoding these molecules in A. nidulans, with a similar cluster organization and gene order which indicated a high degree of certainty that these set of BGCs are responsible for encoding the biosynthesis the metabolites listed in Table 3.
These BGCs (Table 3) share a high identity and, in all but one instance (Figure S7), and the same gene order between the two strains. However, when we examined the gene cluster boundaries, adjacent genes, and genome loci of these BGCs between A. pachycristatus and A. nidulans, we unexpectedly found that the corresponding BGCs were located at different genome loci, even though they shared identical boundary genes in almost all cases. Thus, even in close sister species, the secondary metabolic gene clusters are inherited as independent units during the course of evolutionary time.
3. Materials and Methods
3.1. Construction of Gene Deletion and Marker-Recycling Strains
The basic strategy for gene disruption experiments and protoplast transformation methods were previously described in detail [12]. We disrupted the Apc.pyrG gene (orotidine-5′-phosphate decarboxylase, MK689400, AN6157 homologue) from NRRL 11440 by protoplast transformation to generate uracil-uridine auxotrophic strains [12]. To facilitate high recombination rates in the modified strain, the DNA double-strand break repair enzyme Apc.nkuA (ATP-dependent DNA helicase II, MK689401, AN7753 homolog) was inactivated by insertion of the selective marker pyr4 from Neurospora crassa. This strain was designated wt1. Neither substitution of the native Apc.pyrG gene with pyr4, or disruption of Apc.nkuA appeared to affect colony morphology, sporulation, or secondary metabolism production [12].
For gene disruption, about 1 kb upstream and 1 kb downstream DNA exchange fragments of Apc.stcA or Apc.mcrA genes were fused into each end of afpyrG cassette by overlap PCR by using PrimeSTAR GXL DNA polymerase. Fusion DNA fragments were purified by QIAquick Gel Extraction Kit before use for protoplast transformation. After protoplast transformation of wt2 (for stcA deletion) or wt4 (for mcrA deletion) (Table S3) strains, single-colonies were picked up and screened on MMS selection plates without uracil and uridine for at least three generations and then verified by diagnostic PCR (Figures S8 and S10). For pyrG marker recycling, about 1 kb upstream and 1 kb downstream DNA exchange fragments of Apc.stcA gene were cloned from wild type strain and fused together by overlap PCR. Purified fusion DNA fragments were used for protoplast transformation of wt3 strain. Positive transformants were screened on YAS medium supplemented with 1.0 mg/mL 5-FOA, 9 mM uracil and 10 mM uridine and verified by diagnostic PCR (Figure S9). Primers used for gene deletion, marker recycling, and diagnostic PCR are listed in Table S5. Thus, the wt3 strain is identical to wt1 strain, except that Apc.stcA was inactivated, thus abolishing sterigmatocystin production.
3.2. Growth and Sample Preparation
The medium for routine growth and sporulation was yeast extract medium (YAG, 1 L deionized H2O), 5 g yeast extract, 10 g dextrose, 1 mL trace elements (1000 × stock), 2 mL vitamin mix (500 × stock), 10 mL 1 M MgSO4 ·7H2O with or without 1.5% agar. To produce metabolites, strains were grown in 250-mL flasks containing 50 mL SMY medium (Bacto neopeptone 10 g, maltose 40 g, yeast extract 10 g per 1000 ml of deionized H2O). Cultures were maintained for 10 days at 24 °C and 220 rpm. For the extraction procedure, an equal volume of ethyl acetate was added to the culture and the extraction carried out overnight, with agitation. The extract was vacuum-filtered, and the organic phase separated. A 5.0-mL aliquot was placed in a clean glass flask and evaporated to dryness at 37 °C under gentle airflow. The dried extract was reconstituted in 0.25 mL of acetonitrile, diluted with 0.25 mL of ultra-pure water, filtered through 0.22 µm RC membrane and submitted to chromatographic analyses.
3.3. HPLC-MS Analysis
HPLC-MS data were acquired on an Agilent 1260 HPLC equipped with a diode array detector (DAD) and coupled to an Agilent 6120 single quadrupole mass spectrometer (MS), with an Ace Equivalence 5 C18, 4.6 × 150 mm, 5 μm column kept at 30 °C. The pump used a binary solvent system consisting of 0.1% aqueous formic acid (solvent A) and, 0.1% formic acid in acetonitrile (ACN) (solvent B) with the following gradient: 20%−100% B for 22 min, and maintaining 100%B for 4 min, 1.0 mL.min−1. The chromatographic profiles were monitored by wavelength scanning from 190 to 400 nm and by positive and negative ESI-MS from m/z 160–1500. The injection volume was 10 µL.
3.4. UPLC-HRMS Analysis
Extracts were analyzed with a Waters Acquity I-Class UPLC with DAD detector and equipped with an Acquity UPLC BEH C18 (1.7 um, 2.1 × 50 mm) at 40 °C, with flow rate of 0.5 mL.min−1. The following gradient was applied using 0.1% aqueous acetic acid (solvent A) and 0.1% acetic acid in ACN (solvent B): at 0 min., 5% B, maintained for 2 min, 14 min, 95% B. The column eluent was directed to a Thermo Orbitrap mass spectrometer, operating in the 100–1000 mass range, using as heat temp 375 °C, sheath gas flow rate 45, Aux Gas Flow Rate 10, Sweep Gas Flow Rate 3, I Spray Voltage 4.10 kV, Capillary Temp 320 °C, S-Lens RF Level 55%. The injection volume was 2 µL.
3.5. Metabolic Profiling, Molecular Networking and Compound Dereplication
Chromatograms were analyzed with Thermo Xcalibur software for the identification of peaks, measurements of individual peak height, and accurate MS1 m/z.
Raw data from UPLC–HRMS/MS were converted to mzXML using ProteoWizard (version 3.0.10051, Vanderbilt University, USA). All mzXML data were uploaded to the Global Natural Products Social Networking (GNPS) website (https://gnps.ucsd.edu/ProteoSAFe/static/gnps-splash.jsp). A molecular network was created using the online workflow at the GNPS. The data were filtered by removing all MS/MS peak\s within +/− 17 Da of the precursor m/z. MS/MS spectra were window filtered by choosing only the top six peaks in the +/− 50 Da window throughout the spectrum. The data was then clustered with MS-Cluster with a parent mass tolerance of 0.02 Da and a MS/MS fragment ion tolerance of 0.02 Da to create consensus spectra. Furthermore, consensus spectra that contained less than one spectra were discarded. A network was then created where edges were filtered to have a cosine score above 0.7 and more than four matching peaks. Edges between two nodes were kept in the network if, and only if, each of the nodes appeared in each other’s respective top 10 most similar nodes. The spectra in the network were then searched against the GNPS spectral libraries. The library spectra were filtered in the same manner as the input data. All matches kept between network spectra and library spectra were required to have a score above 0.7 and at least six matched peaks. Analog searching was enabled against the library with a maximum mass shift of 0.02 Da [4]. The metabolic network was built using Cytoscape 3.7.0 [35]. The job ID is ID = 5b9e80c94f054e0d91f194be81594019.
Dereplication of known molecules was enabled by matching to hits in GNPS database and aided by manually searching other databases such as the Natural Product Atlas (www.npatlas.org/) and the Dictionary of Natural Products (dnp.chemnetbase.com/). Moreover, due to the genetic similarities of A. pachycristatus and A. nidulans secondary metabolism, previous works describing A. nidulans metabolites were used as proxy set [28,31,36,37,38,39,40,41,42,43].
3.6. Comparison of Secondary Metabolic Biosynthestic Gene Clusters of A. pachycristatus NRRL 11440 and A. nidulans FGSC A4
The genetically encoded BGCs of A. pachycristatus NRRL 11440 were predicted by submitting the unannotated scaffold sequences for antiSMASH analysis (https://fungismash.secondarymetabolites.org/) [44,45] and by reciprocal BLAST searches of predicted proteins and annotated scaffolds with sequences of previously determined core genes and gene clusters from A. nidulans [37,38,42,43,46,47,48] followed by manual correction of some incorrectly predicted ORFs. Orthologous biosynthetic gene clusters between A. pachycristatus NRRL 11440 and A. nidulans FGSC A4 were aligned and illustrated using Easyfig [49] to determine gene identity (%) and microsynteny.
4. Conclusions
By referencing an extensively explored and annotated model organism as a proxy, we were able to efficiently dereplicate several gene clusters and classes of previously undescribed metabolites from mutant strains of A. pachycristatus. Collectively, the results of this study confirm that A. pachycristatus shares roughly two-thirds of secondary biosynthetic gene clusters and metabolites with that of A. nidulans. However, it still produces or has the potential to produce a large set of yet unknown secondary metabolites and biosynthetic genes that can be further explored as a potential source for the discovery of novel molecules. Moreover, we confirmed the possibility of expanding the observed metabolome of A. pachycristatus through the disruption of global regulator genes. The metabolic effects of gene disruptions in A. pachycristataus (laeA, veA, mcrA) approximately paralleled reported effects in A. nidulans. These results provide additional evidence that these gene disruptions can be a useful tool to enhance and vary overall secondary metabolite expression and enabling the discovery of silent and cryptic metabolites.
Supplementary Materials
The following are available online at https://www.mdpi.com/1420-3049/25/4/913/s1, Figure S1. Morphological variations in Aspergillus pachycristatus mutants grown on agar media. Figure S2. Extracted ion chromatogram (EIC+ = m/z 1042–1044) from LC-MS analysis of Aspergillus pachycristatus strains in SYM media highlighting the production of echinocandin B [M + H − H2O]+ ion in all tested media. Figure S3. A) MS1; B) MS2 and C) Proposed fragmentation pathway of N,N’,N’’-Triacetylfusarinine C. Figure S4. A) MS1; B) MS2 and C) Proposed fragmentation pathway of N,N’,N’’-Triacetylfusarinine C analog containing an extra methylene. Blue fragments indicates m/z similar to those observed in TAFC, while orange m/z represent fragments containing an increased 14 Da. Figure S5. Chromatogram and MS1 spectra showing other possible analogs of N,N’,N’’-Triacetylfusarinine C (22, 23) containing extra methylene units, when compared to N,N’,N’’-Triacetylfusarinine C (21) and their putative structure. Figure S6. MS1 and MS2 HRMS spectra of fellutamides: (a) Antibiotic 1656G (10); (b) Antibiotic 3127 (11); (c) Fellutamide B (12); (d) Fellutamide C (13); (e) Fellutamide derivative 1 (25); (f) Fellutamide derivative 2 (26); (g) Fellutamide derivative 3 (27); (h) Fellutamide derivative 4 (28). Figure S7. Microsynteny comparisons of emericellamide, fellutamide, aspercryptin, aspernidine A, orsellinic acid/9775 and sterigmatocystin BGCs between A. pachycristatus NRRL 11440 and A. nidulans FGSC A4. Figure S8. Construction of DNA fusion fragments for apc.stcA deletion and diagnostic PCR. A. Schematic diagram of Apc.stcA disruption by insertion of Af.pyrG as selective marker gene by homologous recombination. B. Agarose gel images of PCR products of construction of DNA fusion fragments for protoplast transformation. C. Schematic diagram of verification of △Apc.stcA transformants. D. Agarose gel images of PCR products for detection of Apc.stcA deletion of eight independent transformants. Figure S9. Recycling of Af.pyrG marker gene from △Apc.stcA mutant. A. Schematic diagram of Af.pyrG deletion from Apc.stcA locus and verification of Af.pyrG recycling transformants. B. Agarose gel images of PCR products of construction of DNA fusion fragments for protoplast transformation and PCR products for detection of Af.pyrG deletion of 10 independent transformants Figure S10. Construction of DNA fusion fragments for ΔApc.mcrA deletion and diagnostic PCR. A. Schematic diagram of Apc.mcrA disruption by insertion of Af.pyrG as selective marker gene by homologous recombination. B. Agarose gel images of PCR products of construction of DNA fusion fragments for protoplast transformation. C. Schematic diagram of verification of △Apc.mcrA transformants. D. Agarose gel images of PCR products for detection of Apc.mcrA deletion transformants. Table S1. HRMS error for diagnostic MS and MS2 ions for fellutamides. Table S2. Qualitative evaluation of the presence of detected metabolites in the extracts of A. pachycristatus strains. Table S4. Correspondence of secondary metabolic BGCs between strains A. pachycristatus NRRL 11440 and A. nidulans FGSC A4. Table S5. Primers used in this study.
Author Contributions
Conceptualization, B.P., L.N., G.B.; methodology, B.P., L.N., G.B.; investigation, B.P., L.N., Y.J., G.B.; resources, Y.J., Z.A., G.B.; data curation, B.P., L.N.; writing—original draft preparation, B.P., L.N., G.B.; writing—review and editing, B.P., L.N., G.B.; supervision, Z.A., G.B.; funding acquisition, Z.A., G.B. All authors have read and agreed to the published version of the manuscript.
Funding
Portions of this work were supported by Cidara Therapeutics.
Acknowledgments
The authors are grateful to Cidara Therapeutics for the supporting with the genome sequencing of Aspergillus pachycristatus NRRL 11440.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Tormo, J.R.; Asensio, F.J.; Bills, G.F. Manipulating filamentous fungus chemical phenotypes by growth on nutritional arrays. Methods Mol. Biol. 2012, 944, 59–78. [Google Scholar] [CrossRef] [PubMed]
- Bouslimani, A.; Sanchez, L.M.; Garg, N.; Dorrestein, P.C. Mass spectrometry of natural products: current, emerging and future technologies. Nat. Prod. Rep. 2014, 31, 718–729. [Google Scholar] [CrossRef] [PubMed]
- El-Elimat, T.; Figueroa, M.; Ehrmann, B.M.; Cech, N.B.; Pearce, C.J.; Oberlies, N.H. High-resolution MS, MS/MS, and UV database of fungal secondary metabolites as a dereplication protocol for bioactive natural products. J. Nat. Prod. 2013, 76, 1709–1716. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.J.; Frisvad, J.C.; Sun, B.D.; Varga, J.; Kocsube, S.; Dijksterhuis, J.; Kim, D.H.; Hong, S.B.; Houbraken, J.; Samson, R.A. Aspergillus section Nidulantes (formerly Emericella): Polyphasic taxonomy, chemistry and biology. Stud. Mycol. 2016, 84, 1–118. [Google Scholar] [CrossRef] [PubMed]
- Tóth, V.; Nagy, C.T.; Miskei, M.; Pócsi, I.; Emri, T. Polyphasic characterization of “Aspergillus nidulans var. roseus” ATCC 58397. Folia. Microbiol. (Praha) 2011, 56, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Boeck, L.D. New antibiotics: Antifungals from Aspergillus. In Frontiers in Industrial Mycology; Leatham, G., Ed.; Chapman & Hall: New York, NY, USA, 1992; pp. 54–65. [Google Scholar]
- Boeck, L.D.; Kastner, R.E. Method of producing the A-30912 antibiotics. U.S. Patent 4,288,549, 21 January 1981. [Google Scholar]
- Emri, T.; Majoros, L.; Toth, V.; Pocsi, I. Echinocandins: Production and applications. Appl. Microbiol. Biotechnol. 2013, 97, 3267–3284. [Google Scholar] [CrossRef]
- Andersen, M.R.; Nielsen, J.B.; Klitgaard, A.; Petersen, L.M.; Zachariasen, M.; Hansen, T.J.; Blicher, L.H.; Gotfredsen, C.H.; Larsen, T.O.; Nielsen, K.F.; et al. Accurate prediction of secondary metabolite gene clusters in filamentous fungi. Proc. Natl. Acad. Sci. USA 2013, 110, E99–E107. [Google Scholar] [CrossRef]
- De Vries, R.P.; Riley, R.; Wiebenga, A.; Aguilar-Osorio, G.; Amillis, S.; Uchima, C.A.; Anderluh, G.; Asadollahi, M.; Askin, M.; Barry, K.; et al. Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol. 2017, 18, 28. [Google Scholar] [CrossRef]
- Lan, N.; Yue, Q.; An, Z.; Bills, G.F. Apc.LaeA and Apc.VeA of the velvet complex govern secondary metabolism and morphological development in the echinocandin-producing fungus Aspergillus pachycristatus. J. Ind. Microbiol. Biotechnol. 2019. [Google Scholar] [CrossRef]
- Bayram, O.; Braus, G.H. Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins. FEMS Microbiol. Rev. 2012, 36, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Bok, J.W.; Keller, N.P. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot Cell 2004, 3, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Bok, J.W.; Soukup, A.A.; Chadwick, E.; Chiang, Y.M.; Wang, C.C.; Keller, N.P. VeA and MvlA repression of the cryptic orsellinic acid gene cluster in Aspergillus nidulans involves histone 3 acetylation. Mol. Microbiol. 2013, 89, 963–974. [Google Scholar] [CrossRef] [PubMed]
- Kale, S.P.; Milde, L.; Trapp, M.K.; Frisvad, J.C.; Keller, N.P.; Bok, J.W. Requirement of LaeA for secondary metabolism and sclerotial production in Aspergillus flavus. Fungal. Genet. Biol. 2008, 45, 1422–1429. [Google Scholar] [CrossRef] [PubMed]
- Lind, A.L.; Smith, T.D.; Saterlee, T.; Calvo, A.M.; Rokas, A. Regulation of secondary metabolism by the velvet complex is temperature-responsive in Aspergillus. G3 Genes Genomes Genetics 2016, 6, 4023–4033. [Google Scholar] [CrossRef] [PubMed]
- Oakley, C.E.; Ahuja, M.; Sun, W.W.; Entwistle, R.; Akashi, T.; Yaegashi, J.; Guo, C.J.; Cerqueira, G.C.; Russo Wortman, J.; Wang, C.C.; et al. Discovery of McrA, a master regulator of Aspergillus secondary metabolism. Mol. Microbiol. 2017, 103, 347–365. [Google Scholar] [CrossRef] [PubMed]
- De la Cruz, M.; Martin, J.; Gonzalez-Menendez, V.; Perez-Victoria, I.; Moreno, C.; Tormo, J.R.; El Aouad, N.; Guarro, J.; Vicente, F.; Reyes, F.; et al. Chemical and physical modulation of antibiotic activity in emericella species. Chem. Biodivers 2012, 9, 1095–1113. [Google Scholar] [CrossRef]
- Schrettl, M.; Bignell, E.; Kragl, C.; Sabiha, Y.; Loss, O.; Eisendle, M.; Wallner, A.; Arst, H.N., Jr.; Haynes, K.; Haas, H. Distinct roles for intra- and extracellular siderophores during Aspergillus fumigatus infection. PLoS Pathog. 2007, 3, 1195–1207. [Google Scholar] [CrossRef]
- De Lorenzo, V.; Bindereif, A.; Paw, B.H.; Neilands, J.B. Aerobactin biosynthesis and transport genes of plasmid ColV-K30 in Escherichia coli K-12. J. Bacteriol. 1986, 165, 570–578. [Google Scholar] [CrossRef]
- Martin, J.D.; Ito, Y.; Homann, V.V.; Haygood, M.G.; Butler, A. Structure and membrane affinity of new amphiphilic siderophores produced by Ochrobactrum sp. SP18. J. Biol. Inorg. Chem. 2006, 11, 633–641. [Google Scholar] [CrossRef]
- McMahon, M.D.; Rush, J.S.; Thomas, M.G. Analyses of MbtB, MbtE, and MbtF suggest revisions to the mycobactin biosynthesis pathway in Mycobacterium tuberculosis. J. Bacteriol. 2012, 194, 2809–2818. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Guo, Y.; Lu, Y.; Wang, B.; Sun, J.; Zhang, H.; Wang, H. Chemistry and biology of siderophores from narine microbes. Mar. Drugs 2019, 17, 562. [Google Scholar] [CrossRef] [PubMed]
- Haas, H. Fungal siderophore metabolism with a focus on Aspergillus fumigatus. Nat. Prod. Rep. 2014, 31, 1266–1276. [Google Scholar] [CrossRef] [PubMed]
- Hilder, R.C.; Kong, X. Chemistry and biology of siderophores. Nat. Products Rep. 2010, 27, 637–657. [Google Scholar] [CrossRef]
- Renshaw, J.C.; Robson, G.D.; Trinci, A.P.J.; Wiebe, M.G.; Livens, F.R.; Collison, D.; Taylor, R.J. Fungal siderophores: Structures, functions and applications. Mycol. Res. 2002, 106, 1123–1142. [Google Scholar] [CrossRef]
- Lee, Y.M.; Dang, H.T.; Hong, J.; Lee, C.O.; Bae, K.S.; Kim, D.K.; Jung, J.H. A cytotoxic lipopeptide from the sponge-derived fungus Aspergillus versicolor. Bull. Korean Chem. Soc. 2010, 31, 205–208. [Google Scholar] [CrossRef]
- Lee, Y.M.; Dang, H.T.; Li, J.; Zhang, P.; Hong, J.; Lee, C.O.; Jung, J.H. A cytotoxic fellutamide analogue from the sponge-derived fungus Aspergillus versicolor. Bull. Korean Chem. Soc. 2011, 32, 3817–3820. [Google Scholar] [CrossRef]
- Shigemori, H.; Wakuri, S.; Yazawa, K.; Nakamura, T.; Sasaki, T.; Kobayashi, J.I. Fellutamides A and B, cytotoxic peptides from a marine fish-possessing fungus Penicillium fellutanum. Tetrahedron 1991, 47, 8529–8534. [Google Scholar] [CrossRef]
- Xu, D.; Ondeyka, J.; Harris, G.H.; Zink, D.; Kahn, J.N.; Wang, H.; Bills, G.; Platas, G.; Wang, W.; Szewczak, A.A.; et al. Isolation, structure, and biological activities of fellutamides C and D from an undescribed Metulocladosporiella (Chaetothyriales) using the genome-wide Candida albicans fitness test. J. Nat. Prod. 2011, 74, 1721–1730. [Google Scholar] [CrossRef]
- Wu, C.J.; Li, C.W.; Cui, C.B. Seven new and two known lipopeptides as well as five known polyketides: The activated production of silent metabolites in a marine-derived fungus by chemical mutagenesis strategy using diethyl sulphate. Mar. Drugs 2014, 12, 1815–1838. [Google Scholar] [CrossRef]
- Bok, J.W.; Hoffmeister, D.; Maggio-Hall, L.A.; Murillo, R.; Glasner, J.D.; Keller, N.P. Genomic mining for Aspergillus natural products. Chem. Biol. 2006, 13, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Mead, M.E.; Knowles, S.L.; Raja, H.A.; Beattie, S.R.; Kowalski, C.H.; Steenwyk, J.L.; Silva, L.P.; Chiaratto, J.; Ries, L.N.A.; Goldman, G.H.; et al. Characterizing the pathogenic, genomic, and chemical traits of Aspergillus fischeri, a close relative of the major human fungal pathogen Aspergillus fumigatus. mSphere 2019, 4. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Albright, J.C.; Henke, M.T.; Soukup, A.A.; McClure, R.A.; Thomson, R.J.; Keller, N.P.; Kelleher, N.L. Large-scale metabolomics reveals a complex response of Aspergillus nidulans to epigenetic perturbation. ACS Chem. Biol. 2015, 10, 1535–1541. [Google Scholar] [CrossRef]
- Chiang, Y.M.; Szewczyk, E.; Nayak, T.; Davidson, A.D.; Sanchez, J.F.; Lo, H.C.; Ho, W.Y.; Simityan, H.; Kuo, E.; Praseuth, A.; et al. Molecular genetic mining of the Aspergillus secondary metabolome: discovery of the emericellamide biosynthetic pathway. Chem. Biol. 2008, 15, 527–532. [Google Scholar] [CrossRef]
- Henke, M.T.; Soukup, A.A.; Goering, A.W.; McClure, R.A.; Thomson, R.J.; Keller, N.P.; Kelleher, N.L. New aspercryptins, lipopeptide natural products, revealed by HDAC inhibition in Aspergillus nidulans. ACS Chem. Biol. 2016, 11, 2117–2123. [Google Scholar] [CrossRef]
- Nielsen, K.F.; Larsen, T.O. The importance of mass spectrometric dereplication in fungal secondary metabolite analysis. Front. Microbiol. 2015, 6, 71. [Google Scholar] [CrossRef]
- Oh, D.C.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Induced production of emericellamides A and B from the marine-derived fungus Emericella sp. in competing co-culture. J. Nat. Prod. 2007, 70, 515–520. [Google Scholar] [CrossRef]
- Scherlach, K.; Schuemann, J.; Dahse, H.M.; Hertweck, C. Aspernidine A and B, prenylated isoindolinone alkaloids from the model fungus Aspergillus nidulans. J. Antibiot. (Tokyo) 2010, 63, 375–377. [Google Scholar] [CrossRef]
- Yaegashi, J.; Praseuth, M.B.; Tyan, S.W.; Sanchez, J.F.; Entwistle, R.; Chiang, Y.M.; Oakley, B.R.; Wang, C.C. Molecular genetic characterization of the biosynthesis cluster of a prenylated isoindolinone alkaloid aspernidine A in Aspergillus nidulans. Org. Lett. 2013, 15, 2862–2865. [Google Scholar] [CrossRef]
- Yeh, H.H.; Ahuja, M.; Chiang, Y.M.; Oakley, C.E.; Moore, S.; Yoon, O.; Hajovsky, H.; Bok, J.W.; Keller, N.P.; Wang, C.C.; et al. Resistance gene-guided genome mining: Serial promoter exchanges in Aspergillus nidulans reveal the biosynthetic athway for fellutamide B, a proteasome inhibitor. ACS Chem. Biol. 2016, 11, 2275–2284. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Wolf, T.; Chevrette, M.G.; Lu, X.; Schwalen, C.J.; Kautsar, S.A.; Suarez Duran, H.G.; de Los Santos, E.L.C.; Kim, H.U.; Nave, M.; et al. antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic. Acids. Res. 2017, 45, W36–W41. [Google Scholar] [CrossRef] [PubMed]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Muller, R.; Wohlleben, W.; et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic. Acids. Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.W.; Yu, J.H.; Kelkar, H.S.; Fernandes, M.; Nesbitt, T.C.; Keller, N.P.; Adams, T.H.; Leonard, T.J. Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 1996, 93, 1418–1422. [Google Scholar] [CrossRef] [PubMed]
- David, H.; Özçelik, I.Ş.; Hofmann, G.; Nielsen, J. Analysis of Aspergillus nidulans metabolism at the genome-scale. BMC Genomics 2008, 9. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, J.F.; Chiang, Y.M.; Szewczyk, E.; Davidson, A.D.; Ahuja, M.; Elizabeth Oakley, C.; Woo Bok, J.; Keller, N.; Oakley, B.R.; Wang, C.C. Molecular genetic analysis of the orsellinic acid/F9775 gene cluster of Aspergillus nidulans. Mol. Biosyst. 2010, 6, 587–593. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
Sample Availability: Samples of the crude extracts are available from the authors. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).