Relationship Between G-Quadruplex Sequence Composition in Viruses and Their Hosts
Abstract
1. Introduction
2. Results and Discussion
2.1. G-Quadruplex Metrics in Viral Genomes
2.2. Thermodynamically Stable G4 Motifs are Enriched in Viral Genomes
2.3. The PQS Loop Composition Within the Herpesviridae Family of Viruses and Their Host are Correlated
2.4. G4 Motifs in Viral Genomes Overlap Hosts’ Transcription Factor Binding Sites
2.5. High Prevalence of Telomere-Like PQSs across Herpesviridae Infecting Vertebrates
3. Materials and Methods
3.1. Genome Assembly Retrieval
3.2. Genome Metrics
3.3. G-Quadruplex Motif Identification and Loop Composition Analysis
3.4. Putative Quadruplex Sequence Analysis in Eukaryote Genomes
- 18 mammals (Minke whale balAcu1, Marmoset calJac3, Dog canFam3, Green monkey chlSab2, Kangaroo rat dipOrd1, Wallaby macEug2, Crab-eating macaque macFas5, Mouse lemur micMur2, Mouse mm10, Gibbon nomLeu3, Bushbaby otoGar3, Baboon papAnu2, Orangutan ponAbe2, Rhesus macaque rheMac8, Golden snub-nosed monkey rhiRox1, Squirrel monkey saiBol1, Tarsier tarSyr2, Tree shrew tupBel1);
- 9 birds/reptiles (American alligator allMis1, Chicken galGal5, Painted turtle chrPic1, Garter snake thaSir1, Lizard anoCar2, Zebra finch taeGut2, Medium ground finch geoFor1, Turkey melGal5, Budgerigar melUnd1);
- 8 fish (Elephant shark calMil1, Zebrafish danRer11, Fugu fr3, Stickleback gasAcu1, Coelacanth latCha1, Medaka oryLat2, Lamprey petMar2, Tetraodon tetNig2);
- 3 amphibians (Tibetan frog nanPar1, African clawed frog xenLae2, Xenopus tropicalis xenTro7);
- 9 invertebrates (Apis mellifera apiMel3, Caenorhabditis elegans ce11, Caenorhabditis japonica caeJap1, Caenorhabditis brenneri caePb2, Caenorhabditis remanei caeRem3, Caenorhabditis briggsae cb3, Ciona intestinalis ci3, Drosophila melanogaster dm6, Pristionchus pacificus priPac1).
3.5. Loop Composition Analysis in Herpesviruses
3.6. Statistics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Gellert, M.; Lipsett, M.N.; Davies, DR. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. USA 1962, 48, 2013–2018. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Halder, K.; Halder, R.; Yadav, V.K.; Rawal, P.; Thakur, R.K.; Mohd, F.; Sharma, A.; Chowdhury, S. Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J. Med. Chem. 2008, 51, 5641–5649. [Google Scholar] [CrossRef]
- Du, Z.; Zhao, Y.; Li, N. Genome-wide analysis reveals regulatory role of G4 DNA in gene transcription. Genome Res. 2008, 18, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Bugaut, A.; Balasubramanian, S. 5′-UTR RNA G-quadruplexes: Translation regulation and targeting. Nucleic Acids Res. 2012, 40, 4727–4741. [Google Scholar] [CrossRef]
- Huppert, J.L.; Balasubramanian, S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005, 33, 2908–2916. [Google Scholar] [CrossRef] [PubMed]
- Bedrat, A.; Lacroix, L.; Mergny, J.L. Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 2016, 44, 1746–1759. [Google Scholar] [CrossRef]
- Maizels, N.; Gray, L.T. The G4 genome. PLoS Genet. 2013, 9, e1003468. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, D.; Lipps, H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43, 8627–8637. [Google Scholar] [CrossRef] [PubMed]
- Kwok, C.K.; Merrick, C.J. G-Quadruplexes: Prediction, Characterization, and Biological Application. Trends Biotechnol. 2017, 35, 997–1013. [Google Scholar] [CrossRef] [PubMed]
- Todd, A.K.; Johnston, M.; Neidle, S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005, 33, 2901–2907. [Google Scholar] [CrossRef] [PubMed]
- Métifiot, M.; Amrane, S.; Litvak, S.; Andreola, M.-L. G-quadruplexes in viruses: Function and potential therapeutic applications. Nucleic Acids Res. 2014, 42, 12352–12366. [Google Scholar] [CrossRef]
- Lavezzo, E.; Berselli, M.; Frasson, I.; Perrone, R.; Palu, G.; Brazzale, A.R.; Richter, S.N.; Toppo, S. G-quadruplex forming sequences in the genome of all known human viruses: A comprehensive guide. PLoS Comput Biol. 2018, 14, e1006675. [Google Scholar] [CrossRef]
- Harris, L.M.; Merrick, C.J. G-quadruplexes in pathogens: A common route to virulence control? PLoS Pathog. 2015, 11, e1004562. [Google Scholar] [CrossRef]
- Ravichandran, S.; Kim, Y.E.; Bansal, V.; Ghosh, A.; Hur, J.; Subramani, V.K.; Pradhan, S.; Lee, M.K.; Kim, K.K.; Ahn, J.H. Genome-wide analysis of regulatory G-quadruplexes affecting gene expression in human cytomegalovirus. PLoS Pathog. 2018, 14, e1007334. [Google Scholar] [CrossRef] [PubMed]
- Ruggiero, E.; Richter, S.N. G-quadruplexes and G-quadruplex ligands: targets and tools in antiviral therapy. Nucleic Acids Res. 2018, 46, 3270–3283. [Google Scholar] [CrossRef]
- Sundquist, W.I.; Heaphy, S. Evidence for interstrand quadruplex formation in the dimerization of human immunodeficiency virus 1 genomic RNA. Proc. Natl. Acad. Sci. USA 1993, 90, 3393–3397. [Google Scholar] [CrossRef] [PubMed]
- Perrone, R.; Nadai, M.; Poe, J.A.; Frasson, I.; Palumbo, M.; Palù, G.; Smithgall, T.E.; Richter, S.N. Formation of a unique cluster of G-quadruplex structures in the HIV-1 Nef coding region: Implications for antiviral activity. PLoS ONE 2013, 8, e73121. [Google Scholar] [CrossRef]
- Perrone, R.; Nadai, M.; Frasson, I.; Poe, J.A.; Butovskaya, E.; Smithgall, T.E.; Palumbo, M.; Palù, G.; Richter, S.N. A dynamic G-quadruplex region regulates the HIV-1 long terminal repeat promoter. J. Med. Chem. 2013, 56, 6521–6530. [Google Scholar] [CrossRef]
- Amrane, S.; Kerkour, A.; Bedrat, A.; Vialet, B.; Andreola, M.L.; Mergny, J.L. Topology of a DNA G-quadruplex structure formed in the HIV-1 promoter: A potential target for anti-HIV drug development. J. Am. Chem Soc. 2014, 136, 5249–5252. [Google Scholar] [CrossRef] [PubMed]
- Krafčíková, P.; Demkovičová, E.; Halaganová, A.; Víglaský, V. Putative HIV and SIV G-Quadruplex Sequences in Coding and Noncoding Regions Can Form G-Quadruplexes. J. Nucleic Acids. 2017, 2017, 6513720. [Google Scholar] [CrossRef]
- Norseen, J.; Johnson, F.B.; Lieberman, P.M. Role for G-quadruplex RNA binding by Epstein-Barr virus nuclear antigen 1 in DNA replication and metaphase chromosome attachment. J. Virol. 2009, 83, 10336–10346. [Google Scholar] [CrossRef] [PubMed]
- Murat, P.; Zhong, J.; Lekieffre, L.; Cowieson, N.P.; Clancy, J.L.; Preiss, T.; Balasubramanian, S.; Khanna, R.; Tellam, J. G-quadruplexes regulate Epstein-Barr virus-encoded nuclear antigen 1 mRNA translation. Nat. Chem. Biol. 2014, 10, 5358–6410. [Google Scholar] [CrossRef] [PubMed]
- Tluckova, K.; Marusic, M.; Tothova, P.; Bauer, L.; Sket, P.; Plavec, J.; Viglasky, V. Human papillomavirus G-quadruplexes. Biochemistry 2013, 52, 7207–7216. [Google Scholar] [CrossRef] [PubMed]
- Marušič, M.; Hošnjak, L.; Krafčikova, P.; Poljak, M.; Viglasky, V.; Plavec, J. The effect of single nucleotide polymorphisms in G-rich regions of high-risk human papillomaviruses on structural diversity of DNA. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1229–1236. [Google Scholar] [CrossRef] [PubMed]
- Zahin, M.; Dean, W.L.; Ghim, S.J.; Joh, J.; Gray, R.D.; Khanal, S.; Bossart, G.D.; Mignucci-Giannoni, A.A.; Rouchka, E.C.; Jenson, A.B. Identification of G-quadruplex forming sequences in three manatee papillomaviruses. PLoS ONE 2018, 13, e0195625. [Google Scholar] [CrossRef] [PubMed]
- Artusi, S.; Nadai, M.; Perrone, R.; Biasolo, M.A.; Palù, G.; Flamand, L.; Calistri, A.; Richter, S.N. The Herpes Simplex Virus-1 genome contains multiple clusters of repeated G-quadruplex: Implications for the antiviral activity of a G-quadruplex ligand. Antiviral Res. 2015, 118, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Biswas, B.; Kumari, P.; Vivekanandan, P. Pac1 Signals of Human Herpesviruses Contain a Highly Conserved G-Quadruplex Motif. ACS Infect. Dis. 2018, 4, 744–751. [Google Scholar] [CrossRef] [PubMed]
- Lyonnais, S.; Gorelick, R.J.; Mergny, J.L.; Le Cam, E.; Mirambeau, G. G-quartets direct assembly of HIV-1 nucleocapsid protein along single-stranded DNA. Nucleic Acids Res. 2003, 31, 5754–5763. [Google Scholar] [CrossRef]
- Musumeci, D.; Riccardi, C.; Montesarchio, D. G-Quadruplex Forming Oligonucleotides as Anti-HIV Agents. Molecules 2015, 20, 17511–17532. [Google Scholar] [CrossRef] [PubMed]
- González, V.M.; Martín, M.E.; Fernández, G.; García-Sacristán, A. Use of Aptamers as Diagnostics Tools and Antiviral Agents for Human Viruses. Pharmaceuticals (Basel) 2016, 9, 78. [Google Scholar] [CrossRef]
- Risitano, A.; Fox, K.R. Influence of loop size on the stability of intramolecular DNA quadruplexes. Nucleic Acids Res. 2004, 32, 2598–2606. [Google Scholar] [CrossRef] [PubMed]
- Rachwal, P.A.; Brown, T.; Fox, K.R. Effect of G-tract length on the topology and stability of intramolecular DNA quadruplexes. Biochimie 2008, 90, 686–696. [Google Scholar] [CrossRef]
- Guédin, A.; De Cian, A.; Gros, J.; Lacroix, L.; Mergny, J.L. Sequence effects in single-base loops for quadruplexes. Biochemistry 2007, 46, 3036–3044. [Google Scholar] [CrossRef] [PubMed]
- Guédin, A.; Gros, J.; Alberti, P.; Mergny, J.L. How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 2010, 38, 7858–7868. [Google Scholar] [CrossRef]
- Piazza, A.; Adrian, M.; Samazan, F.; Heddi, B.; Hamon, F.; Serero, A.; Lopes, J.; Teulade-Fichou, M.P.; Phan, A.T.; Nicolas, A. Short loop length and high thermal stability determine genomic instability induced by G-quadruplex-forming minisatellites. EMBO J. 2015, 34, 1718–1734. [Google Scholar] [CrossRef]
- Puig Lombardi, E.; Holmes, A.; Verga, D.; Teulade-Fichou, M.P.; Nicolas, A.; Londoño-Vallejo, A. Thermodynamically stable and genetically unstable G-quadruplexes are depleted in genomes across species. Nucleic Acids Res. 2019. [accepted]. [Google Scholar]
- Siddiqui-Jain, A.; Grand, C.L.; Bearss, D.J.; Hurley, L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA 2002, 99, 11593–11598. [Google Scholar] [CrossRef]
- Sun, D.; Guo, K.; Rusche, J.J.; Hurley, L.H. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005, 33, 6070–6080. [Google Scholar] [CrossRef]
- De Armond, R.; Wood, S.; Sun, D.; Hurley, L.H.; Ebbinghaus, S.W. Evidence for the presence of a guanine quadruplex forming region within a polypurine tract of the hypoxia inducible factor 1alpha promoter. Biochemistry 2005, 44, 16341–16350. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Dexheimer, T.S.; Chen, D.; Carver, M.; Ambrus, A.; Jones, R.A.; Yang, D. An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J. Am. Chem. Soc. 2006, 128, 1096–1098. [Google Scholar] [CrossRef]
- Fernando, H.; Reszka, A.P.; Huppert, J.; Ladame, S.; Rankin, S.; Venkitaraman, A.R.; Neidle, S.; Balasubramanian, S. A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry 2006, 45, 7854–7860. [Google Scholar] [CrossRef] [PubMed]
- Kropp, K.A.; Angulo, A.; Ghazal, P. Viral enhancer mimicry of host innate-immune promoters. PLoS Pathog. 2014, 10, e1003804. [Google Scholar] [CrossRef]
- Stano, M.; Beke, G.; Klucar, L. viruSITE-integrated database for viral genomics. Database (Oxford) 2016, 2016, baw162. [Google Scholar] [CrossRef]
- Marsico, G.; Chambers, V.S.; Sahakyan, A.B.; McCauley, P.; Boutell, J.M.; Di Antonio, M.; Balasubramanian, S. Whole genome experimental maps of DNA G-quadruplexes in multiple species. Nucleic Acids Res. 2019, 47, 3862–3874. [Google Scholar] [CrossRef] [PubMed]
- Jaubert, C.; Bedrat, A.; Bartolucci, L.; Di Primo, C.; Ventura, M.; Mergny, J.L.; Amrane, S.; Andreola, M.L. RNA synthesis is modulated by G-quadruplex formation in Hepatitis C virus negative RNA strand. Sci. Rep. 2018, 8, 8120. [Google Scholar]
- Ding, Y.; Fleming, A.M.; Burrows, C.J. Case studies on potential G-quadruplex-forming sequences from the bacterial orders Deinococcales and Thermales derived from a survey of published genomes. Sci Rep. 2018, 8, 15679. [Google Scholar] [CrossRef]
- Davison, A.J.; Eberle, R.; Ehlers, B.; Hayward, G.S.; McGeoch, D.J.; Minson, A.C.; Pellett, P.E.; Roizman, B.; Studdert, M.J.; Thiry, E. The Order Herpesvirales. Arch. Virol. 2009, 154, 171–177. [Google Scholar] [CrossRef]
- Todd, A.K.; Neidle, S. The relationship of potential G-quadruplex sequences in cis-upstream regions of the human genome to SP1-binding elements. Nucleic Acids Res. 2008, 36, 2700–2704. [Google Scholar] [CrossRef]
- Khalil, M.I.; Ruyechan, W.T.; Hay, J.; Arvin, A. Differential effects of Sp cellular transcription factors on viral promoter activation by varicella-zoster virus (VZV) IE62 protein. Virology 2015, 485, 47–57. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tatarowicz, W.A.; Martin, C.E.; Pekosz, A.S.; Madden, S.L.; Rauscher, F.J., 3rd.; Chiang, S.Y.; Beerman, T.A.; Fraser, N.W. Repression of the HSV-1 latency-associated transcript (LAT) promoter by the early growth response (EGR) proteins: Involvement of a binding site immediately downstream of the TATA box. J. Neurovirol. 1997, 3, 212–224. [Google Scholar] [CrossRef] [PubMed]
- Satkunanathan, S.; Thorpe, R.; Zhao, Y. The function of DNA binding protein nucleophosmin in AAV replication. Virology 2017, 510, 46–54. [Google Scholar] [CrossRef]
- Parkinson, G.N.; Lee, M.P.; Neidle, S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 2002, 417, 876–880. [Google Scholar] [CrossRef]
- Pantry, S.N.; Medveczky, P.G. Latency, Integration, and Reactivation of Human Herpesvirus-6. Viruses 2017, 9, 194. [Google Scholar] [CrossRef]
- Gilbert-Girard, S.; Gravel, A.; Artusi, S.; Richter, S.N.; Wallaschek, N.; Kaufer, B.B.; Flamand, L. Stabilization of telomere G-quadruplexes interferes with human herpesvirus 6A chromosomal integration. J. Virol. 2017, 91, e402–e417. [Google Scholar] [CrossRef] [PubMed]
- Wallaschek, N.; Sanyal, A.; Pirzer, F.; Gravel, A.; Mori, Y.; Flamand, L.; Kaufer, B.B. The Telomeric Repeats of Human Herpesvirus 6A (HHV-6A) Are Required for Efficient Virus Integration. PLoS Pathog. 2016, 12, e1005666. [Google Scholar] [CrossRef]
- McPherson, M.C.; Cheng, H.H.; Smith, J.M.; Delany, M.E. Vaccination and Host Marek’s Disease-Resistance Genotype Significantly Reduce Oncogenic Gallid alphaherpesvirus 2 Telomere Integration in Host Birds. Cytogenet Genome Res. 2018, 156, 204–214. [Google Scholar] [CrossRef] [PubMed]
- Mihara, T.; Nishimura, Y.; Shimizu, Y.; Nishiyama, H.; Yoshikawa, G.; Uehara, H.; Hingamp, P.; Goto, S.; Ogata, H. Linking virus genomes with host taxonomy. Viruses 2016, 8, 66. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Thomas-Chollier, M.; Sand, O.; Turatsinze, J.V.; Janky, R.; Defrance, M.; Vervisch, E.; Brohée, S.; van Helden, J. RSAT: Regulatory sequence analysis tools. Nucleic Acids Res. 2008, 36, W119–W127. [Google Scholar] [CrossRef]
- Khan, A.; Fornes, O.; Stigliani, A.; Gheorghe, M.; Castro-Mondragon, J.A.; van der Lee, R.; Bessy, A.; Chèneby, J.; Kulkarni, S.R.; Tan, G. JASPAR 2018: Update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018, 46, D260–D266. [Google Scholar] [CrossRef]
- Huppert, J.L.; Balasubramanian, S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007, 35, 406–413. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |
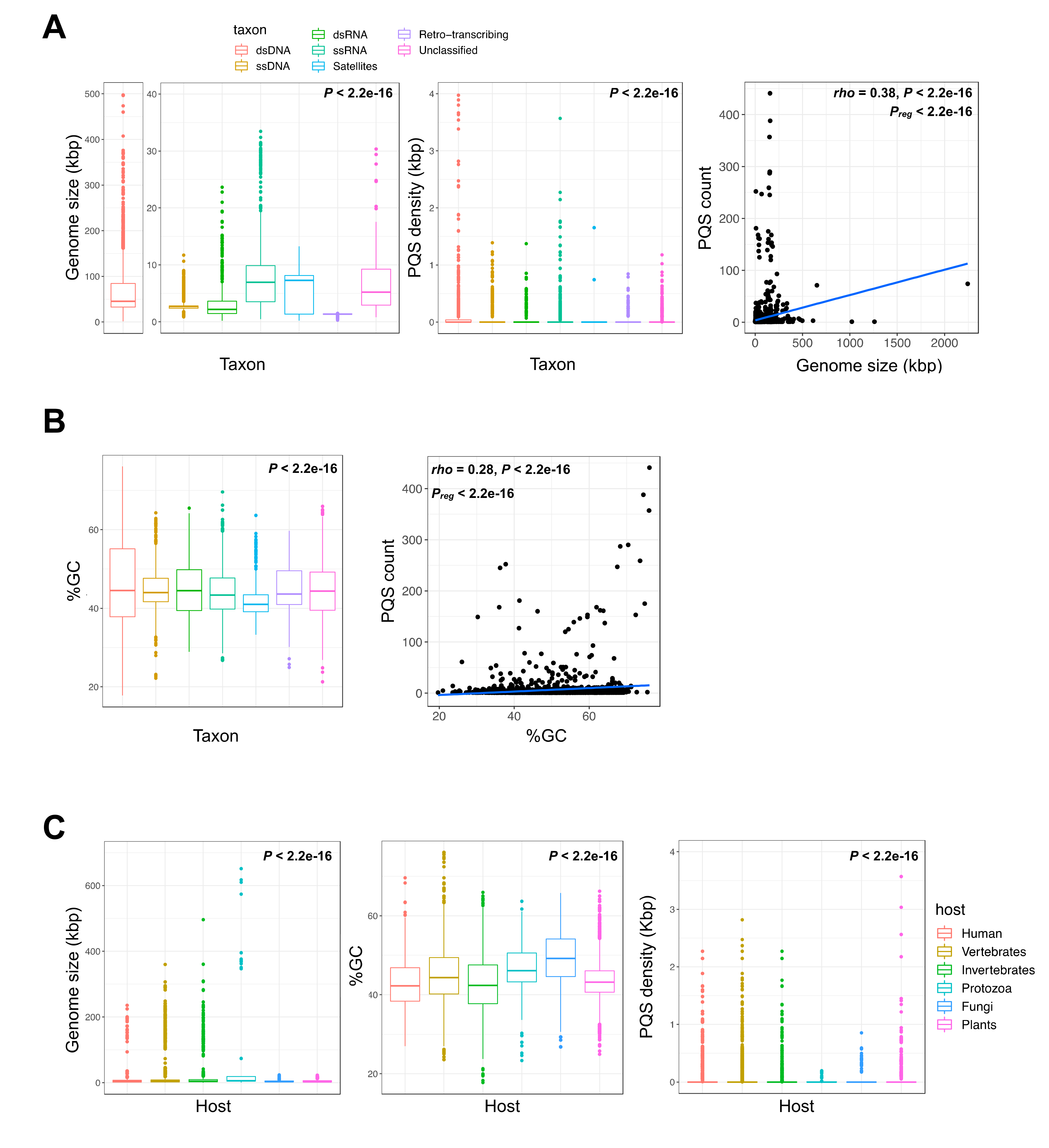
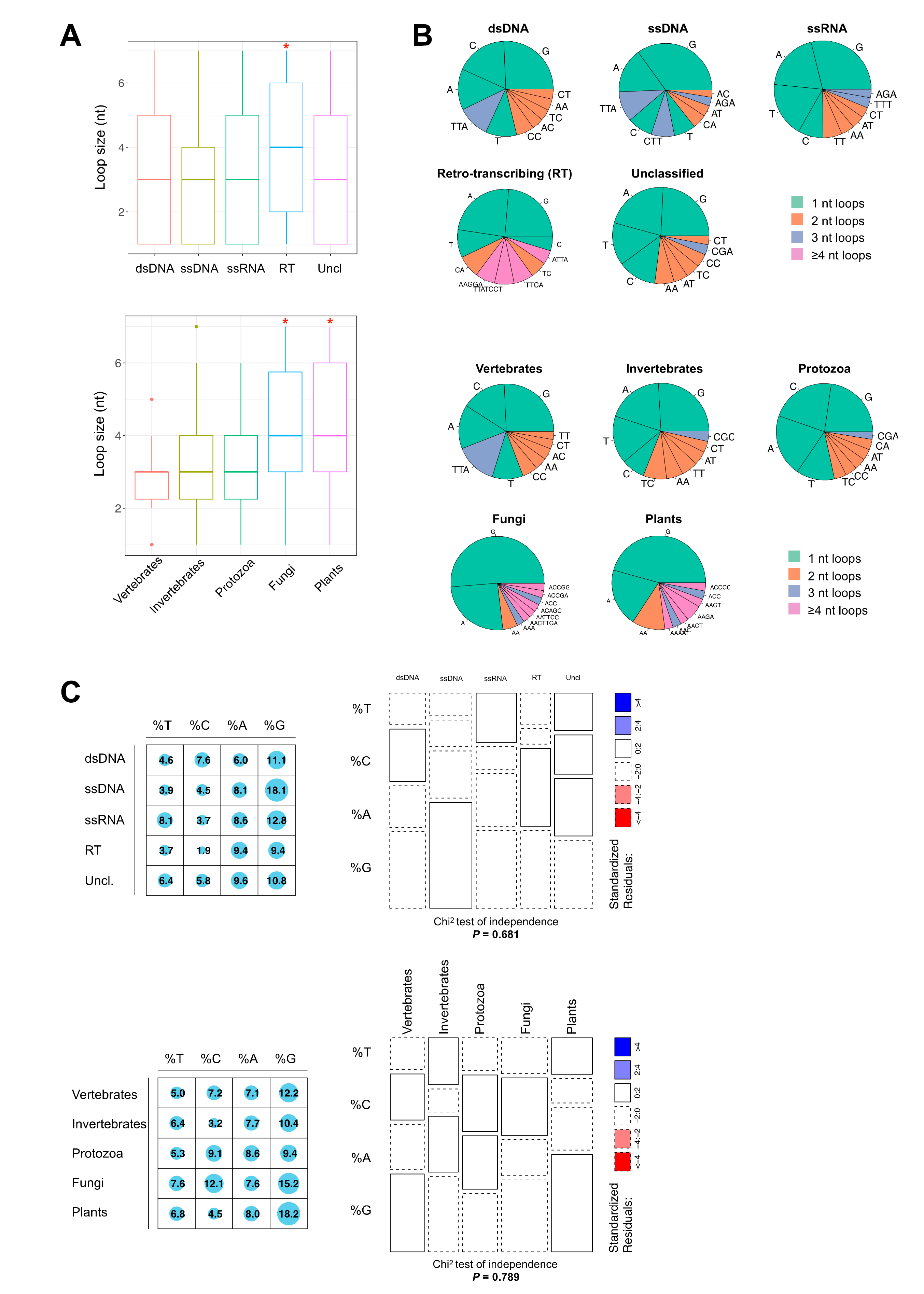
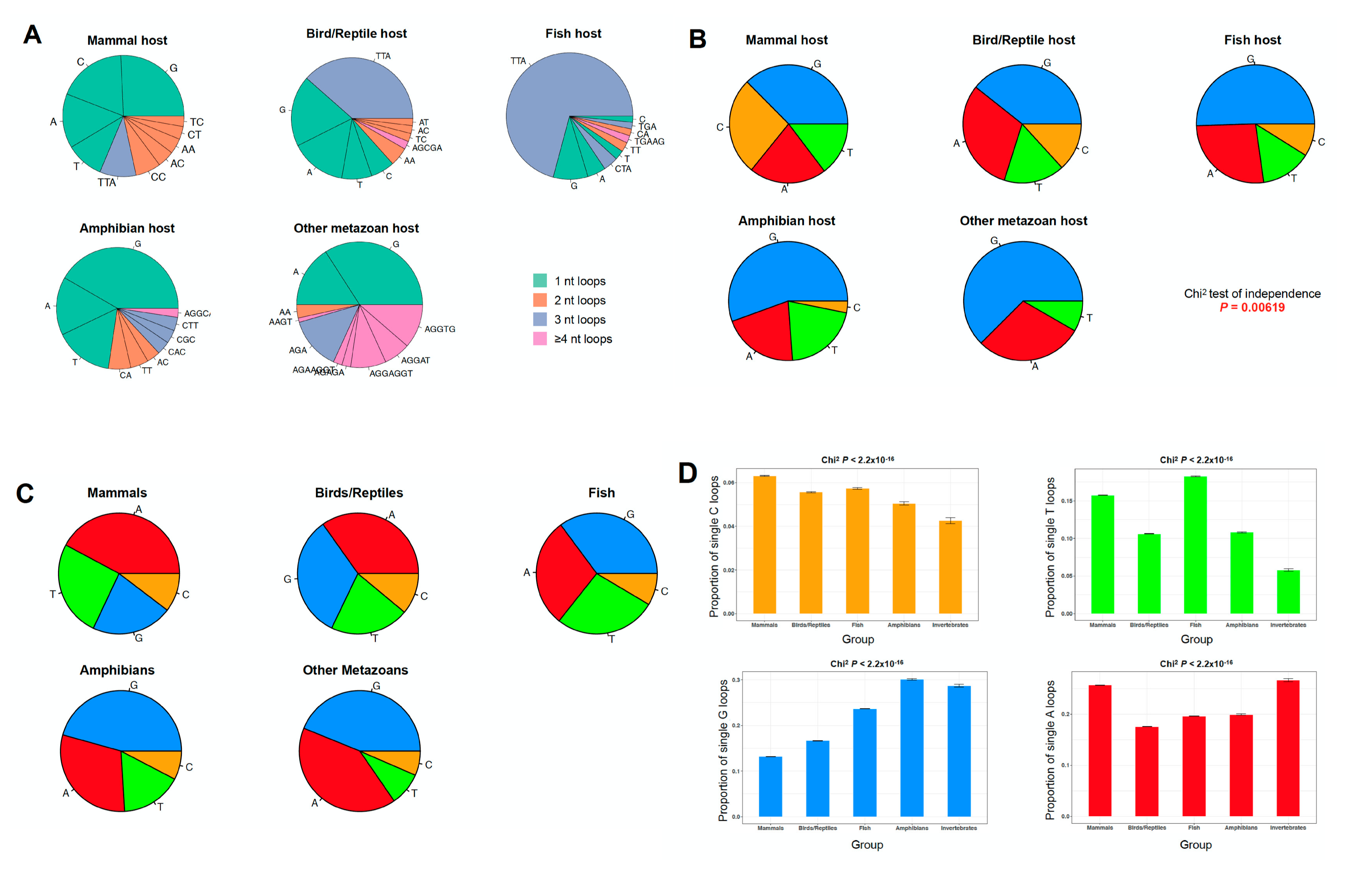


| Taxon | Assemblies | Median % GC | Median Genome Size (Base Pairs) | Total PQS Count | Mean PQS Density 1 |
|---|---|---|---|---|---|
| dsDNA | 2758 | 44.5 | 45,531 | 11,315 | 0.083 |
| dsRNA | 301 | 44.5 | 2178 | 11 | 0.018 |
| RT 2 | 153 | 43.6 | 7743 | 85 | 0.074 |
| Satellites | 227 | 41.0 | 1348 | 2 | 0.011 |
| ssDNA | 988 | 44.0 | 2707 | 102 | 0.058 |
| ssRNA | 1784 | 43.4 | 6944 | 553 | 0.036 |
| Unclassified | 1158 | 44.6 | 4492 | 206 | 0.024 |
| Organism | Median % GC | Genome Size (Mb) | Total PQS Count 1 | Mean PQS Density 2 |
|---|---|---|---|---|
| Human | 37.8 | 3095.69 | 434,272 | 0.140 |
| Mouse | 42.6 | 2730.87 | 327,452 | 0.120 |
| Zebrafish | 36.8 | 1371.72 | 25,677 | 0.019 |
| Drosophila melanogaster | 42.1 | 143.73 | 5262 | 0.037 |
| Caenorhabditis elegans | 35.4 | 100.29 | 1561 | 0.016 |
| Saccharomyces cerevisiae | 38.4 | 12.16 | 7 | 0.001 |
| Leishmania major | 59.6 | 32.86 | 7913 | 0.241 |
| Trypanosoma brucei | 46.8 | 35.83 | 635 | 0.018 |
| Plasmodium falciparum | 19.6 | 23.33 | 51 | 0.002 |
| Arabidopsis thaliana | 36.1 | 119.67 | 338 | 0.003 |
| Rhodobacter sphaeroides | 68.8 | 4.64 | 5 | 0.001 |
| E. coli | 50.8 | 4.6 | 109 | 0.024 |
| Host | Assemblies | Median % GC | Median Genome Size (bp) | Total PQS Count | Mean PQS Density 1 |
|---|---|---|---|---|---|
| Vertebrates | 2769 | 44.4 | 5079 | 7945 | 0.082 |
| Human 2 | 1144 | 42.3 | 4325 | 1410 | 0.076 |
| Invertebrates | 2930 | 42.4 | 4534 | 442 | 0.024 |
| Protozoa | 61 | 46.1 | 6038 | 618 | 0.024 |
| Fungi | 292 | 49.2 | 3147 | 41 | 0.039 |
| Plants | 2484 | 43.2 | 2759 | 262 | 0.027 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Puig Lombardi, E.; Londoño-Vallejo, A.; Nicolas, A. Relationship Between G-Quadruplex Sequence Composition in Viruses and Their Hosts. Molecules 2019, 24, 1942. https://doi.org/10.3390/molecules24101942
Puig Lombardi E, Londoño-Vallejo A, Nicolas A. Relationship Between G-Quadruplex Sequence Composition in Viruses and Their Hosts. Molecules. 2019; 24(10):1942. https://doi.org/10.3390/molecules24101942
Chicago/Turabian StylePuig Lombardi, Emilia, Arturo Londoño-Vallejo, and Alain Nicolas. 2019. "Relationship Between G-Quadruplex Sequence Composition in Viruses and Their Hosts" Molecules 24, no. 10: 1942. https://doi.org/10.3390/molecules24101942
APA StylePuig Lombardi, E., Londoño-Vallejo, A., & Nicolas, A. (2019). Relationship Between G-Quadruplex Sequence Composition in Viruses and Their Hosts. Molecules, 24(10), 1942. https://doi.org/10.3390/molecules24101942






