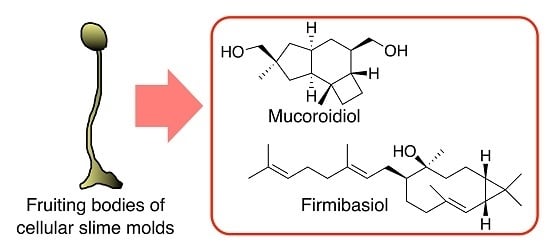
Two New Terpenes Isolated from Dictyostelium Cellular Slime Molds
Abstract
1. Introduction
2. Results
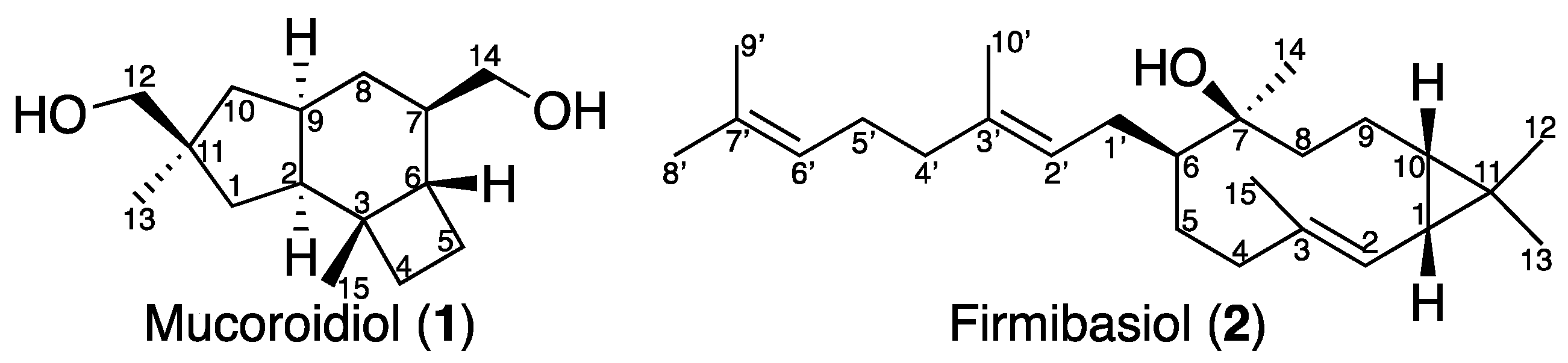
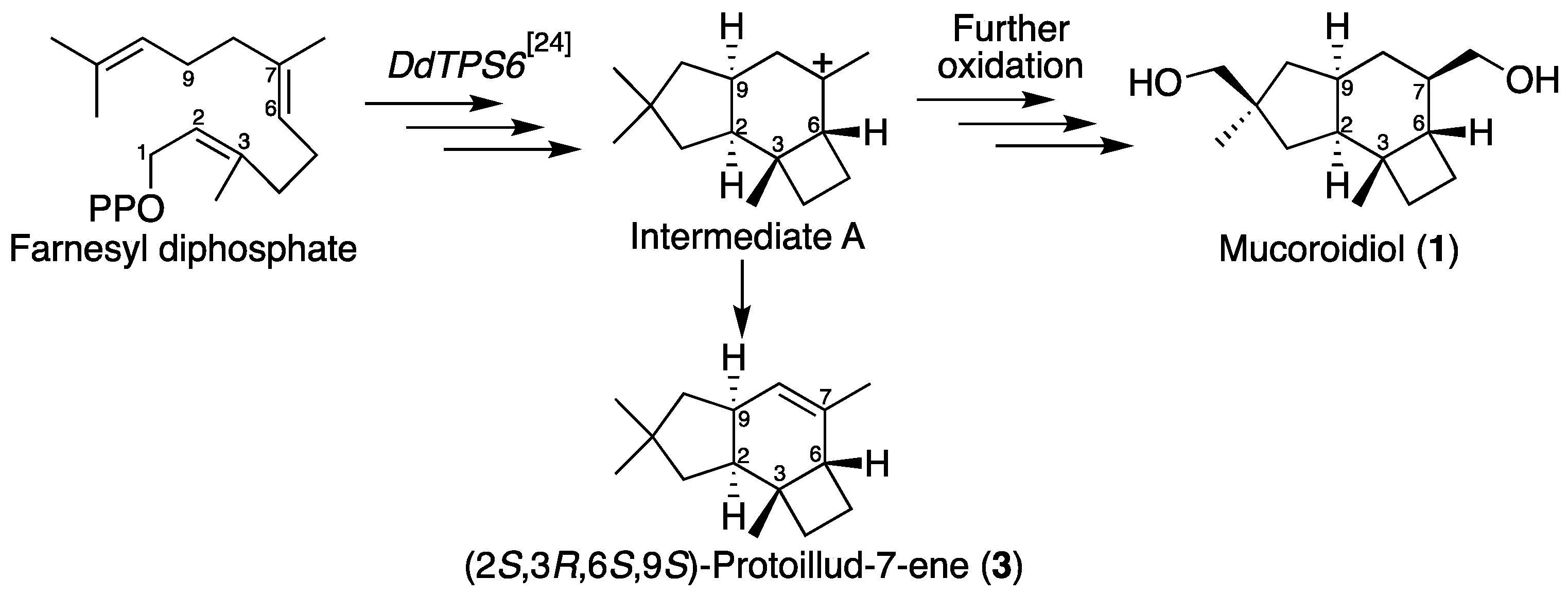
2.1. Isolation and Structural Elucidation of Mucoroidiol
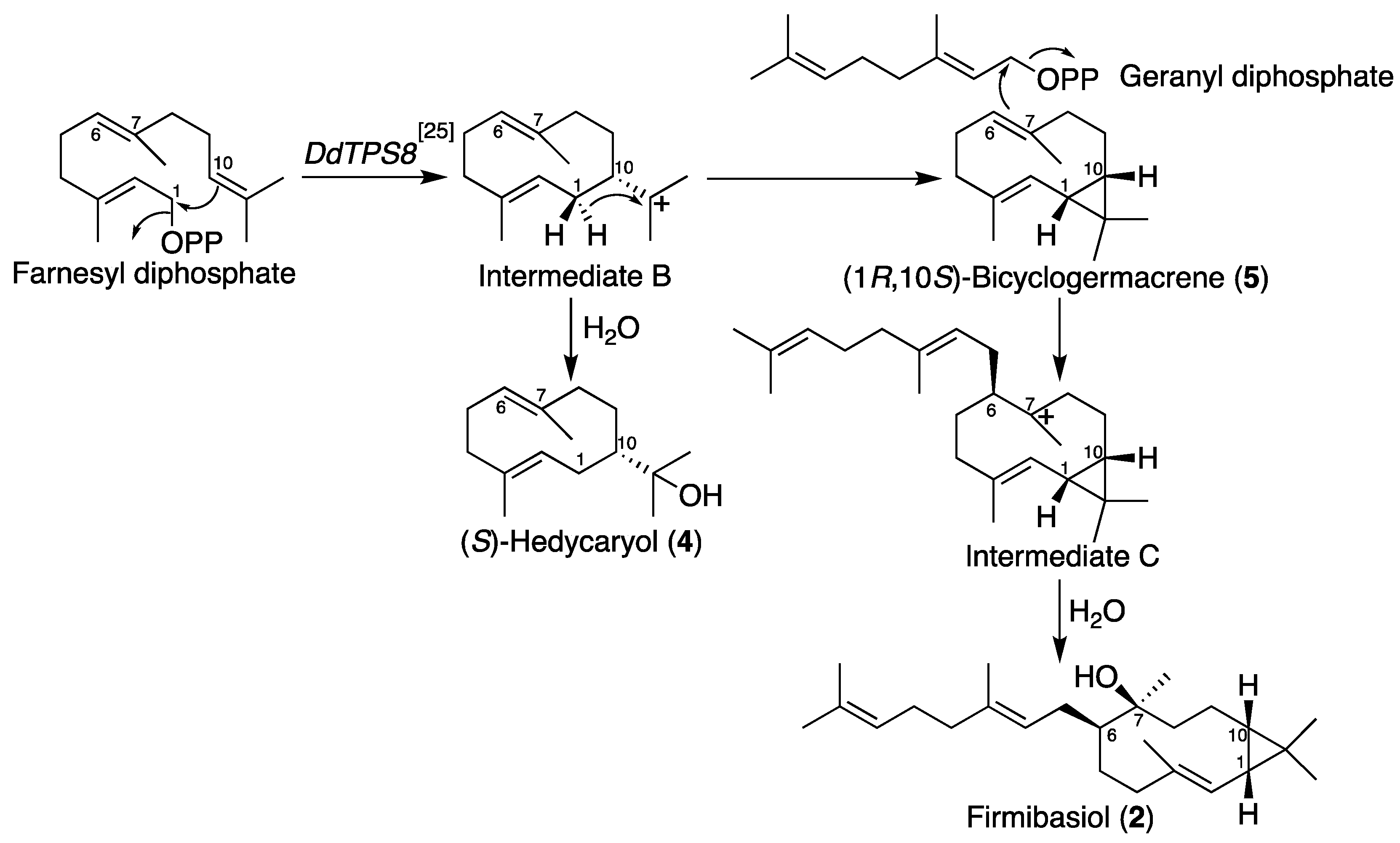
2.2. Isolation and Structural Elucidation of Firmibasiol
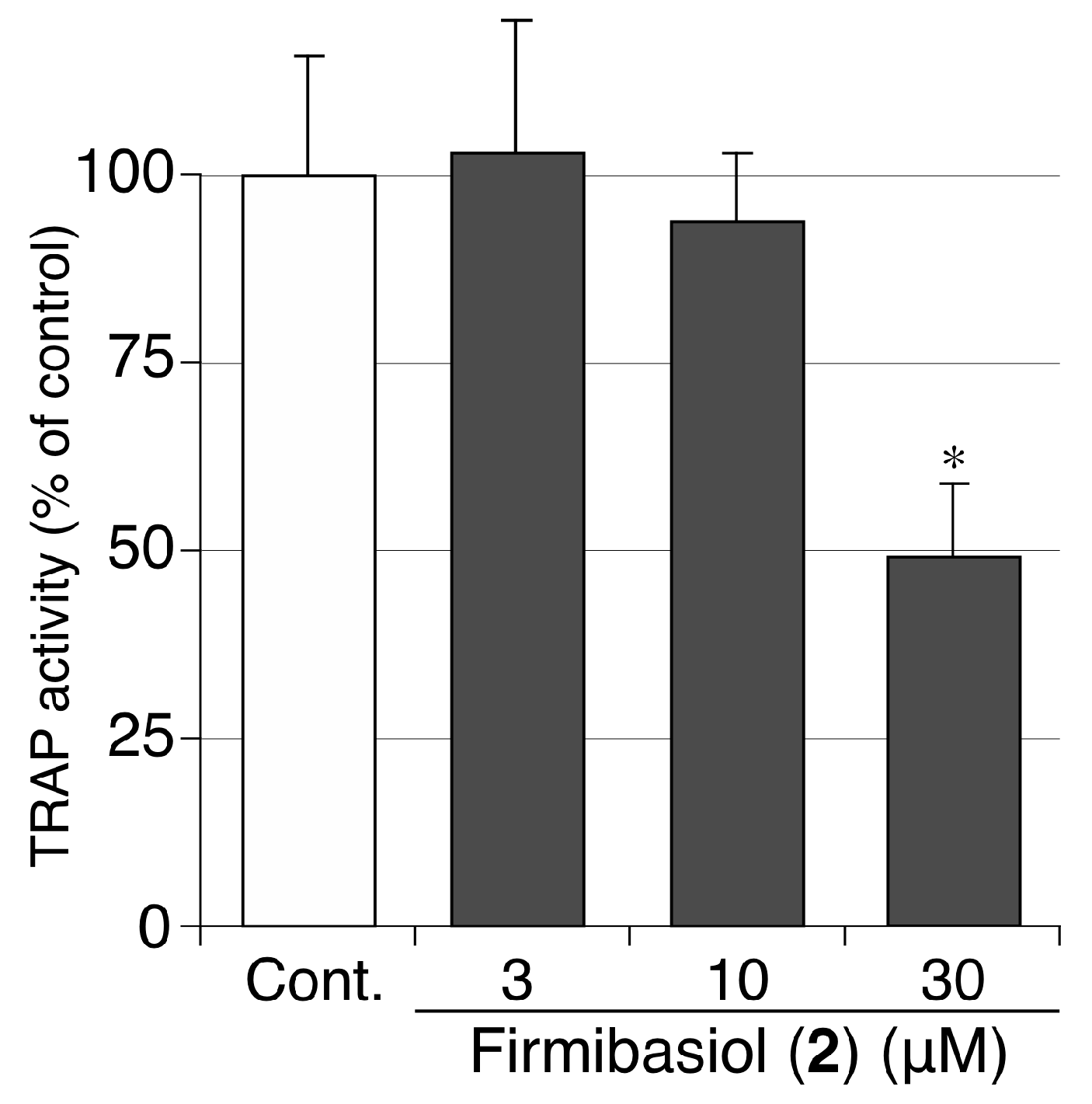
2.3. Biological Activity of Compounds 1 and 2
3. Discussion
4. Materials and Methods
4.1. General Methods
4.2. Organism and Culture Conditions
4.3. Isolation of Mucoroidiol (1)
4.4. Isolation of Firmibasiol (2)
4.5. Cell Proliferation Assay
4.6. Measurement of Minimum Inhibitory Concentration (MIC)
4.7. Osteoclast Differentiation Experiments
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Li, J.W.-H.; Vederas, J.C. Drug Discovery and Natural Products: End of an Era or an Endless Frontier? Science 2009, 325, 161–165. [Google Scholar] [CrossRef] [PubMed]
- Wolfender, J.-L.; Queiroz, E.F. New Approaches for Studying the Chemical Diversity of Natural Resources and the Bioactivity of their Constituents. CHIMIA 2012, 66, 324–329. [Google Scholar] [CrossRef] [PubMed]
- Eichinger, L.; Pachebat, J.; Glockner, G.; Rajandream, M.-A.; Sucgang, R.; Berriman, M.; Song, J.; Olsen, R.; Szafranski, K.; Xu, Q.; et al. The genome of the social amoeba Dictyostelium discoideum. Nature 2005, 435, 43–57. [Google Scholar] [CrossRef] [PubMed]
- Adl, S.; Simpson, A.G.B.; Lane, C.E.; Lukeš, J.; Bass, D.; Bowser, S.S.; Brown, M.W.; Burki, F.; Dunthorn, M.; Hampl, V.; et al. The Revised Classification of Eukaryotes. J. Eukaryot. Microbiol. 2012, 59, 429–493. [Google Scholar] [CrossRef] [PubMed]
- Firtel, R.A.; Meili, R. Dictyostelium: A model for regulated cell movement during morphogenesis. Curr. Opin. Genet. Dev. 2000, 10, 421–427. [Google Scholar] [CrossRef]
- Calvo-Garrido, J.; Carilla-Latorre, S.; Kubohara, Y.; Santos-Rodrigo, N.; Mesquita, A.; Soldati, T.; Golstein, P.; Escalante, R. Autophagy in Dictyostelium: Genes and pathways, cell death and infection. Autophagy 2010, 6, 686–701. [Google Scholar] [CrossRef]
- Nichols, J.M.E.; Veltman, D.; Kay, R.R. Chemotaxis of a model organism: Progress with Dictyostelium. Curr. Opin. Cell Boil. 2015, 36, 7–12. [Google Scholar] [CrossRef]
- Stuelten, C.; Parent, C.A.; Montell, D.J. Cell motility in cancer invasion and metastasis: Insights from simple model organisms. Nat. Rev. Cancer 2018, 18, 296–312. [Google Scholar] [CrossRef]
- Annesley, S.J.; Fisher, P.R. Dictyostelium discoideum–a model for many reasons. Mol. Cell. Biochem. 2009, 329, 73–91. [Google Scholar] [CrossRef]
- Kubohara, Y.; Kikuchi, H. Dictyostelium: An Important Source of Structural and Functional Diversity in Drug Discovery. Cells 2018, 8, 6. [Google Scholar] [CrossRef]
- Takaya, Y.; Kikuchi, H.; Terui, Y.; Komiya, J.; Furukawa, K.-I.; Seya, K.; Motomura, S.; Ito, A.; Oshima, Y. Novel Acyl α-Pyronoids, Dictyopyrone A, B, and C, from Dictyostelium Cellular Slime Molds. J. Org. Chem. 2000, 65, 985–989. [Google Scholar] [CrossRef]
- Kikuchi, H.; Nakamura, K.; Kubohara, Y.; Gokan, N.; Hosaka, K.; Maeda, Y.; Oshima, Y. Dihydrodictyopyrone A and C: New Members of Dictyopyrone Family Isolated from Dictyostelium Cellular Slime Molds. Tetrahedron Lett. 2007, 48, 5905–5909. [Google Scholar] [CrossRef]
- Nguyen, V.H.; Kikuchi, H.; Sasaki, H.; Iizumi, K.; Kubohara, Y.; Oshima, Y. Production of novel bispyrone metabolites in the cellular slime mold Dictyostelium giganteum induced by zinc(II) ion. Tetrahedron 2017, 73, 583–588. [Google Scholar] [CrossRef]
- Kikuchi, H.; Saito, Y.; Komiya, J.; Takaya, Y.; Honma, S.; Nakahata, N.; Ito, A.; Oshima, Y. Furanodictine A and B: Amino Sugar Analogues Produced by Cellular Slime Molds Dictyostelium discoideum Showing Neuronal Differentiation Activity. J. Org. Chem. 2001, 66, 6982–6987. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, H.; Saito, Y.; Sekiya, J.; Okano, Y.; Saito, M.; Nakahata, N.; Kubohara, Y.; Oshima, Y. Isolation and Synthesis of a New Aromatic Compound, Brefelamide, from Dictyostelium Cellular Slime Molds and Its Inhibitory Effect on the Proliferation of Astrocytoma Cells. J. Org. Chem. 2005, 70, 8854–8858. [Google Scholar] [CrossRef]
- Kikuchi, H.; Ishiko, S.; Nakamura, K.; Kubohara, Y.; Oshima, Y. Novel prenylated and geranylated aromatic compounds isolated from Polysphondylium cellular slime molds. Tetrahedron 2010, 66, 6000–6007. [Google Scholar] [CrossRef]
- Kikuchi, H.; Matsuo, Y.; Katou, Y.; Kubohara, Y.; Oshima, Y. Isolation, synthesis, and biological activity of biphenyl and m-terphenyl-type compounds from Dictyostelium cellular slime molds. Tetrahedron 2012, 68, 8884–8889. [Google Scholar] [CrossRef]
- Kikuchi, H.; Ito, I.; Takahashi, K.; Ishigaki, H.; Iizumi, K.; Kubohara, Y.; Oshima, Y. Isolation, Synthesis, and Biological Activity of Chlorinated Alkylresorcinols from Dictyostelium Cellular Slime Molds. J. Nat. Prod. 2017, 80, 2716–2722. [Google Scholar] [CrossRef]
- Zhang, J.; Yamada, O.; Kida, S.; Matsushita, Y.; Murase, S.; Hattori, T.; Kubohara, Y.; Kikuchi, H.; Oshima, Y. Identification of brefelamide as a novel inhibitor of osteopontin that suppresses invasion of A549 lung cancer cells. Oncol. Rep. 2016, 36, 2357–2364. [Google Scholar] [CrossRef]
- Bai, G.; Matsuba, T.; Kikuchi, H.; Chagan-Yasutan, H.; Motoda, H.; Ozuru, R.; Yamada, O.; Oshima, Y.; Hattori, T. Inhibition of inflammatory-molecule synthesis in THP-1 cells stimulated with phorbol 12-myristate 13-acetate by brefelamide derivatives. Int. Immunopharmacol. 2019, 75, 105831. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yamada, O.; Kida, S.; Murase, S.; Hattori, T.; Oshima, Y.; Kikuchi, H. Downregulation of PD-L1 via amide analogues of brefelamide: Alternatives to antibody-based cancer immunotherapy. Exp. Ther. Med. 2020, 19, 3150–3158. [Google Scholar] [CrossRef] [PubMed]
- Kubohara, Y.; Okamoto, K. Specific Induction by Zinc of Dictyostelium Stalk Cell Differentiation. Exp. Cell Res. 1994, 214, 367–372. [Google Scholar] [CrossRef] [PubMed]
- Rabe, P.; Rinkel, J.; Nubbemeyer, B.; Köllner, T.G.; Chen, F.; Dickschat, J.S. Terpene Cyclases from Social Amoebae. Angew. Chem. Int. Ed. 2016, 55, 15420–15423. [Google Scholar] [CrossRef]
- Chen, X.; Luck, K.; Rabe, P.; Dinh, C.Q.; Shaulsky, G.; Nelson, D.R.; Gershenzon, J.; Dickschat, J.S.; Köllner, T.G.; Chen, F. A terpene synthase-cytochrome P450 cluster in Dictyostelium discoideum produces a novel trisnorsesquiterpene. eLife 2019, 8. [Google Scholar] [CrossRef]
- Raola, V.K.; Chakraborty, K. Two rare antioxidative prenylated terpenoids from loop-root Asiatic mangrove Rhizophora mucronata (Family Rhizophoraceae) and their activity against pro-inflammatory cyclooxygenases and lipoxidase. Nat. Prod. Res. 2017, 31, 418–427. [Google Scholar] [CrossRef]
- Asagiri, M.; Takayanagi, H. The molecular understanding of osteoclast differentiation. Bone 2007, 40, 251–264. [Google Scholar] [CrossRef]
- Boyle, W.J.; Simonet, W.S.; Lacey, D.L. Osteoclast differentiation and activation. Nature 2003, 423, 337–342. [Google Scholar] [CrossRef]
- Chen, X.; Köllner, T.G.; Jia, Q.; Norris, A.; Santhanam, B.; Rabe, P.; Dickschat, J.S.; Shaulsky, G.; Gershenzon, J.; Chen, F. Terpene synthase genes in eukaryotes beyond plants and fungi: Occurrence in social amoebae. Proc. Natl. Acad. Sci. USA 2016, 113, 12132–12137. [Google Scholar] [CrossRef]
- Rinkel, J.; Rabe, P.; Chen, X.; Köllner, T.G.; Chen, F.; Dickschat, J.S. Mechanisms of the Diterpene Cyclases β-Pinacene Synthase from Dictyostelium discoideum and Hydropyrene Synthase from Streptomyces clavuligerus. Chem. Eur. J. 2017, 23, 10501–10505. [Google Scholar] [CrossRef]
- Kubohara, Y.; Kikuchi, H.; Matsuo, Y.; Oshima, Y.; Homma, Y. Mitochondria Are the Target Organelle of Differentiation-Inducing Factor-3, an Anti-Tumor Agent Isolated from Dictyostelium discoideum. PLoS ONE 2013, 8, e72118. [Google Scholar] [CrossRef]
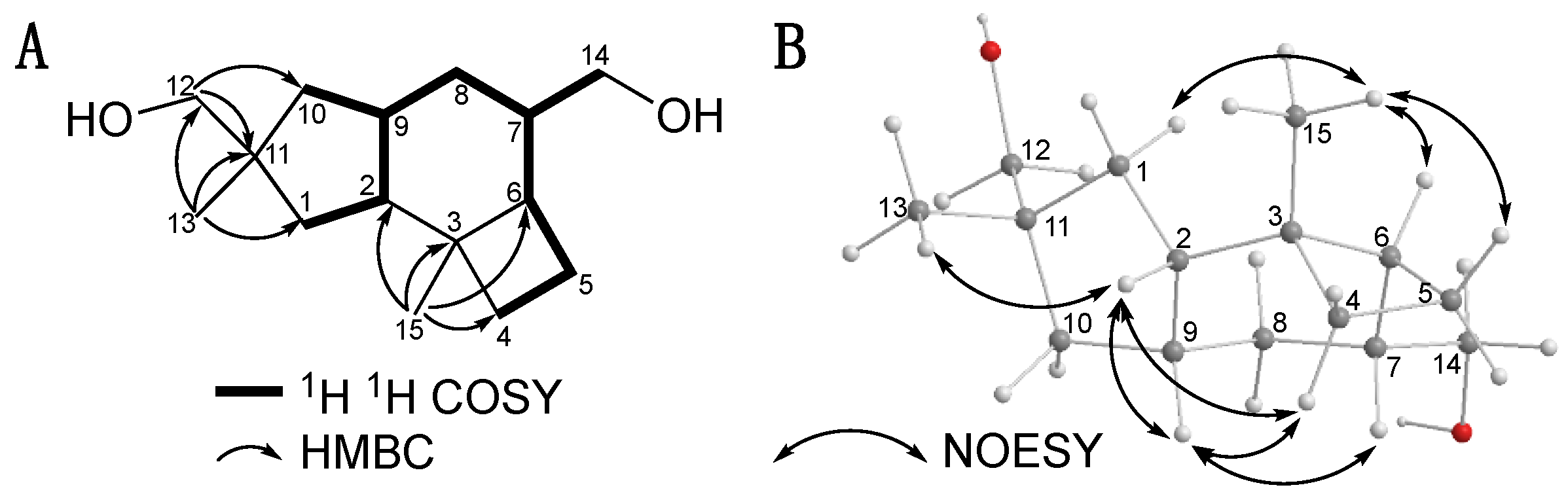
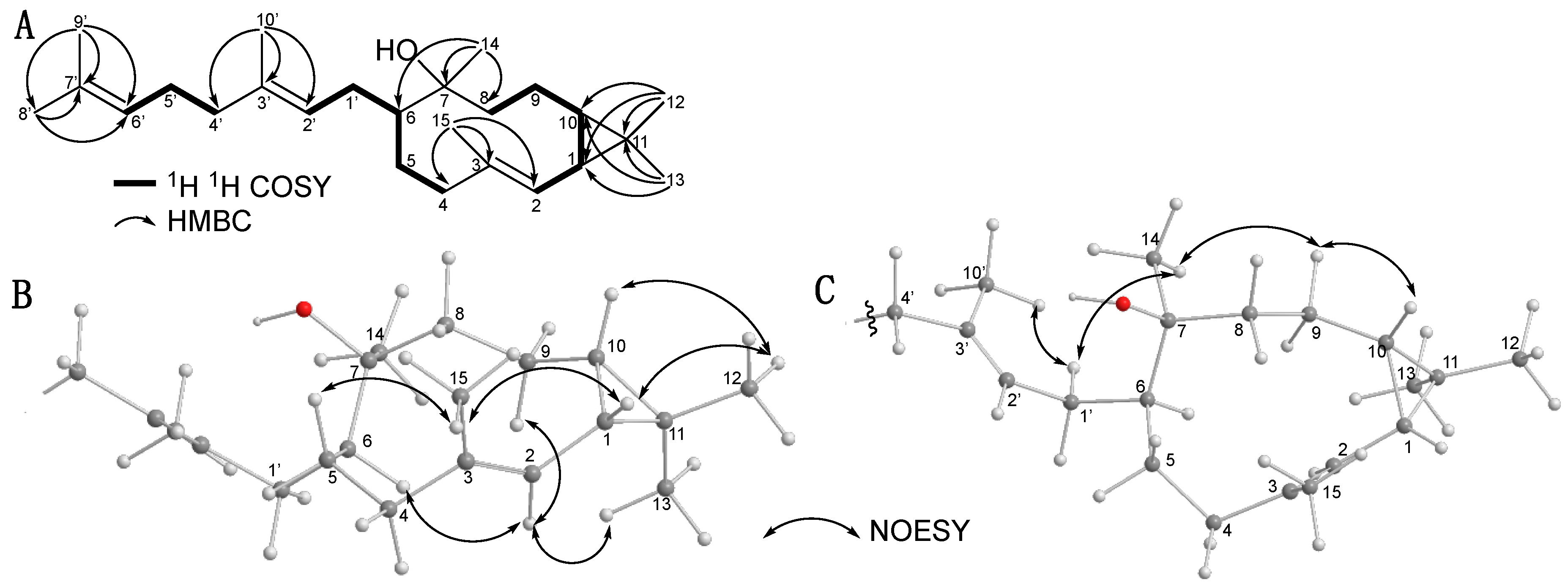
| 13C | (DEPT) | 1H | |
|---|---|---|---|
| 1 | 37.0 | CH2 | 1.23 (1H, dd, J = 12.5, 6.5 Hz) |
| 1.32–1.37 (1H, m) | |||
| 2 | 45.6 | CH | 1.84 (1H, dt, J = 12.9, 6.5 Hz) |
| 3 | 39.0 | C | |
| 4 | 31.7 | CH2 | 1.42–1.47 (1H, m) |
| 2.03 (1H, q, J = 9.4 Hz) | |||
| 5 | 24.4 | CH2 | 1.33–1.37 (1H, m) |
| 2.14–2.21 (1H, m) | |||
| 6 | 40.2 | CH | 1.33–1.39 (1H, m) |
| 7 | 44.7 | CH | 1.60–1.63 (1H, m) |
| 8 | 30.3 | CH2 | 0.78 (1H, q, J = 12.9 Hz) |
| 1.35–1.41 (1H, m) | |||
| 9 | 38.4 | CH | 2.10–2.16 (1H, m) |
| 10 | 42.8 | CH2 | 1.30–1.35 (1H, m) |
| 1.59 (1H, dd, J = 13.5, 7.8 Hz) | |||
| 11 | 41.7 | C | |
| 12 | 73.0 | CH2 | 3.34 (1H, d, J = 10.6 Hz) |
| 3.37 (1H, d, J = 10.6 Hz) | |||
| 13 | 26.5 | CH3 | 0.99 (3H, s) |
| 14 | 67.2 | CH2 | 3.32–3.37 (1H, m) |
| 3.51 (1H, dd, J = 10.5, 4.8 Hz) | |||
| 15 | 26.3 | CH3 | 1.11 (3H, s) |
| 13C | (DEPT) | 1H | |
|---|---|---|---|
| 1 | 25.1 | CH | 1.28 (1H, t, J = 9.0 Hz) |
| 2 | 121.0 | CH | 4.88 (1H, d, J = 9.0 Hz) |
| 3 | 137.2 | C | |
| 4 | 40.5 | CH2 | 1.80 (1H, t, J = 12.0 Hz) |
| 2.07–2.11 (1H, m) | |||
| 5 | 26.7 | CH2 | 1.18–1.25 (1H, m) |
| 1.44–1.51 (1H, m) | |||
| 6 | 42.3 | CH | 1.41–1.45 (1H, m) |
| 7 | 76.0 | C | |
| 8 | 38.3 | CH2 | 1.18–1.27 (2H, m) |
| 9 | 21.3 | CH2 | 0.58 (1H, q, J = 12.0 Hz) |
| 1.50–1.54 (1H, m) | |||
| 10 | 27.8 | CH | 0.66 (1H, ddd, J = 12.0, 9.0, 3.6 Hz) |
| 11 | 20.1 | C | |
| 12 | 28.8 | CH3 | 1.04 (3H, s) |
| 13 | 15.1 | CH3 | 1.11 (3H, s) |
| 14 | 25.8 | CH3 | 1.07 (3H, s) |
| 15 | 17.3 | CH3 | 1.73 (3H, s) |
| 1′ | 30.2 | CH2 | 2.08–2.14 (2H, m) |
| 2′ | 123.1 | CH | 5.27 (1H, t, J = 7.2 Hz) |
| 3′ | 136.2 | C | |
| 4′ | 40.0 | CH2 | 2.03 (2H, t, J = 7.2 Hz) |
| 5′ | 26.8 | CH2 | 2.05–2.10 (2H, m) |
| 6′ | 124.3 | CH | 5.01 (1H, tq, J = 7.2, 1.2 Hz) |
| 7′ | 131.5 | C | |
| 8′ | 25.7 | CH3 | 1.68 (3H, d, J = 7.2, 1.2 Hz) |
| 9′ | 17.7 | CH3 | 1.60 (3H, s) |
| 10′ | 16.3 | CH3 | 1.63 (3H, s) |
| IC50 (μM) vs. | MIC (μM) vs. | |||
|---|---|---|---|---|
| HeLa | S. aureus (MSSA) | S. aureus (MRSA) | E. coli | |
| Mucoroidiol (1) | >40 | >100 | >100 | >100 |
| Firmibasiol (2) | >40 | >100 | >100 | >100 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sasaki, H.; Kubohara, Y.; Ishigaki, H.; Takahashi, K.; Eguchi, H.; Sugawara, A.; Oshima, Y.; Kikuchi, H. Two New Terpenes Isolated from Dictyostelium Cellular Slime Molds. Molecules 2020, 25, 2895. https://doi.org/10.3390/molecules25122895
Sasaki H, Kubohara Y, Ishigaki H, Takahashi K, Eguchi H, Sugawara A, Oshima Y, Kikuchi H. Two New Terpenes Isolated from Dictyostelium Cellular Slime Molds. Molecules. 2020; 25(12):2895. https://doi.org/10.3390/molecules25122895
Chicago/Turabian StyleSasaki, Hitomi, Yuzuru Kubohara, Hirotaka Ishigaki, Katsunori Takahashi, Hiromi Eguchi, Akihiro Sugawara, Yoshiteru Oshima, and Haruhisa Kikuchi. 2020. "Two New Terpenes Isolated from Dictyostelium Cellular Slime Molds" Molecules 25, no. 12: 2895. https://doi.org/10.3390/molecules25122895
APA StyleSasaki, H., Kubohara, Y., Ishigaki, H., Takahashi, K., Eguchi, H., Sugawara, A., Oshima, Y., & Kikuchi, H. (2020). Two New Terpenes Isolated from Dictyostelium Cellular Slime Molds. Molecules, 25(12), 2895. https://doi.org/10.3390/molecules25122895