The Presence and Localization of G-Quadruplex Forming Sequences in the Domain of Bacteria
Abstract
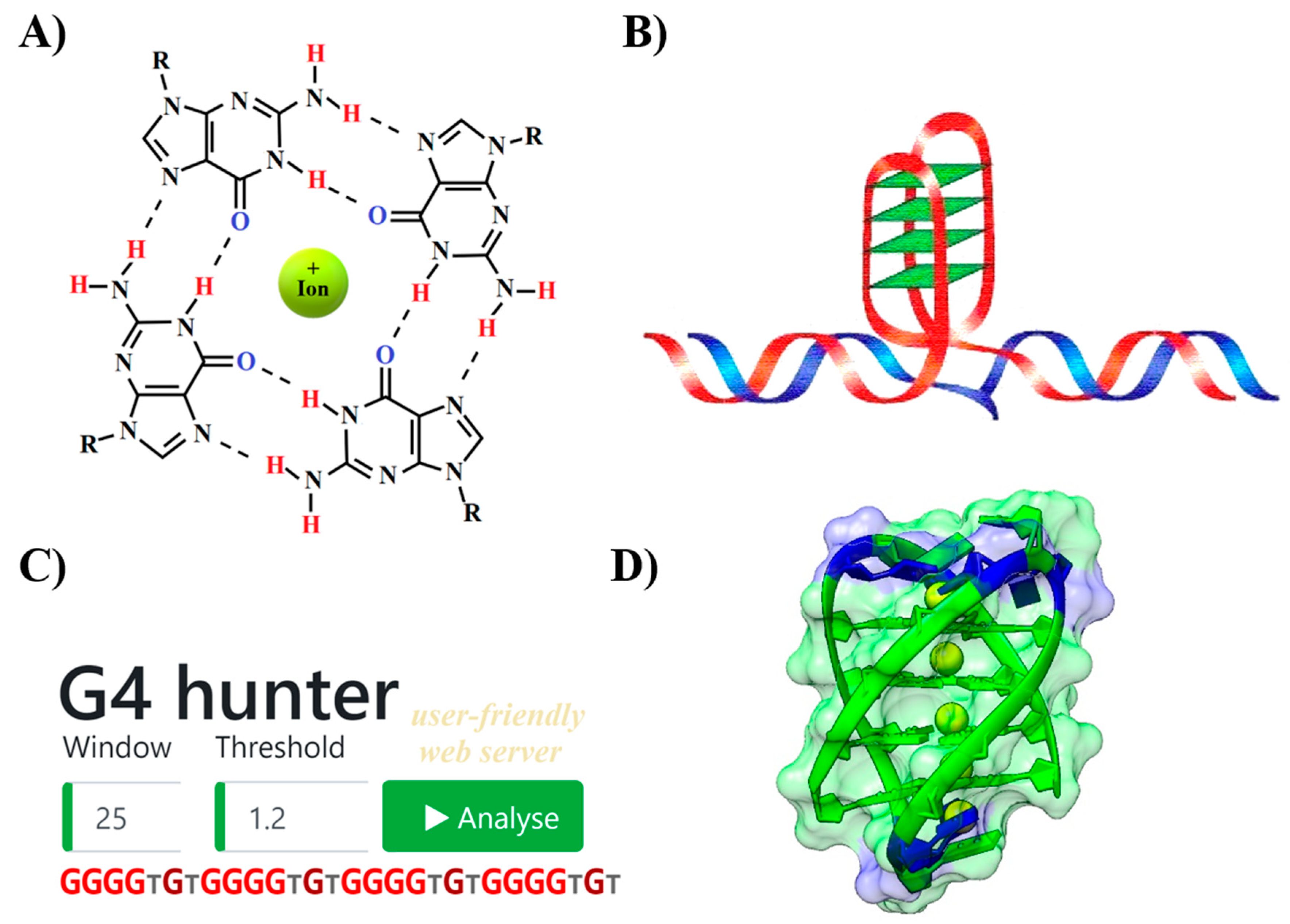
1. Introduction
2. Results
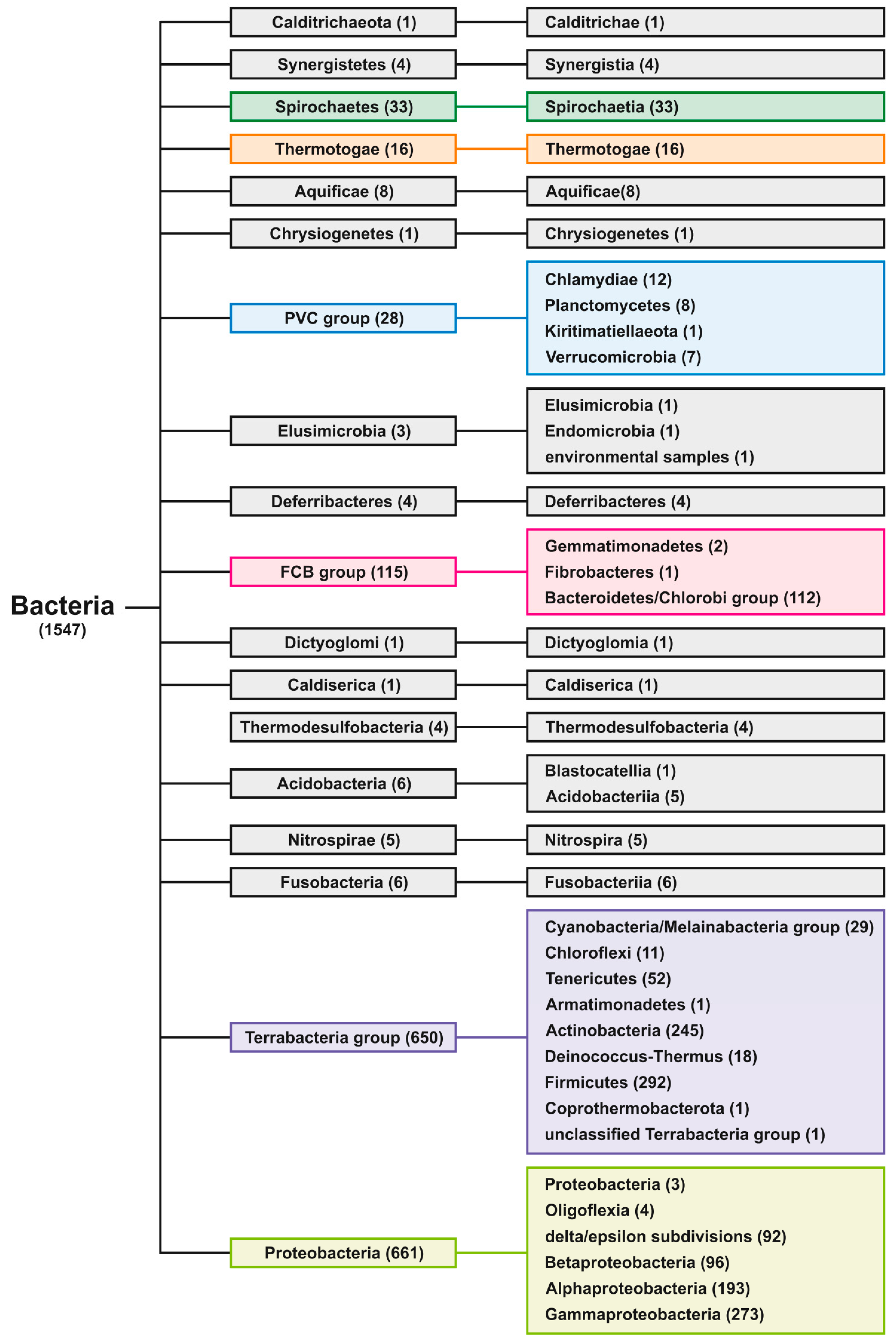
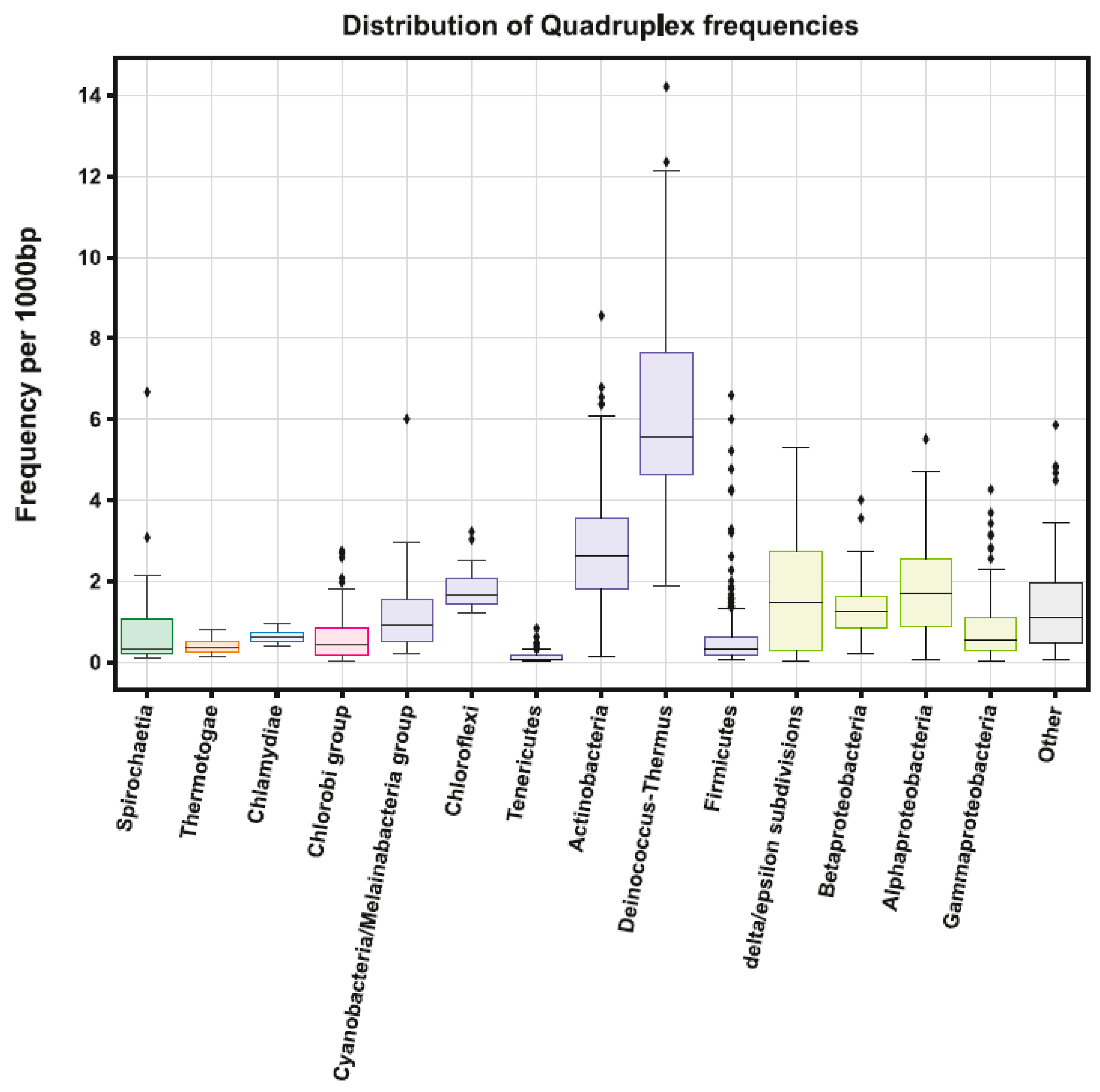
2.1. Variation in Frequency for G4-forming Sequences in Bacteria
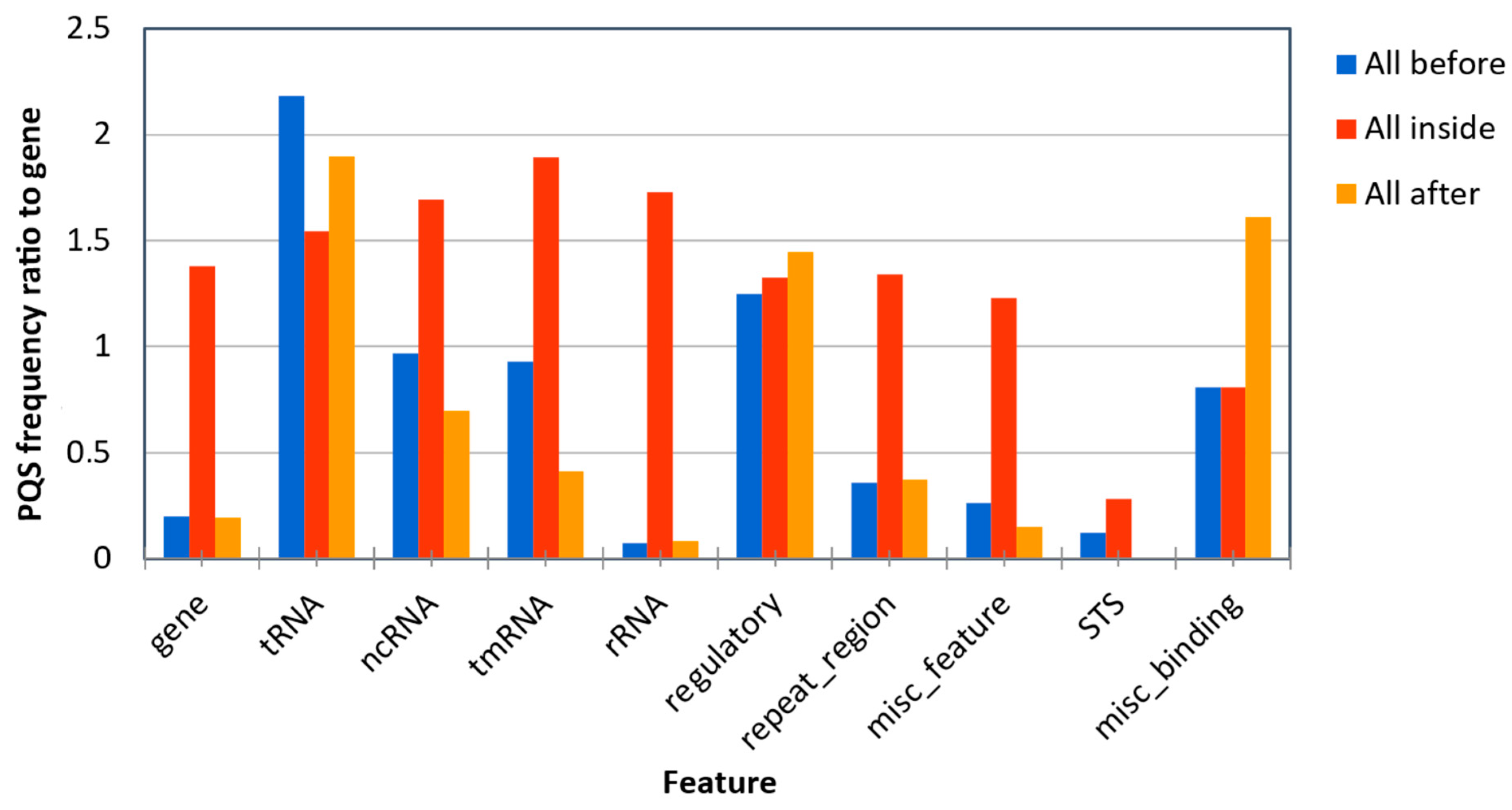
2.2. Localization of PQS in Genomes
3. Discussion
4. Methods
4.1. Selection of DNA Sequences
4.2. Process of Analysis
4.3. Analysis of Putative G4 Sequences Around Annotated NCBI Features
4.4. Phylogenetic Tree Construction
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Watson, J.D.; Crick, F.H. Molecular structure of nucleic acids. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef]
- Szlachta, K.; Thys, R.G.; Atkin, N.D.; Pierce, L.C.T.; Bekiranov, S.; Wang, Y.-H. Alternative DNA secondary structure formation affects RNA polymerase II promoter-proximal pausing in human. Genome Biol. 2018, 19, 89. [Google Scholar] [CrossRef] [PubMed]
- Brázda, V.; Laister, R.C.; Jagelská, E.B.; Arrowsmith, C. Cruciform structures are a common DNA feature important for regulating biological processes. BMC Mol. Biol. 2011, 12, 33. [Google Scholar] [CrossRef]
- Sun, Z.-Y.; Wang, X.-N.; Cheng, S.-Q.; Su, X.-X.; Ou, T.-M. Developing Novel G-Quadruplex Ligands: From Interaction with Nucleic Acids to Interfering with Nucleic Acid–Protein Interaction. Molecules 2019, 24, 396. [Google Scholar] [CrossRef]
- Nelson, L.D.; Bender, C.; Mannsperger, H.; Buergy, D.; Kambakamba, P.; Mudduluru, G.; Korf, U.; Hughes, D.; Van Dyke, M.W.; Allgayer, H. Triplex DNA-binding proteins are associated with clinical outcomes revealed by proteomic measurements in patients with colorectal cancer. Mol. Cancer 2012, 11, 38. [Google Scholar] [CrossRef] [PubMed]
- Gellert, M.; Lipsett, M.N.; Davies, D.R. Helix Formation by Guanylic acid. Proc. Natl. Acad. Sci. USA 1962, 48, 2013–2018. [Google Scholar] [CrossRef]
- Harkness, R.W.; Mittermaier, A.K. G-quadruplex dynamics. Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 1544–1554. [Google Scholar] [CrossRef]
- Siddiqui-Jain, A.; Grand, C.L.; Bearss, D.J.; Hurley, L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA 2002, 99, 11593–11598. [Google Scholar] [CrossRef]
- Lee, S.C.; Zhang, J.; Strom, J.; Yang, D.; Dinh, T.N.; Kappeler, K.; Chen, Q.M. G-Quadruplex in the NRF2 mRNA 5′ Untranslated Region Regulates De Novo NRF2 Protein Translation under Oxidative Stress. Mol. Cell. Biol. 2016, 37, e00122-16. [Google Scholar] [CrossRef] [PubMed]
- Endoh, T.; Kawasaki, Y.; Sugimoto, N. Stability of RNA quadruplex in open reading frame determines proteolysis of human estrogen receptor α. Nucleic Acids Res. 2013, 41, 6222–6231. [Google Scholar] [CrossRef] [PubMed]
- Lam, E.Y.N.; Beraldi, D.; Tannahill, D.; Balasubramanian, S. G-quadruplex structures are stable and detectable in human genomic DNA. Nat. Commun. 2013, 4, 1796. [Google Scholar] [CrossRef]
- Long, X.; Stone, M.D. Kinetic Partitioning Modulates Human Telomere DNA G-Quadruplex StructuralPolymorphism. PLoS ONE 2013, 8, e83420. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Thompson, B.; Cathers, B.E.; Salazar, M.; Kerwin, S.M.; Trent, J.O.; Jenkins, T.C.; Neidle, S.; Hurley, L.H. Inhibition of human telomerase by a G-Quadruplex-Interactive compound. J. Med. Chem. 1997, 40, 2113–2116. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-S.; Carmena, M.; Liskovykh, M.; Peat, E.; Kim, J.-H.; Oshimura, M.; Masumoto, H.; Teulade-Fichou, M.-P.; Pommier, Y.; Earnshaw, W.C.; et al. Systematic Analysis of Compounds Specifically Targeting Telomeres and Telomerase for Clinical Implications in Cancer Therapy. Cancer Res. 2018, 78, 6282–6296. [Google Scholar] [CrossRef]
- Dickerhoff, J.; Onel, B.; Chen, L.; Chen, Y.; Yang, D. Solution Structure of a MYC Promoter G-Quadruplex with 1:6:1 Loop Length. ACS Omega 2019, 4, 2533–2539. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Hurley, L.H.; Neidle, S. Targeting G-quadruplexes in gene promoters: A novel anticancer strategy? Nat. Rev. Drug Discov. 2011, 10, 261–275. [Google Scholar] [CrossRef]
- Cimino-Reale, G.; Zaffaroni, N.; Folini, M. Emerging Role of G-quadruplex DNA as Target in Anticancer Therapy. Curr. Pharm. Design 2016, 22, 6612–6624. [Google Scholar] [CrossRef]
- Asamitsu, S.; Obata, S.; Yu, Z.; Bando, T.; Sugiyama, H. Recent Progress of Targeted G-Quadruplex-Preferred Ligands Toward Cancer Therapy. Molecules 2019, 24, 429. [Google Scholar] [CrossRef]
- Yoshida, W.; Saikyo, H.; Nakabayashi, K.; Yoshioka, H.; Bay, D.H.; Iida, K.; Kawai, T.; Hata, K.; Ikebukuro, K.; Nagasawa, K.; et al. Identification of G-quadruplex clusters by high-throughput sequencing of whole-genome amplified products with a G-quadruplex ligand. Sci. Rep. 2018, 8, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Brázda, V.; Hároníková, L.; Liao, J.C.C.; Fojta, M. DNA and RNA Quadruplex-Binding Proteins. Int. J. Mol. Sci. 2014, 15, 17493–17517. [Google Scholar] [CrossRef]
- Mishra, S.K.; Tawani, A.; Mishra, A.; Kumar, A. G4IPDB: A database for G-quadruplex structure forming nucleic acid interacting proteins. Sci. Rep. 2016, 6, 38144. [Google Scholar] [CrossRef] [PubMed]
- Brázda, V.; Cerveň, J.; Bartas, M.; Mikysková, N.; Coufal, J.; Pečinka, P. The amino acid composition of quadruplex binding proteins reveals a shared motif and predicts new potential quadruplex interactors. Molecules 2018, 23, 2341. [Google Scholar] [CrossRef]
- Patro, L.P.P.; Kumar, A.; Kolimi, N.; Rathinavelan, T. 3D-NuS: A web server for automated modeling and visualization of non-canonical 3-dimensional nucleic acid structures. J. Mol. Biol. 2017, 429, 2438–2448. [Google Scholar] [CrossRef]
- Huppert, J.L.; Balasubramanian, S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005, 33, 2908–2916. [Google Scholar] [CrossRef] [PubMed]
- Eddy, J.; Maizels, N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006, 34, 3887–3896. [Google Scholar] [CrossRef]
- Bedrat, A.; Lacroix, L.; Mergny, J.L. Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 2016, 44, 1746–1759. [Google Scholar] [CrossRef]
- diCenzo, G.C.; Finan, T.M. The Divided Bacterial Genome: Structure, Function, and Evolution. Microbiol. Mol. Biol. Rev. 2017, 81, e00019-17. [Google Scholar] [CrossRef]
- Yadav, V.K.; Abraham, J.K.; Mani, P.; Kulshrestha, R.; Chowdhury, S. QuadBase: Genome-wide database of G4 DNA—Occurrence and conservation in human, chimpanzee, mouse and rat promoters and 146 microbes. Nucleic Acids Res. 2008, 36, D381–D385. [Google Scholar] [CrossRef]
- König, S.L.B.; Huppert, J.L.; Sigel, R.K.O.; Evans, A.C. Distance-dependent duplex DNA destabilization proximal to G-quadruplex/i-motif sequences. Nucleic Acids Res. 2013, 41, 7453–7461. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.K.; Jain, N.; Shankar, U.; Tawani, A.; Sharma, T.K.; Kumar, A. Characterization of highly conserved G-quadruplex motifs as potential drug targets in Streptococcus pneumoniae. Sci. Rep. 2019, 9, 1791. [Google Scholar] [CrossRef]
- Rawal, P.; Kummarasetti, V.B.R.; Ravindran, J.; Kumar, N.; Halder, K.; Sharma, R.; Mukerji, M.; Das, S.K.; Chowdhury, S. Genome-wide prediction of G4 DNA as regulatory motifs: Role in Escherichia coli global regulation. Genome Res. 2006, 16, 644–655. [Google Scholar] [CrossRef]
- Neidle, S. The structures of quadruplex nucleic acids and their drug complexes. Curr. Opin. Struct. Biol. 2009, 19, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Saranathan, N.; Vivekanandan, P. G-Quadruplexes: More than just a kink in microbial genomes. Trends Microbiol. 2018, 27, 148–163. [Google Scholar] [CrossRef]
- Kaplan, O.I.; Berber, B.; Hekim, N.; Doluca, O. G-quadruplex prediction in E. coli genome reveals a conserved putative G-quadruplex-Hairpin-Duplex switch. Nucleic Acids Res. 2016, 44, 9083–9095. [Google Scholar] [PubMed]
- Brocchieri, L. The GC Content of Bacterial Genomes. J. Phylogenet. Evolut. Biol. 2013, 2, 1–3. [Google Scholar] [CrossRef]
- Ding, Y.; Fleming, A.M.; Burrows, C.J. Case studies on potential G-quadruplex-forming sequences from the bacterial orders Deinococcales and Thermales derived from a survey of published genomes. Sci. Rep. 2018, 8, 15679. [Google Scholar] [CrossRef]
- Brumm, P.J.; Monsma, S.; Keough, B.; Jasinovica, S.; Ferguson, E.; Schoenfeld, T.; Lodes, M.; Mead, D.A. Complete Genome Sequence of Thermus aquaticus Y51MC23. PLoS ONE 2015, 10, e0138674. [Google Scholar] [CrossRef]
- Waller, Z.A.E.; Pinchbeck, B.J.; Buguth, B.S.; Meadows, T.G.; Richardson, D.J.; Gates, A.J. Control of bacterial nitrate assimilation by stabilization of G-quadruplex DNA. Chem. Commun. 2016, 52, 13511–13514. [Google Scholar] [CrossRef] [PubMed]
- Čechová, J.; Lýsek, J.; Bartas, M.; Brázda, V. Complex analyses of inverted repeats in mitochondrial genomes revealed their importance and variability. Bioinformatics 2018, 34, 1081–1085. [Google Scholar] [CrossRef]
- Brázda, V.; Kolomazník, J.; Lýsek, J.; Hároníková, L.; Coufal, J.; Št’astný, J. Palindrome analyser—A new web-based server for predicting and evaluating inverted repeats in nucleotide sequences. Biochem. Biophys. Res. Commun. 2016, 478, 1739–1745. [Google Scholar] [CrossRef]
- Brázda, V.; Lýsek, J.; Bartas, M.; Fojta, M. Complex Analyses of Short Inverted Repeats in All Sequenced Chloroplast DNAs. BioMed Res. Int. 2018, 2018, 1097018. [Google Scholar] [CrossRef]
- Ruggiero, E.; Richter, S.N. G-quadruplexes and G-quadruplex ligands: Targets and tools in antiviral therapy. Nucleic Acids Res. 2018, 46, 3270–3283. [Google Scholar] [CrossRef]
- Chen, B.-J.; Wu, Y.-L.; Tanaka, Y.; Zhang, W. Small Molecules Targeting c-Myc Oncogene: Promising Anti-Cancer Therapeutics. Int. J. Biol. Sci. 2014, 10, 1084–1096. [Google Scholar] [CrossRef] [PubMed]
- Belmonte-Reche, E.; Martínez-García, M.; Guédin, A.; Zuffo, M.; Arévalo-Ruiz, M.; Doria, F.; Campos-Salinas, J.; Maynadier, M.; López-Rubio, J.J.; Freccero, M.; et al. G-Quadruplex Identification in the Genome of Protozoan Parasites Points to Naphthalene Diimide Ligands as New Antiparasitic Agents. J. Med. Chem. 2018, 61, 1231–1240. [Google Scholar] [CrossRef]
- Li, F.; Mulyana, Y.; Feterl, M.; Warner, J.M.; Collins, J.G.; Keene, F.R. The antimicrobial activity of inert oligonuclear polypyridylruthenium(II) complexes against pathogenic bacteria, including MRSA. Dalton Trans. 2011, 40, 5032–5038. [Google Scholar] [CrossRef]
- Li, F.; Grant Collins, J.; Richard Keene, F. Ruthenium complexes as antimicrobial agents. Chem. Soc. Rev. 2015, 44, 2529–2542. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Chen, X.; Wu, J.; Wang, J.; Ji, L.; Chao, H. Dinuclear Ruthenium(II) Complexes That Induce and Stabilise G-Quadruplex DNA. Chem. Eur. J. 2015, 21, 4008–4020. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, D.; Huang, J.; Deng, M.; Zhang, M.; Zhou, X. High fluorescence selectivity and visual detection of G-quadruplex structures by a novel dinuclear ruthenium complex. Chem. Commun. 2010, 46, 743–745. [Google Scholar] [CrossRef] [PubMed]
- Wilson, T.; Williamson, M.P.; Thomas, J.A. Differentiating quadruplexes: Binding preferences of a luminescent dinuclear ruthenium (II) complex with four-stranded DNA structures. Org. Biomol. Chem. 2010, 8, 2617–2621. [Google Scholar] [CrossRef] [PubMed]
- Codd, G.A.; Lindsay, J.; Young, F.M.; Morrison, L.F.; Metcalf, J.S. Harmful cyanobacteria. In Harmful Cyanobacteria; Springer: Dordrecht, The Netherlands, 2005; pp. 1–23. [Google Scholar]
- Perrone, R.; Lavezzo, E.; Riello, E.; Manganelli, R.; Palù, G.; Toppo, S.; Provvedi, R.; Richter, S.N. Mapping and characterization of G-quadruplexes in Mycobacterium tuberculosis gene promoter regions. Sci. Rep. 2017, 7, 5743. [Google Scholar] [CrossRef] [PubMed]
- Lavezzo, E.; Berselli, M.; Frasson, I.; Perrone, R.; Palù, G.; Brazzale, A.R.; Richter, S.N.; Toppo, S. G-quadruplex forming sequences in the genome of all known human viruses: A comprehensive guide. PLOS Comput. Biol. 2018, 14, e1006675. [Google Scholar] [CrossRef]
- Beaume, N.; Pathak, R.; Yadav, V.K.; Kota, S.; Misra, H.S.; Gautam, H.K.; Chowdhury, S. Genome-wide study predicts promoter-G4 DNA motifs regulate selective functions in bacteria: Radioresistance of D. radiodurans involves G4 DNA-mediated regulation. Nucleic Acids Res. 2013, 41, 76–89. [Google Scholar] [CrossRef]
- Kota, S.; Dhamodharan, V.; Pradeepkumar, P.I.; Misra, H.S. G-quadruplex forming structural motifs in the genome of Deinococcus radiodurans and their regulatory roles in promoter functions. Appl. Microbiol. Biotechnol. 2015, 99, 9761–9769. [Google Scholar] [CrossRef]
- Repoila, F.; Darfeuille, F. Small regulatory non-coding RNAs in bacteria: Physiology and mechanistic aspects. Biol. Cell 2009, 101, 117–131. [Google Scholar] [CrossRef] [PubMed]
- Brázda, V.; Kolomazník, J.; Lýsek, J.; Bartas, M.; Fojta, M.; Št’astný, J.; Mergny, J.-L. G4Hunter web application: A web server for G-quadruplex prediction. Bioinformatics 2019, btz087. [Google Scholar] [CrossRef]
- Sayers, E.W.; Agarwala, R.; Bolton, E.E.; Brister, J.R.; Canese, K.; Clark, K.; Connor, R.; Fiorini, N.; Funk, K.; Hefferon, T.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2019, 47, D23–D28. [Google Scholar] [CrossRef]
- Federhen, S. The NCBI taxonomy database. Nucleic Acids Res. 2011, 40, D136–D143. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
Sample Availability: Not available. |
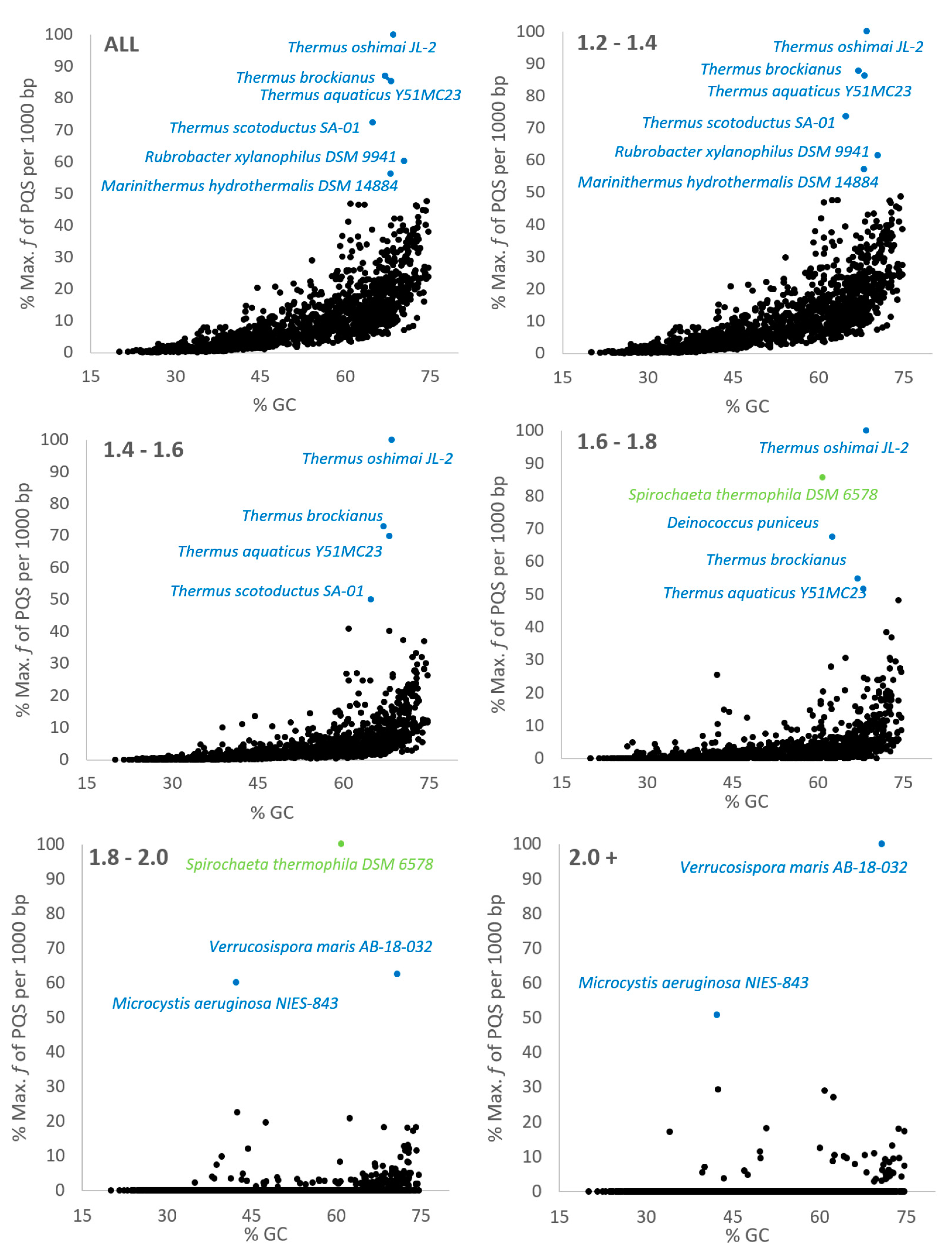
| Interval of G4Hunter Score | Number of PQS in Dataset | PQS Frequency per 1000 bp |
|---|---|---|
| 1.2–1.4 | 9,009,593 | 1.315033 |
| 1.4–1.6 | 180,395 | 0.025058 |
| 1.6–1.8 | 11,779 | 0.00155 |
| 1.8–2.0 | 511 | 0.000055 |
| 2.0–more | 86 | 0.000009 |
| Domain | Seq | Median | Short | Long | GC% | PQS | Mean f | Min f | Max f |
| Bacteria | 1627 | 3,307,820 | 83,026 | 13,033,779 | 50.6 | 9,202,364 | 1.342 | 0.013 | 14.213 |
| Group | Seq | Median | Short | Long | GC% | PQS | Mean f | Min f | Max f |
| Spirochaetes | 38 | 2,646,038 | 277,655 | 4,653,970 | 39.7 | 87,109 | 0.809 | 0.079 | 6.668 |
| Thermotogae | 16 | 2,150,379 | 1,884,562 | 2,974,229 | 39.1 | 13,617 | 0.395 | 0.149 | 0.812 |
| PVC group | 28 | 2,917,407 | 1,041,170 | 9,629,675 | 50.7 | 198,358 | 1.646 | 0.388 | 4.802 |
| FCB group | 117 | 3,914,632 | 605,745 | 9,127,347 | 42.3 | 302,949 | 0.608 | 0.013 | 2.746 |
| Terrabacteria | 659 | 3,018,755 | 91,776 | 11,936,683 | 50.4 | 4,766,517 | 1.601 | 0.016 | 14.213 |
| Proteobacteria | 724 | 3,551,512 | 83,026 | 13,033,779 | 53.4 | 3,688,101 | 1.276 | 0.025 | 5.507 |
| Other | 45 | 2,157,835 | 1,012,010 | 6,237,577 | 44.3 | 145,713 | 1.103 | 0.062 | 5.855 |
| Subgroup | Seq | Median | Short | Long | GC% | PQS | Mean f | Min f | Max f |
| Spirochaetia | 38 | 2,646,038 | 277,655 | 4,653,970 | 39.7 | 87,109 | 0.809 | 0.079 | 6.668 |
| Thermotogae | 16 | 2,150,379 | 1,884,562 | 2,974,229 | 39.1 | 13,617 | 0.395 | 0.149 | 0.812 |
| Chlamydiae | 12 | 1,168,953 | 1,041,170 | 3,072,383 | 40.3 | 12,453 | 0.646 | 0.388 | 0.957 |
| Bacteroidetes/Chlorobi | 114 | 3,878,527 | 605,745 | 912,7347 | 41.9 | 282,516 | 0.585 | 0.013 | 2.746 |
| Cyanobacteria/Melainab. | 29 | 5,315,554 | 1,657,990 | 9,673,108 | 42.6 | 193,894 | 1.247 | 0.201 | 6.004 |
| Chloroflexi | 12 | 2,333,610 | 1,252,731 | 5,723,298 | 60 | 62,688 | 1.89 | 1.223 | 3.222 |
| Tenericutes | 52 | 981,001 | 564,395 | 1,877,792 | 28 | 6460 | 0.136 | 0.016 | 0.834 |
| Actinobacteria | 246 | 3,960,961 | 775,354 | 11,936,683 | 66.2 | 3,590,884 | 2.821 | 0.143 | 8.556 |
| Deinococcus-Thermus | 18 | 2,895,913 | 2,035,182 | 3,881,839 | 66.8 | 311,949 | 6.626 | 1.885 | 14.213 |
| Firmicutes | 298 | 2,835,823 | 91,776 | 8,739,048 | 40.8 | 579,740 | 0.56 | 0.064 | 6.587 |
| delta/epsilon subdiv. | 92 | 3,136,746 | 1,457,619 | 13,033,779 | 50 | 807,281 | 1.681 | 0.034 | 5.282 |
| Betaproteobacteria | 110 | 3,763,620 | 820,037 | 6,987,670 | 60.6 | 585,984 | 1.306 | 0.195 | 4.007 |
| Alphaproteobacteria | 213 | 3,424,964 | 83,026 | 9,207,384 | 61.5 | 126,134 | 1.764 | 0.051 | 5.507 |
| Gammaproteobacteria | 302 | 3,777,066 | 298,471 | 7,783,862 | 48.8 | 31,686 | 0.799 | 0.025 | 4.264 |
| other | 75 | 2,406,157 | 1,012,010 | 9,629,675 | 48.4 | 432,683 | 1.406 | 0.0616 | 5.855 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bartas, M.; Čutová, M.; Brázda, V.; Kaura, P.; Šťastný, J.; Kolomazník, J.; Coufal, J.; Goswami, P.; Červeň, J.; Pečinka, P. The Presence and Localization of G-Quadruplex Forming Sequences in the Domain of Bacteria. Molecules 2019, 24, 1711. https://doi.org/10.3390/molecules24091711
Bartas M, Čutová M, Brázda V, Kaura P, Šťastný J, Kolomazník J, Coufal J, Goswami P, Červeň J, Pečinka P. The Presence and Localization of G-Quadruplex Forming Sequences in the Domain of Bacteria. Molecules. 2019; 24(9):1711. https://doi.org/10.3390/molecules24091711
Chicago/Turabian StyleBartas, Martin, Michaela Čutová, Václav Brázda, Patrik Kaura, Jiří Šťastný, Jan Kolomazník, Jan Coufal, Pratik Goswami, Jiří Červeň, and Petr Pečinka. 2019. "The Presence and Localization of G-Quadruplex Forming Sequences in the Domain of Bacteria" Molecules 24, no. 9: 1711. https://doi.org/10.3390/molecules24091711
APA StyleBartas, M., Čutová, M., Brázda, V., Kaura, P., Šťastný, J., Kolomazník, J., Coufal, J., Goswami, P., Červeň, J., & Pečinka, P. (2019). The Presence and Localization of G-Quadruplex Forming Sequences in the Domain of Bacteria. Molecules, 24(9), 1711. https://doi.org/10.3390/molecules24091711









