A Novel G-Quadruplex Binding Protein in Yeast—Slx9
Abstract
1. Introduction
2. Results
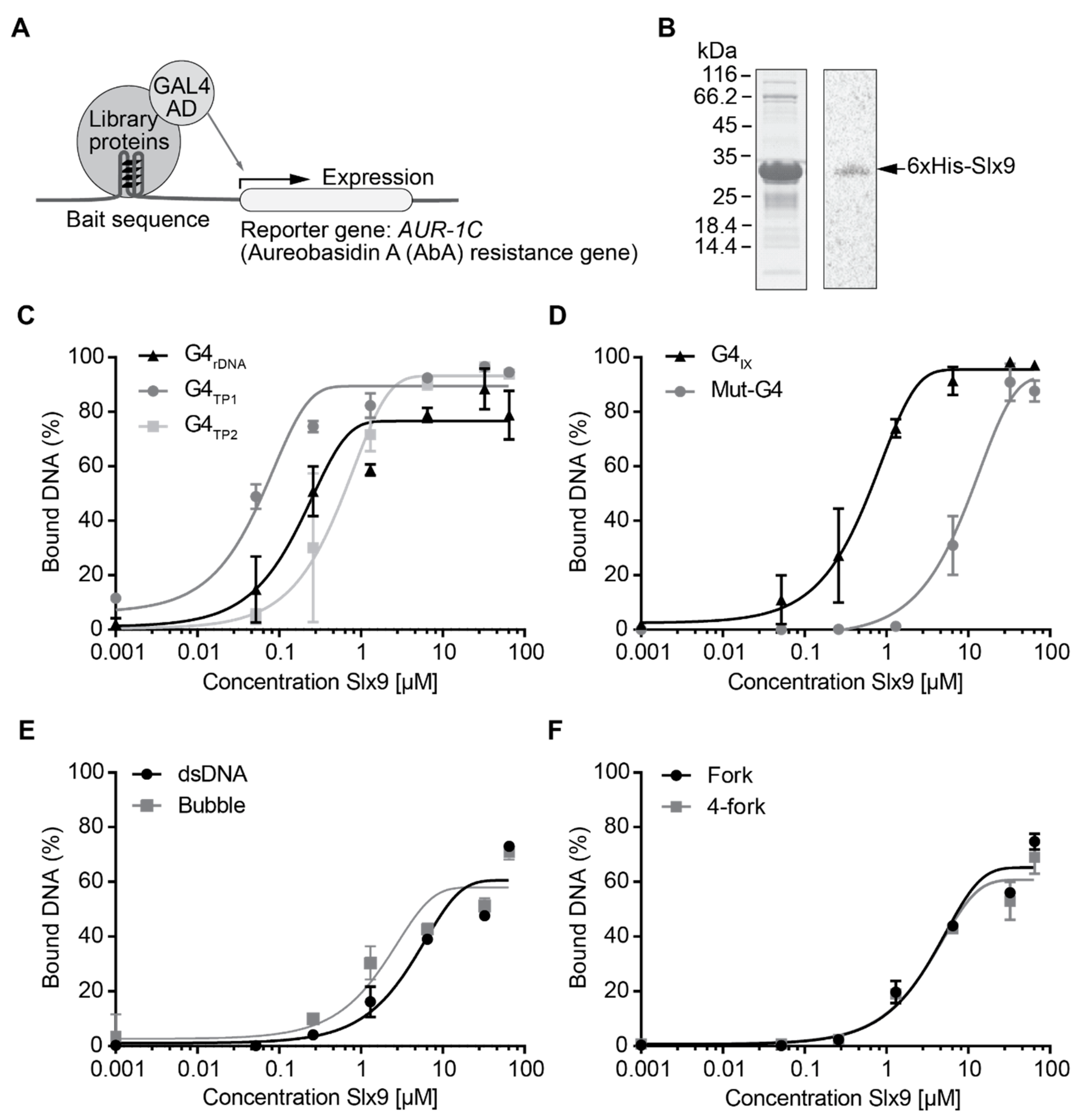
2.1. Identification of Slx9 as a Novel G4-Binding Protein in Vivo
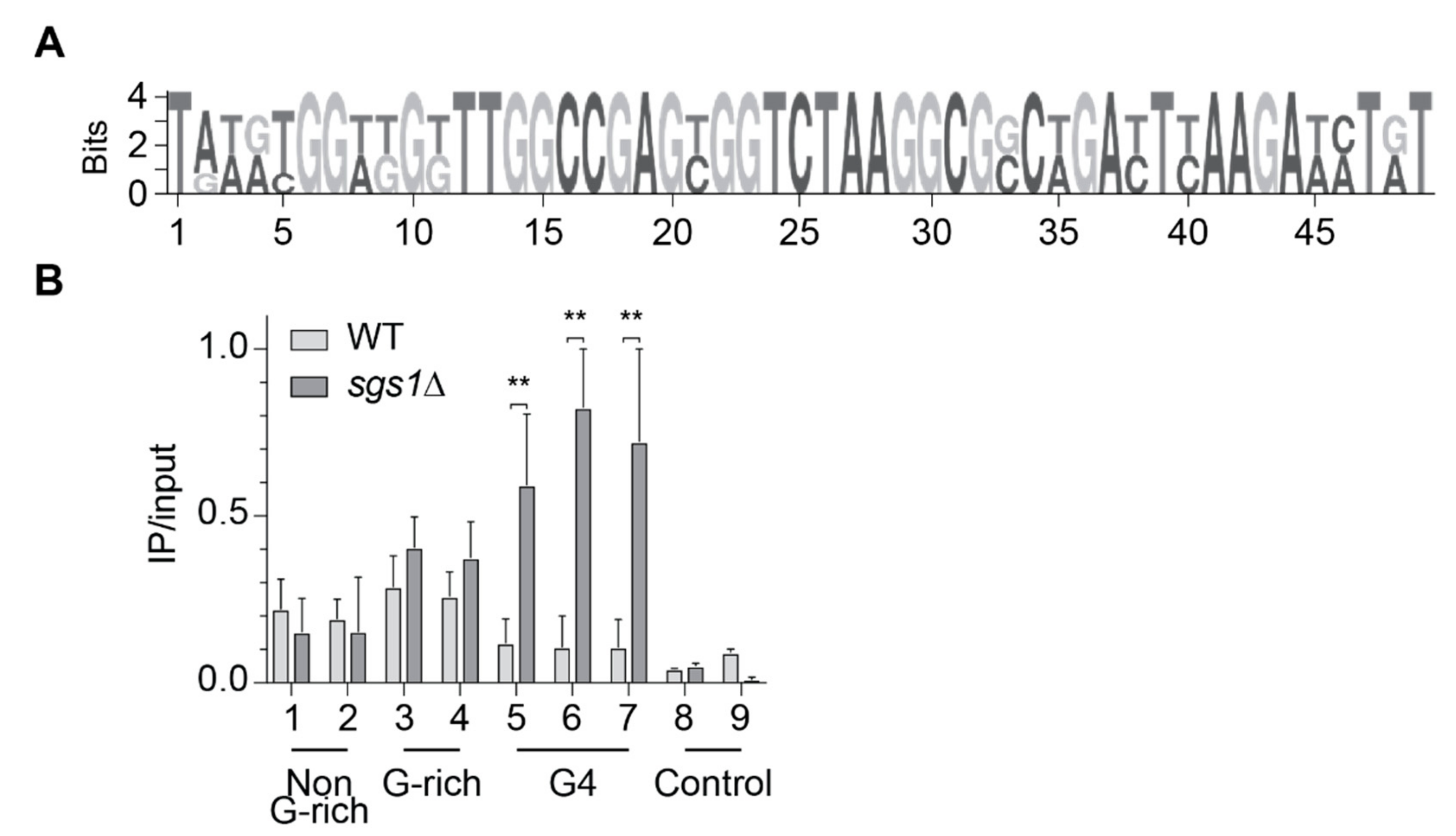
2.2. Slx9 Binds to G-Rich Regions Genome-Wide
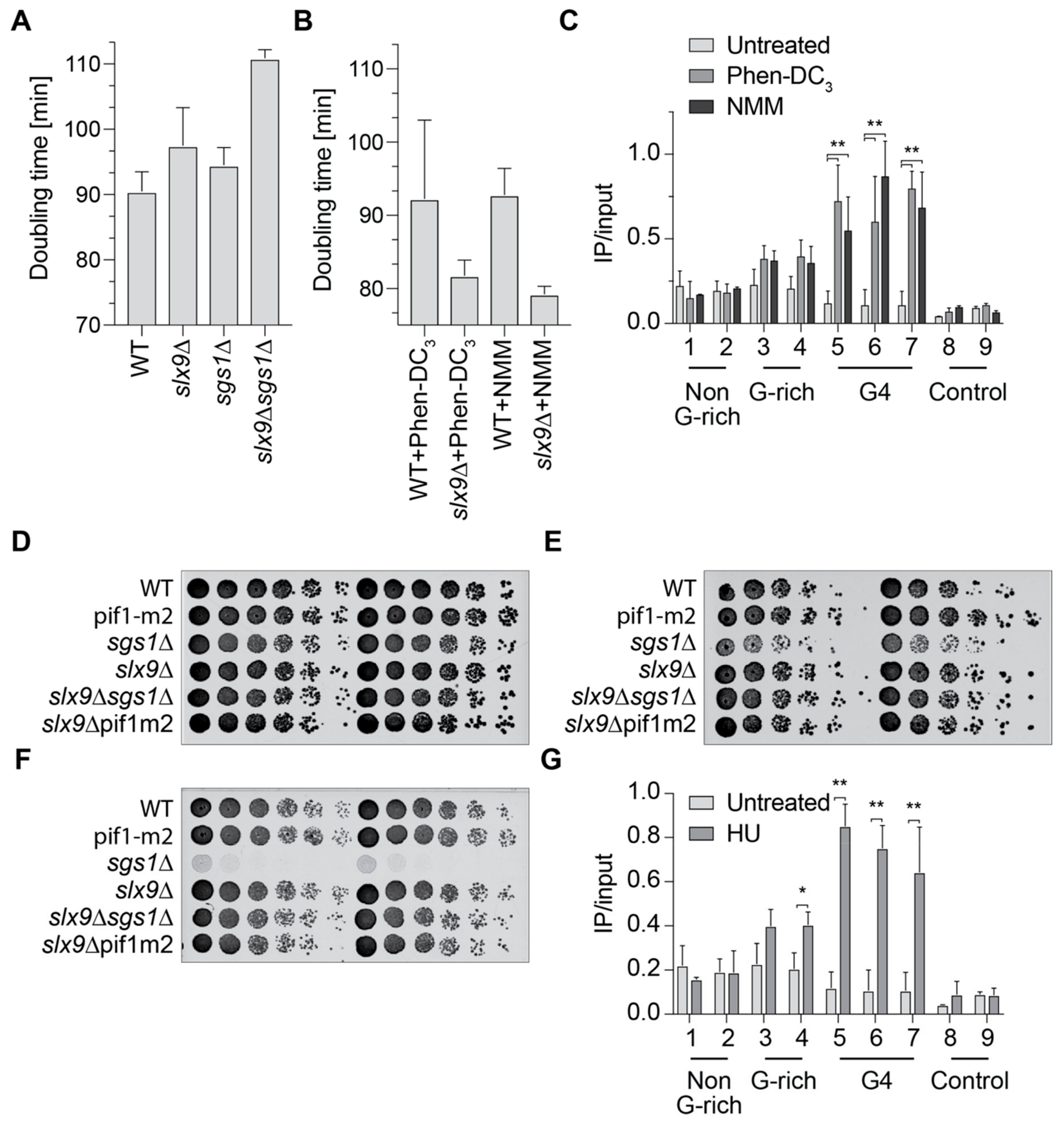
2.3. Slx9 Binds in a Sgs1-Dependent Manner to G4 Motifs
2.4. Slx9 Recognizes G4 Structures That Are Stabilized In Vivo
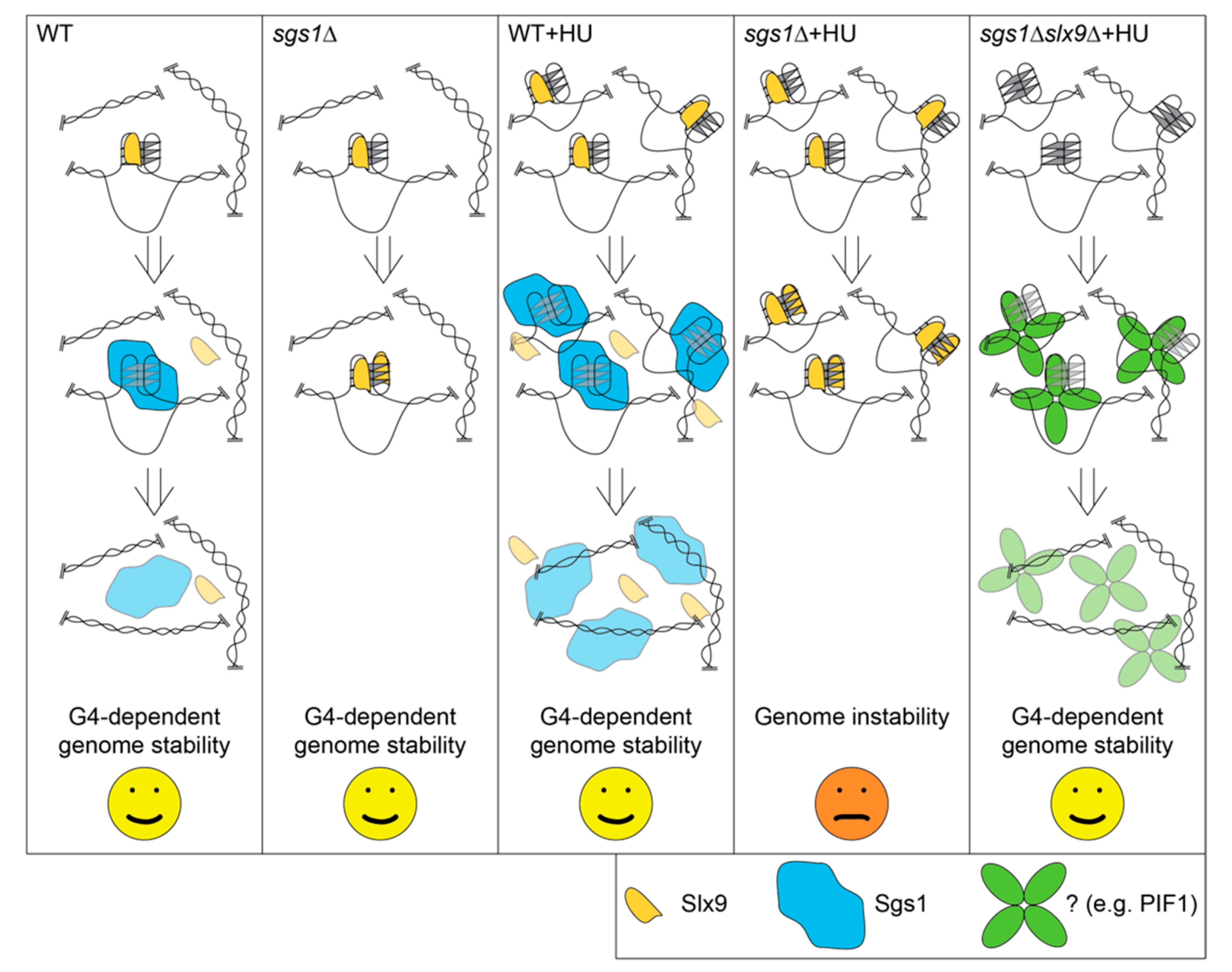
2.5. Slx9 Binding to G4 Structures Affects the “Repair” of These Structures
3. Discussion
4. Materials and Methods
4.1. Yeast One-Hybrid Screen
4.2. Cloning, Expression, and Purification of Slx9
4.3. In Vitro Folding and Analysis of G4 Structures and Annealing of Control DNA Structures
4.4. Binding Studies
4.5. ChIP-Seq -and ChIP Plus qPCR Analysis
4.6. Growth Assay
4.7. Spot Assay
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bikard, D.; Loot, C.; Baharoglu, Z.; Mazel, D. Folded DNA in Action: Hairpin Formation and Biological Functions in Prokaryotes. Microbiol. Mol. Biol. Rev. 2010, 74, 570–588. [Google Scholar] [CrossRef]
- Bochman, M.L.; Paeschke, K.; Zakian, V.A. DNA Secondary Structures: Stability and Function of G-Quadruplex Structures. Nat. Rev. Genet. 2012, 13, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Doherty, E.A.; Doudna, J.A. Ribozyme Structures and Mechanisms. Annu. Rev. Biochem. 2000, 69, 597–615. [Google Scholar] [CrossRef]
- Mirkin, S.M. Discovery of Alternative DNA Structures: A Heroic Decade (1979–1989). Front. Biosci. 2008, 13, 1064–1071. [Google Scholar] [CrossRef]
- Rhodes, D.; Lipps, H.J. G-Quadruplexes and Their Regulatory Roles in Biology. Nucleic Acids Res. 2015, 43, 8627–8837. [Google Scholar] [CrossRef]
- Chambers, V.S.; Marsico, G.; Boutell, J.M.; di Antonio, M.; Smith, G.P.; Balasubramanian, S. High-Throughput Sequencing of DNA G-Quadruplex Structures in the Human Genome. Nat. Biotechnol. 2015, 33, 877–881. [Google Scholar] [CrossRef] [PubMed]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative Visualization of DNA G-Quadruplex Structures in Human Cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef]
- Paeschke, K.; Capra, J.A.; Zakian, V.A. DNA Replication through G-Quadruplex Motifs Is Promoted by the Saccharomyces Cerevisiae Pif1 DNA Helicase. Cell 2011, 145, 678–691. [Google Scholar] [CrossRef] [PubMed]
- Juranek, S.A.; Paeschke, K. Cell Cycle Regulation of G-Quadruplex DNA Structures at Telomeres. Curr. Pharm. Des. 2012, 18, 1867–1872. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui-Jain, A.; Grand, C.L.; Bearss, D.J.; Hurley, L.H. Direct Evidence for a G-Quadruplex in a Promoter Region and Its Targeting with a Small Molecule to Repress C-Myc Transcription. Proc. Natl. Acad. Sci. USA 2002, 99, 11593–11598. [Google Scholar] [CrossRef]
- Wolfe, A.L.; Singh, K.; Zhong, Y.; Drewe, P.; Rajasekhar, V.K.; Sanghvi, V.R.; Mavrakis, K.J.; Jiang, M.; Roderick, J.E.; Van der Meulen, J.; et al. Rna G-Quadruplexes Cause Eif4a-Dependent Oncogene Translation in Cancer. Nature 2014, 513, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Besnard, E.; Babled, A.; Lapasset, L.; Milhavet, O.; Parrinello, H.; Dantec, C.; Marin, J.M.; Lemaitre, J.M. Unraveling Cell Type-Specific and Reprogrammable Human Replication Origin Signatures Associated with G-Quadruplex Consensus Motifs. Nat Struct. Mol. Biol. 2012, 19, 837–844. [Google Scholar] [CrossRef] [PubMed]
- Paeschke, K.; Simonsson, T.; Postberg, J.; Rhodes, D.; Lipps, H.J. Telomere End-Binding Proteins Control the Formation of G-Quadruplex DNA Structures in Vivo. Nat. Struct. Mol. Biol. 2005, 12, 847–854. [Google Scholar] [CrossRef]
- Hershman, S.G.; Chen, Q.; Lee, J.Y.; Kozak, M.L.; Yue, P.; Wang, L.S.; Johnson, F.B. Genomic Distribution and Functional Analyses of Potential G-Quadruplex-Forming Sequences in Saccharomyces Cerevisiae. Nucleic Acids Res. 2008, 36, 144–156. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Hurley, L.H.; Neidle, S. Targeting G-Quadruplexes in Gene Promoters: A Novel Anticancer Strategy? Nat. Rev. Drug Discov. 2011, 10, 261–275. [Google Scholar] [CrossRef] [PubMed]
- Huppert, J.L.; Balasubramanian, S. G-Quadruplexes in Promoters Throughout the Human Genome. Nucleic Acids Res. 2007, 35, 406–413. [Google Scholar] [CrossRef]
- Capra, J.A.; Paeschke, K.; Singh, M.; Zakian, V.A. G-Quadruplex DNA Sequences Are Evolutionarily Conserved and Associated with Distinct Genomic Features in Saccharomyces Cerevisiae. PLoS Comput. Biol. 2010, 6. [Google Scholar] [CrossRef] [PubMed]
- Simonsson, T.; Pecinka, P.; Kubista, M. DNA Tetraplex Formation in the Control Region of C-Myc. Nucleic Acids Res. 1998, 26, 1167–1172. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, O.; Bourdoncle, A.; Boule, J.B.; Brosh, R.M., Jr.; Mergny, J.L. G-Quadruplexes and Helicases. Nucleic Acids Res. 2016, 44, 1989–2006. [Google Scholar] [CrossRef]
- Bharti, S.K.; Sommers, J.A.; Zhou, J.; Kaplan, D.L.; Spelbrink, J.N.; Mergny, J.L.; Brosh, R.M., Jr. DNA Sequences Proximal to Human Mitochondrial DNA Deletion Breakpoints Prevalent in Human Disease Form G-Quadruplexes, a Class of DNA Structures Inefficiently Unwound by the Mitochondrial Replicative Twinkle Helicase. J. Biol. Chem. 2014, 289, 29975–29993. [Google Scholar] [CrossRef]
- Cheung, I.; Schertzer, M.; Rose, A.; Lansdorp, P.M. Disruption of Dog-1 in Caenorhabditis Elegans Triggers Deletions Upstream of Guanine-Rich DNA. Nat. Genet. 2002, 31, 405–409. [Google Scholar] [CrossRef]
- Tarsounas, M.; Tijsterman, M. Genomes and G-Quadruplexes: For Better or for Worse. J. Mol. Biol. 2013, 425, 4782–4789. [Google Scholar] [CrossRef]
- Sauer, M.; Paeschke, K. G-Quadruplex Unwinding Helicases and Their Function in Vivo. Biochem. Soc. Trans 2017, 45, 1173–1182. [Google Scholar] [CrossRef]
- Brazda, V.; Haronikova, L.; Liao, J.C.; Fojta, M. DNA and Rna Quadruplex-Binding Proteins. Int. J. Mol. Sci. 2014, 15, 17493–17517. [Google Scholar] [CrossRef]
- Maizels, N. G4-Associated Human Diseases. EMBO Rep. 2015, 16, 910–922. [Google Scholar] [CrossRef]
- Paeschke, K.; Bochman, M.L.; Garcia, P.D.; Cejka, P.; Friedman, K.L.; Kowalczykowski, S.C.; Zakian, V.A. Pif1 Family Helicases Suppress Genome Instability at G-Quadruplex Motifs. Nature 2013, 497, 458–462. [Google Scholar] [CrossRef]
- Smith, J.S.; Chen, Q.; Yatsunyk, L.A.; Nicoludis, J.M.; Garcia, M.S.; Kranaster, R.; Balasubramanian, S.; Monchaud, D.; Teulade-Fichou, M.P.; Abramowitz, L.; et al. Rudimentary G-Quadruplex-Based Telomere Capping in Saccharomyces Cerevisiae. Nat. Struct. Mol. Biol. 2011, 18, 478–485. [Google Scholar] [CrossRef]
- Wanzek, K.; Schwindt, E.; Capra, J.; Paeschke, K. Mms1 Binds to G-Rich Regions in Saccharomyces Cerevisiae and Influences Replication and Genome Stability. Nucleic Acids Res. 2017, 45, 7796–7806. [Google Scholar] [CrossRef][Green Version]
- Ooi, S.L.; Shoemaker, D.D.; Boeke, J.D. DNA Helicase Gene Interaction Network Defined Using Synthetic Lethality Analyzed by Microarray. Nat. Genet. 2003, 35, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Chung, W.H.; Shim, E.Y.; Lee, S.E.; Ira, G. Sgs1 Helicase and Two Nucleases Dna2 and Exo1 Resect DNA Double-Strand Break Ends. Cell 2008, 134, 981–994. [Google Scholar] [CrossRef]
- Sun, H.; Bennett, R.J.; Maizels, N. The Saccharomyces Cerevisiae Sgs1 Helicase Efficiently Unwinds G-G Paired Dnas. Nucleic Acids Res. 1999, 27, 1978–1984. [Google Scholar] [CrossRef]
- Papadopoulou, C.; Guilbaud, G.; Schiavone, D.; Sale, J.E. Nucleotide Pool Depletion Induces G-Quadruplex-Dependent Perturbation of Gene Expression. Cell Rep. 2015, 13, 2491–2503. [Google Scholar] [CrossRef]
- Bachrati, C.Z.; Hickson, I.D. Analysis of the DNA Unwinding Activity of Recq Family Helicases. Methods Enzymol. 2006, 409, 86–100. [Google Scholar]
- Mohaghegh, P.; Karow, J.K.; Brosh, R.M., Jr.; Bohr, V.A.; Hickson, I.D. The Bloom’s and Werner’s Syndrome Proteins Are DNA Structure-Specific Helicases. Nucleic Acids Res. 2001, 29, 2843–2849. [Google Scholar] [CrossRef]
- Wong, I.; Lohman, T.M. A Double-Filter Method for Nitrocellulose-Filter Binding: Application to Protein-Nucleic Acid Interactions. Proc. Natl. Acad. Sci. USA 1993, 90, 5428–5432. [Google Scholar] [CrossRef]
- Zhang, W.; Duhr, S.; Baaske, P.; Laue, E. Microscale Thermophoresis for the Assessment of Nuclear Protein-Binding Affinities. Methods Mol. Biol. 2014, 1094, 269–276. [Google Scholar]
- Steinmetz, E.J.; Warren, C.L.; Kuehner, J.N.; Panbehi, B.; Ansari, A.Z.; Brow, D.A. Genome-Wide Distribution of Yeast Rna Polymerase Ii and Its Control by Sen1 Helicase. Mol. Cell 2006, 24, 735–746. [Google Scholar] [CrossRef]
- Szilard, R.K.; Jacques, P.E.; Laramee, L.; Cheng, B.; Galicia, S.; Bataille, A.R.; Yeung, M.; Mendez, M.; Bergeron, M.; Robert, F.; et al. Systematic Identification of Fragile Sites Via Genome-Wide Location Analysis of Gamma-H2ax. Nat. Struct. Mol. Biol. 2010, 17, 299–305. [Google Scholar] [CrossRef]
- Guedin, A.; Gros, J.; Alberti, P.; Mergny, J.L. How Long Is Too Long? Effects of Loop Size on G-Quadruplex Stability. Nucleic Acids Res. 2010, 38, 7858–7868. [Google Scholar] [CrossRef]
- Mukundan, V.T.; Phan, A.T. Bulges in G-Quadruplexes: Broadening the Definition of G-Quadruplex-Forming Sequences. J. Am. Chem. Soc. 2013, 135, 5017–5028. [Google Scholar] [CrossRef]
- Tippana, R.; Xiao, W.; Myong, S. G-Quadruplex Conformation and Dynamics Are Determined by Loop Length and Sequence. Nucleic Acids Res. 2014, 42, 8106–8114. [Google Scholar] [CrossRef]
- Agrawal, P.; Lin, C.; Mathad, R.I.; Carver, M.; Yang, D. The Major G-Quadruplex Formed in the Human Bcl-2 Proximal Promoter Adopts a Parallel Structure with a 13-Nt Loop in K+ Solution. J. Am. Chem. Soc. 2014, 136, 1750–1753. [Google Scholar] [CrossRef]
- Piazza, A.; Boule, J.B.; Lopes, J.; Mingo, K.; Largy, E.; Teulade-Fichou, M.P.; Nicolas, A. Genetic Instability Triggered by G-Quadruplex Interacting Phen-Dc Compounds in Saccharomyces Cerevisiae. Nucleic Acids Res. 2010, 38, 4337–4348. [Google Scholar] [CrossRef]
- Saffi, J.; Pereira, V.R.; Henriques, J.A. Importance of the Sgs1 Helicase Activity in DNA Repair of Saccharomyces Cerevisiae. Curr. Genet. 2000, 37, 75–78. [Google Scholar] [CrossRef]
- Mullen, J.R.; Kaliraman, V.; Ibrahim, S.S.; Brill, S.J. Requirement for Three Novel Protein Complexes in the Absence of the Sgs1 DNA Helicase in Saccharomyces Cerevisiae. Genetics 2001, 157, 103–118. [Google Scholar]
- Frei, C.; Gasser, S.M. The Yeast Sgs1p Helicase Acts Upstream of Rad53p in the DNA Replication Checkpoint and Colocalizes with Rad53p in S-Phase-Specific Foci. Genes Dev. 2000, 14, 81–96. [Google Scholar]
- Gangloff, S.; Soustelle, C.; Fabre, F. Homologous Recombination Is Responsible for Cell Death in the Absence of the Sgs1 and Srs2 Helicases. Nat. Genet. 2000, 25, 192–194. [Google Scholar] [CrossRef]
- Putnam, C.D.; Kolodner, R.D. Determination of Gross Chromosomal Rearrangement Rates. Cold Spring Harb. Protoc. 2010, 2010. [Google Scholar] [CrossRef]
- Chen, C.; Kolodner, R.D. Gross Chromosomal Rearrangements in Saccharomyces Cerevisiae Replication and Recombination Defective Mutants. Nat. Genet. 1999, 23, 81–85. [Google Scholar] [CrossRef]
- Hall, B.M.; Ma, C.X.; Liang, P.; Singh, K.K. Fluctuation Analysis Calculator: A Web Tool for the Determination of Mutation Rate Using Luria-Delbruck Fluctuation Analysis. Bioinformatics 2009, 25, 1564–1565. [Google Scholar] [CrossRef]
- Bianchi, V.; Pontis, E.; Reichard, P. Changes of Deoxyribonucleoside Triphosphate Pools Induced by Hydroxyurea and Their Relation to DNA Synthesis. J. Biol. Chem. 1986, 261, 16037–16042. [Google Scholar]
- Koc, A.; Wheeler, L.J.; Mathews, C.K.; Merrill, G.F. Hydroxyurea Arrests DNA Replication by a Mechanism That Preserves Basal Dntp Pools. J. Biol. Chem. 2004, 279, 223–230. [Google Scholar] [CrossRef]
- Bax, R.; Raue, H.A.; Vos, J.C. Slx9p Facilitates Efficient Its1 Processing of Pre-Rrna in Saccharomyces Cerevisiae. RNA 2006, 12, 2005–2013. [Google Scholar] [CrossRef][Green Version]
- Kruisselbrink, E.; Guryev, V.; Brouwer, K.; Pontier, D.B.; Cuppen, E.; Tijsterman, M. Mutagenic Capacity of Endogenous G4 DNA Underlies Genome Instability in Fancj-Defective C. Elegans. Curr. Biol. 2008, 18, 900–905. [Google Scholar] [CrossRef]
- Lopes, J.; Piazza, A.; Bermejo, R.; Kriegsman, B.; Colosio, A.; Teulade-Fichou, M.P.; Foiani, M.; Nicolas, A. G-Quadruplex-Induced Instability During Leading-Strand Replication. EMBO J. 2011, 30, 4033–4046. [Google Scholar] [CrossRef]
- Lemmens, B.; van Schendel, R.; Tijsterman, M. Mutagenic Consequences of a Single G-Quadruplex Demonstrate Mitotic Inheritance of DNA Replication Fork Barriers. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef]
- London, T.B.; Barber, L.J.; Mosedale, G.; Kelly, G.P.; Balasubramanian, S.; Hickson, I.D.; Boulton, S.J.; Hiom, K. Fancj Is a Structure-Specific DNA Helicase Associated with the Maintenance of Genomic G/C Tracts. J. Biol. Chem. 2008, 283, 36132–36139. [Google Scholar] [CrossRef]
- Maniatis, T.; Fritsch, E.F.; Sambrook, J. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor: New York, NY, USA, 1982. [Google Scholar]
- Azvolinsky, A.; Giresi, P.G.; Lieb, J.D.; Zakian, V.A. Highly Transcribed Rna Polymerase Ii Genes Are Impediments to Replication Fork Progression in Saccharomyces Cerevisiae. Mol. Cell 2009, 34, 722–734. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and Memory-Efficient Alignment of Short DNA Sequences to the Human Genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-Based Analysis of Chip-Seq (Macs). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. Meme Suite: Tools for Motif Discovery and Searching. Nucleic Acids Res. 2009, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Longtine, M.S.; McKenzie, A., 3rd; Demarini, D.J.; Shah, N.G.; Wach, A.; Brachat, A.; Philippsen, P.; Pringle, J.R. Additional Modules for Versatile and Economical Pcr-Based Gene Deletion and Modification in Saccharomyces Cerevisiae. Yeast 1998, 14, 953–961. [Google Scholar] [CrossRef]
Sample Availability: Samples of the yeast strains are available from the authors on request. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Götz, S.; Pandey, S.; Bartsch, S.; Juranek, S.; Paeschke, K. A Novel G-Quadruplex Binding Protein in Yeast—Slx9. Molecules 2019, 24, 1774. https://doi.org/10.3390/molecules24091774
Götz S, Pandey S, Bartsch S, Juranek S, Paeschke K. A Novel G-Quadruplex Binding Protein in Yeast—Slx9. Molecules. 2019; 24(9):1774. https://doi.org/10.3390/molecules24091774
Chicago/Turabian StyleGötz, Silvia, Satyaprakash Pandey, Sabrina Bartsch, Stefan Juranek, and Katrin Paeschke. 2019. "A Novel G-Quadruplex Binding Protein in Yeast—Slx9" Molecules 24, no. 9: 1774. https://doi.org/10.3390/molecules24091774
APA StyleGötz, S., Pandey, S., Bartsch, S., Juranek, S., & Paeschke, K. (2019). A Novel G-Quadruplex Binding Protein in Yeast—Slx9. Molecules, 24(9), 1774. https://doi.org/10.3390/molecules24091774





